Abstract
GPR88 is an orphan G-protein coupled receptor originally characterized as a striatal-enriched transcript and is a potential target for neuropsychiatric disorders. At present, gene knockout studies in the mouse have essentially focused on striatal-related functions and a comprehensive knowledge of GPR88 protein distribution and function in the brain is still lacking. Here, we first created Gpr88-Venus knock-in mice expressing a functional fluorescent receptor to fine-map GPR88 localization in the brain. The receptor protein was detected in neuronal soma, fibers and primary cilia depending on the brain region, and remarkably, whole-brain mapping revealed a yet unreported layer-4 cortical lamination pattern specifically in sensory processing areas. The unique GPR88 barrel pattern in L4 of the somatosensory cortex appeared three days after birth and persisted into adulthood, suggesting a potential function for GPR88 in sensory integration. We next examined Gpr88 knockout mice for cortical structure and behavioral responses in sensory tasks. Magnetic resonance imaging of live mice revealed abnormally high fractional anisotropy, predominant in somatosensory cortex and caudate putamen, indicating significant microstructural alterations in these GPR88-enriched areas. Further, behavioral analysis showed delayed responses in somatosensory-, visual- and olfactory-dependent tasks, demonstrating a role for GPR88 in the integration rather than perception of sensory stimuli. In conclusion, our data show for the first time a prominent role for GPR88 in multisensory processing. Because sensory integration is disrupted in many psychiatric diseases, our study definitely positions GPR88 as a target to treat mental disorders perhaps via activity on cortical sensory networks.
Keywords: Orphan G protein-coupled receptor, Gpr88, knock-in and knockout mice, Gpr88-Venus fluorescent protein, layer 4 cortex, primary cilia
Introduction
Gpr88 is a neural orphan G protein-coupled receptor (GPCR) originally described as a striatal-enriched transcript (Mizushima et al. 2000; Logue et al. 2009; Ghate et al. 2007; Massart et al. 2009; Van Waes et al. 2011) regulated by pharmacological treatment, including antidepressants (Conti et al. 2007), mood stabilizers (Ogden et al. 2004; Brandish et al. 2005) or drugs of abuse (Le Merrer et al. 2012; Befort et al. 2008). Two synthetic agonists are proposed, Compound 19 not tested in vivo (Dzierba et al. 2015) and 2-PCCA (Jin et al. 2014; Bi 2013; Jin et al. 2017) with unclear in vivo effects (Li et al. 2013), hence our current knowledge of GPR88 function essentially stems from studies using mice lacking the Gpr88 gene (Gpr88-/- mice). Analyses of Gpr88-/- mice have primarily focused on striatal function, and report altered dopamine signaling (Logue et al. 2009), increased medium spiny neuron (MSN) excitability (Quintana et al. 2012), altered locomotor responses to dopamine ligands (Quintana et al. 2012) and amphetamine (Logue et al. 2009) that was also observed upon local Gpr88 silencing in the rat ventral striatum (Ingallinesi et al. 2015), reduced motor coordination and increased stereotypies (Meirsman et al. 2016a; Quintana et al. 2012). In addition to these striatal-related phenotypes, Gpr88-/- mice showed impaired associative and procedural learning (Meirsman et al. 2016a; Quintana et al. 2012), sensorimotor gating deficits (Logue et al. 2009) and dysfunctional emotional responses leading to risk-taking behavior (Meirsman et al. 2016a; Meirsman et al. 2016b), consistent with the notion that receptor activity broadly influences brain function. Human genetic studies showed association of GPR88 with bipolar disorder and psychoses (Del Zompo et al. 2014; Hodges et al. 2006; Ogden et al. 2004) and a GPR88 variant was linked to a familial developmental disorder characterized by childhood chorea, learning disabilities and speech retardation (Alkufri et al. 2016). Animal and human data therefore concur to indicate that GPR88 is both a disease gene and a potential therapeutic target for neuropsychiatry.
In-depth analysis of Gpr88 expression is critical to further understand receptor function in brain development and disease. Previous reports have mapped Gpr88 mRNA distribution in rodents. In the striatum, the Gpr88 transcript is homogenously expressed early-on (E16), then develops a patchy pattern, and finally shows dense expression in MSNs from both direct and indirect pathways (Massart et al. 2016). The Gpr88 transcript is also detected in the cortex, where expression level is about 50-fold lower compared to striatum (Ghate et al. 2007; Logue et al. 2009). There, Gpr88 mRNA is observed in the cortical plate from embryonic days, and progressively migrates to neocortical layers with adult expression in layers 2-6 (Massart et al. 2016). In the adult, Gpr88 mRNA is also detected at low levels in central amygdala (Becker et al. 2008; Befort et al. 2008), septum and hypothalamus (Massart et al. 2016). Expression of the GPR88 receptor protein has been more difficult to address, however one group reported GPR88 immunoreactivity in both striatum and cortex, with differential subcellular localization (Massart et al. 2009; Massart et al. 2016).
Here, we first substantiate the mouse as a valid model to study Gpr88 function with comparable mouse and human Gpr88 transcript expression patterns in the brain. To fine-map distribution of the GPR88 protein, we employed our unique approach (Erbs et al. 2014; Pradhan et al. 2011), which we used previously to study delta (Faget et al. 2012; Pradhan et al. 2009; Pradhan et al. 2010; Rezai et al. 2012; Scherrer et al. 2006) and mu (Erbs et al. 2015; Gardon et al. 2014) opioid receptors and report the successful generation of Gpr88-Venus mice expressing a fully functional and visible receptor, and describe the detailed receptor distribution in mutant mice, throughout the brain and within neuronal compartments. Receptor mapping revealed a yet unreported layer 4-enriched protein expression in the sensory cortex. Further, diffusion magnetic resonance imaging (MRI) analysis revealed deficits in cortical microstructure of Gpr88-/- mice and behavioral studies showed impaired sensory responses, demonstrating for the first time a prominent role of this orphan receptor in cortical sensory processing.
Materials and Methods
Animals
Gpr88Venus/Venus mice were generated at the Institut Clinique de la Souris (Illkirch-Graffenstaden, France). Gpr88-/- mice were generated as previously reported (Meirsman et al. 2016a). Male Gpr88-/- and Gpr88+/+ mice aged 8 to 10 weeks or male and female Gpr88+/+, Gpr88Venus/+, Gpr88Venus/Venus mice aged 8-12 weeks were bred in-house at the Neurophenotyping Center of the McGill University/Douglas Hospital Research Institute. Animals were group-housed and maintained on a 12-hour light/dark cycle (lights on at 8:00 AM) at a controlled temperature (22°C ± 1°C). Food and water were available ad libitum unless otherwise stated.
Gpr88Venus/Venus mice expressing the Gpr88 receptor fused at its C terminus to a YFP variant, Venus, an improved thermostable and photo-stable YFP variant (Nagai et al. 2002) were generated by homologous recombination. In brief, the Venus cDNA was introduced into exon 2 of the Gpr88 receptor gene via a targeting construct in which the Gpr88 stop codon has been replaced by a Gly-Ser-Ile-Ala-Thr- linker sequence followed by Venus encoding cDNA. Following, a neomycin resistance gene flanked by FRT sites was transfected into embryonic stem (ES) cells. Two independent homologous recombinants were electroporated with an FLP recombinase expressing plasmid to excise the neomycin gene and microinjected into C57Bl/6N blastocysts. F1 heterozygous progenies were obtained from chimeras crossed with C57Bl/6N (Charles River Laboratories, Wilmington, MA, USA) mice. Gpr88Venus/+ mice were intercrossed to generate Gpr88Venus/Venus mice which are fertile and develop normally. All experiments were analyzed blind to genotypes. All experimental procedures were performed in accordance with the guidelines of the Canadian Council of Animal Care, and all animal procedures were approved by the McGill University/Douglas Hospital Animal Care Committee.
Human Brain Samples
Postmortem human brain tissues were obtained from the Quebec Suicide Brain Bank, part of the Douglas – Bell Canada Brain Bank (Douglas Mental Health University Institute, Montreal, Quebec, Canada). The six healthy subjects had died suddenly from accidental or natural causes. Dissections were performed on fresh frozen 0.5 cm-thick coronal sections with the guidance of a human brain atlas (Mai et al. 2008). Samples were obtained from ten different brain regions: orbital frontal cortex (OFC; BA11), prefrontal cortex (PFC; BA9-10), motor cortex (MoCtx; BA4), somatosensory cortex (SSCtx; BA1,2,3), nucleus accumbens (ACB), caudate putamen (CP), thalamus (Th), substantia nigra (SN), midbrain (MB) and cerebellum (Cer). Ethical approval for this study was obtained from the Institutional Review Board of the Douglas Mental Health University Institute.
Human RNA Isolation and Integrity Analysis
Total RNA was isolated using NucleoZol (Macherey-Nagel, Düren, Germany). RNA quantity and quality were determined spectrophotometrically by NanoDrop ND2000 (ThermoFisher, Waltham, MA, USA). RNA integrity numbers (RIN) were obtained using automated electrophoresis by the 2200 TapeStation system (Agilent, Santa Clara, CA, USA). All samples that had a RIN higher than 5 were considered good quality RNA samples, and samples with a RIN higher than 8 were considered as perfect (Fleige and Pfaffl 2006). Samples with very low RNA concentrations and integrity (RIN <4) were excluded from the real-time quantitative polymerase chain reaction (RT-qPCR) analysis (n = 6 by ten dissected regions).
Mouse RNA Sample Preparation and Isolation
Mice in both RT-qPCR studies, were euthanized by cervical dislocation and brains were immediately extracted and placed into a coronal matrix with 1 mm intervals (ASI instruments, Warren, MI, USA). Brain regions were taken from both hemispheres as two (1 mm) punches for orbital frontal cortex (OFC), motor cortex (MoCtx), somatosensory cortex (SSCtx), hypothalamus (Hy), nucleus accumbens (ACB), caudate putamen (CP), thalamus (Th), substantia nigra (SN), midbrain (MB) and cerebellum (Cer) and one punch centered between two hemispheres (1 mm) for prefrontal cortex (PFC). Smaller regions, central amygdala (CEA), basal lateral amygdala (BLA) were taken as 0.5 mm punches, from both hemispheres. Dissected tissue was immediately frozen on dry ice and stored at -80°C. TRIzol (Invitrogen) reagent was added to the sample directly and mechanically dissociated to yield a homogenate. Extraction proceeded according to the manufacture's protocol and Dr. Gentle (Takara, Kusatsu, Japan) was added as a precipitate carrier. Total RNA was re-suspended in RNase DNase free water (ThermoFisher). RNA quantity and quality was evaluated spectrophotometrically by NanoDrop ND2000 (ThermoFisher).
RT-qPCR
RT-qPCR was performed as described previously (Meirsman et al. 2016a) with some modifications. Briefly, 400 ng of RNA was reverse transcribed using the M-MLV Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The twice diluted cDNA was subjected to 45 cycles of amplification by using LightCycler 480 SYBR I Green Master Mix (Roche, Basel, Switzerland) on the LightCycler 480 II Real-Time PCR System (Roche). cDNA samples were loaded in triplicate with 2 μl cDNA and 10 μl final volume. A no-template control (NTC) reaction, with just water, was included to check for non-specific amplification. Relative fold changes were calculated by the comparative Ct method (2˄-ΔΔCT) (Livak and Schmittgen 2001). Normalization was done to housekeeping genes by subtraction of the average triplicate CT values for each sample (ΔCT). For human versus wild-type mouse gene expression study, the ΔΔCT was calculated by subtracting ΔCT of the highest Gpr88 expressing brain region, CP, from each of the 9 other regions per individual. Thereafter, the fold change calculation was presented as a percent of CP (% of CP). The housekeeping genes used for normalization for human gene expression studies were human B2M and RPL19, for mouse gene expression studies Tubb2a was used. In the second gene expression study, Gpr88-Venus mice (2˄-ΔΔCT) values were in function of Gpr88+/+ mice. The housekeeping genes used for normalization were B2m and Tubb2a.
| Table of RT-qPCR Primers | ||
| Gene | Forward | Reverse |
| Venus | CACATGAAGCAGCACGACTT | CATTGTGGGCGTTGTAGTTG |
| MmGpr88 | CGAGGAGGTATTTCCTGGCATC | ACTTGCCTGCTTGATTTGGG |
| MmTubb2a | CTGGCACTTACCACGGAGAT | GTCTGAAGATCTGCCCGAAG |
| MmB2m | TGGTGCTTGTCTCACTGACC | GTATGTTCGGCTTCCCATTC |
| HsGPR88 | TGGCCAGAATGACCAACTCC | TTTCGGAAGGACGACACGAG |
| HsB2M | TGTCTTTCAGCAAGGACTGGT | GCTTACATGTCTCGATCCCA |
| HsRPL19 | CCGAGCGAGCTCTTTCCTTTC | TGGCATTGGCGATTTCATTGG |
Plasmids for the in vitro Characterization of GPR88-Venus
Mouse GPR88 coding sequence was subcloned into pIRES vector (Invitrogen). The Venus tag sequence was added to the C-terminus of GPR88 construct, through a short 5 amino acid linker and protein sequence was optimized by addition of a signal peptide at the N-terminal domain. Plasmids encoding all the different human G protein subunits used in this study were purchased from the Missouri University of Science and Technology (www.cdna.org). GFP10 tag sequence was linked to the C-terminus of GRK2 through a 11-amino acid linker and GRK2-GFP10 was subcloned into pcDNA3.1 vector. Gγ3 coding sequence was fused in frame to the humanized Renilla luciferase II (RlucII) and subcloned into pcDNA3.1 vector. The coding sequence of all mouse G protein subunits were subcloned into pcDNA3.1 vector.
Cell Culture and Transfection
For BRET assays, HEK-293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin (Wisent) in a 37°C humidified incubator with a 5% CO2 atmosphere. Two days prior to experiments, 35,000 cells were distributed in poly-ornithine (Sigma-Aldrich) coated 96-well white culture plates (PerkinElmer) and transfected with plasmids encoding for GPR88 or GPR88-Venus mouse receptor, mouse Gβ1, human Gγ3-RlucII and human GRK2-GFP10 along with the indicated mouse or human Gαi subunit using polyethylenimine 25 kD linear (PEI; Polysciences) as a transfecting agent (3:1 PEI/DNA ratio).
Functional characterization of GPR88-Venus by BRET assay
For BRET assays, HEK-293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin (Wisent, Saint-Jean-Baptiste, Canada) in a 37°C humidified incubator with a 5% CO2 atmosphere. Two days prior to experiments, 35,000 cells were distributed in poly-ornithine (Sigma-Aldrich, St. Louis, MI, USA) coated 96-well white culture plates (PerkinElmer, Waltham, MA, USA) and transfected with plasmids encoding for GPR88 or GPR88-Venus mouse receptor, mouse Gβ1, human Gγ3-RlucII and human GRK2-GFP10 along with the indicated mouse or human Gαi subunit using polyethylenimine 25 kD linear (PEI; Polysciences, Warrington, PA, USA) as a transfecting agent (3:1 PEI/DNA ratio). Transfected cells were washed twice with phosphate-buffered saline (PBS) then incubated 1 hour in Tyrode buffer (137 mM NaCI, 0.9 mM KCI, 1 mM MgCI2, 1 .9 mM NaHCO3, 3.6 mM NaH2PO4, 25 mM HEPES, 5.5 mM Glucose and 1 mM CaCI2, pH 7.4). Cells were then treated with the indicated concentrations of ligands (Compound 19 or 2-PCCA) or vehicle for the indicated times. For the generation of the dose response curve, Compound 19 was dissolved in the presence of 0.1 % Pluronic® F-127 (ThermoFisher). We monitored G protein activation using a similar previously reported BRET-based (Masuho et al. 2015). We measured BRET between Gγ3-RlucII (BRET donor) and GRK2-GFP10 (BRET acceptor), after addition of the luciferase substrate, coelenterazine 400a (NanoLight Technologies, Pinetop, AZ, USA) using a Mithras™ LB940 Multimode Microplate Reader (Berthold Technologies, Bad Wildbad, Germany) equipped with the energy donor filter (400 nm ± 70 nm) and energy acceptor (515 ± 20 nm). BRET2 values are obtained by calculating the ratio of the light emitted by GFP (515 nm) over the light emitted by Rluc (400 nm). Data were collected using the MicroWin 2000 software (Berthold Technologies). Data are expressed as mean of three independent experiments ± SEM and significance was tested using a one-way or a 2-way ANOVA with Dunnett or Bonferroni post hoc adjustment for multiple comparisons, respectively, as appropriate, using Prism 6.0 software (GraphPad Software, Inc). A value of P<0.05 was considered significant.
[35S]-GTPγS Binding in Mouse Tissues
The assay was performed on membrane preparations from dissected whole striata and performed as described previously (Pradhan et al., 2009; Erbs et al., 2015; Meirsman et al., 2016a). To evaluate GPR88-Venus receptor function, striata were dissected rapidly following mouse cervical dislocation and placed on dry ice and stored at -80°C until use. Striata from 3 mice, male or female equally balanced, were pooled together for a single membrane preparation and this was repeated twice for each genotype. Results are expressed by the mean average of all preparations. Membranes were prepared by homogenizing tissue in 0.25M Sucrose (ThermoFisher) with mechanical dissociation for 30 seconds. Centrifugation followed at 2500 rpm, 4°C for 10 min. Samples were diluted in TMEN (50mM TrisHCl (pH 7.4), 3mM MgCl2, 0.2mM EGTA, 100mM NaCl) and ultracentrifugation (Optimax-XP, Beckman Coultier, Brea, CA, USA) proceeded at 40,000 g, 4°C for 40 minutes with a MLA-55 rotor (Beckman). The membrane pellet was re-suspended in 0.32M sucrose by 10 strokes with a glass tissue grinder. Protein concentration was determined by the Bradford assay (Bio-Rad, Hercules, CA, USA) with a standard curve of BSA (ThermoFisher) and 3 dilutions of each sample. The membrane preparations were diluted to 0.1 mg/ml and stored at -80°C in aliquots of 500 μl. For each assay, 5 μg of protein was used per well. Samples were incubated with and without ligands, for 1 hour at 25°C in assay buffer containing 5 mM GDP and 0.1 nM [35S] GTPγS. Bound radioactivity was quantified by a liquid scintillation counter, TopCount (Perkin Elmer). Non-specific binding is determined by binding in the presence of 10 μM GTPγS and basal activity was determined in the absence of agonist. Calculations and sigmoidal dose-response binding curves were done using GraphPad PRISM 6 (GraphPad Software, Inc, San Diego, CA, USA).
Tissue Preparation for Immunohistochemistry
Mice were anesthetized with i.p. injections of 100 μl/100 g of a cocktail containing Ketamine/ Xylazine/ Acépromazine. Male and female mice were intracardially perfused with 10 ml of 1× PBS pH 7.4 (Life Technologies) followed by 50 ml of 4% paraformaldehyde or PFA (Cedarlane, Burlington, Ontario, Canada) in 1× PBS pH 7.4. For P0 and P3, heads were severed and brains were dissected in 1× PBS pH 7.4. P0, P3 and adult brains were extracted into 10 ml 4% PFA/1× PBS, and left for overnight. Cryoprotection in 30% sucrose (ThermoFisher) 1× PBS pH 7.4 was carried out at 4°C until brains were sunk. Brains were embedded on dry ice in O.C.T. (Sakura Finetek, Alphen aan den Rijn, Netherlands) and stored at -80°C until processing. A cryostat (Leica, Wetzlar, Germany) set at -20°C was used to section tissue at 30 μm for adult and 50 μm for P0 and P3. Tissue slices were kept free floating in 1× PBS pH 7.4 until staining procedure. For all staining the procedure was as follows, slices were taken from 2 wells approximately 300 μm apart and permeabilized and washed in PBS-T (1× PBS pH 7.4, 0.1% Triton X-100) followed by a one hour incubation with agitation in blocking buffer (1× PBS, 3% NGS, 0.2% Triton X-100). Tissue was incubated with primary antibody diluted in blocking buffer at 4°C overnight with gentle agitation. Tissue was thoroughly washed in PBS-T. Secondary antibodies, Goat Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies, Carlsbad, CA, USA), were diluted in blocking buffer and incubated with tissue sections for two hours at room temperature with gentle agitation. Following another washing step, tissue was mounted onto slides (ThermoFisher) and sealed with coverslips (ThermoFisher) in Moviol (Sigma-Aldrich) and left to dry overnight at room temperature with short term storage at 4°C and long term at -20°C. DAPI (ThermoFisher) was added in the final washing step with 1× PBS pH 7.4 or together with Moviol. Slides were scanned on the Olympus VS120 (Olympus Corporation, Shinjuku, Tokyo, Japan) with a 10× objective. For confocal microscopy imaging, an Olympus FV1200, oil immersion 60× objective, was used to take Z-stack images of 0.4 μm steps and the projection of maximum intensity images are shown.
| Antibody | Purpose | Dilution | Maker |
| (ch) Anti-Venus (GFP) | Venus | 1:2000 | Novus NB100-1614 |
| (rb) Anti-Venus (GFP) | Venus | 1:2000 | Life Technologies A111222 |
| (rb) Anti-Arl13b | Primary Cilia | 1:500 | PTG-Lab 17711-1-AP |
| (rb) Anti-CDP(Cux1) | Cortical layer 2-4 | 1:50 | Santa Cruz sc-13024 |
| (rt) Anti-Ctip2 | Cortical layer 5-6 | 1:750 | Abcam ab18465 |
| (rb) Anti-NeuN | Neurons | 1:2000 | CST 24307S |
| (ch) Anti-GFAP | Glia | 1:2000 | Abcam ab4674 |
Mapping GPR88
Slide scanner images of coronal brain slices from Gpr88Venus/Venus (n=3), Gpr88Venus/+ mice (n=3) and Gpr88+/+ mice (n=3) were examined. Annotations of GPR88-Venus expression were performed on 20 cortical areas and 54 brain anatomical sub-regions, previously identified using the Allen Brain Interactive Atlas Viewer (http://atlas.brain-map.org/). In the cortex, the level of GPR88 fluorescence were characterized according to layer. First, all sections were scanned by an Olympus VS120 with a 10× objective and analyzed in detail using OlyVIA 2.8 software (Olympus) and VS-Desktop 2.8 (Olympus), at 5× to 30× magnification. The level of GPR88-Venus expression amplified by Anti-Venus antibody (red) were scored by comparing the fluorescence signal between Gpr88+/+ corresponding to background level, and Gpr88Venus/+ or Gpr88Venus/Venus slices for each brain region of interest. A scale of four levels of fluorescence was used to determine GPR88-Venus expression at each region (Supplementary Figure 4). Data for each brain area were pooled to generate a final score. For confocal microscopy analysis of GPR88-Venus subcellular localization, Olympus FV1200, oil immersion 60× objective lens was used to examine the intrinsic signal. Four different patterns of expression were distinguished: soma, fiber, both cell and fiber or primary cilia.
DT-MRI Methods
Animals
DT-MRI was performed on two groups (n=14 / group) of 7 – 8 weeks old live adult male mice (74.9% C57B/6J, 25% 129/SvPas, 0.05% FVB/N, 0.05% SJL/J): Gpr88+/+ – control mice (Gpr88+/+: n = 14) and Gpr88-/- group (Gpr88-/-: n = 14) respectively.
MRI Data Acquisition and Processing
Mouse brain DT-MRI data was acquired with a 7T small bore animal scanner (Biospec 70/20, Bruker, Germany) and a mouse head adapted CryoCoil (MRI CryoProbe, Bruker, Germany). Mice were anesthetized via Isoflurane (∼1.5 Vol%) and high angular resolution diffusion imaging (HARDI) was performed on respiration triggering using 4-shot Diffusion Tensor Imaging - Echo Planar Imaging (DTI-EPI) sequence (TE/TR = 27 ms/3750 ms), with diffusion gradients applied along 30 non-colinear directions, b factor of 1000 s/mm2, diffusion gradient's duration (δ) of 4 ms and separation (Δ) of 10 ms respectively. 25 axial slices of 0.5 mm thickness were acquired with a FOV of 1.5 × 1.2 cm2 and an acquisition matrix of 160 × 128 resulting in an image resolution of 94 × 94 μm2. Brain fractional anisotropy (FA) maps were generated and after spatial normalization and co-registration on the Allen Mouse Brain Atlas were further used to evaluate the brain microstructural modifications. Statistical significance was evaluated using two-sample t-test (P<0.05, FWE corrected).
Behavioral Experiments
Home Recognition Test
To assess the olfactory function, a home recognition test adapted from Metz and Schwab was performed (Metz and Schwab 2004). A conditioned place preference apparatus (Harvard Apparatus, Holliston, MA, USA) was fitted with a compartment containing home litter and the other compartment containing new litter. Each animal was placed in a neutral area between the two compartments with equal access to both compartments. Four trials were carried out alternating the litter between left and right compartments to avoid side or pattern bias. The time to decide to enter a compartment and the compartment chosen (home or new litter) as well as the duration of time spent in each compartment was measured with a cut-off at 1 minute.
Sticky Paper Test
The somatosensory response to a tactile stimulus was assessed with the sticky paper test (Metz and Schwab 2004; Schallert and Whishaw 1984). Adhesive tape (0.7 × 1.1 cm) was applied to the palmar side of the hind-limb. The mouse was then placed in a Plexiglas box (40 × 40 cm) and the latency of the first reaction to the stimulus (paw lifting, sniffing, biting, or removal) with a limit of 180 seconds was recorded. Four trials with alternation between left and right sides was performed.
Visual Cliff
A particle board box measuring 50 cm × 50 cm was built with a first horizontal plane connected to a second horizontal plane at a lower level by a vertical drop of 50 cm. A black and white pattern was glued on to accentuate the visual cliff effect and a Plexiglas floor was placed on top of the upper horizontal plane extending over the vertical drop. To reduce reflections on the Plexiglas, white particle board was used for the box walls. Equal illuminations for both sides of the visual cliff apparatus was applied. Each animal was placed on the center of the ridge of the visual cliff apparatus, facing the cliff and the side on which the mouse stepped and latency to move off of the ridge was recorded for 10 trials.
Odor Discrimination
Odor discrimination was adapted to test habituation and dishabituation to odors (Arbuckle et al. 2015). Odors were prepared by diluting 100 μl of almond, lemon or anise with 10 ml of water prior to the test outside of the testing room. The odorant tubes were kept outside the test room and gloves were changed between each mouse to avoid social odor contamination. Mice were placed in individual home cages with a wire lid on the top and no bottle of water and allowed to acclimate to the cage for 45 minutes. For each mouse, 50 μl of the tested odorant was pipetted onto the tip of a cotton swab in the following order: water, almond, lemon, anise and each odor was consecutively presented three times for 2 minutes each trial. Trials were separated by 1 minute intervals. The cumulative active sniffing time defined by the nose oriented towards the odorant tip (∼2 cm or closer) were recorded for each 2-minute presentation. The dishabituation to an odor was calculated by subtracting a former odors' 3rd trial from the following odor's first trial; for example - water trial 3 (W3) was subtracted from almond trial 1 (Al1) to determine dishabituation from water to almond.
Data Analysis
Data is presented as mean per group value standard error of the mean (SEM) and analyzed using Student's t-test or analysis of variance (ANOVA) when appropriate. Post-hoc analysis tests were done using Bonferroni multiple comparison test. The criterion for statistical significance was P<0.05. All statistics were done using GraphPad PRISM 6 (GraphPad Software, Inc).
Results
Gpr88 transcript distribution is similar in the human and mouse brain
In humans, the Gpr88 transcript was reportedly expressed at high levels in both the caudate nucleus and putamen, and faintly in the medulla (Mizushima et al. 2000). We first tested whether Gpr88 mRNA could be detected in other regions of the human brain, and if the pattern is similar to the mouse brain. Using RT-qPCR, we quantified Gpr88 expression on dissected post-mortem brain tissue from healthy human subjects (Quebec Suicide Brain Bank, see Methods) and commercial (C57Bl/6N) mice, across 10 regions of the brain (Figure 1A-B). In both species, the Gpr88 transcript was highly expressed in the caudate putamen (CP) and nucleus accumbens (ACB) and moderately expressed in the cortex compared to striatum. In cortical areas, the transcript was observed at similar levels in somatosensory cortex (SSCtx) and prefrontal cortex (PFC), and low in the motor cortex (MoCtx). The Gpr88 transcript was also low in the midbrain (Mb) and thalamus (Th), and undetectable in substantia nigra (SN) and cerebellum (Cer) for both species. Together, in both human and mouse brains, Gpr88 expression is strongest in striatum, moderate in the forebrain, and virtually absent in mid- and hindbrains. Comparable Gpr88 mRNA distribution therefore, indicates that the mouse is an appropriate translational model to examine Gpr88 function.
Figure 1.
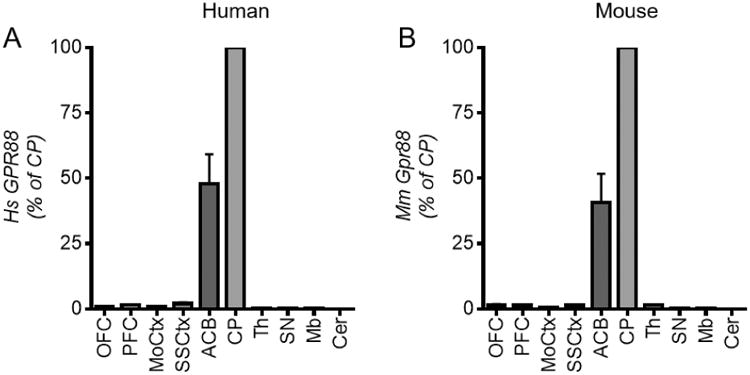
Gpr88 mRNA expression profile is similar in the human and mouse brain. RT-qPCR was performed using mRNA samples from 10 brain regions for (A) healthy human subjects (n=6) and (B) C57/Bl6N mice (n=7). ΔΔCT values were normalized using B2M and RPL19 (human) and B2m and Tubb2a (mouse) as the housekeeping genes, and gene expression is expressed as a percentage of Gpr88 expression level in the CP. Abbreviations- Orbital frontal cortex (OFC), prefrontal cortex (PFC), motor cortex (MoCtx), somatosensory cortex (SSCtx), nucleus accumbens (ACB), caudate putamen (CP), thalamus (Th), substantia nigra (SN), midbrain (Mb) and cerebellum (Cer) (statistical analysis not shown here for clarity, was done by One-Way ANOVA with Bonferroni post-hoc test, see Supplementary Table I).
In vitro and in vivo molecular characterization of Gpr88-Venus
Functionality of the GPR88-Venus C-terminal fusion protein was first tested by a bioluminescence resonance energy transfer (BRET) assay in HEK-293 cells (Figure 2A). Stimulation of GPR88-Venus with Compound 19, a GPR88 agonist (Dzierba et al. 2015), produced a similar Gαi2 activation response as compared to native GPR88, including comparable potency (EC50 = 1.2 μM vs 1.9 μM, respectively) and efficacy (Emax = 97.5 ± 2.5 vs 100 ± 4.7, respectively) (Figure 2B). A profiling of GPR88 activation by Compound 19 or 2-PCCA, (Dzierba et al. 2015; Jin et al. 2014) on 13 human and 7 mouse G proteins showed highly similar signaling profiles for the two receptor constructs, with preferred activation of both mouse and human Gαi1 and Gαi2 isoforms (Supplementary Figures 1A and B). These results strongly suggest that the Venus fusion does not impair signaling properties of the receptor.
Figure 2.
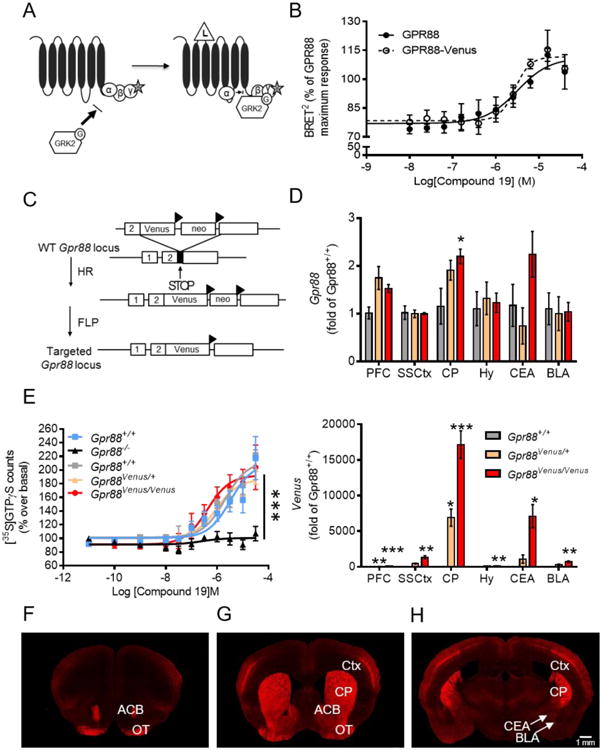
Characterization of Gpr88-Venus. (A, B) GPR88 signaling in transfected cells. (A) Schematic representation of the BRET-based biosensor assay used to monitor ligand-induced G-protein activation. Upon ligand (L) binding to the receptor, the Gα subunit dissociates from the βγ-RlucII (γ-R, donor BRET2 sensor) dimer allowing GRK2-GFP10 (GRK2-G, acceptor BRET2) sensor recruitment to the βγ dimer, leading to an increase in BRET2 signal. (B) GPR88-mediated activation of mouse Gαi2 protein. HEK-293 cells were co-transfected with GPR88 or GPR88-Venus mouse receptor construct, Gβ1, Gγ3-RlucII (donor BRET2 sensor), GRK2-GFP10 (acceptor BRET2 sensor) and Gαi2 subunit, and stimulated 15 minutes with increasing concentrations of Compound 19. Potency values are expressed as LogEC50 ± SEM (EC50 μM). Emax values are expressed as mean % of maximal GPR88 response (mean ± SEM, 3 replicate experiments). (C-H) Characterization of the Gpr88-Venus knock-in mouse line. (C) Schematic diagram of the gene targeting strategy to create Gpr88-Venus mice. The Venus cDNA was inserted in the Gpr88 gene coding exon (exon 2) upstream from the STOP codon to produce a C-terminally-tagged GPR88 receptor protein. Triangles denote FLP sites flanking the Neo cassette, which was ultimately deleted. (D) Quantification of Gpr88 mRNA expression (top) by RT-qPCR in prefrontal cortex (PFC), somatosensory cortex (SSCtx), caudate putamen (CP), hypothalamus (Hy), central extended amygdala (CEA) and basal lateral amygdala (BLA) shows no modification of Gpr88 transcript level between Gpr88Venus/Venus, Gpr88Venus/+ and Gpr88+/+ littermates, with the exception of slightly higher expression in CP. A subtle increase of mRNA level was also observed in our earlier work (Erbs et al. 2015; Scherrer et al. 2006), perhaps reflecting increased mRNA stability. The Venus transcript (bottom) is detected in homozygous and heterozygous (about half) but not control knock-in mice for all the brain regions (n=3-6; One-way ANOVA, Bonferroni post-hoc, see Supplementary Table I). (E) Compound 19 induced [35S]-GTPγS binding is indistinguishable across the three genotypes, and another group of control, indicating that the Venus fusion does not alter GPR88-mediated G protein activation (Two-way analysis of variance, Gpr88+/+ vs. Gpr88-/- interaction of genotype and drug effect, Bonferroni post-hoc test, see Supplementary Table I). (F-H) Brain sections from Gpr88Venus/Venus mice stained with the Venus antibody show Gpr88 expression pattern at the level of (F-G) olfactory tubercule (OT) and nucleus accumbens (ACB), as well as (G-H) caudate putamen (CP), cortex (Ctx). (H) GPR88 expression is visible in central extended amygdala (CEA) but not basal lateral amygdala (BLA), see white arrows. GPR88-Venus expression is similar to previously reported in situ hybridization studies (Ghate et al., 2007; Becker et al., 2008; Befort et al., 2008; Logue et al., 2009; Massart et al., 2009; Massart et al., 2016) and one immunohistochemistry study (Massart et al., 2016) (Scale bar 1 mm). Data are presented as mean ± SEM: *P<0.05; **P<0.01, ***P<0.001.
We next engineered a knock-in mouse line that expresses GPR88 fused to Venus (Nagai et al. 2002) in place of the endogenous receptor by a targeting strategy (Figure 2C) similar to our previous work (Erbs et al. 2015; Scherrer et al. 2006), and we obtained animals heterozygous (Gpr88Venus/+) and homozygous (Gpr88Venus/Venus) for the mutation, which we characterized at molecular and cellular levels.
We first examined whether insertion of the Venus cDNA into the Gpr88 gene locus modifies Gpr88 gene transcription using RT-qPCR (Figure 2D, top). Gpr88 mRNA level did not significantly differ across the three genotypes in PFC, SSCtx, hypothalamus (Hy), central extended amygdala (CEA) or basal lateral amygdala (BLA), and was slightly higher for Gpr88Venus/Venus mice, in the CP. We also quantified the Venus mRNA expression and found that, as expected, transcript level was half for Gpr88Venus/+ compared to Gpr88Venus/Venus and undetectable in Gpr88+/+ mice (Figure 2D, bottom).
Next, we evaluated functionality of the modified receptor in the [35S]-GTPγS binding assay, which quantifies G protein activation in response to a receptor agonist in native tissues (Figure 2E and Supplementary Table 1). As in our previous report (Meirsman et al. 2016a), Compound 19 (Dzierba et al. 2015) elicited strong [35S]-GTPγS binding in striatal membranes from Gpr88+/+ mice, and no response was observed in mice lacking Gpr88 (Gpr88-/-) demonstrating GPR88 selectivity of the agonist. Further, activity measures from Gpr88Venus/Venus mice showed [35S]-GTPγS binding similar to Gpr88Venus/+ and Gpr88+/+ littermates The Gpr88Venus/Venus curve was left-shifted, perhaps related to the slight increase of mRNA transcript in GPR88-Venus CP, but overall there was no statistical significant difference across genotypes (Supplementary Table 1). The Venus fusion therefore does not modify signaling properties of the GPR88 receptor in mouse tissues, at least in this assay.
We finally examined GPR88-Venus protein expression at macroscopic level. In brain regions with known enriched Gpr88 expression, intrinsic GPR88-Venus fluorescent signal was easily detectable (Figure 2 F-H). Signal amplification using the Venus antibody improved the signal-noise ratio and further increased the signal (Supplementary Figure 2).
Developmental expression of GPR88-Venus at P0, P3 and in the adult
In the rat, Gpr88 mRNA was reported to change throughout development (Massart et al. 2016; Van Waes et al. 2011). We therefore examined GPR88-Venus expression in Gpr88Venus/Venus mice, here after called Gpr88-Venus mice, at three time points including P0, P3 and P56 (adult). Representative sections are shown in Figure 3. The CP showed highest GPR88-Venus signal at all developmental stages with increasing homogeneity, from patches at P0 and P3 to matrix compartments in adult. In the neocortex, the lamination pattern evident by GPR88-Venus was different across all three developmental time points. In the P0 neocortex, GPR88-Venus was present in layer 1 (L1) and layer 6b (L6b), and virtually undetectable in the other layers. At P3, GPR88-Venus decreased but was still detectable in L1 and L6b, and appeared in layer 4 (L4) and less-so in layer 5 (L5) and 6a (L6a). In the adult cortex, GPR88-Venus was primarily enriched at L4, with low expression in layer 2/3 (L2/3), sparse detection in L5-L6a, and discrete populations of neurons remaining in L1 and L6b. At both P3 and adult, the GPR88-Venus signals observed, formed a barrel pattern in SSCtx L4, also known as the barrel cortex (Van der Loos and Woolsey 1973), further identified in the adult by tangential slice preparations (Supplementary Figure 3). Finally, GPR88-Venus expression was observed as soon as P0 in ACB, OT and CeA but not BLA, and the pattern was similar across developmental stages (data not shown).
Figure 3.
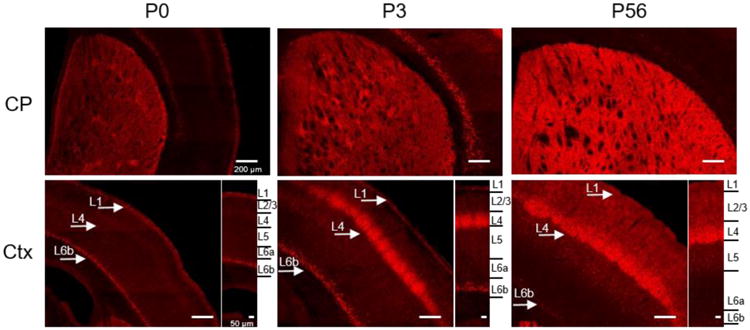
Developmental GPR88 expression at postnatal stage: P0, P3 and P56 (adult). Gpr88Venus/Venus mouse tissue sections were prepared for each time point, and the GPR88-Venus intrinsic signal was amplified (red). Representative images show the heterogeneous expression localized in striatal patches at P0 and P3 while expression in the adult is stronger and homogenously distributed across all caudate putamen (CP) compartments. The cortical expression of GPR88 (Ctx) transitions from layer 1 and 6b to layer 4 between P0 and P3, finally in adult predominantly in layer 2-4 and scant in layer 5-6b. Scale bar is 200 μm and 50 μm for layer view in the cortex. Arrows point at layer 1, 4 and 6b on the sections, and right insets show cortical layer details.
GPR88-Venus layer 4 pattern is regionally restricted to the sensory cortex
We then mapped GPR88-Venus expression in the entire adult brain and semi-quantified the degree of expression (Supplementary Figure 4) in 20 cortical and 54 sub-cortical regions (Table I). We found prominent GPR88 expression in OT, ACB, CP, parasubiculum (PAR) and in the cortex (detailed below). Areas that showed moderate expression outside of the cortex include, olfactory areas (cortical amygdalar (COA), piriform amygdalar (PAA), CEA and intercalated amygdalar nucleus (IA) and substantia nigra. Low expression of GPR88-Venus was also detected in discreet regions of the olfactory bulb, hippocampus, cortical subplate, septum, hypothalamus, pallidum and thalamus. No GPR88-Venus signal could be detected in BLA, medulla, pons and Cer (Table I).
Table I.
Complete table of GPR88-Venus expression.
| Olfactory bulb | Subcortical Areas | ||
|---|---|---|---|
| Accessory | Striatum | ||
| Glomerular | - | Anterior amygdalar area | + |
| Granular | - | Caudoputamen | +++ |
| Mitral | - | Central amygdalar nucleus | + |
| Main | Intercalated amygdalar nucleus | ++ | |
| Glomerular | + | Lateral septal complex | + |
| Granular | - | Medial amygdalar nucleus | + |
| Inner plexiform | - | Nucleus Accumbens | +++ |
| Mitral | - | Olfactory Tubercle | +++ |
| Outer plexiform | - | Olfactory Area | |
| Anterior olfactory | + | Cortical amygdalar area | ++ |
| Cortex | Piriform-amygdalar area | ++ | |
| Agranular Insular | Pallidum | ||
| I | + | Bed nuclei stria terminalis | - |
| II | + | Caudal | - |
| III | + | Dorsal | + |
| V | - | Medial | - |
| VI | - | Ventral | + |
| VIb | - | Cortical Subplate | |
| Anterior Cingulate | Basolateral amygdalar nucleus | - | |
| I | + | Basomedial Amygdalar nucleus | + |
| II | + | Claustrum | - |
| III | + | Endopiriform nucleus | + |
| V | - | Posterior amygdalar nucleus | - |
| VI | - | Hippocampus | |
| VIb | - | CA1 | - |
| Auditory | CA2 | - | |
| I | ++ | CA3 | - |
| II | ++ | Dentate Gyrus | + |
| III | ++ | Parasubiculum | +++ |
| IV | ++ | Subiculum | - |
| V | - | Thalamus | |
| VI | - | Anteromedial nucleus, dorsal part | - |
| VIb | + | Lateral posterior nucleus | - |
| Ectorhinal | Lateral habenula | + | |
| I | ++ | Medial habenula | + |
| II | ++ | Posterior complex | - |
| III | ++ | Submedial nucleus | + |
| V | - | Ventral anterior-lateral complex | + |
| VI | - | Ventral posterolateral nucleus | + |
| VIb | + | Ventral posteromedial nucleus | + |
| Entorhinal, lateral part | Hypothalamus | ||
| I | + | Ventromedial hypothalamic nucleus | + |
| II | + | Midbrain, Behavioral | |
| III | + | Dorsal raphe | - |
| IV | - | Midbrain raphe | - |
| V | - | Midbrain, Motor | |
| VI | - | Edinger-Westphal nucleus | - |
| VIb | + | Periaqueductal gray | - |
| Gustatory | Substantia Nigra | ++ | |
| I | ++ | Ventral Tegmental Area | - |
| II | ++ | Hindbrain | |
| III | ++ | Medulla | - |
| IV | +++ | Pons | - |
| V | + | Cerebellum | - |
| VI | - | ||
| VIb | + | ||
| Infralimbic | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | - | ||
| VI | - | ||
| Motor Area Primary | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | - | ||
| VI | - | ||
| Motor Area Secondary | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | - | ||
| VI | - | ||
| Orbital Area Lateral | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | - | ||
| VI | - | ||
| VIb | - | ||
| Orbital Area Medial | |||
| I | + | ||
| II | + | ||
| III | - | ||
| V | - | ||
| VI | - | ||
| Orbital Area Ventrolateral | |||
| I | + | ||
| II | ++ | ||
| III | ++ | ||
| V | + | ||
| VI | + | ||
| VIb | + | ||
| Perirhinal | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | - | ||
| VI | - | ||
| VIb | + | ||
| Parietal Association | |||
| I | ++ | ||
| II | ++ | ||
| III | ++ | ||
| IV | ++ | ||
| V | - | ||
| VI | - | ||
| VIb | + | ||
| Prelimbic | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | - | ||
| VI | - | ||
| Retrosplenial | |||
| I | + | ||
| II | + | ||
| III | + | ||
| V | ++ | ||
| VI | - | ||
| Somatosensory | |||
| I | ++ | ||
| II | ++ | ||
| III | ++ | ||
| IV | +++ | ||
| V | + | ||
| VI | - | ||
| VIb | + | ||
| Temporal Association | |||
| I | ++ | ||
| II | ++ | ||
| III | ++ | ||
| IV | - | ||
| V | - | ||
| VI | - | ||
| VIb | + | ||
| Visceral Area | |||
| I | ++ | ||
| II | ++ | ||
| III | ++ | ||
| IV | +++ | ||
| V | - | ||
| VI | - | ||
| VIb | + | ||
| Visual Area | |||
| I | ++ | ||
| II | ++ | ||
| III | ++ | ||
| IV | ++ | ||
| V | - | ||
| VI | - | ||
| VIb | + |
Adult (P56) GPR88-Venus expression categorized according to the Allen Mouse Brain atlas, and ranked in terms of degree of expression: Absent (-), Low (+), Moderate (++) or High (+++).
Fine-mapping cortical GPR88-Venus revealed the highest expression in L4 with a pattern of enrichment across sensory cortical regions (Supplementary Figure 5A). Prominent SSCtx GPR88-Venus signal extended into gustatory (GU) and visceral (VISC) sensory areas (Figure 4A). Moderate expression in L4 was also detected in other adjacent sensory areas, including auditory (AUD), parietal (PTL), and visual (VIS). Sensory cortical regions that show moderate expression in other layers, L1 and L2/3, (ectorhinal or ECT) and L5 (retrosplenial or RSP). Finally, low expression was observed in other cortical regions, entorhinal (ENT), perirhinal (PERI), agranular insular, PFC and MoCtx (Table I). The cortical areas with high or moderate GPR88-Venus signal are graphically represented in Figure 4B. We next confirmed the L4-restricted GPR88-Venus fluorescent signal in the SSCtx using the CUX1 antibody, a known marker of L1-4 (Molyneaux et al. 2007; Nieto et al. 2004) (Figure 4C). In addition, sparse GPR88-Venus-positive cells in L5 and L6 overlapped with CTIP2 (Figure 4D), a deep cortical layer marker (Arlotta et al. 2005).
Figure 4.
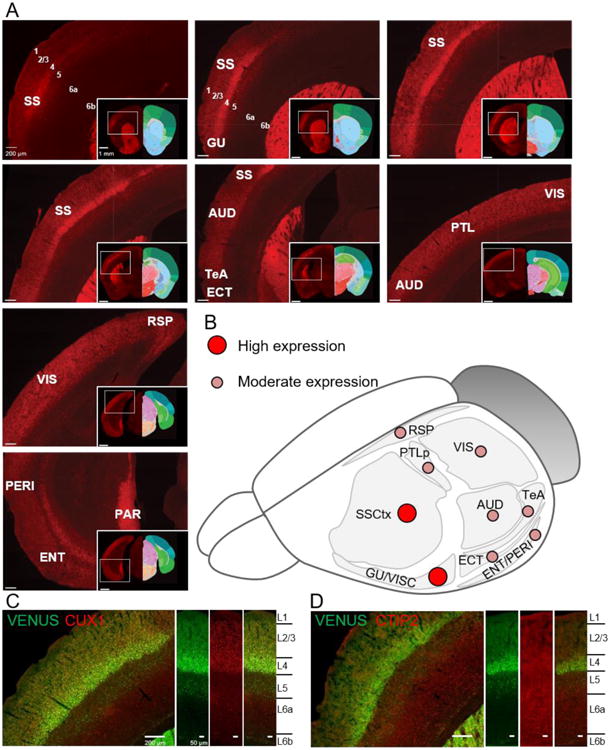
Mapping GPR88-Venus expression throughout the cortex. (A) Shown is the progression of strong GPR88-Venus expression along the rostro-caudal axis, beginning with somatosensory cortex (SSCtx) layer 4 (L4) and contiguous gustatory (GU) to the visceral cortex (VISC). Moderate expression continues into auditory cortex (AUD), temporal association area (TeA) and ectorhinal cortex (ECT). Finally, parietal (PTL), visual (VIS) and retrosplenial (RSP) cortex express GPR88-Venus moderately and expression in the perirhinal (PERI), entorhinal (ENT) is low in contrast to the high expression in parasubiculum (PAR) (Scale bar is 200 μm and 1 mm for the insets). Allen Mouse Brain atlas reference is included in the inset (Image credit: Allen Institute.). (B) An overview illustrates the high and moderate distribution of GPR88-Venus in above cortical areas. (C) CUX1, a layer 2-4 marker co-labels with GPR88-Venus at the L4 of the SSCtx. (D) CTIP2, a layer 5-6 marker, largely remains distinct from GPR88-Venus. Scale bar is 200 μm and 50 μm for cortical columns.
Altogether, the complete mapping of GPR88-Venus refines our knowledge of GPR88 receptor distribution in the mouse brain and, in addition to the well-known massive receptor expression throughout the striatum, reveals strong cortical GPR88-Venus expression confined essentially to layer 4 of sensory processing areas.
GPR88-Venus is localized in different neuronal compartments across the brain
GPR88 is expressed in neurons as demonstrated by high co-localization with NeuN, a neuronal marker, but not GFAP, a glia marker (Figure 5). We next examined GPR88-Venus subcellular localization with confocal imaging. Depending on the brain region, the GPR88-Venus signal was detected in neuronal cell bodies, fibers, or both with or without primary cilia localization (Figure 6A). Intrinsic GPR88-Venus was localized to soma in SSCtx, fibers in SN and shell of ACB, or both soma and fibers in OT, CP, IA and COA.
Figure 5.
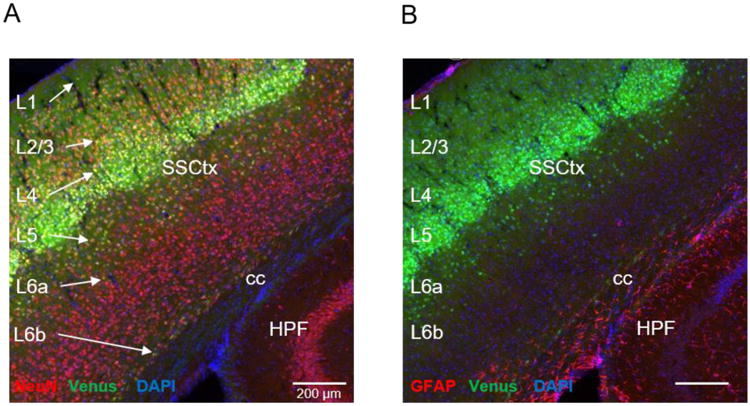
GPR88-Venus is expressed in neurons and not glia. Tissue sections of 30 μmwere prepared from Gpr88Venus/Venus mice, immunostained with Anti-Venus antibody (green), DAPI to stain nuclei (blue) and either Anti-NeuN, neuronal nuclei (red) (A), or Anti-GFAP to detect glia (red) (B). GPR88-Venus signal is observed (see white arrows) as colocalized with NeuN (yellow) in L1-6b of (A), but not with GFAP in (B). All images, scale bar is 200 μm. Annotations; Somatosensory cortex (SSCtx), corpus callosum (cc) and hippocampal formation (HPF).
Figure 6.
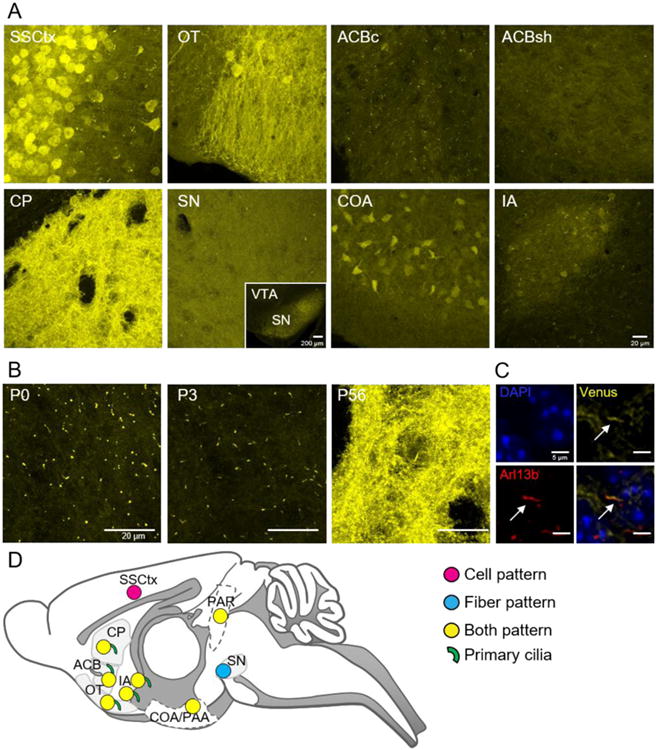
Mapping GPR88-Venus subcellular expression. (A) Confocal images show intrinsic GPR88-Venus, in the somatosensory cortex (SSCtx), was localized primarily to the soma, and was virtually undetectable at the level of processes or primary cilia (Supplementary Figure 6). The olfactory tubercle (OT) features GPR88-Venus expression throughout the neuron and neural processes. In the nucleus accumbens core (ACBc) GPR88-Venus was remarkably localized to primary cilia only, while a predominantly diffuse fiber-like pattern can be seen in the shell (ACBsh) with few primary cilia. In the caudate putamen (CP), GPR88-Venus is seen throughout the neuron, at the cell membrane and in neural processes (Supplementary Movie 1) as well as in primary cilia. The substantia nigra (SN), distinctively contains GPR88-Venus fibers perhaps demonstrating GPR88-Venus in terminals, with no detectable cellular or ciliary signal. The olfactory area, COA, exhibited well-defined soma expression and fibers without primary cilia localization. Finally, in the intercalated amygdala (IA), a cilia-restricted GPR88 expression pattern was observed with scant cellular and fiber localization (scale bar 20 μm, inset 200 μm). (B) In the CP, GPR88-Venus is observed essentially in primary cilia at P0 and P3, contrasting with the densely packed expression in other cellular compartments in the adult (P56) that masks the ciliary expression (scale bar 20 μm). (C) Adult brain section at the level of CP. Triple immunolabeling for cell nuclei (DAPI, blue), GPR88-Venus (yellow) and Arl13b (red), a marker of primary cilia, aids in discriminating GPR88-Venus localized in primary cilia from the dense receptor expression in cell bodies and fibers (scale bar 5 μm). (D) Overview map of GPR88-Venus subcellular pattern summarizes the distribution in cell bodies, fibers, both or primary cilia throughout brain regions.
A previous study reported that FLAG-GPR88, overexpressed in cultured striatal neurons, is targeted mainly to primary cilia (Marley et al. 2013), an antenna-like feature found on most cells including neurons (Chen et al. 2016). We report here for the first time that endogenous GPR88 is indeed localized in primary cilia of neurons, in a regional manner, at core of ACB, CP, and IA but not seen in areas such as SSCtx (Supplementary Figure 6). We next examined the primary cilia pattern of GPR88-Venus along development, and found that in CP, the fluorescent signal was restricted to primary cilia only at P0 and P3, contrasting with the broad cell/ fiber/cilia pattern seen in the adult (Figure 6B and Supplementary Movie 1-3), suggesting that a primary cilia-specific expression at early stages further extends to all neuronal compartments in the adult. Co-localization with the cilia marker Arl13b (Caspary et al. 2016) confirmed the subcellular expression of GPR88-Venus in the cilia compartment of adult CP neurons (Figure 6C). The subcellular distribution of GPR88-Venus across the brain is graphically summarized in Figure 6D. Altogether, the diverse subcellular pattern of GPR88-Venus demonstrates differential GPR88 targeting within neuronal compartments.
Mice lacking GPR88 show altered brain microstructure in the cortex and dorsal striatum
The high density of GPR88 in both striatum and cortex, and the evolving distribution pattern of GPR88 in the cortex during development, prompted us to probe microstructural integrity in the brain of mice lacking the Gpr88 gene. Our previous histological analysis already demonstrated altered spine densities and dendritic length in several brain areas of Gpr88-/- mice, suggesting that Gpr88 contributes to establish neuronal morphology (Meirsman et al. 2016a). Here we used non-invasive magnetic resonance imaging (Harsan et al. 2013) to compare brain microstructure structure in live Gpr88-/- mice and their controls. We performed parametric mapping of diffusion tensor indices quantifying the extent (mean diffusivity), directionality (axial and radial diffusivity indices) and anisotropy (fractional anisotropy – FA) of the water diffusion processes. Among these parameters, FA is generally accepted as a sensitive measure of microstructural modifications. We found abnormally high FA values (voxel-wise statistical group comparison between Gpr88-/- and Gpr88+/+ groups, statistical significance threshold P<0.05, family wise error - FWE corrected) in several brain areas of the Gpr88-/- mice (Figure 7). Most notable differences were observed at the level of SSCtx, and CP, corresponding to areas of richest GPR88-Venus density. Gpr88 gene activity therefore considerably contributes to shape the brain microstructure, with a prominent impact on the somatosensory cortex, and further analysis of functional connectivity confirms these findings (Arefin et al. In revision).
Figure 7.
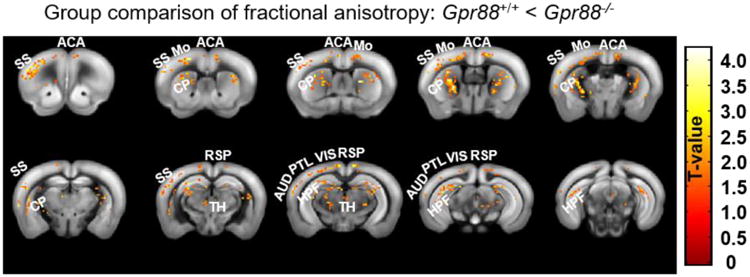
Structural Magnetic Resonance Imaging of live Gpr88-/- mice. The figure shows significant alterations of fractional anisotropy (FA) in mutant mice. Strong modification of FA was primarily in SSCtx and CP of Gpr88-/- mice compared to Gpr88+/+ mice. The panel shows statistical significance overlaid on T2-weighted anatomical brain slices with higher FA in mutants compared to controls. Corresponding T-value is shown on the right color scale (positive correlations from 0 to +4.5: red to yellow). Statistical significance was evaluated using two-sample t-test (P<0.05, FWE corrected). Abbreviations are as follows: somatosensory cortex (SS), anterior cingulate area (ACA), Motor cortex (Mo), caudate putamen (CP), retrosplenial cortex (RSP), thalamus (TH), auditory cortex (AUD), parietal cortex (PTL), visual cortex (VIS), hippocampal formation (HPF).
Mice lacking GPR88 show impaired sensory-task performance
Previous behavioral studies of Gpr88-/- mice have highlighted deficits mainly related to striatal function, and deficits in sensorimotor gating (Logue et al. 2009; Meirsman et al. 2017), however responses of these mice to sensory stimuli have not been examined otherwise. We therefore designed a test battery to challenge the integration of olfactory, somatosensory and visual sensory responses, all related to the remarkable receptor expression pattern in the cortex (Figure 8A).
Figure 8.
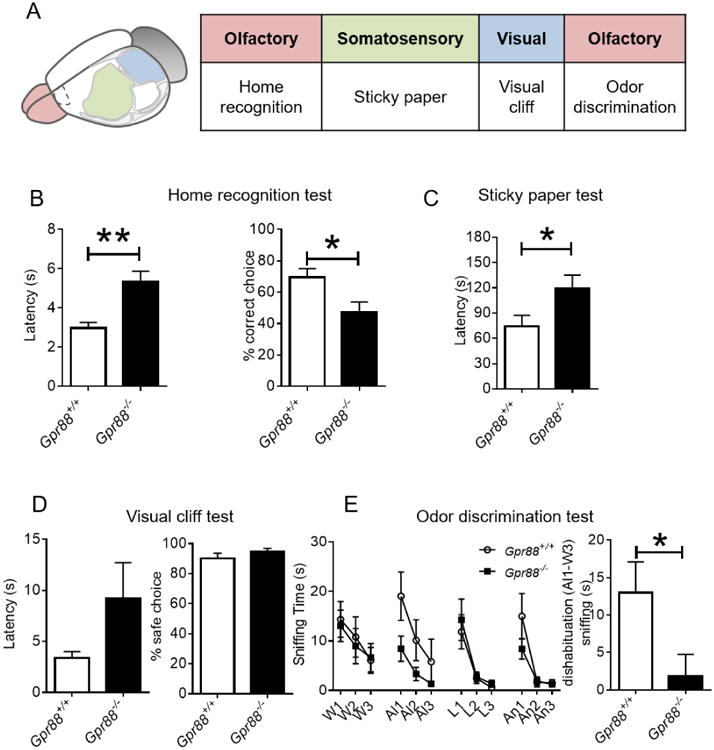
Gpr88-/- mice demonstrate impaired performance in multisensorimotor tasks. (A) Sensory behavior scheme (B) In the home recognition test Gpr88-/- mice performed lower in home recognition and took significantly longer time to decide which compartment to enter. (C) Somatosensory function is impaired in Gpr88-/- mice, which measured longer latency to notice adhesive on hind-paw compared to Gpr88+/+ mice in the sticky paper test. (D) The visual cliff test demonstrates Gpr88-/- mice have normal vision with a tendency of longer latency to move to the safe choice. (E) Odor discrimination task, Gpr88-/- mice perform similar to Gpr88+/+ in habituation to new odors but display significant difference in dishabituation to odors. Gpr88+/+ (n=9), Gpr88-/- (n=9). Significance is denoted as * if P<0.05 or ** if P<0.01 for genotype unpaired t-test, complete analysis see Supplementary Table I.
We first evaluated olfactory processing with a home recognition test (Metz and Schwab 2004). To do so, Gpr88+/+ and Gpr88-/- mice were placed in a central compartment with equal access to two identical compartments containing either home cage or neutral litter. Gpr88-/- mice had a longer latency to the first entry of any compartment (Figure 8B; t(16) =3.717, P<0.01) and fewer correct choices (t(16) =2.596, P<0.05) in comparison to Gpr88+/+. These data suggest Gpr88 mediates some aspects of olfactory processing. Second, we evaluated the contribution of Gpr88 in somatosensory processing with the sticky paper test (Metz and Schwab 2004; Liguz-Lecznar et al. 2014). Gpr88-/- mice showed a significantly higher latency to try and remove the tape (Figure 8C; t(16) =2.171, P<0.05). Furthermore, two Gpr88-/- mice persistently reached the 180 seconds' cut-off without removing the tape in all trials whereas all Gpr88+/+ mice succeeded in removing the tape in at least one trial. This result indicates a role for Gpr88 in tactile sensation.
We next examined the role of Gpr88 in depth perception and visual information processing by submitting mice to the visual cliff test (Fox 1965). In this test, Gpr88-/- mice had a trend to take longer time to move-off the ridge (Figure 8D; t(16) =1.616, P=0.13) in comparison to Gpr88+/+ mice. The safe choice was selected for both groups at a similar rate. These results suggest that Gpr88 does not contribute to visual acuity but is perhaps implicated in visual information processing.
The home recognition test suggested modified olfactory perception or processing in Gpr88-/- mice, therefore we next determined whether Gpr88-/- mice are able to detect and differentiate odors with the odor discrimination test (Arbuckle et al. 2015; Yang and Crawley 2009). There was no significant genotype effect, suggesting that both groups were able to discriminate novel odors (Figure 8E; odor effect two-way ANOVA F (11, 176) = 8.072, P<0.001). However, the ability for mice to dishabituate from water to almond was significantly reduced for Gpr88-/- as compared to Gpr88+/+ mice (Figure 8E; t(16) =2.214, P<0.05). This impairment was not observed with ensuing odor dishabituation, suggesting that Gpr88 deficiency likely does not contribute directly to olfactory sensation but rather may impact the processing of odor information.
Together the data suggest that primary sensory perception (olfaction and vision) is intact, but the loss of Gpr88 contributes to delayed behavioral responses that require sensory processing.
Discussion
GPR88 is an orphan GPCR with strongly conserved mRNA expression in human and mouse striatum, making this receptor an obvious target for intervention in striatal dysfunction-related disorders. However, to allow better positioning of GPR88 as a target for neuropsychiatric disorders, and facilitate drug design and development, a comprehensive knowledge of GPR88 protein distribution in the brain is necessary. To address this gap, we extended our knock-in fluorescent receptor approach (Erbs et al. 2015; Pradhan et al. 2009; Scherrer et al. 2006) by the creation of Gpr88-Venus mice. Mutant animals show physiological receptor mRNA levels, native receptor signaling activity and an overall fluorescence distribution consistent with previous reports (Massart et al. 2009; Massart et al. 2016). Protein expression is concordant with mRNA transcript levels reported in both rodent and human studies (Becker et al. 2008; Befort et al. 2008; Conti et al. 2007; Ghate et al. 2007; Hodges et al. 2006; Logue et al. 2009; Mizushima et al. 2000; Van Waes et al. 2011; Vassilatis et al. 2003; Quintana et al. 2012) suggesting that GPR88 is mainly localized at its mRNA expression sites. Our extensive mapping of GPR88-Venus protein in the sensory cortex layer 4 prompted us to examine Gpr88-/- mice for brain microstructure and behaviors relevant to sensory processing. Our data show altered connection patterns in both striatal and somatosensory cortical areas of Gpr88-/- mice, and delayed responses in sensory processing tasks. Thus, beyond its known role in the regulation of striatal function, GPR88 is a key player in the integration of sensory information.
Methods to study distribution of GPCR proteins are scarce. With our approach (Scherrer et al. 2006; Erbs et al. 2015), we have an advantage to examine receptor distribution at cellular and subcellular levels (Erbs et al. 2014; Pradhan et al. 2011; Gardon et al. 2014; Rezai et al. 2012; Scherrer et al. 2009), providing a novel and comprehensive view of receptor expression throughout brain cells and networks (Pradhan et al. 2009; Faget et al. 2012). Massart et al., engineered a polyclonal antibody to examine GPR88 expression across development and in the adult rat (Massart et al. 2009; Massart et al. 2016). In the striatum of E21 rats, Massart et al., reported enrichment of GPR88 in patch compartments (Massart et al. 2016). We also found this pattern of GPR88 at mouse P0 and P3 and observed a loss of this distinction in the adult as GPR88 density increases. Neurons in the patch compartment appear earlier than the matrix (Fishell and van der Kooy 1989; Song and Harlan 1994), therefore our data suggests that GPR88 is expressed in both early neurons and later migrating neurons that form the striatal matrix. Cortical GPR88 was described as transitioning from high expression in the cortical plate to L5 and L6 at the time of birth (Massart et al. 2016), and finally high in L2-4 of the adult rat brain. We find GPR88-Venus only in L1 and L6b at P0, emerging in L4 while still evident in L1 and L6b at P3, and finally prominent in L4, low in L1-6b in the adult mouse. Differences in observation of GPR88 expression during cortical development may be due to species difference or distinct detection sensitivity. Altogether however, both studies show a dynamic GPR88 expression pattern, suggesting a potential role for GPR88 in cortical migration and lamination with likely lasting effects into adulthood. This hypothesis is concordant with our MRI-based analysis showing significant microstructural reshaping of cortical connectivity in the brain of adult Gpr88-/-. In the future, temporally-controlled genetic inactivation of Gpr88 will be required to dissociate developmental from tonic roles of GPR88.
Our approach uniquely reveals variable cell/fiber/primary cilia localization across the brain. Intriguingly, we observed GPR88-Venus fluorescence in primary cilia. A previous report showed that overexpressed GPR88 in primary striatal neurons is mainly targeted to the primary cilia (Marley et al. 2013). Here, for the first time, we demonstrate that endogenous GPR88 is indeed transported to this compartment, and this phenomenon is observable during development, as well as in several brain regions. Other GPCRs show expression enriched to primary cilia (Berbari et al. 2008; Badgandi et al. 2017), where signaling activity of the receptor may differ from somatic signaling (Marley et al. 2013). Overall, the biological significance of primary cilia localization remains an open question in GPCR research, but a role in neuronal cell fate and migration during development as well the dendritic refinement and synaptic integration of adult-born neurons has been suggested (Guemez-Gamboa et al. 2014; Kumamoto et al. 2012). GPR88 may serve as a model receptor to investigate implications of cilia-specific signaling in brain structure, development and function.
Our study uncovers a novel role of GPR88 in multisensory processing. Our data suggest that tactile, visual and olfactory perception is intact however, behavioral responses in multisensory processing tasks are slower, (delay in removing sticky paper, delayed time to choose home and trend to delay moving to the safe side in visual cliff). Thus, sensory deficits were observable when sensory information requires filtration or “gating” for subsequent behavioral outcome, and we propose that GPR88 plays a role at the level of circuits involved in the integration and processing of sensory information, downstream of primary perception as our own work detects no modification of both acoustic or visual startle response in Gpr88-/- mice (Meirsman et al. 2017). Furthermore, this conclusion is consistent with the observation that Gpr88-/- mice show intact startle response but impaired inhibition of the startle response (Logue et al. 2009), implicating modification of sensory gating rather than perception. Importantly, the major role of GPR88 in somatosensory processing evidenced in this study, may influence all the behavioral phenotypes of Gpr88-/- mice that have been reported to date, including prepulse inhibition (Logue et al. 2009), activity responses (hyperactivity, impaired motor coordination), increased risk taking and impaired cue-based learning which have all been ascribed to a lack of Gpr88 regulated actions on MSNs in the striatum (Logue et al. 2009; Meirsman et al. 2016a; Meirsman et al. 2016b; Quintana et al. 2012).
GPR88-regulated sensory processing may operate through several mechanisms. First, GPR88 activity may contribute to neural wiring during development. This is supported by our observation with MRI, of microstructural changes in cortical and striatal areas, which likely impacts connectivity and may impinge on corticostriatal circuit function. Second, tonic GPR88 activity in the adult may regulate sensory processing at both striatal and cortical levels, as previous reports showed that striatal neurons do respond to multimodal sensory inputs (Brown et al. 1996; Nagy et al. 2005; Schulz et al. 2009; Wilson et al. 1983), lesions in cortical regions and their corresponding striatal areas produce a similar effect (Divac et al. 1967). Interestingly, recordings of striatal neurons in recipient zones of sensory, or visual cortex identified by axonal tracing, provided strong evidence for a cortical origin of striatal sensory responses (Reig and Silberberg 2014; Wilson 2014). Third, impaired sensory processing in Gpr88-/- mice may arise from dysfunction of cortico-thalamic loops that are essential for gating of sensory input (Feldmeyer 2012; Zembrzycki et al. 2013; Niell 2015). GPR88 is prominently expressed in glutamatergic barrels of L4 and sporadically other cortical layers that are belonging to thalamic-cortical circuitry (Feldmeyer 2012; Maier et al. 1999). The importance of L4 in barrel cortex function and associated sensorimotor behavior (Ferezou et al. 2007; Manita et al. 2015; Schneider et al. 2014; Zhang et al. 2014; Guo et al. 2014), and L6 projections to the thalamus are known to filter and feedback sensory inputs to the cortex (Crandall et al. 2015; Cudeiro and Sillito 2006). GPR88 acting at these terminals may reduce thalamic inputs to the cortex contributing to filter sensory information. Ultimately, circuit-targeted studies will be necessary to determine the exact site(s) of GPR88-mediated sensory processing.
The integration of sensory information is disrupted in many psychiatric diseases (Powell et al. 2012). The ability to discern critical sensory information from unimportant sensory stimuli is necessary for survival, and dysfunctional sensory gating is a hallmark in schizophrenia (Braff et al. 2001; Swerdlow et al. 2008), and one feature of bipolar disease (Perry et al. 2001), Huntington's disease (Swerdlow et al. 1995) and panic disorder (Ludewig et al. 2002). This study reinforces the notion that GPR88 is a promising target to treat a number of mental disorders, perhaps also via activity on sensory processing networks.
Supplementary Material
Supplementary Figure 1. Profiling GPR88-mediated G protein activation. (A-B) G protein activation profile of GPR88 (left) versus GPR88-Venus (right) following 10-minute stimulation with 31.6 μM Compound 19 (A) or 2-PCCA (B). HEK-293 cells were co-transfected with GPR88 or GPR88-Venus, Gβ1, Gγ3-RlucII (donor BRET2 sensor), GRK2-GFP10 (acceptor BRET2 sensor) and either of the indicated human (h) (grey) or mouse (m) (red) Gα subunits. Mock is a condition where cells were not co-transfected with any Gα subunit and shows GPR88-mediated activation of endogenous G proteins. Data are expressed as mean ± SEM, *P<0.05, ** P <0.01, *** P<0.001 vs Mock condition, 3-4 replicate experiments.
Supplementary Figure 2. (A-B) Immunostaining of 30 μm sections from Gpr88-Venus (A) Intrinsic (i) GPR88-Venus signal is shown in yellow and (B) amplified (a) GPR88-Venus is displayed in red. (Scale bar is 1 mm and 200 μm for insets).
Supplementary Figure 3. Gpr88-Venus is enriched in the mouse barrel cortex. Perfused brains of adult Gpr88-Venus mice were sliced at 30 μm on a cryostat tangentially. Gpr88-Venus was amplified (a) in red and the intrinsic signal (i) can be seen in yellow, DAPI was used to stain nuclei. Annotated barrels are indicated on the merge image (Maier et al. 1999; Strominger and Woolsey 1987; Petersen 2007) Medial (M), rostral (R). All images, scale bar is 200um.
Supplementary Figure 4. Representative images for fluorescence scale used to fine-map GPR88-Venus expression: 0, not detectable; 1, weak expression; 2, moderate expression; 3, dense.
Supplementary Figure 5. Expanded cortical GPR88-Venus expression shows the discrete pattern of sensory cortical GPR88 in the layer 4. In this panel 30 μm sections with ∼300μm separation were stained with Anti-Venus to amplify the Venus signal in red and imaged with an Olympus slide scanner from approximately Bregma 1.94 mm to -5.52 mm. Scale bar is 1 mm.
Supplementary Figure 6. GPR88-Venus localizes to the soma in somatosensory neurons. Confocal microscopy Z-stack projection image shown here to demonstrate GPR88-Venus in the somatosensory cortex L4 is not prominently localized to primary cilia. GPR88-Venus was amplified with an antibody shown here in yellow and the primary cilia marker, Arl13b, can be seen in red, DAPI was used to stain nuclei. White arrows indicate primary cilia observable at this focal plane. All images, scale bar is 5 μm.
Supplementary Movies 1-3. GPR88-Venus Confocal Z-stack movies of images of the CP in P56 (Movie 1), P0 (Movie 2) and P3 (Movie 3). 30 μm tissue sections imaged in Venus channel (intrinsic) 60× oil immersion objective with 0.4 μm step size.
Supplementary Table for Figure 1A, Human GPR88 mRNA: One-Way ANOVA (P value <0.0001, ****)
Supplementary Table for Figure 1B, Mouse Gpr88 mRNA: One-Way ANOVA (P value <0.0001, ****)
Supplementary Table for Figure 2D (top), Mouse Gpr88 mRNA One-Way ANOVA
Supplementary Table for Figure 2D (bottom), Mouse Venus mRNA One-Way ANOVA
Supplementary Table for Figure 2E, [35S]-GTPγS Assay: (Gpr88+/+ vs. Gpr88-/-), Two-Way ANOVA
Supplementary Table for Figure 2E: (Gpr88+/+ vs. Gpr88-/-), Bonferroni's multiple comparisons test
Supplementary Table for Figure 2E, [35S]-GTPγS Assay: (Gpr88+/+ vs. Gpr88Venus/+ vs. Gpr88Venus/Venus), Two-Way ANOVA
Supplementary Table for Figure 8. Sensory Behavior Tests: (Gpr88+/+ vs. Gpr88-/-)
Acknowledgments
Financial Disclosure: This work was supported by CQDM/Region Alsace/EU to BLK & MB, the US National Institute of Health (National Institute of Drug Abuse Grant No. 05010 to BLK and National Institute on Alcohol Abuse and Alcoholism, Grant No. 16658 to BLK), the Canada Fund for Innovation and the Canada Research Chairs to BLK and MB, and the Canadian Institute for Health Research (Grant no. MOP-10501 to MB).
We acknowledge Josée Prud'homme for preparation of human tissue specimens and the Douglas – Bell Canada Brain Bank that is supported by the Quebec Suicide Research Network of the Fonds de Recherche du Québec - Santé (FRQS) and by the Douglas Institute Foundation. The present study used the services of the Molecular and Cellular Microscopy Platform at the Douglas Hospital and Research Center. We would like to thank Monique Lagace for helpful discussions on the use of BRET biosensors. We also thank the mouse clinic institute (Illkirch, France) for mouse generation. We thank Aude Villemain, Eujin Kim and Aimee Lee Luco for animal care and genotyping. We thank the staff at the animal facility of the Neurophenotyping Center Douglas Hospital Research Center for the housing and maintenance of the animals.
Footnotes
Conception and design of experiments were performed by ATE, JB, MS, LH, BLK, MB. Acquisition of data were performed by ATE, MS, JB, SW, TMA, CC, FG. Analysis of data were contributed by ATE, MS, FG, JB, TMA and ED. Design of tools or reagents were performed by CL, MH and VL. The original manuscript was written by ATE, ED and BLK. Revising and editing of the manuscript was done by ATE, BLK, ED, MS, TMA, LH, MB.
Conflict of interest: The authors report no potential conflict of interest.
References
- Alkufri F, Shaag A, Abu-Libdeh B, Elpeleg O. Deleterious mutation in GPR88 is associated with chorea, speech delay, and learning disabilities. Neurol Genet. 2016;2(3):e64. doi: 10.1212/NXG.0000000000000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle EP, Smith GD, Gomez MC, Lugo JN. Testing for odor discrimination and habituation in mice. J Vis Exp. 2015;(99):e52615. doi: 10.3791/52615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefin TM, Mechling AE, Meirsman C, Bienert T, Hubner N, Lee H, Ben-Hamida S, Ehrlich AT, Roquet D, Hennig J, Elverfeldt DV, Kieffer BL, Harsan L. Remodeling of Sensorimotor Brain Connectivity in Gpr88 deficient mice. Brain Connectivity. doi: 10.1089/brain.2017.0486. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45(2):207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Badgandi HB, Hwang SH, Shimada IS, Loriot E, Mukhopadhyay S. Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J Cell Biol. 2017;216(3):743–760. doi: 10.1083/jcb.201607095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JA, Befort K, Blad C, Filliol D, Ghate A, Dembele D, Thibault C, Koch M, Muller J, Lardenois A, Poch O, Kieffer BL. Transcriptome analysis identifies genes with enriched expression in the mouse central extended amygdala. Neuroscience. 2008;156(4):950–965. doi: 10.1016/j.neuroscience.2008.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Befort K, Filliol D, Ghate A, Darcq E, Matifas A, Muller J, Lardenois A, Thibault C, Dembele D, Le Merrer J, Becker JA, Poch O, Kieffer BL. Mu-opioid receptor activation induces transcriptional plasticity in the central extended amygdala. Eur J Neurosci. 2008;27(11):2973–2984. doi: 10.1111/j.1460-9568.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19(4):1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, P, NJ, US, Dzierba Carolyn Diane, Middletown, CT, US, Bronson Joanne J, Durham, CT, US, Fink Cynthia, Lebanon, NJ, US, Green Michael, Easton, PA, US, Kimball David, East Windsor, NJ, US, Macor John E, Gullford, CT, US, Kwon Soojin, Zhang Yulian, Zipp Greg. Modulators of G protein-coupled receptor 88. United States Patent 2013
- Braff DL, Geyer MA, Light GA, Sprock J, Perry W, Cadenhead KS, Swerdlow NR. Impact of prepulse characteristics on the detection of sensorimotor gating deficits in schizophrenia. Schizophr Res. 2001;49(1-2):171–178. doi: 10.1016/s0920-9964(00)00139-0. [DOI] [PubMed] [Google Scholar]
- Brandish PE, Su M, Holder DJ, Hodor P, Szumiloski J, Kleinhanz RR, Forbes JE, McWhorter ME, Duenwald SJ, Parrish ML, Na S, Liu Y, Phillips RL, Renger JJ, Sankaranarayanan S, Simon AJ, Scolnick EM. Regulation of gene expression by lithium and depletion of inositol in slices of adult rat cortex. Neuron. 2005;45(6):861–872. doi: 10.1016/j.neuron.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Brown LL, Hand PJ, Divac I. Representation of a single vibrissa in the rat neostriatum: peaks of energy metabolism reveal a distributed functional module. Neuroscience. 1996;75(3):717–728. doi: 10.1016/0306-4522(96)00310-7. [DOI] [PubMed] [Google Scholar]
- Caspary T, Marazziti D, Berbari NF. Methods for Visualization of Neuronal Cilia. Methods Mol Biol. 2016;1454:203–214. doi: 10.1007/978-1-4939-3789-9_13. [DOI] [PubMed] [Google Scholar]
- Chen X, Luo J, Leng Y, Yang Y, Zweifel LS, Palmiter RD, Storm DR. Ablation of Type III Adenylyl Cyclase in Mice Causes Reduced Neuronal Activity, Altered Sleep Pattern, and Depression-like Phenotypes. Biol Psychiatry. 2016;80(11):836–848. doi: 10.1016/j.biopsych.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti B, Maier R, Barr AM, Morale MC, Lu X, Sanna PP, Bilbe G, Hoyer D, Bartfai T. Region-specific transcriptional changes following the three antidepressant treatments electro convulsive therapy, sleep deprivation and fluoxetine. Mol Psychiatry. 2007;12(2):167–189. doi: 10.1038/sj.mp.4001897. [DOI] [PubMed] [Google Scholar]
- Crandall SR, Cruikshank SJ, Connors BW. A corticothalamic switch: controlling the thalamus with dynamic synapses. Neuron. 2015;86(3):768–782. doi: 10.1016/j.neuron.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudeiro J, Sillito AM. Looking back: corticothalamic feedback and early visual processing. Trends Neurosci. 2006;29(6):298–306. doi: 10.1016/j.tins.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Del Zompo M, Deleuze JF, Chillotti C, Cousin E, Niehaus D, Ebstein RP, Ardau R, Mace S, Warnich L, Mujahed M, Severino G, Dib C, Jordaan E, Murad I, Soubigou S, Koen L, Bannoura I, Rocher C, Laurent C, Derock M, Faucon Biguet N, Mallet J, Meloni R. Association study in three different populations between the GPR88 gene and major psychoses. Mol Genet Genomic Med. 2014;2(2):152–159. doi: 10.1002/mgg3.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divac I, Rosvold HE, Szwarcbart MK. Behavioral effects of selective ablation of the caudate nucleus. J Comp Physiol Psychol. 1967;63(2):184–190. doi: 10.1037/h0024348. [DOI] [PubMed] [Google Scholar]
- Dzierba CD, Bi Y, Dasgupta B, Hartz RA, Ahuja V, Cianchetta G, Kumi G, Dong L, Aleem S, Fink C, Garcia Y, Green M, Han J, Kwon S, Qiao Y, Wang J, Zhang Y, Liu Y, Zipp G, Liang Z, Burford N, Ferrante M, Bertekap R, Lewis M, Cacace A, Grace J, Wilson A, Nouraldeen A, Westphal R, Kimball D, Carson K, Bronson JJ, Macor JE. Design, synthesis, and evaluation of phenylglycinols and phenyl amines asagonists of GPR88. Bioorg Med Chem Lett. 2015;25(7):1448–1452. doi: 10.1016/j.bmcl.2015.01.036. [DOI] [PubMed] [Google Scholar]
- Erbs E, Faget L, Scherrer G, Matifas A, Filliol D, Vonesch JL, Koch M, Kessler P, Hentsch D, Birling MC, Koutsourakis M, Vasseur L, Veinante P, Kieffer BL, Massotte D. A mu-delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct Funct. 2015;220(2):677–702. doi: 10.1007/s00429-014-0717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs E, Faget L, Veinante P, Kieffer BL, Massotte D. In vivo neuronal co-expression of mu and delta opioid receptors uncovers new therapeutic perspectives. Receptors Clin Investig. 2014;1(5) doi: 10.14800/rci.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget L, Erbs E, Le Merrer J, Scherrer G, Matifas A, Benturquia N, Noble F, Decossas M, Koch M, Kessler P, Vonesch JL, Schwab Y, Kieffer BL, Massotte D. In vivo visualization of delta opioid receptors upon physiological activation uncovers a distinct internalization profile. J Neurosci. 2012;32(21):7301–7310. doi: 10.1523/JNEUROSCI.0185-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D. Excitatory neuronal connectivity in the barrel cortex. Front Neuroanat. 2012;6:24. doi: 10.3389/fnana.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferezou I, Haiss F, Gentet LJ, Aronoff R, Weber B, Petersen CC. Spatiotemporal dynamics of cortical sensorimotor integration in behaving mice. Neuron. 2007;56(5):907–923. doi: 10.1016/j.neuron.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Fishell G, van der Kooy D. Pattern formation in the striatum: developmental changes in the distribution of striatonigral projections. Brain Res Dev Brain Res. 1989;45(2):239–255. doi: 10.1016/0165-3806(89)90042-4. [DOI] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27(2-3):126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fox MW. The visual cliff test for the study of visual depth perception in the mouse. Anim Behav. 1965;13(2):232–233. doi: 10.1016/0003-3472(65)90040-0. [DOI] [PubMed] [Google Scholar]
- Gardon O, Faget L, Chu Sin Chung P, Matifas A, Massotte D, Kieffer BL. Expression of mu opioid receptor in dorsal diencephalic conduction system: newinsights for the medial habenula. Neuroscience. 2014;277:595–609. doi: 10.1016/j.neuroscience.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghate A, Befort K, Becker JA, Filliol D, Bole-Feysot C, Demebele D, Jost B, Koch M, Kieffer BL. Identification of novel striatal genes by expression profiling in adult mouse brain. Neuroscience. 2007;146(3):1182–1192. doi: 10.1016/j.neuroscience.2007.02.040. [DOI] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Coufal NG, Gleeson JG. Primary cilia in the developing and mature brain. Neuron. 2014;82(3):511–521. doi: 10.1016/j.neuron.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZV, Li N, Huber D, Ophir E, Gutnisky D, Ting JT, Feng G, Svoboda K. Flow of cortical activity underlying a tactile decision in mice. Neuron. 2014;81(1):179–194. doi: 10.1016/j.neuron.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsan LA, David C, Reisert M, Schnell S, Hennig J, von Elverfeldt D, Staiger JF. Mapping remodeling of thalamocortical projections in the living reeler mouse brain by diffusion tractography. Proc Natl Acad Sci U S A. 2013;110(19):E1797–1806. doi: 10.1073/pnas.1218330110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L, Luthi-Carter R. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Ingallinesi M, Le Bouil L, Biguet NF, Thi AD, Mannoury la Cour C, Millan MJ, Ravassard P, Mallet J, Meloni R. Local inactivation of Gpr88 in the nucleus accumbens attenuates behavioral deficits elicited by the neonatal administration of phencyclidine in rats. Mol Psychiatry. 2015;20(8):951–958. doi: 10.1038/mp.2014.92. [DOI] [PubMed] [Google Scholar]
- Jin C, Decker AM, Huang XP, Gilmour BP, Blough BE, Roth BL, Hu Y, Gill JB, Zhang XP. Synthesis, pharmacological characterization, and structure-activity relationship studies of small molecular agonists for the orphan GPR88 receptor. ACS Chem Neurosci. 2014;5(7):576–587. doi: 10.1021/cn500082p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Decker AM, Langston TL. Design, synthesis and pharmacological evaluation of 4-hydroxyphenylglycine and 4-hydroxyphenylglycinol derivatives as GPR88 agonists. Bioorg Med Chem. 2017;25(2):805–812. doi: 10.1016/j.bmc.2016.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto N, Gu Y, Wang J, Janoschka S, Takemaru K, Levine J, Ge S. A role for primary cilia in glutamatergic synaptic integration of adult-born neurons. Nat Neurosci. 2012;15(3):399–405. S391. doi: 10.1038/nn.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J, Befort K, Gardon O, Filliol D, Darcq E, Dembele D, Becker JA, Kieffer BL. Protracted abstinence from distinct drugs of abuse shows regulation of a common gene network. Addict Biol. 2012;17(1):1–12. doi: 10.1111/j.1369-1600.2011.00365.x. [DOI] [PubMed] [Google Scholar]
- Li JX, Thorn DA, Jin C. The GPR88 receptor agonist 2-PCCA does not alter the behavioral effects of methamphetamine in rats. Eur J Pharmacol. 2013;698(1-3):272–277. doi: 10.1016/j.ejphar.2012.10.037. [DOI] [PubMed] [Google Scholar]
- Liguz-Lecznar M, Zakrzewska R, Daniszewska K, Kossut M. Functional assessment of sensory functions after photothrombotic stroke in the barrel field of mice. Behav Brain Res. 2014;261:202–209. doi: 10.1016/j.bbr.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Logue SF, Grauer SM, Paulsen J, Graf R, Taylor N, Sung MA, Zhang L, Hughes Z, Pulito VL, Liu F, Rosenzweig-Lipson S, Brandon NJ, Marquis KL, Bates B, Pausch M. The orphan GPCR, GPR88, modulates function of the striatal dopamine system: a possible therapeutic target for psychiatric disorders? Mol Cell Neurosci. 2009;42(4):438–447. doi: 10.1016/j.mcn.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Ludewig K, Geyer MA, Etzensberger M, Vollenweider FX. Stability of the acoustic startle reflex, prepulse inhibition, and habituation in schizophrenia. Schizophr Res. 2002;55(1-2):129–137. doi: 10.1016/s0920-9964(01)00198-0. [DOI] [PubMed] [Google Scholar]
- Mai JK, Voss T, Paxinos G. Atlas of the human brain. 3rd. Elsevier/Academic Press; Amsterdam ; Boston: 2008. [Google Scholar]
- Maier DL, Mani S, Donovan SL, Soppet D, Tessarollo L, McCasland JS, Meiri KF. Disrupted cortical map and absence of cortical barrels in growth-associated protein (GAP)-43 knockout mice. Proc Natl Acad Sci U S A. 1999;96(16):9397–9402. doi: 10.1073/pnas.96.16.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manita S, Suzuki T, Homma C, Matsumoto T, Odagawa M, Yamada K, Ota K, Matsubara C, Inutsuka A, Sato M, Ohkura M, Yamanaka A, Yanagawa Y, Nakai J, Hayashi Y, Larkum ME, Murayama M. A Top-Down Cortical Circuit for Accurate Sensory Perception. Neuron. 2015;86(5):1304–1316. doi: 10.1016/j.neuron.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Marley A, Choy RW, von Zastrow M. GPR88 reveals a discrete function of primary cilia as selective insulators of GPCR cross-talk. PLoS One. 2013;8(8):e70857. doi: 10.1371/journal.pone.0070857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R, Guilloux JP, Mignon V, Sokoloff P, Diaz J. Striatal GPR88 expression is confined to the whole projection neuron population and is regulated by dopaminergic and glutamatergic afferents. Eur J Neurosci. 2009;30(3):397–414. doi: 10.1111/j.1460-9568.2009.06842.x. [DOI] [PubMed] [Google Scholar]
- Massart R, Mignon V, Stanic J, Munoz-Tello P, Becker JA, Kieffer BL, Darmon M, Sokoloff P, Diaz J. Developmental and adult expression patterns of the G-protein-coupled receptor GPR88 in the rat: Establishment of a dual nuclear-cytoplasmic localization. J Comp Neurol. 2016;524(14):2776–2802. doi: 10.1002/cne.23991. [DOI] [PubMed] [Google Scholar]
- Masuho I, Ostrovskaya O, Kramer GM, Jones CD, Xie K, Martemyanov KA. Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci Signal. 2015;8(405):ra123. doi: 10.1126/scisignal.aab4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirsman AC, de Kerchove d'Exaerde A, Kieffer BL, Ouagazzal AM. GPR88 in A2A receptor-expressing neurons modulates locomotor response to dopamine agonists but not sensorimotor gating. Eur J Neurosci. 2017;46(4):2026–2034. doi: 10.1111/ejn.13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirsman AC, Le Merrer J, Pellissier LP, Diaz J, Clesse D, Kieffer BL, Becker JA. Mice Lacking GPR88 Show Motor Deficit, Improved Spatial Learning, and Low Anxiety Reversed by Delta Opioid Antagonist. Biol Psychiatry. 2016a;79(11):917–927. doi: 10.1016/j.biopsych.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirsman AC, Robe A, de Kerchove d'Exaerde A, Kieffer BL. GPR88 in A2AR Neurons Enhances Anxiety-Like Behaviors. eNeuro. 2016b;3(4) doi: 10.1523/ENEURO.0202-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz GA, Schwab ME. Behavioral characterization in a comprehensive mouse test battery reveals motor and sensory impairments in growth-associated protein-43 null mutant mice. Neuroscience. 2004;129(3):563–574. doi: 10.1016/j.neuroscience.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Mizushima K, Miyamoto Y, Tsukahara F, Hirai M, Sakaki Y, Ito T. A novel G-protein-coupled receptor gene expressed in striatum. Genomics. 2000;69(3):314–321. doi: 10.1006/geno.2000.6340. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nagy A, Paroczy Z, Norita M, Benedek G. Multisensory responses and receptive field properties of neurons in the substantia nigra and in the caudate nucleus. Eur J Neurosci. 2005;22(2):419–424. doi: 10.1111/j.1460-9568.2005.04211.x. [DOI] [PubMed] [Google Scholar]
- Niell CM. Cell types, circuits, and receptive fields in the mouse visual cortex. Annu Rev Neurosci. 2015;38:413–431. doi: 10.1146/annurev-neuro-071714-033807. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upperlayers II-IV of the cerebral cortex. J Comp Neurol. 2004;479(2):168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, Kuczenski R, Niculescu AB. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9(11):1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Perry W, Minassian A, Feifel D, Braff DL. Sensorimotor gating deficits in bipolar disorder patients with acute psychotic mania. Biol Psychiatry. 2001;50(6):418–424. doi: 10.1016/s0006-3223(01)01184-2. [DOI] [PubMed] [Google Scholar]
- Powell SB, Weber M, Geyer MA. Genetic models of sensorimotor gating: relevance to neuropsychiatric disorders. Curr Top Behav Neurosci. 2012;12:251–318. doi: 10.1007/7854_2011_195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan AA, Becker JA, Scherrer G, Tryoen-Toth P, Filliol D, Matifas A, Massotte D, Gaveriaux-Ruff C, Kieffer BL. In vivo delta opioid receptor internalization controlsbehavioral effects of agonists. PLoS One. 2009;4(5):e5425. doi: 10.1371/journal.pone.0005425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan AA, Befort K, Nozaki C, Gaveriaux-Ruff C, Kieffer BL. The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol Sci. 2011;32(10):581–590. doi: 10.1016/j.tips.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan AA, Walwyn W, Nozaki C, Filliol D, Erbs E, Matifas A, Evans C, Kieffer BL. Ligand-directed trafficking of the delta-opioid receptor in vivo: two paths toward analgesic tolerance. J Neurosci. 2010;30(49):16459–16468. doi: 10.1523/JNEUROSCI.3748-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana A, Sanz E, Wang W, Storey GP, Guler AD, Wanat MJ, Roller BA, La Torre A, Amieux PS, McKnight GS, Bamford NS, Palmiter RD. Lack of GPR88 enhances medium spiny neuron activity and alters motor- and cue-dependent behaviors. Nat Neurosci. 2012;15(11):1547–1555. doi: 10.1038/nn.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig R, Silberberg G. Multisensory integration in the mouse striatum. Neuron. 2014;83(5):1200–1212. doi: 10.1016/j.neuron.2014.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezai X, Faget L, Bednarek E, Schwab Y, Kieffer BL, Massotte D. Mouse delta opioid receptors are located on presynaptic afferents to hippocampal pyramidal cells. Cell Mol Neurobiol. 2012;32(4):509–516. doi: 10.1007/s10571-011-9791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallert T, Whishaw IQ. Bilateral cutaneous stimulation of the somatosensory system in hemidecorticate rats. Behav Neurosci. 1984;98(3):518–540. doi: 10.1037//0735-7044.98.3.518. [DOI] [PubMed] [Google Scholar]
- Scherrer G, Imamachi N, Cao YQ, Contet C, Mennicken F, O'Donnell D, Kieffer BL, Basbaum AI. Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell. 2009;137(6):1148–1159. doi: 10.1016/j.cell.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer G, Tryoen-Toth P, Filliol D, Matifas A, Laustriat D, Cao YQ, Basbaum AI, Dierich A, Vonesh JL, Gaveriaux-Ruff C, Kieffer BL. Knockin mice expressing fluorescent delta-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc Natl Acad Sci U S A. 2006;103(25):9691–9696. doi: 10.1073/pnas.0603359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DM, Nelson A, Mooney R. A synaptic and circuit basis for corollary discharge in the auditory cortex. Nature. 2014;513(7517):189–194. doi: 10.1038/nature13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JM, Redgrave P, Mehring C, Aertsen A, Clements KM, Wickens JR, Reynolds JN. Short-latency activation of striatal spiny neurons via subcortical visual pathways. J Neurosci. 2009;29(19):6336–6347. doi: 10.1523/JNEUROSCI.4815-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song DD, Harlan RE. Genesis and migration patterns of neurons forming the patch and matrix compartments of the rat striatum. Brain Res Dev Brain Res. 1994;83(2):233–245. doi: 10.1016/0165-3806(94)00144-8. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Lipska BK, Weinberger DR, Braff DL, Jaskiw GE, Geyer MA. Increased sensitivity to the sensorimotor gating-disruptive effects of apomorphine after lesions of medial prefrontal cortex or ventral hippocampus in adult rats. Psychopharmacology (Berl) 1995;122(1):27–34. doi: 10.1007/BF02246438. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Weber M, Qu Y, Light GA, Braff DL. Realistic expectations ofprepulse inhibition in translational models for schizophrenia research. Psychopharmacology (Berl) 2008;199(3):331–388. doi: 10.1007/s00213-008-1072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Loos H, Woolsey TA. Somatosensory cortex: structural alterations following early injury to sense organs. Science. 1973;179(4071):395–398. doi: 10.1126/science.179.4071.395. [DOI] [PubMed] [Google Scholar]
- Van Waes V, Tseng KY, Steiner H. GPR88 - a putative signaling molecule predominantly expressed in the striatum: Cellular localization and developmental regulation. Basal Ganglia. 2011;1(2):83–89. doi: 10.1016/j.baga.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003;100(8):4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ. The sensory striatum. Neuron. 2014;83(5):999–1001. doi: 10.1016/j.neuron.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Wilson JS, Hull CD, Buchwald NA. Intracellular studies of the convergence of sensory input on caudate neurons of cat. Brain Res. 1983;270(2):197–208. doi: 10.1016/0006-8993(83)90593-0. [DOI] [PubMed] [Google Scholar]
- Yang M, Crawley JN. Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci. 2009;Chapter 8(Unit 8):24. doi: 10.1002/0471142301.ns0824s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zembrzycki A, Chou SJ, Ashery-Padan R, Stoykova A, O'Leary DD. Sensory cortex limits cortical maps and drives top-down plasticity in thalamocortical circuits. Nat Neurosci. 2013;16(8):1060–1067. doi: 10.1038/nn.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xu M, Kamigaki T, Hoang Do JP, Chang WC, Jenvay S, Miyamichi K, Luo L, Dan Y. Selective attention. Long-range and local circuits for top-down modulation of visual cortex processing. Science. 2014;345(6197):660–665. doi: 10.1126/science.1254126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Profiling GPR88-mediated G protein activation. (A-B) G protein activation profile of GPR88 (left) versus GPR88-Venus (right) following 10-minute stimulation with 31.6 μM Compound 19 (A) or 2-PCCA (B). HEK-293 cells were co-transfected with GPR88 or GPR88-Venus, Gβ1, Gγ3-RlucII (donor BRET2 sensor), GRK2-GFP10 (acceptor BRET2 sensor) and either of the indicated human (h) (grey) or mouse (m) (red) Gα subunits. Mock is a condition where cells were not co-transfected with any Gα subunit and shows GPR88-mediated activation of endogenous G proteins. Data are expressed as mean ± SEM, *P<0.05, ** P <0.01, *** P<0.001 vs Mock condition, 3-4 replicate experiments.
Supplementary Figure 2. (A-B) Immunostaining of 30 μm sections from Gpr88-Venus (A) Intrinsic (i) GPR88-Venus signal is shown in yellow and (B) amplified (a) GPR88-Venus is displayed in red. (Scale bar is 1 mm and 200 μm for insets).
Supplementary Figure 3. Gpr88-Venus is enriched in the mouse barrel cortex. Perfused brains of adult Gpr88-Venus mice were sliced at 30 μm on a cryostat tangentially. Gpr88-Venus was amplified (a) in red and the intrinsic signal (i) can be seen in yellow, DAPI was used to stain nuclei. Annotated barrels are indicated on the merge image (Maier et al. 1999; Strominger and Woolsey 1987; Petersen 2007) Medial (M), rostral (R). All images, scale bar is 200um.
Supplementary Figure 4. Representative images for fluorescence scale used to fine-map GPR88-Venus expression: 0, not detectable; 1, weak expression; 2, moderate expression; 3, dense.
Supplementary Figure 5. Expanded cortical GPR88-Venus expression shows the discrete pattern of sensory cortical GPR88 in the layer 4. In this panel 30 μm sections with ∼300μm separation were stained with Anti-Venus to amplify the Venus signal in red and imaged with an Olympus slide scanner from approximately Bregma 1.94 mm to -5.52 mm. Scale bar is 1 mm.
Supplementary Figure 6. GPR88-Venus localizes to the soma in somatosensory neurons. Confocal microscopy Z-stack projection image shown here to demonstrate GPR88-Venus in the somatosensory cortex L4 is not prominently localized to primary cilia. GPR88-Venus was amplified with an antibody shown here in yellow and the primary cilia marker, Arl13b, can be seen in red, DAPI was used to stain nuclei. White arrows indicate primary cilia observable at this focal plane. All images, scale bar is 5 μm.
Supplementary Movies 1-3. GPR88-Venus Confocal Z-stack movies of images of the CP in P56 (Movie 1), P0 (Movie 2) and P3 (Movie 3). 30 μm tissue sections imaged in Venus channel (intrinsic) 60× oil immersion objective with 0.4 μm step size.
Supplementary Table for Figure 1A, Human GPR88 mRNA: One-Way ANOVA (P value <0.0001, ****)
Supplementary Table for Figure 1B, Mouse Gpr88 mRNA: One-Way ANOVA (P value <0.0001, ****)
Supplementary Table for Figure 2D (top), Mouse Gpr88 mRNA One-Way ANOVA
Supplementary Table for Figure 2D (bottom), Mouse Venus mRNA One-Way ANOVA
Supplementary Table for Figure 2E, [35S]-GTPγS Assay: (Gpr88+/+ vs. Gpr88-/-), Two-Way ANOVA
Supplementary Table for Figure 2E: (Gpr88+/+ vs. Gpr88-/-), Bonferroni's multiple comparisons test
Supplementary Table for Figure 2E, [35S]-GTPγS Assay: (Gpr88+/+ vs. Gpr88Venus/+ vs. Gpr88Venus/Venus), Two-Way ANOVA
Supplementary Table for Figure 8. Sensory Behavior Tests: (Gpr88+/+ vs. Gpr88-/-)


