Abstract
Ubiquitin confined within nanodroplets was irradiated with a variable-power CO2 laser. Mass spectrometry analysis shows evidence for a protein “melting”-like transition within droplets prior to solvent evaporation and ion formation. Ion mobility spectrometry reveals that structures associated with early steps of denaturation are trapped because of short droplet lifetimes.
While irradiating small (~0.05 to 1.0 μm dia.) droplets containing individual ubiquitin molecules with 10.6 μm light from a CO2 laser, we find that it is possible to induce “melting” transitions and trap early structures associated with denaturation. Mass spectrometry (MS) techniques show that upon ramping the laser power (from 0 to 20 W), the distributions shift from low charge state ([M+7H]7+ and [M+8H]8+) to high charge state ([M+9H]9+ to [M+13H]13+) species, indicating that the protein has undergone a melting-like transition within the droplet, prior to solvent evaporation and ion formation.
This transition is sensitive to solution pH, suggesting that the droplet behaves as a bulk solution at equilibrium. However, ion mobility spectrometry (IMS) measurements of conformer distributions within charge states reveal that after irradiative heating, rapid, droplet evaporation1 traps structures associated with initial stages of denaturation, before the equilibrium of denatured states is established. In their classic 1954 paper,2 “Conformation changes of proteins,” Lumry & Eyring began by stating that “[t]he term protein denaturation even in its original meaning included all those reactions destroying the solubility of native proteins and has since acquired so many other meanings as to become virtually useless.” The ability to denature proteins in droplets and directly examine the structures, stabilities, and dynamics of how ensembles of equilibrium melted states are formed complements traditional spectroscopic methods and provides a rare glimpse into the complex processes that are hidden within cooperative transitions. Our approach may be useful in studying conformation-specific unfolding pathways and enzyme inactivation mechanisms such as those reported by Fernandez-Lafuente and coworkers.3
We illustrate these phenomena with two examples involving ubiquitin, a small, 76-residue protein, because several populations of structures (the native β-grasp fold, extended helical A-state, and unstructured U-state) have been characterized in the condensed phase4,5 as well as in vacuo.6–8 First, we show that the A-state product of thermal denaturation9 is not produced at high laser powers for the [M+8H]8+ ion; apparently the complex rearrangement associated with unfolding the native β-grasp structure and subsequent assembly of the hydrogen bond networks associated with the extended helices between Gln40 – Arg72 that make up the A-state5 is impossible during the short lifetime of the droplet. Second, we show that the ubiquitin [M+11H]11+ denatured product initially emerges in a conformation that favors a cis-configured Glu18–Pro19 bond; the mixture of denatured cis- and trans-configurations observed at equilibrium evolves over longer times.9 This is remarkable since the native structure has a trans-configured Glu18–Pro19 bond;4 thus, denaturation to form [M+11H]11+ initiates through a trans→cis Glu18–Pro19 isomerization, a process that is often rate limiting;10 the unfolded trans-configured structure observed in equilibrium9 must involve a relatively complex mechanism, e.g., it might arise from a subsequent cis→trans Glu18–Pro19 isomerization step.
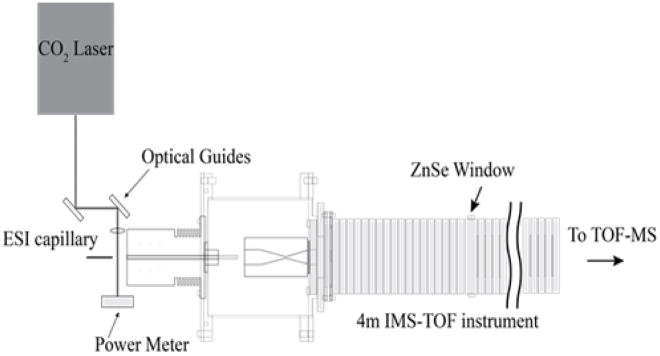
Figure 1 shows the laser-droplet activation source used in these studies. In this configuration, droplets, produced by electrospray ionization (ESI), pass through an orthogonal IR-laser beam prior to evaporation and ion formation. Changes in protein conformation upon irradiating the droplets are monitored by nested IMS-MS techniques, as described elsewhere.11
Figure 1.
Schematic diagram of the instrument. Droplets diameters are estimated to be ~0.05 and 1.0 μm when produced from small ~1.0 and 20 μm dia. ESI capillary emitters (see Experimental section in the Supplementary Information). For 10 μM ubiquitin solutions, we estimate that only one in three of the droplets contains a protein molecule. After their formation by electrospray, droplets pass through a CO2 laser beam focused at the immediate entrance to the instrument orifice. Activation in this region may induce structural changes in the protein which leads to changes in the protein charge state distribution and ion structures. This instrument is also equipped with a ZnSe window in the middle of the drift tube, which allows ions of a known mobility to be excited with 10.6 μm radiation. A series of control experiments in which the laser is focused through the drift tube shows that gaseous protein ions are not activated in the absence of solvent at the laser powers used (See Supplementary Information). This approach has similarities with an elegant “laser spray” technique, in which 10.6 μm light from an IR laser was focused into the metal-capillary tip of an ESI source to heat the bulk solution inside the capillary (see ref. 12 for details).
Figure 2 shows equilibrium melting data (from ref. 9) obtained by thermally heating a bulk ubiquitin solution (aqueous, pH 3). When electrosprayed at 25 °C, the charge state distribution favors [M+7H]7+. As the solution temperature is increased, a new distribution of highly charged ions centered around [M+11H]11+ emerges. This transition is consistent with thermally unfolding the native ubiquitin protein; following earlier pioneering ESI-MS studies as a function of solution temperature,13 a sigmoidal two-state model fit to the weighted average charge state yields a melting temperature (Tm) of 71 ± 2 °C, in agreement with literature values.4
Figure 2.
(top) Average charge state as a function of solution temperature (squares) (from ref. 9) and laser power (open circles) for ubiquitin in aqueous solution at pH 3. Solid lines show the best fit of the data assuming a two-state model with Tm = 71 ± 2 °C and Tp =10.4 ± 0.3 W. Insets show representative mass spectra at different solution temperatures and laser powers. Upon blocking the radiation, a melted charge state distribution immediately returns to the room temperature distribution, indicating that laser excitation heats only the droplets. The bottom plot shows ubiquitin ions heated in droplets at pH 2.5 (upside down triangles), pH 3.0 (open circles), and pH 4.0 (diamonds) with melting transitions of Tp = 5.8 ± 0.3, 10.4 ± 0.3, and 11.8 ± 0.3 W, respectively.
When droplets containing ubiquitin are exposed to a laser beam, mass spectra (also shown in Figure 2) are indistinguishable from those obtained from heated solutions. At 0 W (i.e., allowing droplets to pass through without laser excitation) the charge state distribution is dominated by [M+7H]7+, as expected. When the droplets pass through a 11 W laser field, highly charged ions (centered around [M+11H]11+) are observed, indicating that a substantial fraction of the protein molecules underwent an unfolding transition. Unfolded structures dominate the spectrum at 17 W. The weighted average charge state distribution for these ions is also sigmoidal in shape and analysis yields a melting power, Tp = 10.4 ± 0.3 W. The similar shapes of the temperature-induced and droplet-IR-irradiated charge state distribution curves were somewhat surprising to us. One might expect irradiation of droplets to induce immediate desolvation, such that solvent is removed before the structure can change – rendering only the native protein charge state in the mass spectrum. The present results require that irradiated droplets survive long enough for the protein to unfold in solution, prior to droplet evaporation and ion formation. But, does the solution environment within the droplet still influence protein stability as it does in the bulk? To explore this, we changed the pH of the solutions used to produce droplets. Figure 2 shows the weighted average charge state curves obtained for ubiquitin confined in droplets produced from pH = 2.5, 3.0, and 4.0 solutions. The midpoint associated with these transitions shifts to higher laser powers with increasing solution pH, corroborating the idea that solution environment within the droplet influences the denaturation transition.
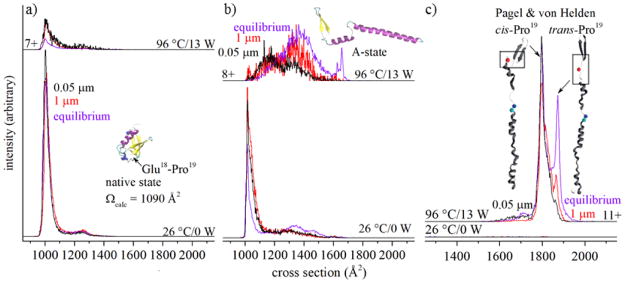
While the MS analysis shows that our intuition – that irradiated droplets might evaporate so quickly that solution transitions might not be observed – was incorrect, IMS measurements reveal that there are differences between droplet-IR-heating and equilibrium melting studies. An ion’s mobility through a buffer gas is related to its shape, or collision cross section.14 Figure 3 shows IMS cross section distributions for [M+7H]7+ recorded at 26 °C, distributions for [M+7H]7+, [M+8H]8+ and [M+11H]11+ formed at a bulk solution temperature of 96 °C, as well as these product ions formed from irradiating droplets with a laser power of 13 W, i.e., in the region where the melting transition appears to be complete. The [M+7H]7+ species shows a single peak centered at Ω = 1010 Å2, consistent with highly folded structures from solutions that favor the native state.9 Upon increasing either the solution temperature, or the laser power used to irradiate droplets, the compact structures decrease in relative abundance. A comparison of the decreases in abundances of the Ω = 1010 Å2 peak, which yielded a value of Tm = 71 ± 2 °C from prior heated solution studies,9 with the present data obtained upon irradiating ubiquitin in droplets shows the decrease in this peak is indistinguishable between these methods. The present data shows that the loss of compact [M+7H]7+ conformers from droplets also behaves as a melting transition.
Figure 3.
Cross section distributions for (a) [M+7H]7+, (b) [M+8H]8+, and (c) [M+11H]11+ ions of ubiquitin at different temperatures (purple) and laser powers, with black and red lines corresponding to structures that form upon irradiating droplets formed from ~1 μm and ~20 μm electrospray emitters, respectively. Structures in (c) are adapted from reference 8.
Cross section distributions for other charge states formed from heating bulk solutions or from laser-droplet activation show marked differences. At 26 °C, the [M+8H]8+ species exhibits a sharp peak at Ω = 1010 Å2. When the bulk solution of ubiquitin is heated, this peak decreases in abundance and several broad features at Ω ~ 1100 – 1550 Å2 increase in relative abundance (from T ~ 45 to 60 °C) and then subsequently decay at temperatures above T ~ 70 °C. The decrease of these signals9 is associated with the systematic increase in the two sharp peaks at Ω = 1635 and 1650 Å2, previously assigned to the ubiquitin A-state,7 a low abundance product of melting. While the A-state is somewhat unexpected, we note that as the temperature is increased from ~25 to 96 °C, the dielectric constant of water decreases from ε = 78 to 56, near the value of ε = 52,15 which is similar to that of 40:60 water:methanol solution that is known to favor the A-state at 26 °C.5,7
As is the case with heating a bulk ubiquitin solution, broad features, centered at Ω([M+8H]8+) ~1160 and ~1350 Å2, are also observed upon laser irradiation of droplets; however, in comparison with heated solution data, these features are shifted to lower cross sections, and it appears that the shapes and intensities of these features are dependent upon on droplet size. When [M+8H]8+ is formed from ~0.05 μm nanodroplets, we observe two broad features at Ω ~1150 and ~1330 Å2, with the Ω ~1150 Å2 more populated than the Ω ~1330 Å2. In contrast, when the [M+8H]8+ products are formed upon irradiation of larger ~1 μm droplets, larger Ω ~1330 Å2 cross section species increase in abundance and the population of Ω ~1150 Å2 ions decreases slightly. The lifetimes of these droplets depend on their initial size, which is related to the nano-electrospray emitter diameter and solution flow rate. Although the exact droplet lifetimes are unknown, previous experiments using theta-glass emitters indicate that a change in emitter tip diameter from 1.5 μm to 0.24 μm results in a linear decrease in the solution flow rate and a reduction in the droplet lifetime from 9 to 1 μs.16 The twenty-fold difference in tip diameters used in these experiments should change the droplet lifetimes by well over an order of magnitude. Presumably the short lifetimes favor [M+8H]8+ products in the relatively compact Ω ~1150 Å2 region. That is, these species are quenched before the Ω ~1330 Å2 species are formed. When given time to reach equilibrium (as is the case for bulk heating studies), the population of the Ω ~1330 Å2 species increases even more.9
An extreme case illustrating the importance of droplet size (lifetime) in producing different structures is observed in the Ω ~1625 to 1675 Å2 region, where A-state structures show two characteristic peaks. Indeed, this doublet unambiguously denotes the presence of A-state species as a product of equilibrium melting in bulk solutions;9 however, we observe no evidence for the A-state upon laser-droplet activation. The inability to form the A-state must be due to the transient nature of a rapidly desolvating droplet, which limits the accessible unfolding products to only those that can form rapidly. The complex changes that are associated with forming the extended regions of helices between Gln40 – Arg72 cannot be carried out during the short lifetime of even the largest 1.0 μm dia. droplets.
Other products of laser-induced transitions of ubiquitin confined in droplets provide additional details about the first steps of melting. Figure 3 also shows the cross section distributions for [M+11H]11+, the dominant charge state produced at high solution temperatures and high laser power. The most notable products of high-temperature solution melting are the two sharp peaks with cross sections of Ω ~1800 and 1875 Å2. While there can be multiple conformations associated with these peaks, Pagel and von Helden used ultraviolet photodissociation of mobility-selected ions to show that the major differences arise from the orientation of Glu18 – Pro19; the Ω ~1800 Å2 species has a cis-configured peptide bond whereas Ω ~1875 Å2 species is in the trans configuration.8 The products formed from 0.05 μm dia. nanodroplets show that the trans conformer is almost completely absent, as indicated by the shoulder at Ω ~1800 Å2. However, the trans-configured species is formed in appreciable amounts upon irradiating 1.0 μm dia. droplets. These results require that an early step in the unfolding of the native trans-configured Glu18–Pro19 species involves isomerization of this peptide bond. Inspection of the native structure reveals that the Glu18–Pro19 residues are in close proximity to the β-sheet formed between Met1–Lys6 and Glu64–Arg72 residues. These interactions stabilize the β-grasp fold around the hydrophobic core. A trans→cis isomerization of Glu18–Pro19 leading to elongated structures disrupts these stabilizing interactions.
Melting of isolated molecules in electrosprayed droplets17 provides the ability to monitor structures, stabilities, and dynamics of species involved in denaturation, and provides a unique way to study structural changes that occur in confined spaces. While the droplets used here are much larger (~0.05 to 1.0 μm dia.) than the length scale of unfolded ubiquitin (10–30 Å) larger proteins (or confinement in smaller droplets) may introduce some fascinating physical behavior associated with confinement.18 In the studies presented here, we showed that the protonation state of protein ions that underwent irradiative heating within nanodroplets reflects that of the bulk, but significant structural changes have not equilibrated, as measured using IMS. The kinetic trapping of structures provides a new view of denaturation by capturing “snapshots” of the unfolding pathway. Further characterization of structures and pathways in a range of environments using state-of-the-art condensed and in vacuo techniques are likely to provide detailed insights about rearrangements that occur upon protein unfolding, reconciling the notion of an “unstructured” denatured state.
Supplementary Material
Acknowledgments
This work is supported in part by funds from the Waters Corporation, the National Institutes of Health R01 GM117207-03 and R01 GM121751-01A1, and the Robert and Marjorie Mann endowment. T.J.E. was supported by the Robert and Marjorie Mann fellowship from Indiana University.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details; Comparison of cross section distributions obtained by IR heating of droplets, IMS-IR-IMS, and tandem IMS techniques. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Lee SW, Freivogel P, Schindler T, Beauchamp J. J Am Chem Soc. 1998;120:11758. [Google Scholar]; Silveira JA, Fort KL, Pierson NA, Clemmer DE, Russell DH. J Am Chem Soc. 2013;135:19147. doi: 10.1021/ja4114193. [DOI] [PubMed] [Google Scholar]
- 2.Lumry R, Eyring H. J Phys Chem. 1954;58:110. [Google Scholar]
- 3.Rodrigues RC, Ortiz C, Berenguer-Murcia A, Torres R, Fernandez-Lafuente R. Chem Soc Rev. 2013;42:6290. doi: 10.1039/c2cs35231a. [DOI] [PubMed] [Google Scholar]; Sanchez A, Cruz J, Rueda N, dos Santos JCS, Torres R, Ortiz C, Villalonga R, Fernández-Lafuente R. RSC Adv. 2016;6:27329. [Google Scholar]
- 4.Lenkinski RE, Chen DM, Glickson JD, Goldstein G. Biochim Biophys Acta. 1977;494:126. doi: 10.1016/0005-2795(77)90140-4. [DOI] [PubMed] [Google Scholar]; Cary PD, King DS, Crane-Robinson C, Bradbury WM, Rabbani A, Goodwin GH, Johns EW. Eur J Biochem. 1980;112:577. doi: 10.1111/j.1432-1033.1980.tb06123.x. [DOI] [PubMed] [Google Scholar]; Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]; Wintrode PL, Makhatadze GI, Privalov PL. Proteins: Struct, Funct, Genet. 1994;18:246. doi: 10.1002/prot.340180305. [DOI] [PubMed] [Google Scholar]; Briggs MS, Roder H. Proc Natl Acad Sci U S A. 1992;89:2017. doi: 10.1073/pnas.89.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jackson SE. Org Biomol Chem. 2006;4:1845. doi: 10.1039/b600829c. [DOI] [PubMed] [Google Scholar]; Kony DB, Hunenberger PH, van Gunsteren WF. Protein Sci. 2007;16:1101. doi: 10.1110/ps.062323407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson KD, Mayer AN. Arch Biochem Biophys. 1986;250:390. doi: 10.1016/0003-9861(86)90741-1. [DOI] [PubMed] [Google Scholar]; Harding MM, Williams DH, Woolfson DN. Biochemistry. 1991;30:3120. doi: 10.1021/bi00226a020. [DOI] [PubMed] [Google Scholar]; Pan Y, Briggs MS. Biochemistry. 1992;31:11405. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]; Stockman BJ, Euvrard A, Scahill TA. J Biomol NMR. 1993;3:285. doi: 10.1007/BF00212515. [DOI] [PubMed] [Google Scholar]; Cox JPL, Evans PA, Packman LC, Williams DH, Woolfson DN. J Mol Biol. 1993;234:483. doi: 10.1006/jmbi.1993.1600. [DOI] [PubMed] [Google Scholar]; Brutscher B, Brüschweiler R, Ernst RR. Biochemistry. 1997;36:13043. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 6.Jockush RA, Schnier PD, Price WD, Strittmatter EF, Demirev PA, Williams ER. Anal Chem. 1997;69:1119. doi: 10.1021/ac960804q. [DOI] [PMC free article] [PubMed] [Google Scholar]; Oh H, Breuker K, Sze KS, Carpenter BK, McLafferty FW. Proc Natl Acad Sci USA. 2002;99:15863. doi: 10.1073/pnas.212643599. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koeniger SL, Merenbloom SI, Sevugarajan S, Clemmer DE. J Am Chem Soc. 2006;128:11713. doi: 10.1021/ja062137g. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen S-H, Russell DH. J Am Soc Mass Spectrom. 2015;26:1433. doi: 10.1007/s13361-015-1191-1. [DOI] [PubMed] [Google Scholar]; Wagner N, Kim D, Russell DH. Anal Chem. 2016;88:5934. doi: 10.1021/acs.analchem.6b00871. [DOI] [PubMed] [Google Scholar]
- 7.Wyttenbach T, Bowers MT. J Phys Chem B. 2011;115:12266. doi: 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]; Shi H, Clemmer DE. J Phys Chem B. 2014;118:3498. doi: 10.1021/jp4097327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warnke S, Baldauf C, Bowers MT, Pagel K, von Helden G. J Am Chem Soc. 2014;136:10308. doi: 10.1021/ja502994b. [DOI] [PubMed] [Google Scholar]
- 9.El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, Russell DH, Clemmer DE. J Am Chem Soc. 2017;139:6306. doi: 10.1021/jacs.7b02774. [DOI] [PubMed] [Google Scholar]
- 10.Jackson SE, Fersht AR. Biochemistry. 1991;30:10436. doi: 10.1021/bi00107a011. [DOI] [PubMed] [Google Scholar]
- 11.Merenbloom SI, Koeniger SL, Valentine SJ, Plasencia MD, Clemmer DE. Anal Chem. 2006;78:2802. doi: 10.1021/ac052208e. [DOI] [PubMed] [Google Scholar]; Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth H, Smith RS, Clemmer DE. Anal Chem. 2006;78:4161. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 12.Kudaka I, Kojima T, Saito S, Hiraoka K. Rapid Commun Mass Spectrom. 2000;14:1588. doi: 10.1002/1097-0231(20000830)14:16<1558::AID-RCM64>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; Takamizawa A, Fujimaki S, Sunner J, Hiraoka K. J Am Soc Mass Spectrom. 2005;16:860. doi: 10.1016/j.jasms.2005.02.012. [DOI] [PubMed] [Google Scholar]; Nakamura M, Takamizawa A, Yamada H, Hiraoka K, Akashi S. Rapid Commun Mass Spectrom. 2007;21:1635. doi: 10.1002/rcm.2995. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury SL, Katta V, Chait BT. J Am Chem Soc. 1990;112:9012. [Google Scholar]; Loo JA, Loo RRO, Udseth HR, Edmonds CG, Smith RD. Rapid Commun Mass Spectrom. 1991;5:101. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]; Mirza UA, Cohen SL, Chait BT. Anal Chem. 1993;65:1. doi: 10.1021/ac00049a003. [DOI] [PubMed] [Google Scholar]; Benesch JLP, Sobott F, Robinson CV. Anal Chem. 2003;75:2208. doi: 10.1021/ac034132x. [DOI] [PubMed] [Google Scholar]; Wang G, Abzalimov RR, Kaltashov IA. Anal Chem. 2011;83:2870. doi: 10.1021/ac200441a. [DOI] [PubMed] [Google Scholar]
- 14.Jarrold MF, Constant VA. Phys Rev Lett. 1991;67:2994. doi: 10.1103/PhysRevLett.67.2994. [DOI] [PubMed] [Google Scholar]; Wyttenbach T, von Helden G, Bowers MT. J Am Soc Mass Spectrom. 1996;118:8355. [Google Scholar]; Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem. 1996;100:16082. [Google Scholar]; Clemmer DE, Jarrold MF. J Mass Spectrom. 1997;32:577. [Google Scholar]; Wyttenbach T, von Helden G, Batka JJ, Carlat D, Bowers MT. J Am Soc Mass Spectrom. 1997;8:275. [Google Scholar]
- 15.Malmberg CG, Maryott AA. J Res Natl Bur Stand. 1956;56:1. [Google Scholar]
- 16.Mortensen DN, Williams ER. J Am Chem Soc. 2016;138:3453. doi: 10.1021/jacs.5b13081. [DOI] [PubMed] [Google Scholar]
- 17.Lee JK, Kim S, Nam HG, Zare RN. Proc Natl Acad Sci U S A. 2015;112:3898. doi: 10.1073/pnas.1503689112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miller CF, Kulyk DS, Kim JW, Badu-Tawiah A. Analyst. 2017;12:2152. doi: 10.1039/c7an00362e. [DOI] [PubMed] [Google Scholar]
- 18.Sakaue T, Raphaël E. Macromoleules. 2006;39:2621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.