Abstract
Due to the growing field of digital pathology, more and more digital histology slides are becoming available. This improves the accessibility, allows teleconsultations from specialized pathologists, improves education, and might give urologist the possibility to review the slides in patient management systems. Moreover, by stacking multiple two-dimensional (2D) digital slides, three-dimensional volumes can be created, allowing improved insight in the growth pattern of a tumor. With the addition of computer-aided diagnosis systems, pathologist can be guided to regions of interest, potentially reducing the workload and interobserver variation. Digital (3D) pathology has the potential to improve dialog between the pathologist and urologist, and, therefore, results in a better treatment selection for urologic patients.
Electronic supplementary material
The online version of this article (10.1007/s00345-018-2202-1) contains supplementary material, which is available to authorized users.
Keywords: Digital pathology, Three-dimensional, Computer-aided diagnosis, Urinary tract pathology
Introduction
Regardless increase of cervical hernias and repetitive strain disorders, the conventional light microscope and the pathologist seem to remain inseparable partners [1]. Yet, recent developments in the field of digital pathology urge many pathology departments to digitize slides, enabling digital visualization (Fig. 1). Clearly, hospitals benefit from digital patient management systems and fully digitized radiology departments and even allow urologist to plan intrarenal surgeries with special 3D software [2]. Likewise, the digitization of pathology specimens will improve accessibility within the hospital and could facilitate multidisciplinary meetings, allowing oversees consultation from specialized pathologists.
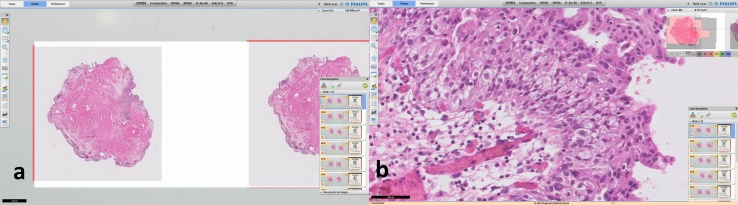
Fig. 1.
Overview of the Philips information management system. a Showing an overview of a case, an en-bloc resection of a bladder tumor; b showing the zoomed in version, focusing on the papillary tissue
Histopathologic analysis using the conventional light microscopy has been the gold standard for cancer detection and grading for decades. In general, of every tissue block arriving at the pathology department, only 1–2 slides per tissue block are assessed [3]. Therefore, it can be difficult for the pathologists to fully understand the growth pattern of a tumor [4]. It can be hypothesized that tumor invasiveness may vary from section to section. Moreover, most tumors are heterogeneous [5]. By assessing a small number of slides, an undersampling error is introduced. A method to sample a whole specimen holds the promise to provide the pathologists with a more accurate understanding of the growth pattern of a tumor [6].
Due to recent developments, two-dimensional (2D) digital microscopy images can be acquired through digital microscope systems. A method to visualize the whole specimen is to cut multiple consecutive 2D sections and create a three-dimensional (3D). Nonetheless, this approach has not been considered by pathologists to examine histology yet, mostly due to the effort and time involved in the preparation of the physical slides.
It is debatable whether the diagnostic accuracy would improve by increasing the number of digital slides. Histopathological examination is already laborious and susceptible for human variation [7]. By presenting even more data within the same time-span, more errors are prone to occur. To reduce the workload and inter-observer variation, computer-aided diagnosis (CAD) systems might be the solution. A CAD system could diminish the workload by automated recognition of suspicious tissue and guide the pathologist in the grading and staging of a tumor.
The current workflow in pathology
The standard workup for a histology specimen is a labor-intensive and time-consuming process (supplementary Figure 1). At the end of this process, the pathologist examines the slides by looking at morphological changes. Using different magnifications, the aggressiveness of tumor cells is graded, and the staging is performed. Over the years, guidelines have been adjusted to improve prognostic information, helping the urologist in their treatment planning. However, histopathology remains notorious for its interobserver variability. In prostate cancer, interobserver studies show an agreement ranging from 10 to 70% when assessing the Gleason score [8]. In 20% of patients, this would have influenced the treatment plan [9]. While the bladder cancer grading system has a large prognostic value, the interobserver agreement is only 60% [10]. Diagnostic accuracy has been seen to improve when assessed by specialized urinary tract pathologist [11].
2D digital pathology
The Food and Drug Administration (FDA) only recently gave approval for the first digital histology slide scanner to be used for diagnostic purposes in the US [12], while it was already being used in various places in Europa and Canada. By optically scanning the histologic glass slide, a 2D ultra-high-resolution digital image is created, a so-called whole slide image (WSI). These WSIs can be visualized on a digital screen, making it possible to examine the image at different magnifications (see Fig. 1). Several studies proved the non-inferiority of WSIs for diagnostic purpose by comparing them with state-of-the-art light microscopy [13].
Even though digital pathology is available, it is not yet broadly implemented in the current clinical practice. Pathologists could be reluctant, since it requires another way of working. As of 2016, the College of American Pathology has issued a set of preliminary guidelines for digital pathology to anticipate the digital era [14]. To incorporate digital pathology, however, the workflow on the pathology department must be adjusted (supplementary Figure 1). It requires investments in WSI scanners, high-performance computers, high-quality color-calibrated monitors, and server solutions for data storage. WSIs consist of an enormous amount of data, depending on the size of the section; the storage size of a WSI can range from 1 to 5 GB for a non-compressed single prostate biopsy. This requires an enormous increase in computational power and network infrastructure.
If digital pathology is implemented correctly, it will lead to reduced costs, e.g., by balancing the workload or tele-consulting of (distant) specialists. For the implementation of digital pathology, it has been estimated that for a large academic institution with 219,000 annual accessions, a shift from conventional to WSIs would save of US$ 18 million over a 5-year period [15]. Table 1 gives a more detailed overview of benefits and barriers for the implementation of digital pathology.
Table 1.
Benefits and barriers of implementing digital pathology
| Benefits whole slide imaging | Barriers for adoption whole slide imaging |
|---|---|
| Accessibility and access by multiple observers | Change in ergonomics |
| Teleconsultations | Need for high-quality scanners |
| Eligible for CAD systems | Need for high-speed network |
| Possibility placing annotations and comments | Large size digital files |
| Sharing slides for research purposes | Costs: hardware, software, information technology support/infrastructure, and maintenance |
| Digital storage | Lack of standards and/or best practice guidelines |
| Portability and flexible work schedules | Scanning artifacts |
| Archiving interesting cases | |
| Enhancing workflow | |
| Integrated into pathology report/patient information system | |
| Pathology education |
Three-dimensional (3D) reconstructions
Digital pathology could accelerate consultations and can replace the bright field microscope for education and clinical conferences [16]. A more recently studied application is the stacking of multiple 2D slides to reconstruct a 3D volume. This enables the pathologist to assess the resected tissue or biopsy as a whole. Orientation on single 2D slides is often difficult due to tangential sectioning and artifacts [17]. A 3D reconstruction can provide improved insight into the architectural features and spatial arrangements with other structures.
However, a major difficulty in the 3D representation is the alignment of the slides, since non-linear deformation occurs during the sampling process of fixating, sectioning, and mounting of the specimen [6, 18–21].
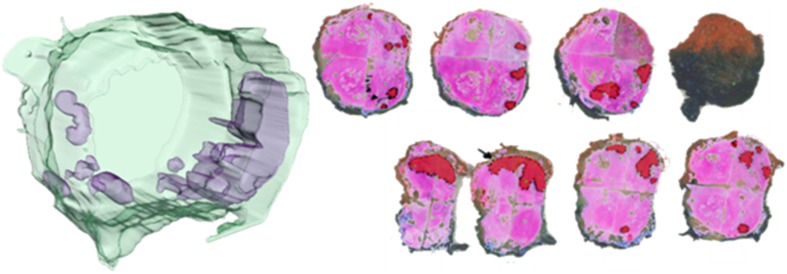
Several studies have focused on the 3D reconstructed histology [6, 18–24]. Boag et al. reconstructed prostate carcinomas out of multiple 2D histological slides and segmented the adenocarcinoma to visualize the architecture of the tumorous glands. They found that Gleason grade 3 glands appear separate from each other on the 2D slides, while the 3D reconstruction showed interconnecting tubules [22]. Muller et al., impart of a correlation study, visualized Gleason grade 3 and 4 tumors in a 3D representation of a whole prostate, using 4 mm spaced WSIs (see Fig. 2) [25, 26].
Fig. 2.
3D reconstruction of a prostatectomy specimen. On the right, the individual whole mount slides are shown in red the manually delineated tumor. From this, the 3D reconstruction on the left is rendered
For the visualization of 3D reconstructions, most studies used manual segmentations of tissue structures. As a next step, Norton et al. showed an automated method for the creation of 3D segmentations of in situ disease of breast tissue [21]. These automated segmentations could alleviate the laborious task of manual segmentations. 3D reconstructions are not solely based on H&E stained tissue. Some studies have shown that registration is also possible with other staining agents or even combined multiple stains or different modalities [6, 18, 19, 23].
The main drawback of these studies is the low out-of-plane resolution of 3D reconstructions. The size of these high-resolution data sets visualizing them in 3D is challenging, especially when looking at a higher magnification. The bias could be that you lose the details which could be of clinical relevance.
Computer-aided diagnosis (CAD)
Several solutions have been sought to handle the increase in workload and to reduce the existing observer variation of pathologists. Since the possibility to digitize histology slides, groups have applied automated pattern recognition software to identify suspicious features. The so-called CAD systems have been introduced to support pathologist in their decision-making [27]. These systems can automatically measure the extent of cancerous area, the grade of the cancer, and generate localized cancer maps (Fig. 3).
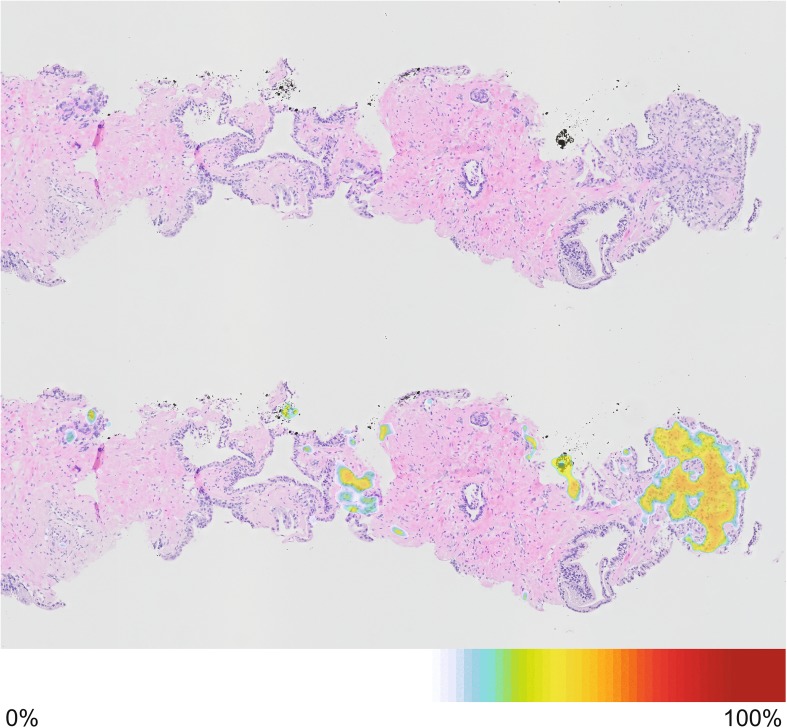
Fig. 3.
Digital prostate needle biopsy. Top image shows the normal H&E stained digital image with on the right and on the left (top) a focus of adenocarcinoma. Lower image shows the heatmap overlay indicating the probability of prostate cancer
Initially, CAD systems used mathematical algorithms based on structural hand-crafted features or variations in pixel intensities [27]. Currently, the majority of CAD systems focus on prostate biopsies and high accuracy levels are found in the detection of prostate cancer [27]. In the grading, however, these systems seem less accurate. An explanation may be the focus on the glandular structures, where the adenocarcinoma originates. In high-grade tumors, there is an absence of glands and loss of differentiation [28] and thus no structures to detect.
More recently, deep learning has become more popular. Deep learning is often used for image recognition; for example, the automated classification of skin lesions in dermatology [29]. By applying a unique set of filters, it is possible to differentiate between preselected groups, for example different tumor grades. Litjens et al. have used deep learning in prostate biopsies and accomplished an accuracy of 93% in differentiating tumorous tissue from benign tissue [30]. Since most interobserver variation exists in the grading and staging of a tumor [7], it would be of great asset to train CAD systems for this application.
The major drawback of these deep learning techniques is the need of large annotated training sets. Preferably, these training sets should be assessed by multiple pathologists to reduce the influence of the interobserver variability. At this moment, there is a lack of multi-institutional validation sets to compare the performance of different CAD systems.
Ideally, a CAD system should be used to guide pathologists to a region of interest or even replace pathologists, certainly with the increased number of slides in 3D reconstructions. By doing so, communication, and thus interpretation, regarding the clinical information and the histopathological input could lead to better understanding of the tumorous tissue.
Future
Currently, the use of histological 3D reconstructions in diagnostics is not feasible for clinical decision-making. Despite the major progress in digital pathology, there are still some hurdles to take. However, a set of challenges are already addressed to enable the future use of 2D and 3D digital pathology.
First, the sample workup for histopathology remains a labor-intensive process. To improve this, Onozato et al. have developed an automated tissue-sectioning machine, automating the sectioning and mounting of the histological sections [31]. At this moment, automated sectioning requires more time than the manual sample workup. However, it is the expectancy that eventually these systems can alleviate the manual work.
Although formalin is still the most common fixative, it may not always be the most optimal. The deleterious effects on DNA and RNA are well known, and proteins are altered by the crosslinking mechanism. An optimal fixative should allow for high-quality histology, preserve sufficient material for analysis using other technical approaches, and desirable, faster than the 1 mm/h penetration rate of formalin [32].
Clearly, 2D and 3D digital pathology is potentially a powerful tool for the pathologist. It has the opportunity to reduce workload, which has been increasing due to increasing number of requested diagnosis and a reduced number of pathologists [33]. It allows pathologists to set a diagnosis from anywhere in the world, and more important, it gives the possibility to consult distant colleagues. These teleconsultations can facilitate the second opinions without the risk-off slide loss. Teleconsultations can be of tremendous value for peripheral hospitals, giving the opportunity to easily consult urinary tract pathologists. Sub-specialization in urinary tract pathology is relatively rare and most pathologists work on a large variety of tissue types. Digital pathology has the potential to amplify sub-specialization and, therefore, limit the variation in diagnosis [34].
Although current CAD systems have high levels of accuracy for the detection of prostate cancer, there is no implementation into clinical practice yet. Most CAD systems are only validated in a single center, while, due to staining differences between the histological slides in different pathology laboratories, staining differences must be incorporated. Therefore, before these CAD systems can be used in daily practice, multi-institutional validation has to be performed by specialized pathologists.
By implementing 3D reconstructions together with CAD systems, pathologists will get a better understanding of the growth pattern of a tumor and can be more easily guided to specific regions of interest. By letting the pathologist focus on only the difficult cases, the workload diminishes.
Finally, by implementing digital pathology into the electronic patient information systems, they are readily available for urologist and, therefore, give easily access during multidisciplinary meetings or for intercollegial consultations. The possible 3D pathology image could be of clinical relevance for urologists. With the ongoing evolution in better imaging technologies, such as MRI and PSMA-PET scan, we hope that integration of these technologies together can help the urologist in optimizing treatment options. Subsequently, with better pathological and radiological mapping of prostate cancer, this could be clinically important in light of possible focal therapy for prostate cancer [35, 36]. Overall, this will result in a better quality of care for the patient.
Conclusion
Digital pathology and 3D reconstructions have the potential to improve dialog between the pathologist and urologist, and, therefore, result in a better treatment selection for urologic patients. For further refinement of analyzing pathology and usage of digital slides, 3D pathology is now on show.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1 a. General pathology workflow. b. workflow with the implementation of digital pathology (TIFF 18958 kb)
Abbreviations
- 2D
Two-dimensional
- 3D
Three-dimensional
- H&E
Hematoxylin and eosin
- CAD
Computer-aided diagnosis
- WSI
Whole slide image
Author contributions
IJ: manuscript writing. ML: manuscript editing. CDSH: manuscript editing. SLM: manuscript editing. HAM: manuscript editing. DMB: manuscript writing. PJZ: manuscript editing.
Funding
I. Jansen and M. Lucas are paid by ITEA3, H. A. Marquering is founder and shareholder of Nico-lab, D. M. de Bruin is founder and shareholder of Offroad Medical.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This project did not involve any human participants or animals.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00345-018-2202-1) contains supplementary material, which is available to authorized users.
Contributor Information
Ilaria Jansen, Phone: +31 (0)205665207, Email: i.jansen@amc.uva.nl.
Marit Lucas, Email: m.lucas@amc.uva.nl.
C. Dilara Savci-Heijink, Email: c.d.savciheijink@amc.uva.nl.
Sybren L. Meijer, Email: s.l.meijer@amc.uva.nl
Henk A. Marquering, Email: h.a.marquering@amc.uva.nl
Daniel M. de Bruin, Email: d.m.debruin@amc.uva.nl
Patricia J. Zondervan, Email: p.j.zondervan@amc.uva.nl
References
- 1.George E. Occupational hazard for pathologists: microscope use and musculoskeletal disorders. Am J Clin Pathol. 2010;133:543–548. doi: 10.1309/AJCPUXDS5KJKRFVW. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Li H, Wu D, et al. Surgical planning and manual image fusion based on 3D model facilitate laparoscopic partial nephrectomy for intrarenal tumors. World J Urol. 2014;32:1493–1499. doi: 10.1007/s00345-013-1222-0. [DOI] [PubMed] [Google Scholar]
- 3.Kohl SK, Lewis SE, Tunnicliffe J, et al. The College of American Pathologists and National Society for Histotechnology workload study. Arch Pathol Lab Med. 2011;135:728–736. doi: 10.5858/2010-0288-CP.1. [DOI] [PubMed] [Google Scholar]
- 4.Sun L, Wang D, Zubovits JT, et al. An improved processing method for breast whole-mount serial sections for three-dimensional histopathology imaging. Am J Clin Pathol. 2009;131:383–392. doi: 10.1309/AJCPVBZZ4IKJHY3U. [DOI] [PubMed] [Google Scholar]
- 5.Moch H, Humphrey P, Ulbright T, Reuter V. WHO classification of tumours of the urinary system and male genital organs. Lyon: International Agency for Research on Cancer; 2016. [Google Scholar]
- 6.Magee D, Song Y, Gilbert S, et al. Histopathology in 3D: from three-dimensional reconstruction to multi-stain and multi-modal analysis. J Pathol Inf. 2015 doi: 10.4103/2153-3539.151890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engers R. Reproducibility and reliability of tumor grading in urological neoplasms. World J Urol. 2007;25:595–605. doi: 10.1007/s00345-007-0209-0. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Urrego PA, Cronin AM, Al-Ahmadie HA, et al. Interobserver and intraobserver reproducibility in digital and routine microscopic assessment of prostate needle biopsies. Hum Pathol. 2011;42:68–74. doi: 10.1016/j.humpath.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Melia J, Moseley R, Ball RY, et al. A UK-based investigation of inter- and intra-observer reproducibility of Gleason grading of prostatic biopsies. Histopathology. 2006;48:644–654. doi: 10.1111/j.1365-2559.2006.02393.x. [DOI] [PubMed] [Google Scholar]
- 10.Babjuk M, Böhle A, Burger M et al (2016) EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur Urol 1–15. 10.1016/j.eururo.2016.05.041 [DOI] [PubMed]
- 11.Mortezavi A, Keller EX, Poyet C, et al. Clinical impact of prostate biopsy undergrading in an academic and community setting. World J Urol. 2016;34:1481–1490. doi: 10.1007/s00345-016-1788-4. [DOI] [PubMed] [Google Scholar]
- 12.Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2016 doi: 10.5858/arpa.2016-0074-OA. [DOI] [PubMed] [Google Scholar]
- 13.Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: a systematic review. Arch Pathol Lab Med. 2017;141:151–161. doi: 10.5858/arpa.2016-0025-RA. [DOI] [PubMed] [Google Scholar]
- 14.Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710–1722. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho J, Ahlers SM, Stratman C, et al. Can digital pathology result in cost savings? A financial projection for digital pathology implementation at a large integrated health care organization. J Pathol Inf. 2014;5:33. doi: 10.4103/2153-3539.139714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantanowitz L, Valenstein PN, Evans AJ, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011 doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng L, Montironi R, Davidson DD, Lopez-Beltran A. Staging and reporting of urothelial carcinoma of the urinary bladder. Mod Pathol. 2009;22:s70–s95. doi: 10.1038/modpathol.2009.1. [DOI] [PubMed] [Google Scholar]
- 18.Roberts N, Magee D, Song Y, et al. Toward routine use of 3D histopathology as a research tool. Am J Pathol. 2012;180:1835–1842. doi: 10.1016/j.ajpath.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y, Treanor D, Bulpitt AJ, Magee DR. 3D reconstruction of multiple stained histology images. J Pathol Inf. 2013 doi: 10.4103/2153-3539.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth ME, Treanor D, Roberts N, et al. Three-dimensional reconstruction of ductal carcinoma in situ with virtual slides. Histopathology. 2015;66:966–973. doi: 10.1111/his.12561. [DOI] [PubMed] [Google Scholar]
- 21.Norton KA, Namazi S, Barnard N, et al. Automated reconstruction algorithm for identification of 3D architectures of cribriform ductal carcinoma in situ. PLoS ONE. 2012;7:1–11. doi: 10.1371/journal.pone.0044011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boag AH, Kennedy LA, Miller MJ. Three-dimensional microscopic image reconstruction of prostatic adenocarcinoma. Arch Pathol Lab Med. 2001;125:562–566. doi: 10.5858/2001-125-0562-TDMIRO. [DOI] [PubMed] [Google Scholar]
- 23.Marchiò C, Sapino A, Arisio R, Bussolati G. A new vision of tubular and tubulo-lobular carcinomas of the breast, as revealed by 3-D modelling. Histopathology. 2006;48:556–562. doi: 10.1111/j.1365-2559.2006.02373.x. [DOI] [PubMed] [Google Scholar]
- 24.Onozato ML, Klepeis VE, Yagi Y, Mino-Kenudson M. A role of three-dimensional (3D)-reconstruction in the classification of lung adenocarcinoma. Anal Cell Pathol. 2012;35:79–84. doi: 10.1155/2012/684751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller BG, de Bruin DM, Brandt MJ, et al. Prostate cancer diagnosis by optical coherence tomography: first results from a needle based optical platform for tissue sampling. J Biophotonics. 2016;9:490–498. doi: 10.1002/jbio.201500252. [DOI] [PubMed] [Google Scholar]
- 26.Muller BG, de Bruin DM, van den Bos W, et al. Prostate cancer diagnosis: the feasibility of needle-based optical coherence tomography. J Med Imaging. 2015;2:037501-1–037501-9. doi: 10.1117/1.JMI.2.3.037501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosquera-Lopez C, Agaian S, Velez-Hoyos A, Thompson I. Computer-aided prostate cancer diagnosis from digitized histopathology: a review on texture-based systems. IEEE Rev Biomed Eng. 2015;8:98–113. doi: 10.1109/RBME.2014.2340401. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen K, Sabata B, Jain AK. Prostate cancer detection: fusion of cytological and textural features. J Pathol Inf. 2011 doi: 10.4103/2153-3539.92030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:1–11. doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onozato ML, Hammond S, Merren M, Yagi Y. Evaluation of a completely automated tissue-sectioning machine for paraffin blocks. J Clin Pathol. 2013;66:151–154. doi: 10.1136/jclinpath-2011-200205. [DOI] [PubMed] [Google Scholar]
- 32.Howat WJ, Wilson BA. Tissue fixation and the effect of molecular fixatives on downstream staining procedures. Methods. 2014;70:12–19. doi: 10.1016/j.ymeth.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(2016) 2016 Physician specialty data report: executive summary. Association of American Medical Colleges. https://www.aamc.org/download/471786/data/2016physicianspecialtydatareportexecutivesummary.pdf. Accessed 30 Jan 2018
- 34.Allsbrook WC, Mangold KA, Johnson MH, et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: urologic pathologists. Hum Pathol. 2001;32:74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 35.Onik G, Barzell W. Transperineal 3D mapping biopsy of the prostate: an essential tool in selecting patients for focal prostate cancer therapy. Urol Oncol Semin Orig Investig. 2008;26:506–510. doi: 10.1016/j.urolonc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Valerio M, Anele C, Charman SC, et al. Transperineal template prostate-mapping biopsies: an evaluation of different protocols in the detection of clinically significant prostate cancer. BJU Int. 2016;118:384–390. doi: 10.1111/bju.13306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 a. General pathology workflow. b. workflow with the implementation of digital pathology (TIFF 18958 kb)