Abstract
Chromobacterium violaceum is a ß-proteobacterium found widely worldwide with important biotechnological properties and is associated to lethal sepsis in immune-depressed individuals. In this work, we report the discover, complete sequence and annotation of a plasmid detected in C. violaceum that has been unnoticed until now. We used DNA single-molecule analysis to confirm that the episome found was a circular molecule and then proceeded with NGS sequencing. After DNA annotation, we found that this extra-chromosomal DNA is probably a defective bacteriophage of approximately 44 kilobases, with 39 ORFs comprising, mostly hypothetical proteins. We also found DNA sequences that ensure proper plasmid replication and partitioning as well as a toxin addiction system. This report sheds light on the biology of this important species, helping us to understand the mechanisms by which C. violaceum endures to several harsh conditions. This discovery could also be a first step in the development of a DNA manipulation tool in this bacterium.
Introduction
Chromobacterium violaceum is a Gram-negative facultative anaerobe bacillus belonging to the Neisseriaceae family1. This free-living ß-proteobacterium reside mainly around tropical and sub-tropical regions. The study of C. violaceum started in the 1970s, focusing on its potential in pharmacology and industry for the production of antibiotics, anti-tumoral substances, biopolymers and others organic compounds (reviewed in refs2–4). C. violaceum is also an opportunistic pathogen that can cause severe infections and lead to sepsis and sometimes death in immuno-depressed individuals5,6.
In 2003, the complete genome of C. violaceum was sequenced and many genes related to stress adaptability were identified. This led to a great number of studies of how the bacterium copes with environmental challenges7–11. Many studies focusing on understanding the mechanisms of quorum sensing in C. violaceum make this organism an important model species12–14.
Despite the great interest in C. violaceum and the sequencing of its entire genome7,8,15, efficient methods to modify its genome are still not developed. For example, a study reported genetic transformation of C. violaceum16 but this methodology proved to be irreproducible by many groups. More recently, a group succeeded in generating mutants in C. violaceum using conjugation17. This method is laborious and mutants often revert. Therefore, there is a demand to develop more efficient tools to conduct genetic studies in C. violaceum.
Here, we used single DNA molecule analysis and next-generation sequencing to identify a plasmid in C. violaceum strain ATCC 12472. The presence of this 44,212 bp plasmid has been unnoticed until now and its characterization may help building a shuttle vector that would greatly facilitate the development of genome engineering tools for C. violaceum.
Experimental Procedures
Plasmid isolation
Four isolated colonies of C. violaceum ATCC 12472 were inoculated in four flasks containing 400 mL of LB medium each for 16–18 h. The cultures were centrifuged at 4 °C, 5 minutes, 7441 × g. The pellets were resuspended with 25 mL of Ressuspension Buffer (50 mM Tris-HCl, 10 mM EDTA, RNAse 100 μg/mL, pH 8.0), and then 25 mL of Lysis Buffer (SDS 1%; 0.2 M NaOH) was added, with 5 minutes of room temperature incubation. The plasmid DNA was precipitated by adding 25 mL of 3 M Potassium Acetate, pH 5.5, followed by centrifugation at 22789 × g, 10 minutes at 4 °C. The supernatant was transferred to a new tube and 0.7 volume of isopropanol was added. After one more step of centrifugation (22789 × g, 10 minutes, 4 °C), the pellets were re-suspended with 1 mL of TE Buffer and one volume of phenol:chloroform was added. After centrifugation (22789 × g, 10 minutes, 4 °C), the aqueous phase was transferred to a new tube and the DNA was precipitated with one volume of isopropanol. Finally, the DNA was washed with 80% Ethanol and the four independent preparations were re-suspended with 500 μL of TE.
In order to certify that our preparation was free of genomic DNA we isolated the band containing the plasmid and digested the agarose using ß-agarase (NEB catalog # - M0392S) according to the manufacturer’s instructions.
Optical analysis of DNA in nanochannels
The optical DNA mapping of the single plasmid molecules were performed as described in ref.18. Using a combination of a DNA fluorescent dye (YOYO-1) and Netropsin, an antibiotic that binds specifically to AT DNA regions, this technique allows the acquisition of DNA barcode images, with dark and bright regions corresponding to AT-rich and GC-rich regions respectively19. In this way, the pattern of the emission intensity reflects the sequence of the DNA molecule, with a resolution on the kilobasepair length scale. The nanofluidic chips were fabricated in fused silica, using conventional techniques, as described in ref.20. All the data was recorded, using a Zeiss AxioObserver. Z1 microscope equipped with a 100× TIRF oil immersion objective (NA = 1.46) from Zeiss and a Photometrics Evolve EMCCD camera.
NGS Sequencing and Assembly
The DNA was quantified using Qubit Fluorometric Quantitation and the quality was checked on an agarose gel. The library was prepared using TruSeq Nano DNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions and then sequenced on the Illumina MiSeq at Fasteris SA. For the base-calling, the CASAVA pipeline 1.8 was used. De novo genomic assembly was made using VELVET v1.2.10 and Burrows-Wheeler Alignment Tool (v0.5.9) for mapping.
Plasmid annotation and comparison
The annotation was made using Glimmer (v3.02b), a software built to find genes in bacteria, archaea and viruses. Bacteria/archaea genetic code and circular topology were chosen. The search for homology of the whole pChV1 sequence was made using the BLASTn program against non-redundant (NR) NCBI database and against a specific bacteriophage database (unclassified bacteriophages – taxid: 12333), also from NCBI. Comparison of the predicted ORFs in genomic databases was made using BLASTx. Hits with more than 50% coverage and with the highest BitScore were picked. Search for tRNAs was made using the online version of tRNAscan-SE v1.21 in default mode. DNA inverted repeated sequences were obtained using Einverted (http://emboss.bioinformatics.nl/cgi-bin/emboss/einverted). The search for palindromic DNA was made using the MEME web-tool21. GC content profile and GC-skew were obtained using GC-Profile22 and GenSkew (http://genskew.csb.univie.ac.at/), respectively.
Data availability
The pChV1 complete sequence is available at GenBank (accession number - MG651603). FASTQ file is also available in the Sequence Read Archive (SRA) repository with accession number SRR6363036.
Results
Identification of an episome in C. Violaceum strain ATCC 1242
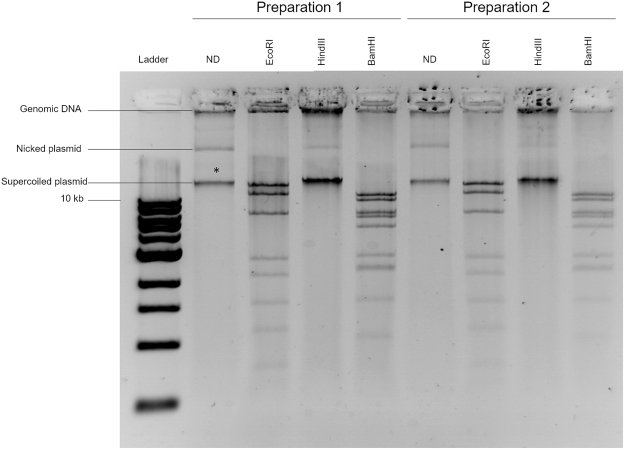
While extracting genomic DNA from C. violaceum strain ATCC 12472 to construct a genomic library, we noticed after agarose gel electrophoresis the recurrence of a DNA species smaller than expected for high molecular weight genomic DNA in our preparations. We hypothesized that this DNA species could be a circular episome. We therefore carried out standard plasmid DNA preparations and analyzed the purified DNA by agarose gel electrophoresis and ethidium bromide staining. As can be seen in lane 2 of Fig. 1, the preparation contained contaminating high molecular weight genomic DNA trapped in the well but also a species with mobility much greater than 10 kb, our putative episome as indicated by a star symbol. A third faster migrating species was also observed. We next performed a restriction enzyme analysis of our preparation with KpnI, BamHI or EcoRI (Fig. 1 lanes 3–5). Consistent with linearization of a circular DNA molecule, digestion with KpnI resulted in a single band and disappearance of genomic DNA, both the DNA trapped in the well and the third species described above, due to characteristic smearing of genomic DNA digestions (Fig. 1 lane 3). Digestion with BamHI or EcoRI resulted instead in defined patterns of discrete DNA fragments (Fig. 1 lanes 4 and 5). We then performed the same analysis using seven different additional C. violaceum strains (Fig. 2). Preparations from strains CVAC02, CVAC05, and CVT8 appeared to contain a putative episome similar to strain ATCC 12472, while preparations from strains CV026, CVT19 and CVRP5 appeared to contain only genomic DNA. The preparation from strain CVT24 also seemed to contain a putative episome species but the result from the restriction analysis is difficult to interpret. Thus, we identified an episome in C. violaceum strain ATCC 12472 and propose to name it pChV1.
Figure 1.
Restriction digestion pattern from two independent preparations of the episome. The asterisk denotes the band corresponding to the plasmid. The restriction enzyme used is mentioned on top of each lane. 0.8% TBE Agarose gel stained with ethidium bromide.
Figure 2.
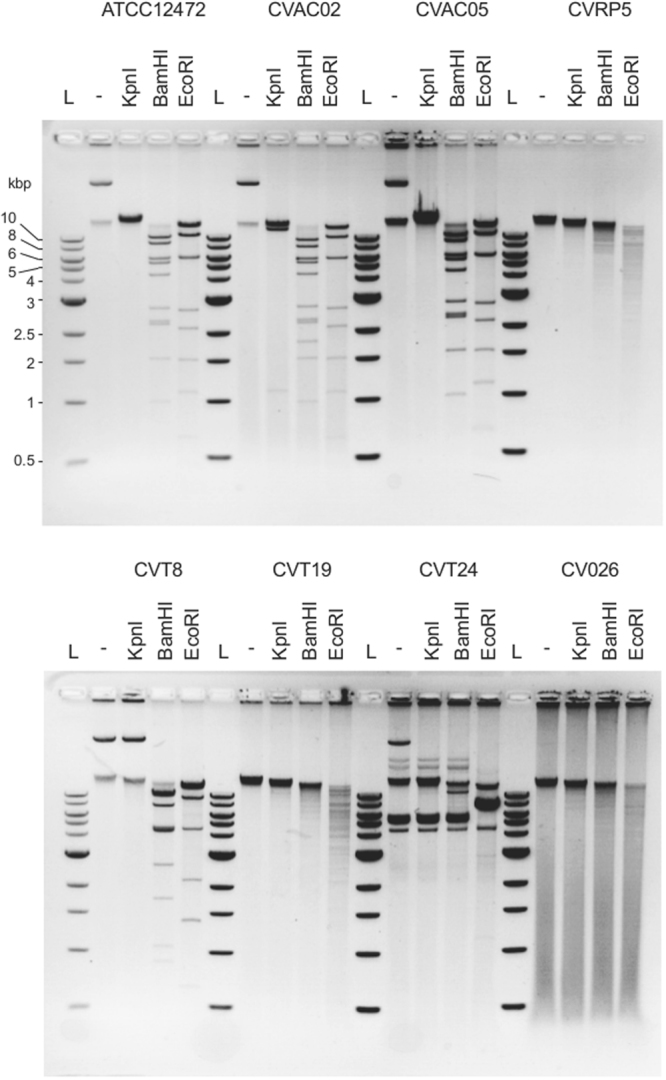
Restriction digestion of extra-chromosomal DNA extracted from eight different C. violaceum strains. Samples were digested with KpnI, BamHI and EcoRI for one hour at 37 °C. “−” reflects non digested samples and L is DNA ladder. 0.8% TBE agarose gel stained with ethidium bromide.
Episome pChV1 is a circular plasmid
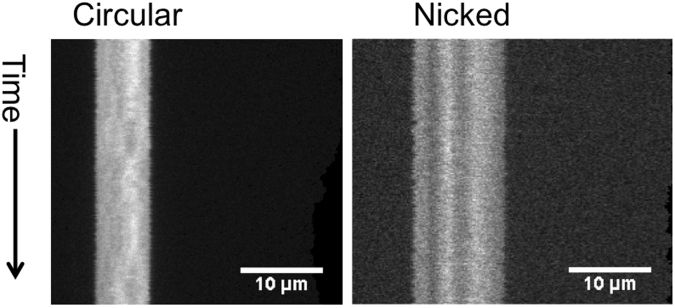
The above restriction enzyme analysis suggested that pChV1 is a circular DNA molecule. To verify this hypothesis the episome DNA was purified after gel electrophoresis (Fig. 1 lane 2, band indicated with a star symbol) and stained with YOYO-1 (a bis-intercalator fluorescent dye) and Netropsin (a minor groove binder of AT-rich sequences of double-stranded DNA) that competes with YOYO-1 intercalation19. Such stained preparations were diluted and injected in nanochannels to observe individual extended episome DNA molecules by fluorescence microscopy (Fig. 3A). Initially, the contour length of individual episome DNA molecules averaged circa 6 μm (Fig. 3A - left kymograph). However, after prolonged illumination, the accumulation of nicks in the DNA molecule induced double strand breakage and linearization of the circular plasmid, evidenced by an increase in contour length (Fig. 3B - right kymograph). Lambda phage DNA (48,502 bp) was used as an internal standard to convert extension from pixels to basepairs. Using a scaling factor of 1.8423 for going from circular to linear extension, we estimated that the longest episome molecule detected was circa 44 kb in size. DNA barcode analysis revealed mostly GC-rich regions with two AT-rich regions of darker signal.
Figure 3.
Kymographs showing the extensions of circular and nicked forms of the episome in nanofluidic channels. Competitive binding was used in order to produce the emission intensity pattern along the linear for of the plasmid.
The pChV1 DNA sequence
The complete sequence of pChV1 revealed a circular element with 44,212 bp with a G + C content of 65.96% (Table 1). 39 Open Reading Frames (ORFs) were found, which comprises 89,66% of the whole plasmid (Fig. 4). From these, 28 are conserved hypothetical proteins and 1 is a hypothetical protein. Comparing the ORFs of the plasmid with other organisms, we observed that 17 (43%) of the ORFs have similarity with ORFs from Pseudogulbenkiania ferrooxidans. No tRNAs genes were found. We also searched for homology with bacteriophages and the BLAST analysis did not give any similarity with any phage genomes.
Table 1.
General features of pChV1.
| Lenght, bp | 44,212 |
| G + C content | 65.96% |
| Total ORFs | 39 |
| Percentage of plasmid sequence constituting coding regions | 89.66% |
| Average ORF lenght, bp | 1017 |
| Number of conserved hypothetical proteins | 28 |
| Number of hypothetical proteins | 1 |
Figure 4.
Map of the pChV1 plasmid. Most of the phage-related genes are present in the same region. The distribution of the plasmid partitioning genes is in accordance to what is seen in the literature. Blue, red and green arrows depict phage, plasmid and hypothetical ORFS respectively.
Plasmid maintenance genes
The plasmid has at least 4 known genes related to plasmid segregation/replication: parA, parB, repA and a gene with RPA domain, involved in plasmid replication initiation (Table 2). parA and parB encode the ParA and ParB proteins, respectively. These proteins are part of the Type I plasmid-partitioning system and are responsible for ensuring the correct propagation of plasmids to daughter cells throughout cell division24. This partitioning system is founded in prophages, plasmids and chromosomes25. RepA is a protein related to plasmid replication and is characteristic of P1 plasmids.
Table 2.
List of ORFs found in pChV1.
| ORF Number | Gene | Length | Domains | Best Hit Identity |
|---|---|---|---|---|
| ORF_01 | Chromosome partitioning protein ParA | 840 | ParA/Soj/Fer4 NifH | 93% |
| ORF_02 | Partitioning protein ParB | 885 | ParB | 96% |
| ORF_03 | Plasmid replication protein RepA | 951 | Plasmid replication initiator protein | 49% |
| ORF_04 | Conserved hypothetical protein | 477 | none | 82% |
| ORF_05 | Conserved hypothetical protein/putative bacteriophage lysis protein | 1029 | COG4623 | 82% |
| ORF_06 | Conserved hypothetical protein | 312 | SlyX | 97% |
| ORF_07 | Conserved hypothetical protein | 756 | Cadherin repeat/Ca2+ binding | 32% |
| ORF_08 | Conserved hypothetical protein | 1422 | none | 44% |
| ORF_09 | Conserved hypothetical protein | 1887 | none | 60% |
| ORF_10 | Conserved hypothetical protein | 363 | none | 99% |
| ORF_11 | Conserved hypothetical protein | 219 | none | 95% |
| ORF_12 | Conserved hypothetical protein | 1167 | DUF4157 | 48% |
| ORF_13 | Toxin | 801 | RhsA | 52% |
| ORF_14 | Conserved hypothetical protein | 708 | none | 63% |
| ORF_15 | Conserved hypothetical protein | 1125 | none | 98% |
| ORF_16 | Conserved Hypothetical protein | 126 | none | 52% |
| ORF_17 | Conserved hypothetical protein | 615 | Transposase | 99% |
| ORF_18 | Hypothetical protein | 726 | none | ND |
| ORF_19 | Conserved hypothetical protein | 891 | DUF4255 | 95% |
| ORF_20 | Conserved hypothetical protein | 1854 | ATPase AAA domain | 72% |
| ORF_21 | Conserved hypothetical protein | 2415 | none | 62% |
| ORF_22 | Conserved hypothetical protein | 453 | none | 78% |
| ORF_23 | Conserved hypothetical protein | 3768 | DUF342 | 95% |
| ORF_24 | Conserved hypothetical protein | 2439 | none | 82% |
| ORF_25 | Conserved hypothetical protein | 993 | ribonuclease e/dihydrolipoamide succyniltransferase | 66% |
| ORF_26 | Conserved hypothetical protein | 3168 | TIGR02243(phage tail-like region) | 77% |
| ORF_27 | Conserved hypothetical protein | 441 | none | 90% |
| ORF_28 | Phage baseplate assembly protein W | 399 | GPW gp25 | 71% |
| ORF_29 | Conserved hypothetical protein | 1593 | Phage Base V | 59% |
| ORF_30 | Conserved hypothetical protein | 729 | Phage Tube | 41% |
| ORF_31 | Conserved Hypothetical protein | 165 | none | 52% |
| ORF_32 | Conserved hypothetical protein | 456 | Phage T4 gp19 | 40% |
| ORF_33 | Phage tail protein | 447 | Phage T4 gp19 | 91% |
| ORF_34 | Phage tail sheath protein | 1416 | COG3497(phage tail sheath protein FI) | 63% |
| ORF_35 | DNA invertase | 624 | mpi/SR ResInv(Recombinase;DNA binding)/HTH Hin like | 97% |
| ORF_36 | Conserved hypothetical protein | 348 | HTH_XRE (transcriptional regulator family)/xenobiotic response | 97% |
| ORF_37 | Toxin HipA | 1350 | HipA/Rna Pol | 90% |
| ORF_38 | Conserved Hypothetical protein | 126 | none | 52% |
| ORF_39 | Plasmid replication initiator protein | 1182 | RPA | 54% |
Structural phage genes
An abundant number of genes related to phage structure are present in the sequence of the plasmid. Genes that codify the baseplate, sheath and tail proteins as well as conserved hypothetical genes with domains related to phage structure are in close proximity in the pChV1 sequence.
Other genes
A DNA invertase (ORF_35), an enzyme that catalyzes site-specific recombination in phages was found. A conserved hypothetical protein (ORF_17) with a transposase domain is also present in the plasmid sequence. Toxins (ORF_13 and ORF_37) that may be related to the toxin-antitoxin (TA) system responsible for assuring the survival only for the cells with a copy of the lisogenyzed phage were also located. Other worthy-mention genes are: conserved hypothetical proteins with Ribonuclease E domain, XRE domain and ATPase AAA domain.
GC profile and GC-skew
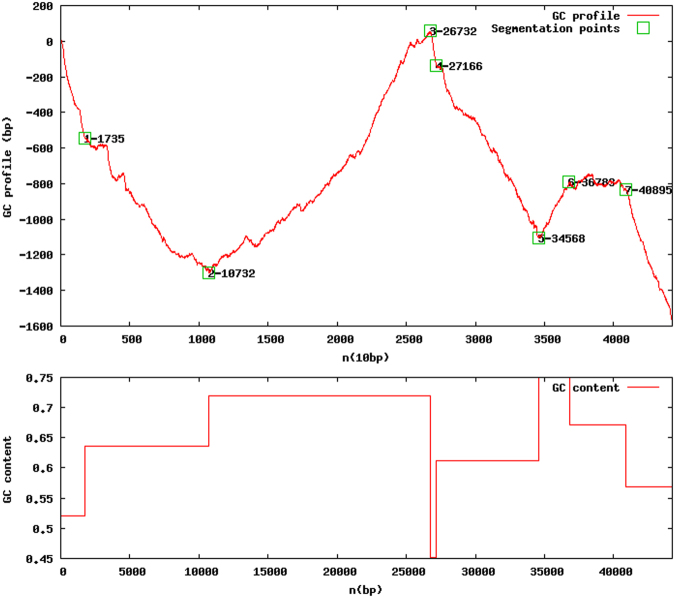
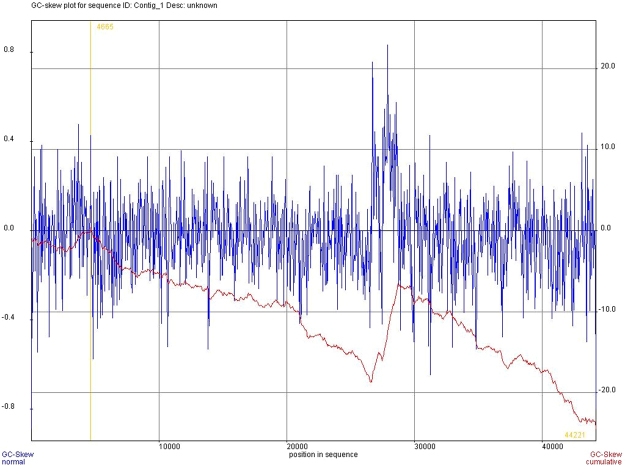
We were able to identify two points in the sequence of pChV1 where the GC content drops when comparing to the whole sequence (Fig. 5). These variations also qualitatively agree with intensity variaitons in the single molecules studies in Fig. 3. This might reflect the presence of two origins of replication that are present in P1-like plasmids, oriR and oriL. GC-skew also helps predicting the location of the leading and lagging strand and cumulative GC-skew values reflect the origin and terminus points of replication26. In our analysis, we can observe throughout the cumulative GC-skew curve, two regions that we could call minimum points that sign the origins of replication oriR and oriL (Fig. 6).
Figure 5.
GC content profile of pChV1. Two points of low GC content are observable in the 1) beginning-end of the plasmid sequence and in the 2) 27000 bp region (second chart, below).
Figure 6.
GC-skew of pChV1. The cumulative GC-skew (red curve) has two decline points, characteristic of origins of replication and might reflect oriR and oriL.
Repeated and palindromic sequences
A 19 bp inverted repeated sequence separated by 1,785 bp was also located and may be involved in the circularization of the phage or other homologous recombination-based process (Table 3). This pair of sequences is located flanking the partitioning related genes parA and parB (Fig. 7). Other inverted repeat sequences with size varying from 23 to 54 bp were also founded although the complementarity between the pair of repeats was not 100% (data not shown). Palindromic sequences located at two distinct sites in the sequence and varying from 7 to 17 bp are also present (Table 3).
Table 3.
Pairs of Inverted repeated and palindromic sequences founded in pChV1.
| Coordinates | Strand | Sequence | Type of sequence |
|---|---|---|---|
| 42355–42373 44177–44159 |
+ + |
TGTAGCAAGTTGCTACACT ACATCGTTCAACGATGTGA |
Inverted repeat sequence |
| 5113–5120 10723–10730 |
+ + |
AAATATTT | Palindrome |
| 44161–44177 42355–42371 |
− + |
TGTAGCAAg/cTTGCTACA | Palindrome |
| 27010–27016 26990–26996 |
− + |
ATAt/aTAT | Palindrome |
| 1608–1615 32309–32316 |
+ + |
TGAATTCA | Palindrome |
| 1465–1478 41121–41134 |
+ − |
TTTTTAACTAAAAA TTTATAGTTAAGAA |
Palindrome |
| 304–312 132–140 |
− + |
TTGAt/aTCAA | Palindrome |
| 34202–34209 33338–33345 |
+ − |
AATTAATT AAGTAATT |
Palindrome |
| 34414–34424 4595–4605 |
+ − |
ATTTGTCATAT | Palindrome |
| 27884–27891 32052–32059 |
− − |
TATTCATA | Palindrome |
Figure 7.
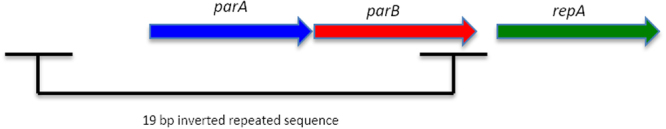
Schematic chart showing the genomic context in which the 19 bp inverted repeated sequence are flanking the partitioning-related genes parA and parB. The diagram is not to scale.
Discussion
The first bacteriophage was discovered in the 1950’s27 and since then, the number and variety of new viruses that infect bacteria has grown considerably, reaching more than 1,300 genome projects according to the NCBI database. While researching the opportunistic pathogen C. violaceum, in genomic preparations we observed an extra-chromosomal DNA of high molecular weight (but lower than it would be if it was genomic DNA). We then isolated and sequenced this putative plasmid which proved to have genes from the P1 bacteriophage/plasmid group.
After the sequencing of C. violaceum7, the presence of four different sequences of prophages (CvP1-4) were observed in the C. violaceum’s genome28. Neither of these is related to the plasmid we report here. Before this, tail-like particles were observed in C. violaceum by electron microscopy although no biological activity was associated to them29,30.
According to the sequence data and annotation, the plasmid founded in C. violaceum could be a P1-like virus due to the presence of genes that encode for structural viral particles. Moreover, genes related to plasmid partitioning and the plasmid initiator protein RepA are strong evidence to classify this plasmid as a P1-like phage. Another hallmark of P1-like phages is the presence of toxin-antitoxin genes that constitute a plasmid addiction system. In pChV1 two ORFs are predicted to be toxin genes (ORFs 13 and 37 with 52% and 90% of identity, respectively) although further studies need to be done to confirm the presence of this system.
From our search for homology, we observed that pChV1 has a nucleotide sequence very different from other phages described so far. This feature hampers the search for phage-related sequences, such as lox sites, incC and incA and others, which are, in general, well conserved between other viruses, but does not exclude the existence of them in pChV1. However, repeated sequences that are founded amongst other phages are also present in pChV1, such as the 19 bp inverted repeated sequence (Fig. 7).
Origins of replication are GC-poor regions and locating them in the plasmid may suggest the locals where replication starts. Although we were not able to predict specific sequences that would correspond to origins of replications in pChV1, the GC content profile and GC-skew showed two regions that might reflect oriR and oriL. oriR, that is used during plasmid maintenance replication, is in the same region as the parA, parB and repA genes. This co-location of a possible origin of replication and the plasmid maintenance genes is observable in pChV1. Conversely, oriL is related to lytic growth and is separated about 9 kb from oriR in P131. We suggest that the second possible origin of replication founded in pChV1 (located approximately at 27 kbp) corresponds to oriL.
One notable feature is that when we aligned the predicted open reading frames using BLASTn (that searches a nucleotide query in a nucleotide database) we obtained no significant result. Conversely, when BLASTx was used (searches a translated nucleotide query in a protein database) we were able to identify genes with high degree of confidence. This means that during evolution this virus accumulated many mutations on its DNA sequence but conserved – to some extent - the amino acid composition of its proteins. For example, pChV1 has many ORFs with more than 90% of identity with other genes found in bacteria (Table 2). When we aligned these same ORFs using BLASTn we did not obtain any significant result.
Besides the presence of phage-related genes and sequences, some essential elements that would make pChV1 a functional P1-phage are still missing31. By the lack of evidence, we cannot conclude if this plasmid is a temperate P1-like phage, or if it is a chimeric DNA, part bacteriophage or plasmid. Moreover, it could be a fragment of DNA that is maintained inside C. violaceum by addiction systems but defective in its capacity of lisogeny. Conversely, the tail-like particles observed in the 1970s29,30 could be an evidence that, under stress, the phage proteins encoded by pChV1 would be produced.
Genetic mobile elements are still important in the field of molecular biology. Beside this, the use of phage-derived systems as tools has allowed genome manipulation of all kind of organisms. In this way, further study of pChV1 would bring new ways to investigate genetic aspects of C. violaceum and maybe other species. Finally, pChV1 with its great number of hypothetical ORFs, is a rich reservoir of unexplored genes that might contribute to our understanding of the mechanisms underlying viral infections and plasmids.
Conclusion
In our work, we discovered an extra-chromosomal DNA – that we named pChV1 - in the opportunistic pathogen Chromobacterium violaceum. This plasmid is present as a low-copy plasmid and has most of its genetic apparatus composed of ORFs with unknown function, making pChV1 an important source of genes to be further explored. More than this, when its biology is better understood, this element can be used in genetic studies in C. violaceum as well as in other organisms.
Acknowledgements
The authors would like to thank UFRN, CNPq, CAPES for financial support. The authors are also grateful to Mauro Modesti who performed Restriction Digestion assay and also hosted D.C.L. in his lab.
Author Contributions
D.C.L. participated in the design of the study, acquisition of data and analysis and interpretation of data and drafted the manuscript. L.K.N. and F.W. performed optical mapping analysis of DNA and interpreted the results. S.R.B.M. contributed to the study conception and design, writing of the manuscript and overall supervision. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Garrity, G. M. & Holt, J. G. In Bergeys Manual of Systematic Bacteriology The Archaea and the Deeply Branching and Phototrophic Bacteria 119–166, 10.1007/978-0-387-21609-6_15 (2001).
- 2.Durán N, et al. MINIREVIEW Violacein: properties and biological activities. Biotechnol. Appl. Biochem. 2007;133:127–133. doi: 10.1042/BA20070115. [DOI] [PubMed] [Google Scholar]
- 3.Durán M, Faljoni-Alario A, Durán N. Chromobacterium violaceum and its important metabolites–review. Folia Microbiol. (Praha). 2010;55:535–47. doi: 10.1007/s12223-010-0088-4. [DOI] [PubMed] [Google Scholar]
- 4.Durán M, et al. Potential applications of violacein: A microbial pigment. Med. Chem. Res. 2012;21:1524–1532. doi: 10.1007/s00044-011-9654-9. [DOI] [Google Scholar]
- 5.Yang C-H, Li Y-H. Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J. Chin. Med. Assoc. 2011;74:435–41. doi: 10.1016/j.jcma.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Luz KG, et al. Chromobacterium violaceum: a fatal case in the northeast of the Brazil. J Bras Patol Med Lab. 2014;50:278–279. [Google Scholar]
- 7.Vasconcelos ATR, et al. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA. 2003;100:11660–5. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baraúna R. Genes (Basel). 2011. a. et al. Proteomics Analysis of the Effects of Cyanate on Chromobacterium violaceum Metabolism; pp. 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lima DC, et al. The influence of iron on the proteomic profile of Chromobacterium violaceum. BMC Microbiol. 2014;14:267. doi: 10.1186/s12866-014-0267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro, D. et al. Proteomic analysis of Chromobacterium violaceum and its adaptability to stress Proteomic analysis of Chromobacterium violaceum and its adaptability to stress. BMC Microbiol. 15 (2015). [DOI] [PMC free article] [PubMed]
- 11.Duarte, F. T. et al. GeLC-MS-based proteomics of Chromobacterium violaceum: comparison of proteome changes elicited by hydrogen peroxide. (2016). [DOI] [PMC free article] [PubMed]
- 12.Zhu, H., He, C.-C. & Chu, Q.-H. Inhibition of quorum sensing in Chromobacterium violaceum by pigments extracted from Auricularia auricular. Lett. Appl. Microbiol. 1–6, 10.1111/j.1472-765X.2011.02993.x (2011). [DOI] [PubMed]
- 13.Chaudhari V, Gosai H, Raval S, Kothari V. Effect of certain natural products and organic solvents on quorum sensing in Chromobacterium violaceum. Asian Pac. J. Trop. Med. 2014;7:S204–S211. doi: 10.1016/S1995-7645(14)60233-9. [DOI] [PubMed] [Google Scholar]
- 14.Skogman, M. E., Kanerva, S., Manner, S., Vuorela, P. M. & Fallarero, A. Flavones as quorum sensing inhibitors identified by a newly optimized screening platform using chromobacterium violaceum as reporter bacteria. Molecules21 (2016). [DOI] [PMC free article] [PubMed]
- 15.Ciprandi A, et al. Proteomic Response to Arsenic Stress in Chromobacterium violaceum. J. Integr. OMICS. 2012;2:69–73. [Google Scholar]
- 16.Gene, A., An, T., Lecula, M. O., Tech, B. I. O. & Broetto, N. L. Stable transformation of Chromobacterium violaceum with a broad-host-range plasmid. Biologia (Bratisl). 450–454, 10.1007/s00253-005-0140-5 (2006). [DOI] [PubMed]
- 17.da Silva Neto JF, Negretto CC, Netto LES. Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a cys-based redox sensor regulated by thioredoxin. PLoS One. 2012;7:e47090. doi: 10.1371/journal.pone.0047090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyberg, L. K. et al. Rapid identification of intact bacterial resistance plasmids via optical mapping of single DNA molecules. Sci. Rep. 6 (2016). [DOI] [PMC free article] [PubMed]
- 19.Nilsson, A. N. et al. Competitive binding-based optical DNA mapping for fast identification of Bacteria - Multi-ligand transfer matrix theory and experimental applications on Escherichia coli. Nucleic Acids Res. 42 (2014). [DOI] [PMC free article] [PubMed]
- 20.Persson F, Tegenfeldt JO. DNA in nanochannels—directly visualizing genomic information. Chem. Soc. Rev. 2010;39:985. doi: 10.1039/b912918a. [DOI] [PubMed] [Google Scholar]
- 21.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 22.Gao, F. & Zhang, C. T. GC-Profile: A web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 34 (2006). [DOI] [PMC free article] [PubMed]
- 23.Alizadehheidari M, et al. Nanoconfined circular and linear DNA: Equilibrium conformations and unfolding kinetics. Macromolecules. 2015;48:871–878. doi: 10.1021/ma5022067. [DOI] [Google Scholar]
- 24.Salje J. Plasmid segregation: how to survive as an extra piece of DNA. Crit. Rev. Biochem. Mol. Biol. 2010;45:296–317. doi: 10.3109/10409238.2010.494657. [DOI] [PubMed] [Google Scholar]
- 25.Pinto UM, Pappas KM, Winans SC. The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 2012;10:755–765. doi: 10.1038/nrmicro2882. [DOI] [PubMed] [Google Scholar]
- 26.Eppinger M, Baar C, Raddatz G, Huson DH, Schuster SC. Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2004;2:872–885. doi: 10.1038/nrmicro1024. [DOI] [PubMed] [Google Scholar]
- 27.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Almeida R, et al. Bacteriophages and insertion sequences of Chromobacterium violaceum ATCC 12472. Genet. Mol. Res. 2004;3:76–84. [PubMed] [Google Scholar]
- 29.Rucinsky TE, Gregory JP, Cota-Robles EH. Organization of bacteriophage tail-like particles in cells of Chromobacterium violaceum. J. Bacteriol. 1972;110:754–757. doi: 10.1128/jb.110.2.754-757.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rucinsky TE, Cota-Robles EH. The intracellular organization of bacteriophage tail-like particles in cells of Chromobacterium violaceum following mitomycin C treatment. J. Ultrastruct. Res. 1973;43:260–269. doi: 10.1016/S0022-5320(73)80038-3. [DOI] [PubMed] [Google Scholar]
- 31.Lobocka, M. B. et al. Genome of Bacteriophage P1. 186 (2004). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The pChV1 complete sequence is available at GenBank (accession number - MG651603). FASTQ file is also available in the Sequence Read Archive (SRA) repository with accession number SRR6363036.