Abstract
Human dihydrolipoamide dehydrogenase (hLADH) is a flavoenzyme component (E3) of the human alpha-ketoglutarate dehydrogenase complex (α-KGDHc) and few other dehydrogenase complexes. Pathogenic mutations of hLADH cause severe metabolic diseases (atypical forms of E3 deficiency) that often escalate to cardiological or neurological presentations and even premature death; the pathologies are generally accompanied by lactic acidosis. hLADH presents a distinct conformation under acidosis (pH 5.5–6.8) with lower physiological activity and the capacity of generating reactive oxygen species (ROS). It has been shown by our laboratory that selected pathogenic mutations, besides lowering the physiological activity of hLADH, significantly stimulate ROS generation by hLADH, especially at lower pH, which might play a role in the pathogenesis of E3-deficiency in respective cases. Previously, we generated by molecular dynamics (MD) simulation the low-pH hLADH structure and analyzed the structural changes induced in this structure by eight of the pathogenic mutations of hLADH. In the absence of high resolution mutant structures these pieces of information are crucial for the mechanistic investigation of the molecular pathogeneses of the hLADH protein. In the present work we analyzed by molecular dynamics simulation the structural changes induced in the low-pH conformation of hLADH by five pathogenic mutations of hLADH; the structures of these disease-causing mutants of hLADH have never been examined before.
Abbreviations: LADH, (dihydro)lipoamide dehydrogenase; PDHc, pyruvate dehydrogenase complex; α-KGDHc, alpha-ketoglutarate dehydrogenase complex; ROS, reactive oxygen species; RMSD, root mean square deviation; FAD, flavin adenine dinucleotide; NAD+/NADH, nicotinamide adenine dinucleotide (oxidized/reduced); MD, molecular dynamics; S.E.M., standard error of the mean; WT, wild-type
Keywords: Lipoamide dehydrogenase, Mutation, Reactive oxygen species, Molecular dynamics
Highlights
-
•
5 disease-causing mutants of hLADH were subjected to MD to reveal structural changes.
-
•
MD simulations were carried out both in vacuum and in water supplemented with ions.
-
•
Functional regions significantly affected by mutation were identified.
-
•
Implicated residues are to be targeted in mechanistic studies of hLADH dysfunction.
1. Introduction
Human dihydrolipoamide dehydrogenase (hLADH) is a flavoenzyme component (E3) of the human alpha-ketoglutarate dehydrogenase complex (hα-KGDHc) and few other dehydrogenase complexes; hLADH oxidizes the E2-bound dihydrolipoate while reducing NAD+ to NADH. In both the forward and reverse reaction, in the latter hα-KGDHc and isolated E3 consume NADH, the enzyme generates superoxide and hydrogen peroxide (ROS; reactive oxygen species) [1], [2], [3], [4]. Excess superoxide and H2O2 generation in mitochondria induces oxidative stress [5], [6]. ROS generation by hα-KGDHc and isolated E3 becomes dominant under pathological conditions at elevated NADH/NAD+ ratios and/or decreased pH (in acidosis) [1], [2]. ROS generation by hα-KGDHc was suggested to be the major source of oxidative stress in senescence/aging, ischemia-reperfusion and neurodegenerative diseases [7], [8], [9], [10], [11].
Pathogenic hLADH mutations result in E3-deficiency in humans [12]. The mutations primarily affect tissues that have high O2 demand, thus E3-deficiency often presents with severe cardiological or neurological symptoms. Clinical complications, which ultimately may lead to premature death, are usually observed already in the neonatal age [12] and are generally accompanied by lactic acidosis. hLADH presents a distinct conformation under acidosis (pH 5.5–6.8) with lower physiological activity; ROS generation activity of hLADH is also accredited to this conformation. Our laboratory recently reported that disease-causing mutations, besides their basal effect of lowering the physiological activity of hLADH, also modulate the ROS-generating capacity of hLADH; four mutations significantly stimulated while few others significantly lowered the rate of ROS generation by hLADH [13]. While elevated ROS generation by a specific mutation is implicated in pathogenesis [14] and could be a target in the treatment [15], elucidation of the mechanism by which other mutations attenuate ROS production by hLADH is potentially also important to reveal the nature of molecular interactions in the hLADH active center crucial for the development of this potentially beneficial effect [16]. Although multiple crystal structures for hLADH have been published, the only report on the low-pH (ROS-generating) conformation of hLADH is a molecular dynamics (MD) study from our own laboratory. In that work the effects of eight disease-causing mutations on the low-pH conformation of hLADH were also analyzed by MD simulations [17].
In the present contribution we analyzed by molecular dynamics simulation the structural changes induced in the low-pH conformation of hLADH by five pathogenic mutations of hLADH; the structures of these disease-causing mutants of hLADH have never been examined before (R447G [18], G101del [19], I445M [20], I12T [12], and I318T [21]). The structural information revealed here are to be used in the mechanistic evaluation of the molecular pathogeneses of these disease-causing hLADH mutants.
2. Methods
The applied methodology was published before [17] and is summarized below with the few modifications detailed.
Structure calculations and manipulations were carried out in XPLOR-NIH [22] and VMD [23] under Linux on a calculation architecture of a Hewlett-Packard supercomputer with a CP4000BL blade technology and AMD Opteron 6174 Magny Cours processors with 12 cores (the total number of cores was 2304); parallelization was not applied. XPLOR-NIH was compiled in-house due to specific adjustments made to the source code.
2.1. Mutant structure building and molecular dynamics simulations
From the eight monomers in the asymmetric unit (pdb code: 1zmc) the functional dimer of A and B was used. After performing initial adjustments to coordinate files and introducing amino acid changes at designated sites, structures for calculations simulating low pH (<6.0) were created by protonating at both nitrogen atoms all the 12 His side chains which are present in each monomer. For simulations in H2O with physiological ionic strength the dimeric holoenzyme was surrounded with a 12 Å water shell supplemented by 150 mM KCl. After an initial stage of calculations comprising conjugate gradient energy minimization steps, atoms were all assigned to a uniform initial distribution of velocities. A heating scheme of gradual steps was applied using an initial temperature of 100 K and a final temperature of 310 K (37 °C; with 10 K increase at a time) permitting 10 ps simulation time at each temperature (10,000 steps with 1 fs integration time). H2O molecules were retained during simulation by the application of a deformable stochastic boundary potential [24]. To secure adequate temperature equilibration during temperature incrementation steps up to 37 °C, a water shell with two layers and Langevin dynamics were applied [25]. The coordinates of 37 °C underwent 2000 steps of energy minimization followed by 2 ns of molecular dynamics simulation with the temperature control switched to temperature coupling [26] (to 310 K); initial velocities were reassigned by a Maxwellian distribution. Molecular dynamics simulations were performed in the Cartesian space applying the third-order finite difference approximation [24]. In water, two parallel experiments were performed and for the initial velocity assignments two different random seeds were applied. As a reference, simulations were repeated in vacuum with a single seed and identical settings except for the final simulation time which was set to 10 ns.
2.2. Analysis of structures
As a first step of analysis, RMS difference curves, relative to the initial structures, as a function of simulation time were generated from the trajectories. RMSD curves were created also for the domain and the 10 amino acid stretch bore the mutation. 10 ps long samples (20 coordinate sets with 1 fs integration time and every 500th step recorded) were collected from the stable parts of the trajectories and averaged. Averaged structures were carried over in case 10% of the protein chain or less was found beyond the solvation sphere after simulation. Structures were energy minimized in two subsequent stages of 2000 steps each, first with planarity restraints and then with constraints imposed on the isoalloxazine ring of FAD. Final structures were validated inside XPLOR-NIH and on the MOLPROBITY server (molprobity.biochem.duke.edu). Mutants were fitted to the wild-type structure through Cα atoms of the dimer and the respective overall RMSD values were calculated. Residue displacement plots were generated after calculating backbone RMSDs on a per residue basis relative to the reference structure. Deviations of greater than 10 Å (in water) or 7 Å (in vacuum) were filtered. Eighty four inter-atomic distances crucial in the hLADH structure were identified (VMD) and measured (XPLOR-NIH) in the FAD/NAD+/NADH or the lipoate binding sites or on the dimerization surface; respective distances in mutants and wt-LADH were compared.
All settings and procedures not detailed here were identical to the settings and procedures applied before in [17].
3. Results
3.1. Analysis of trajectory information
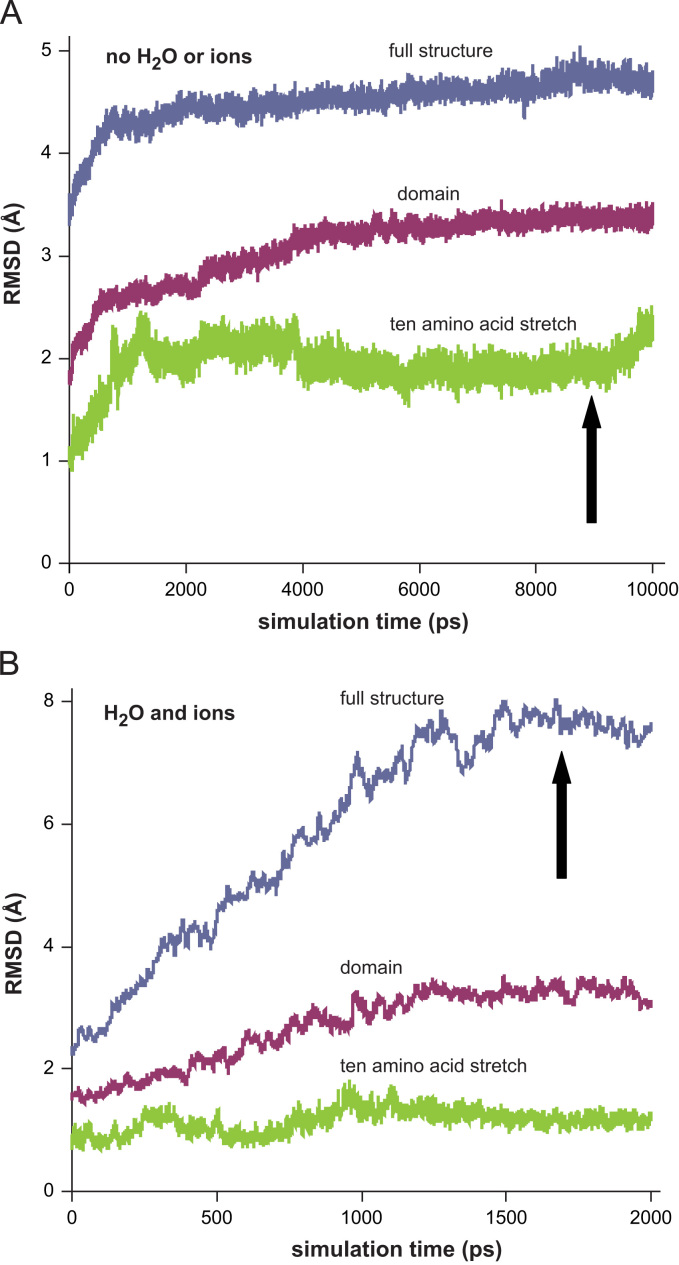
During building mutant structures chirality of the new amino acid was always verified. H2O molecules have not escaped across the boundary wall when simulations were carried out in water. Temperature was found to be very stable during all simulation experiments. At least one or two of the three RMSD curves (full protein, domain, peptide stretch) calculated from trajectories recorded in a vacuum became practically stabilized on or before 2 ns for each mutant (representative data are shown in Fig. 1); flexible loops and/or terminal stretches are the structural elements that may result in drifting RMSD curves. This justified the applied length of the very calculation demanding simulations in water (2 ns). Difference from the regular order of the magnitudes of the three RMSD curves (full protein>domain≥peptide stretch), where the peptide stretch deviated the most as a function of time, was seen only in case of G101del in water (data not shown). A conclusion was drawn that G101del leads to a robust but rather local effect while the other four mutations altered the structure on a more extended scale. All averaged structures calculated from the stable parts of the trajectories showed less than 10% peripheral amino acids outlying beyond the water shell. No energy-minimized protein structures showed any deviation in bond lengths or angles. Only those structures were analyzed further though that possessed at least one FAD with reasonably planar isoalloxazine ring structure. Ramachandran outliers that were not in or entirely borderline to the allowed regions were always ≤1% (Fig. S1).
Fig. 1.
Representative RMSD curves for the MD simulation trajectories of the I445M mutant of the low-pH hLADH structure. RMSDs were calculated for Cαs from both monomers relative to the initial structure in vacuum (A) or in water plus 150 mM KCl (B) (I445M structure #1). The three curves with different colors represent (for A and B) RMSDs calculated using the full structure (blue), the domain carrying the actual mutation (red) or a 10 amino acid stretch that holds the mutation in the middle (green). Arrows designate the points of sampling the trajectories. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Analysis of the mutant structures
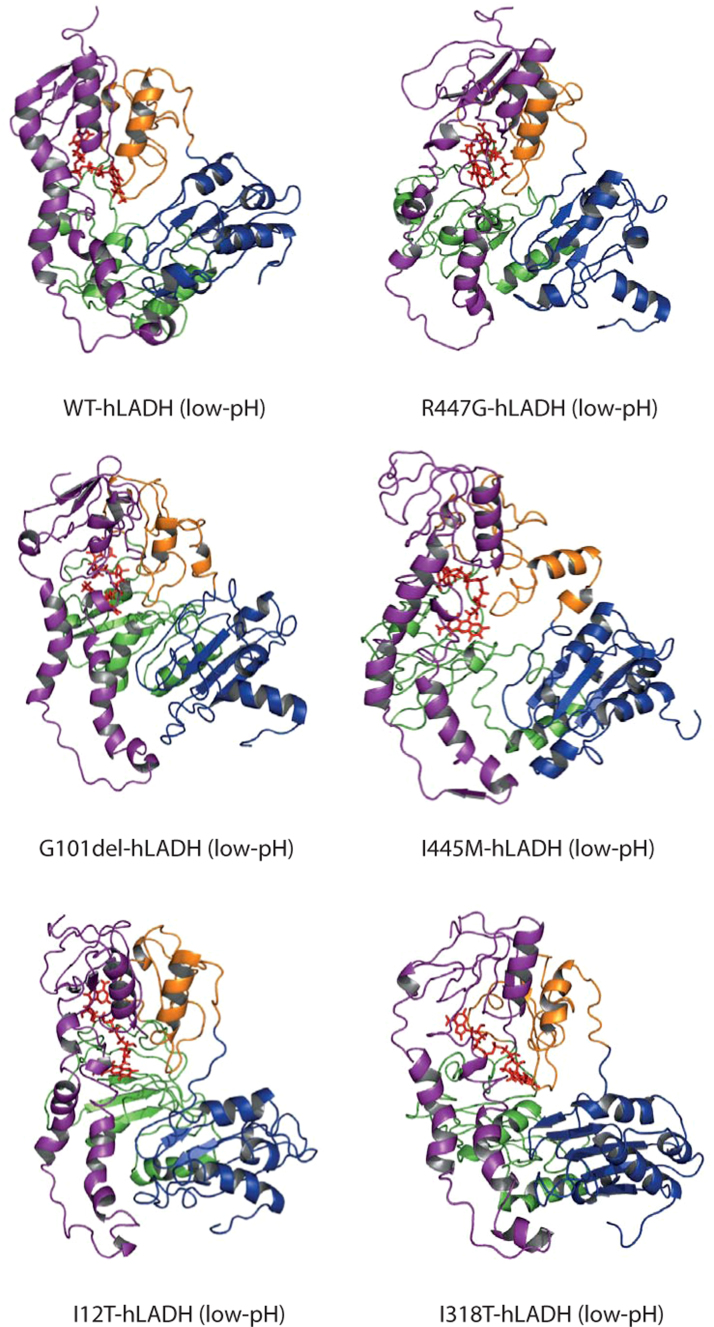
Mutant structures (Fig. 2) were fitted to the wild-type structure (Fig. S2A-B) in both simulation environments and respective full-structure RMSDs were calculated (Table 1). The results show no considerable inter-monomeric displacement for any of the mutants, which is in accordance with our data on the other eight hLADH pathogenic mutants [13], [17]. As calculated in [17], RMSDs greater than 7.51 (in water) or 1.49 Å (in vacuum) provide structures which are significantly different from WT-hLADH; all mutants in Table 1 satisfy this requirement.
Fig. 2.
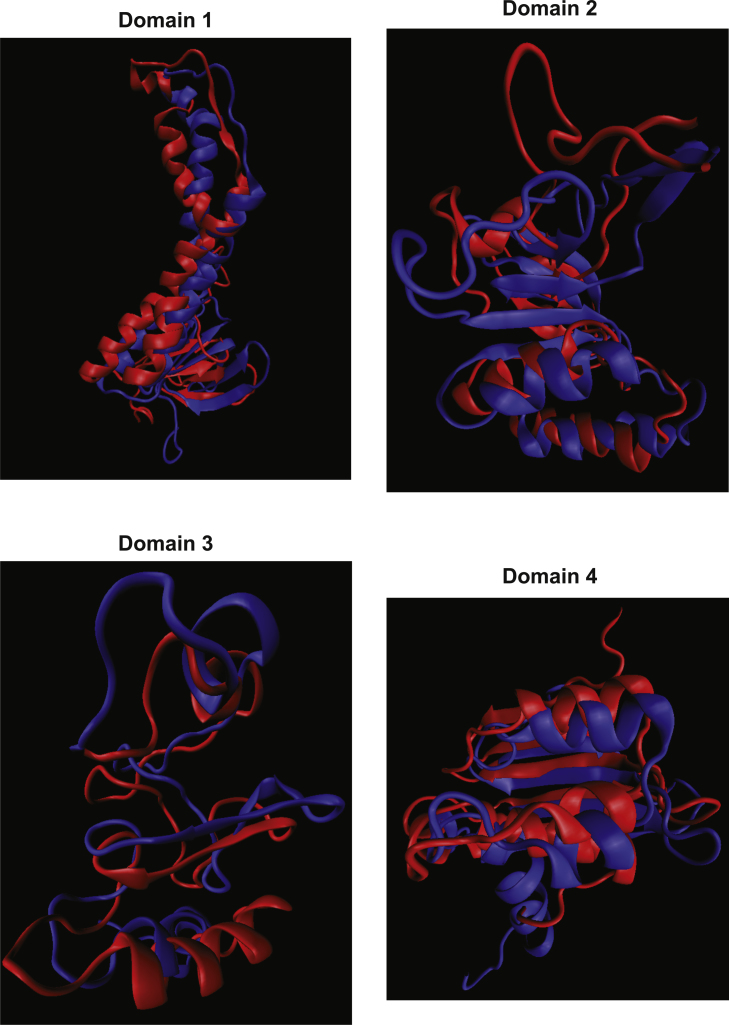
Structural models for wild-type (WT) hLADH and its five pathogenic mutants (structures #1) in their low-pH forms in water plus ions. Only monomer A is shown with different colors for domains: FAD-binding domain (1–149) – purple, NAD+/NADH-binding domain (150–282) – green, central domain (283–350) – orange, and interface domain (351–474) – blue. Structures were created and rendered in Pymol. Representative fittings of I318T- or G101del-hLADH (low-pH) to WT-hLADH (low-pH) can be seen in Fig. S2A,B. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Fitting simulated structures to reference structuresa.
| Structure | RMSD | RMSD | RMSD |
|---|---|---|---|
| (Å, protein) | (Å, FAD-A) | (Å, FAD-B) | |
| R447G #1 | 11.65 | 4.88 | 3.74⁎ |
| R447G #2 | 10.48 | 3.26 | 4.69⁎ |
| G101del #1 | 9.38 | 4.14 | 3.62⁎ |
| G101del #2 | 7.68 | 4.45 | 4.44 |
| I445M #1 | 12.28 | 4.36 | 4.40 |
| I445M #2 | 8.03 | 3.38 | 2.92⁎ |
| I12T #1 | 8.99 | 4.42 | 4.20⁎ |
| I12T #2 | 11.23 | 4.14 | 5.05⁎ |
| I318T #1 | 8.90 | 3.81⁎ | 4.87 |
| I318T #2 | 11.25 | 3.61 | 4.14 |
| R447G | 5.45 | 4.55⁎ | 4.19 |
| G101del | 6.14 | 2.66 | 3.37⁎ |
| I445M | 4.70 | 1.94 | 3.04 |
| I12T | 5.54 | 2.39 | 4.63 |
| I318T | 4.58 | 4.55⁎ | 3.86 |
RMSDs were calculated for Cα atoms from both monomers. Data above the bold separation line are from simulations in water plus ions while under the line are from simulations in vacuum. Asterisks label FADs that were not approved for planar isoalloxazine ring.
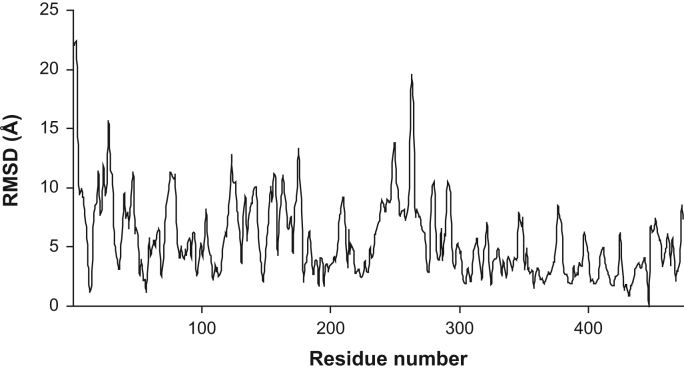
Amino acids that most significantly changed position upon mutation were identified using residue displacement plots (residue-level RMSDs; Fig. 3). Significant displacements were filtered against functionally important residues in the active center, the FAD and NAD+/NADH binding sites, the lipoic moiety binding site and the homodimerization surface (Table 2); in Table 2 the functional locations of the residues are also highlighted so that the primarily affected regions could easily be identified. In Table 2 asterisks label residues that were identified multiple times from the three parallel simulations; a residue marked with two asterisks is the most likely to be displaced in the mutant conformation of the low-pH hLADH structure.
Fig. 3.
Representative residue displacement plot for the simulated R447G mutant structure (#2) in water plus ions relative to the respective low-pH WT-hLADH structure. The plot represents RMS differences as a function of residue number after fitting the respective E3-A monomers. RMSDs are rms-averaged over the backbone atoms of each residue. RMSDs greater than 10 Å (in water plus ions) or 7 Å (in vacuum) are filtered out from plots and collected in Table 2 under the “residue displacement” columns.
Table 2.
Significant amino acid deviations in the low-pH hLADH structure induced by pathogenic mutations identified from residue displacement plots and structure mappinga
| Protein | Residue displacement (water plus ions) | Residue displacement (vacuum) | Structure mapping (water plus ions) | Structure mapping (vacuum) |
|---|---|---|---|---|
| G101del | 209n, 155n | 16f, 17f, 43f, 44f, 48f, 49f, 117f, 118f, 150f, 168f, 280f, 45a, 279n, 357n, 42l, 46l, 99l | 36f, 119f, 280f, 340d–447′d, 444d–438′d⁎, 277n, 51l–452′l | 45a–452′a, 12f, 35f, 99l–392′l |
| I12T | 280f⁎, 155n⁎, 209n, 278n, 279n⁎, 438d, 444d, 447d, 460d | 43f, 44f, 49f, 117f, 150f, 280f⁎, 279n⁎, 42l, 99l, 103l | 36f⁎⁎, 37f⁎, 43f, 118f, 189f, 280f, 283f, 444d–438′d⁎, 460d–333′d, 277n⁎, 51l–453′l | 12f, 36f⁎⁎, 37f⁎, 99l–392′l |
| I318T | 280f, 155n⁎, 279n, 46l, 99l, 103l | 185n, 340d | 13f, 16f–45f, 36f⁎, 37f⁎, 43f⁎, 118f, 119f⁎⁎, 148f, 280f, 359f⁎, 444d–438′d⁎, 460d–333′d, 188n, 277n⁎, 99l–332l, 51l–453′l, 392′l–453′l | 12f, 37f⁎, 117f, 119f⁎⁎, 45l–328l, 99l–392′l |
| I445M | 16fl, 209n, 19l, 46l, 340d | 43f, 150f, 42l, 438d | 16f–45f, 119f⁎, 444d–438′d, 277n⁎, 99l–332l, 51l–453′l | 119f⁎, 45l–328l, 99l–392′l |
| R447G | 48f, 49f⁎, 168f⁎, 280f, 45a, 209n, 210n, 19l, 46l⁎ | 49f⁎, 117f, 149f, 150f, 168f⁎, 326f, 327f, 50a, 279n, 51l | 36f, 37f⁎, 43f, 117f, 118f, 119f⁎, 168f, 280f, 340d–447′d⁎, 277n⁎, 51l–452′l, 51l–453′l, 103l–452′l | 12f, 35f, 37f⁎, 119f⁎, 460d–333′d |
Greater deviations of N- and C-terminal stretches in residue displacement plots were neglected in this table. Residue numbers are based on 474 amino acids (mature protein); in case of G101del, a pseudo residue was included at position 101 during analysis to keep the original numbering scheme for clear comparison of affected amino acids. Residues filtered from displacement plots showed higher than 7 (vacuum) or 10 Å (water plus ions) displacements relative to the respective reference structures and were also previously identified as participants of crucial interactions in the original 1zmc structure (see below). Residues filtered from structure mapping presented with a multiplication factor of higher than 2 for designated atom distances relative to the reference structures. Asterisks label residues positively filtered from multiple analyses (e.g. in vacuum and in water or in both structures in water); the number of asterisks indicates the number of cases a residue was positively filtered (double asterisks equals to three positives). All structures were plotted and residues are presented even when filtered from one analysis (but fulfilled the requirements of the above filters). Subscripts label locations of residues: active center – a, FAD binding site – f, NAD+/NADH binding site – n, lipoic moiety binding site – l, dimer interface – d. Some residues belong to multiple categories. ′ labels the other monomer. For structure mapping FAD atoms are not included in this table, for those see Figs. S3 and S4.
In the crystallographic study of WT-hLADH amino acids in functional vicinity to NAD+(NADH)/FAD or on the homodimerization surface or in the binding channel for lipoate were identified [27]. It is also known which residues participate in the catalytic action [28], [29], [30], [31]. All of this information was used during the analysis of residue displacement plots (see above), but also to map the wild-type structure for functionally representative interatomic distances; 84 such distances (Fig. S3) were selected after the manual inspection of the WT-hLADH structure and screened for in all recorded trajectories. As NAD+/NADH were not included in simulation (as they are substrates), the NAD+/NADH binding site was mapped via the most direct distances to FAD (Fig. S4); FAD stacks close and well-defined to NADH during catalysis. To quantify potential monomerization upon simulation, representative inter-monomeric distances were also specified and determined. Every designated distance was determined both in the mutants and the respective wild-type structures (Fig. 4), and those with significant deviation were listed also in Table 2 under ‘structure mapping’.
Fig. 4.
A representative plot of ratios of specific, functionally significant, interatomic distances for the R447G mutant structure (#2) relative to the respective low-pH WT-hLADH structure in water plus ions. The serial numbers on the abscissa axis refer to the numbering scheme of distances in Fig. S3. Ratios greater than 2 are filtered out from plots and collected in Table 2 under the “structure mapping” columns.
4. Discussion
The structural information revealed here is to be applied in mechanistic studies addressing the elucidation of the molecular pathogeneses of disease-causing mutant hLADH proteins.
We identified, with molecular dynamics simulation, the putative structural changes, including the most deviating amino acids (from residue displacement plots) and interatomic distances (from structure mapping) in functionally critical regions, in human LADH bearing five different pathogenic (disease-causing) amino acid substitutions relative to the wild-type structure. Amino acids and interatomic distances identified with deviations under the filter limits may also possess crucial structural information, nevertheless, our primary goal here was to identify amino acid residues and interatomic distances of structural and functional significance, upon the previously determined crystal structure of WT-hLADH determined at physiological pH, which deviate the most relative to the wild-type structure. These specific amino acid deviations identified in functionally significant regions by molecular dynamics simulations might contribute to the development and dysfunction of low-pH pathogenic mutant hLADH structures and thus their further individual analysis ought to be addressed in future structural and mechanistic studies.
A pathogenic amino acid substitution can cause local and/or distant effects in the structure of a protein. In either case, functional units might be significantly displaced as a whole in the mutant protein, but still kept relatively intact in the internal structure. With residue displacement plots, overall distortions of the structure were tracked (projected to the individual amino acid level) while using structure mapping, all the relevant changes inside the functionally important structural units could be determined. Significantly deviating residues or distances identified in a pathogenic mutant multiple times in different simulation experiments are of special interest. Amino acids affected by the mutations in the low-pH conformation may also contribute to the molecular dysfunction of the physiological hLADH conformation (in terms of loss in physiological hLADH activity); nevertheless, analysis of the low-pH conformation is more justified due to the pathologically relevant usual condition for atypical E3-deficiency (acidosis). Structural alterations induced by the mutations may also result in changes in the ROS-generating behavior of the low-pH conformation of hLADH; for these five pathogenic mutants of hLADH investigated here, there is no data regarding their ROS generation capacities reported.
For evaluation of the structural changes, simulation results obtained in water plus ions ought to be primarily considered. As seen in Table 2 (and from the trajectory deviation curves as well) G101del induced a rather local change (few significant changes in the residue displacement plot and an intensive deviation of the peptide stretch in the trajectory in water plus ions, see Section 3). For the other four mutations, more considerable sets of residues and functional distances were found to be significantly affected, virtually in all regions important for hLADH function (Table 2). As seen in Fig. 2, the overall structure of the FAD prosthetic group was also generally affected by the mutations which might also potentially contribute to dysfunction; the greatest change was detected in R447G-hLADH where the overall shape of FAD was converted to a rather rare U-shaped FAD structure, similar to what was found in DNA-photolyase [32].
Mechanistic validation of the structural information reported here ought to result in new structural restraints and constraints which can then be applied to further refine the simulated structures until relevantly high resolution (NMR, X-ray) structures are determined. NMR failed us to provide adequate signal dispersion due to the size of hLADH proteins (~100 kDa) even at 900 MHz of proton while quality mutant crystals could neither be grown hitherto from any of the pathogenic mutants of hLADH; this is why MD simulation became the method of choice for the structure analysis of these pathogenic forms of hLADH and there is no straightforward technique hence to experimentally validate these simulated structures, yet. The structural data provided by these MD simulation experiments are of low resolution, hence an atomic level evaluation of the structures ought not to be performed at this stage of analysis. These simulated structures will be refined later upon an extensive mechanistic evaluation and hence are only adequate at this stage to serve as preliminary structural information for the informed selection of amino acids in a mechanistic evaluation study. In a mechanistic study, typically implicated amino acids are labeled, modified or substituted and changes in biophysical/biochemical parameters including spectroscopic (e.g. NMR, CD, UV, IR, ESR) and kinetic (like specific activity, Vmax, KM, kcat/KM) information are recorded and evaluated for the elucidation of the structure-based molecular mechanism [1], [13], [33], [34], [35]. With both functional and (refined) structural information in hand for all the 13 known pathogenic mutants of hLADH a plausible attempt can then be made to assess the putative basis of the atomic level relationship between phenotypic properties at the molecular level, like loss of physiological activity and modulation of ROS generation, and these (or all) simulated hLADH structures.
Selective, rationally designed inhibitors against human hLADH mutants presenting with high ROS generation are the future alternatives of the non-selective antioxidant therapy. For this purpose, deep insight into the disease-causing mutant structures in a broader pH range and the relevant molecular mechanisms on the atomic level is inevitable. The present work together with our previous study [17] completes a preliminary structural investigation for all the disease-causing mutants of human LADH reported in the clinical literature until today.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgments
The authors are thankful to Dr. Charles Schwieters (CIT, NIH, Bethesda, MD, USA) for technical assistance in many aspects of computation and to Gareth Ogdon and Daniel Oren (Semmelweis University, Budapest, Hungary) for technical assistance. This work was supported by the Hungarian Academy of Sciences (MTA grant 02001 to A-V.V.; Bolyai Fellowship to A.A.), the Hungarian Scientific Research Fund (OTKA, grant 112230 to A-V. V.) and the Hungarian Brain Research Program (grant KTIA_13_NAP-A-III/6 to A-V.V.).
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.04.006.
Contributor Information
Attila Ambrus, Email: ambrus.attila@med.semmelweis-univ.hu.
Reka Mizsei, Email: mizsei.reka@med.semmelweis-univ.hu.
Vera Adam-Vizi, Email: adam.veronika@med.semmelweis-univ.hu.
Appendix A. Transparency document
Representative Ramachandran plot of the I12T mutant of the low-pH hLADH structure in water plus ions (structure #1). It is apparent that the majority of the outliers are entirely borderline to the allowed regions.
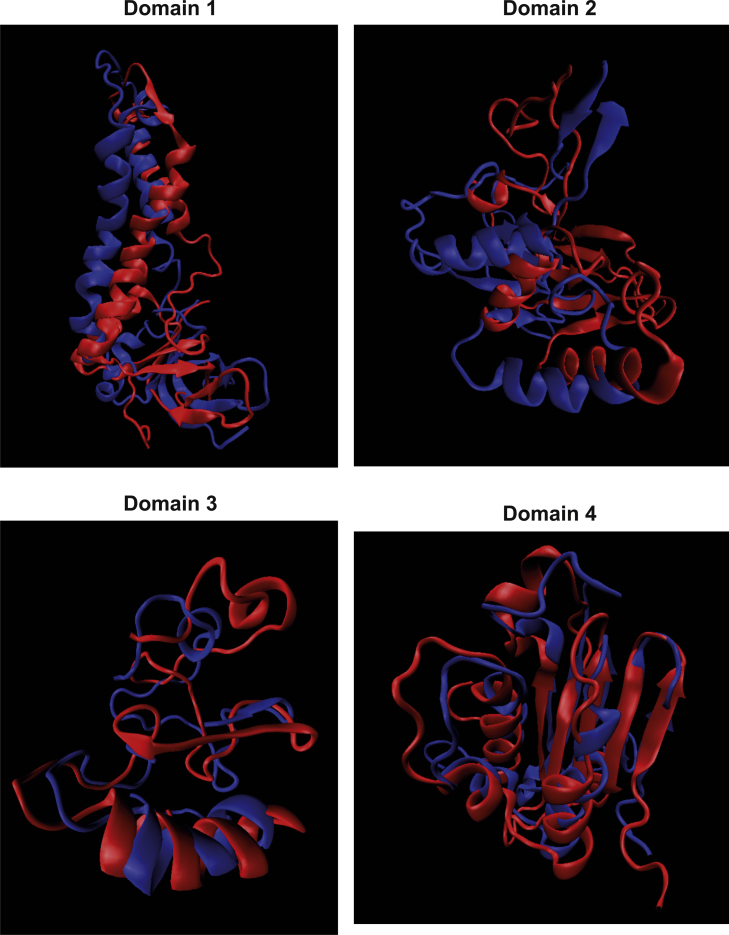
Supplementary Fig. S2A.
Representative fitting of simulation results determined for the I318T mutant of the low-pH hLADH structure (structure #2; blue) and the wild-type low-pH hLADH structure (red) in water plus ions. For clarity, the four domains are presented separately, from only monomer A; FAD-binding domain (1-149, domain 1), NAD+/NADH-binding domain (150-282, domain 2), central domain (283-350, domain 3) and interface domain (351-474, domain 4). Structures were created in VMD.
Supplementary Fig. S2B.
Representative fitting of simulation results determined for the G101del mutant of the low-pH hLADH structure (structure #2; blue) and the wild-type low-pH hLADH structure (red) in water plus ions. For clarity, the four domains are presented separately, from only monomer A; the domain structures are detailed in Fig. S2A. Structures were created in VMD.
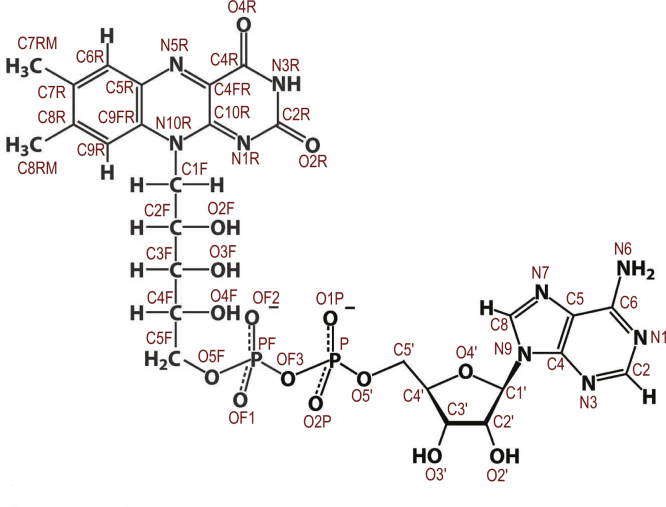
Specific, functionally significant, interatomic distance definitions for structure mapping. Groups of selected atom-based distances represent specific interactions in the hLADH molecule and were grouped in this figure accordingly; the type of interaction each group represents is in the header above each group in parenthesis. All atomic distances were defined after manually inspecting the 1zmc structure in VMD and taking also into consideration the structural information available on hLADH in the PDB database.
Supplementary Fig. S4.
Structure of FAD and its atom labeling. This atom labeling is relevant to Fig. S3.
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
References
- 1.Ambrus A., Tretter L., Adam-Vizi V. Inhibition of the alpha-ketoglutarate dehydrogenase-mediated reactive oxygen species generation by lipoic acid. J. Neurochem. 2009;109:222–229. doi: 10.1111/j.1471-4159.2009.05942.x. [DOI] [PubMed] [Google Scholar]
- 2.Tretter L., Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. J. Neurosci. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bando Y., Aki K. Mechanisms of generation of oxygen radicals and reductive mobilization of ferritin iron by lipoamide dehydrogenase. J. Biochem. 1991;109:450–454. doi: 10.1093/oxfordjournals.jbchem.a123402. [DOI] [PubMed] [Google Scholar]
- 4.Starkov A.A., Fiskum G., Chinopoulos C., Lorenzo B.J., Browne S.E., Patel M.S., Beal M.F. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam-Vizi V., Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol. Sci. 2006;27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Adam-Vizi V. Production of reactive oxygen species in brain mitochondria: contribution by electron transport chain and non-electron transport chain sources. Antioxid. Redox Signal. 2005;7:1140–1149. doi: 10.1089/ars.2005.7.1140. [DOI] [PubMed] [Google Scholar]
- 7.Zundorf G., Kahlert S., Bunik V.I., Reiser G. Alpha-ketoglutarate dehydrogenase contributes to production of reactive oxygen species in glutamate-stimulated hippocampal neurons in situ. Neuroscience. 2009;158:610–616. doi: 10.1016/j.neuroscience.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Tahara E.B., Barros M.H., Oliveira G.A., Netto L.E.S., Kowaltowski A.J. Dihydrolipoyl dehydrogenase as a source of reactive oxygen species inhibited by caloric restriction and involved in Saccharomyces cerevisiae aging. FASEB J. 2007;21:274–283. doi: 10.1096/fj.06-6686com. [DOI] [PubMed] [Google Scholar]
- 9.Adam-Vizi V., Tretter L. The role of mitochondrial dehydrogenases in the generation of oxidative stress. Neurochem. Int. 2013;62:757–763. doi: 10.1016/j.neuint.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Starkov A.A. An update on the role of mitochondrial alpha-ketoglutarate dehydrogenase in oxidative stress. Mol. Cell. Neurosci. 2013;55:13–16. doi: 10.1016/j.mcn.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan C.L., Goncalves R.L., Hey-Mogensen M., Yadava N., Bunik V.I., Brand M.D. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron J.M., Levandovskiy V., MacKay N., Raiman J., Renaud D.L., Clarke J.T.R., Feigenbaum A., Elpeleg O., Robinson B.H. Novel mutations in dihydrolipoamide dehydrogenase deficiency in two cousins with borderline-normal PDH complex activity. Am. J. Med. Genet. A. 2006;140:1542–1552. doi: 10.1002/ajmg.a.31313. [DOI] [PubMed] [Google Scholar]
- 13.Ambrus A., Torocsik B., Tretter L., Ozohanics O., Adam-Vizi V. Stimulation of reactive oxygen species generation by disease-causing mutations of lipoamide dehydrogenase. Hum. Mol. Genet. 2011;20:2984–2995. doi: 10.1093/hmg/ddr202. [DOI] [PubMed] [Google Scholar]
- 14.Vaubel R.A., Rustin P., Isaya G. Mutations in the dimer interface of dihydrolipoamide dehydrogenase promote site-specific oxidative damages in yeast and human cells. J. Biol. Chem. 2011;286:40232–40245. doi: 10.1074/jbc.M111.274415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart S.D., Schauble A., Gupta S., Kennedy A.D., Keppler B.R., Bingham P.M., Zachar Z. A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metab. 2014;2:4. doi: 10.1186/2049-3002-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starkov A.A., Adam-Vizi V. Calcium and mitochondrial reactive oxygen species generation: how to read the facts. J. Alzheim. Dis. 2010;20:S413–S426. doi: 10.3233/JAD-2010-100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrus A., Adam-Vizi V. Molecular dynamics study of the structural basis of dysfunction and the modulation of reactive oxygen species generation by pathogenic mutants of human dihydrolipoamide dehydrogenase. Arch. Biochem. Biophys. 2013;538:145–155. doi: 10.1016/j.abb.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Odievre M.H., Chretien D., Munnich A., Robinson B.H., Dumoulin R., Masmoudi S., Kadhom N., Rötig A., Rustin P., Bonnefont J.P. A novel mutation in the dihydrolipoamide dehydrogenase E3 subunit gene (DLD) resulting in an atypical form of alpha-ketoglutarate dehydrogenase deficiency. Hum. Mutat. 2005;25:323–324. doi: 10.1002/humu.9319. [DOI] [PubMed] [Google Scholar]
- 19.Hong Y.S., Kerr D.S., Liu T.C., Lusk M., Powell B.R., Patel M.S. Deficiency of dihydrolipoamide dehydrogenase due to two mutant alleles (E340K and G101del) – analysis of a family and prenatal testing. Biochim. Biophys. Acta-Mol. Basis Dis. 1997;1362:160–168. doi: 10.1016/s0925-4439(97)00073-2. [DOI] [PubMed] [Google Scholar]
- 20.Quintana E., Pineda M., Font A., Vilaseca M.A., Tort F., Ribes A., Briones P. Dihydrolipoamide dehydrogenase (DLD) deficiency in a Spanish patient with myopathic presentation due to a new mutation in the interface domain. J. Inherit. Metab. Dis. 2010;33:S315–S319. doi: 10.1007/s10545-010-9169-4. [DOI] [PubMed] [Google Scholar]
- 21.Quinonez S.C., Leber S.M., Martin D.M., Thoene J.G., Bedoyan J.K. Leigh syndrome in a girl with a novel DLD mutation causing E3 deficiency. Pediatr. Neurol. 2013;48:67–72. doi: 10.1016/j.pediatrneurol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwieters C.D., Kuszewski J.J., Clore G.M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Mag. Res. Spectrosc. 2006;48:47–62. [Google Scholar]
- 23.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A., Brooks C.L., Karplus M. Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 1984;105:495–500. [Google Scholar]
- 25.Brooks C.L., Karplus M. Deformable stochastic boundaries in molecular dynamics. J. Chem. Phys. 1983;79:6312–6325. [Google Scholar]
- 26.Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 27.Brautigam C.A., Chuang J.L., Tomchick D.R., Machius M., Chuang D.T. Crystal structure of human dihydrolipoamide dehydrogenase: NAD(+)/NADH binding and the structural basis of disease-causing mutations. J. Mol. Biol. 2005;350:543–552. doi: 10.1016/j.jmb.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Liu T.C., Korotchkina L.G., Hyatt S.L., Vettakkorumakankav N.N., Patel M.S. Spectroscopic studies of the characterization of recombinant human dihydrolipoamide dehydrogenase and its side-directed mutants. J. Biol. Chem. 1995;270:15545–15550. doi: 10.1074/jbc.270.26.15545. [DOI] [PubMed] [Google Scholar]
- 29.Kim H., Patel M.S. Characterization of 2 site specifically mutated human dihydrolipoamide dehydrogenases (His-452-]Gln and Glu-457-]Gln) J. Biol. Chem. 1992;267:5128–5132. [PubMed] [Google Scholar]
- 30.Qi F., Pradhan R.K., Dash R.K., Beard D.A. Detailed kinetics and regulation of mammalian 2-oxoglutarate dehydrogenase. BMC Biochem. 2011;12:53. doi: 10.1186/1471-2091-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gazaryan I.G., Krasnikov B.F., Ashby G.A., Thorneley R.N.F., Kristal B.S., Brown A.M. Zinc is a potent inhibitor of thiol oxidoreductase activity and stimulates reactive oxygen species production by lipoamide dehydrogenase. J. Biol. Chem. 2002;277:10064–10072. doi: 10.1074/jbc.M108264200. [DOI] [PubMed] [Google Scholar]
- 32.Hahn J., Michel-Beyerle M.E., Rosch N. Conformation of the flavin adenine dinucleotide cofactor FAD in DNA-photolyase: a molecular dynamics study. J. Mol. Model. 1998;4:73–82. [Google Scholar]
- 33.Ambrus A., Friedrich K., Somogyi A. Oligomerization of nitrophorins. Anal. Biochem. 2006;352:286–295. doi: 10.1016/j.ab.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Ambrus A., Yang D. Diffusion-ordered nuclear magnetic resonance spectroscopy for analysis of DNA secondary structural elements. Anal. Biochem. 2007;367:56–67. doi: 10.1016/j.ab.2007.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemeria N.S., Ambrus A., Patel H., Gerfen G., Adam-Vizi V., Tretter L., Zhou J., Wang J., Jordan F. Human 2-oxoglutarate dehydrogenase complex E1 component forms a thiamin-derived radical by aerobic oxidation of the enamine intermediate. J. Biol. Chem. 2014;289:29859–29873. doi: 10.1074/jbc.M114.591073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative Ramachandran plot of the I12T mutant of the low-pH hLADH structure in water plus ions (structure #1). It is apparent that the majority of the outliers are entirely borderline to the allowed regions.
Specific, functionally significant, interatomic distance definitions for structure mapping. Groups of selected atom-based distances represent specific interactions in the hLADH molecule and were grouped in this figure accordingly; the type of interaction each group represents is in the header above each group in parenthesis. All atomic distances were defined after manually inspecting the 1zmc structure in VMD and taking also into consideration the structural information available on hLADH in the PDB database.
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document