Abstract
In bacteria, the DnaG primase is responsible for synthesis of short RNA primers used to initiate chain extension by replicative DNA polymerase(s) during chromosomal replication. Among the proteins with which Escherichia coli DnaG interacts is the single-stranded DNA-binding protein, SSB. The C-terminal hexapeptide motif of SSB (DDDIPF; SSB-Ct) is highly conserved and is known to engage in essential interactions with many proteins in nucleic acid metabolism, including primase. Here, fragment-based screening by saturation-transfer difference nuclear magnetic resonance (STD-NMR) and surface plasmon resonance assays identified inhibitors of the primase/SSB-Ct interaction. Hits were shown to bind to the SSB-Ct-binding site using 15N–1H HSQC spectra. STD-NMR was used to demonstrate binding of one hit to other SSB-Ct binding partners, confirming the possibility of simultaneous inhibition of multiple protein/SSB interactions. The fragment molecules represent promising scaffolds on which to build to discover new antibacterial compounds.
Keywords: antibacterial agents, fragment-based screening, primase, protein–protein interactions, SSB
1. Introduction
Duplication of chromosomal DNA prior to cell division is a fundamental process in living cells. During initiation of DNA replication in Escherichia coli, DnaB helicase is loaded with the assistance of the helicase loader DnaC onto double-stranded DNA and unwinds it [1,2]. DnaB, through direct physical interaction with DnaG primase, forms the primosome [3], which uses its primase activity to synthesize short RNA primers essential for the function of DNA polymerase III [4,5].
DnaG is a DNA-dependent RNA polymerase [6]. In bacteria it is comprised of three distinct domains: an N-terminal zinc-binding domain (ZBD) responsible for DNA template recognition [7], a central catalytic domain (RNA polymerase domain, RPD) [8,9], and a C-terminal helicase-binding domain (HBD or DnaGC), which is responsible for interaction with DnaB helicase and single-stranded DNA-binding protein (SSB) [10,11,12]. The crystal and solution structures of DnaGC of E. coli were determined by X-ray crystallography as a non-physiological domain-swapped dimer [13] and as a monomer in solution by NMR spectroscopy [14].
SSB protects single-stranded DNA during DNA replication. It is an interaction hub known to bind to more than 14 other proteins involved in various stages of DNA replication, repair, and recombination through a highly conserved C-terminal hexapeptide motif (SSB-Ct, sequence: DDDIPF) [15,16]. SSB’s binding partners include DnaG [10,12], the Pol lll χ subunit [12,17,18,19,20,21,22], the PriA replication restart helicase [23], and exonuclease I [24,25].
The SSB-Ct binding site in DnaGC has been identified by NMR. The binding pocket is formed by basic residues K447, R452, and K518, as well as T450, M451, I455, and L519 [26]. Moreover, the DnaGC point mutants K447A, T450A, R452A, and K518A dramatically attenuate SSB-Ct binding. Mutagenesis and NMR experiments, in particular 15N–1H heteronuclear single-quantum correlation (15N–1H HSQC) experiments suggested that the conserved R452 residue forms a salt bridge with the carboxylic acid of the C-terminal Phe residue of the SSB-Ct, whereas the other positively charged residues around the binding pocket interact with the negatively charged residues of SSB-Ct. The SSB-Ct binding pockets in other SSB-binding proteins have characteristics in common with the binding pocket in DnaGC; e.g., those in ExoI [24], RecO [27], Pol lll χ [22], and PriA [23].
The SSB-Ct binding pockets in some or all of these proteins have been suggested to be very good targets for development of new antibacterial agents because many of the interactions are essential for bacterial survival and resistance to compounds that interfere with multiple interactions could not easily develop by target mutagenesis [16]. This argument depends critically on a single compound mimicking the SSB-Ct peptide sufficiently well that it is able to bind tightly to three or more essential binding pockets that are lined with different residues and thus have structures that are more or less distinct. The observed gross structural similarities among pockets in SSB-Ct binding partners, including the ionic interaction with the C-terminal Phe and the basic rim that interacts with the acidic residues suggest such compounds might exist, but the only useful way to establish this for sure is to quantify the binding to multiple potential targets of compounds selected against one of them.
To begin the process of determining whether SSB-Ct binding pockets are actually suitable targets, we report the use of fragment-based screening (FBS) to find compounds binding to E. coli DnaGC. FBS uses small (<300 Da) compounds called “fragments” as starting points for drug discovery. Several biophysical methods may be used in fragment screening [28]. Here, we report the use of surface plasmon resonance (SPR) and NMR measurements to screen for binders that target the SSB-binding pocket in DnaGC.
2. Results and Discussion
2.1. Screening of Fragment Libraries
An SPR competition assay (example in Figure S1a) was used as the first-pass screen. It identified six small-molecule fragments that competed with immobilized SSB-Ct peptide for binding to a N-terminally truncated DnaG protein missing just the ZBD; we call this protein DnaG-RCD, comprised of the RPD and DnaGC domains. These fragments were validated by saturation-transfer difference (STD) NMR [29], where transient binding of the ligand to the protein is detected by attenuation of the ligand NMR spectrum (example in Figure S1b). In parallel with the confirmation of SPR hits, cocktails encompassing the complete MIPS library of 1140 structurally diverse fragments [30] were independently screened by STD-NMR. Fragments identified in cocktails were re-tested as pure ligands in the STD-NMR assay. The hits identified were ranked according to STD signal intensity. In total, 56 fragments were identified as top hits (rank 3, clear hit, strong intensity difference) and 62 as rank 2 (clear hit, moderate intensity) [30]. From the final STD rank 3 and 2 compounds together (80 compounds), about 50 fragments were identified as “frequent hits” or “PAINS” [30] and were excluded from further screening.
2.2. Validation of Fragment Binding by 2D NMR
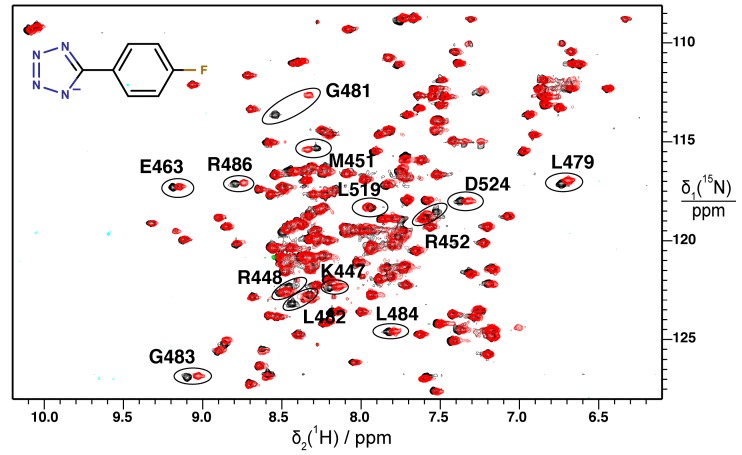
Two-dimensional (2D) 15N–1H HSQC spectra of the uniformly 15N-labeled DnaGC domain were employed to validate STD hits. Buffer conditions were optimized, and no major differences were observed in recorded spectra of the protein alone compared to the original studies [13,14]. We were able to assign some of the missing resonances for residues forming the SSB-Ct binding pocket in the spectrum of the apo-protein reported by Naue et al. [26]; i.e., those of K447, R448, T449, N511, and N565. 15N–1H HSQC spectra were recorded on 0.1 mM solutions of 15N-DnaGC mixed with compounds at 3.3 mM. The compounds were ranked according to the magnitude of the generated weighted chemical shift perturbation (CSP; Section 3.5), and the best four were selected (Figure 1, Figures S2a–c and S3a–d).
Figure 1.
Superimposition of 15N–1H HSQC spectra of DnaGC. The protein spectrum in the absence of fragment in black is compared with its spectrum after addition of fragment 4 (structure shown) in red. Representative assignments of resonances that showed the highest weighted chemical shift perturbation (CSP) (Figure S3d) are shown.
By monitoring the protein chemical shift and peak intensity changes upon addition of compound and mapping the CSP onto the protein surface, the location of the ligand-binding pocket could in each case be identified as the SSB-Ct binding pocket of DnaGC (Figure 2 and Figure S4).
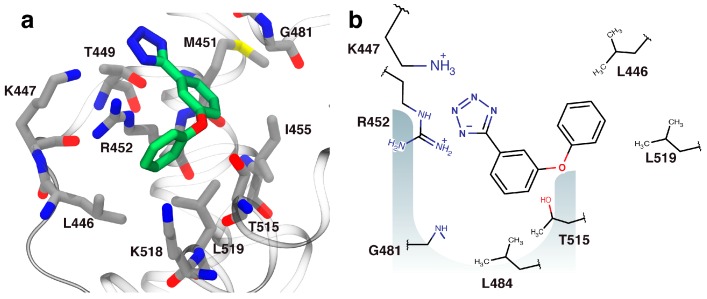
Figure 2.
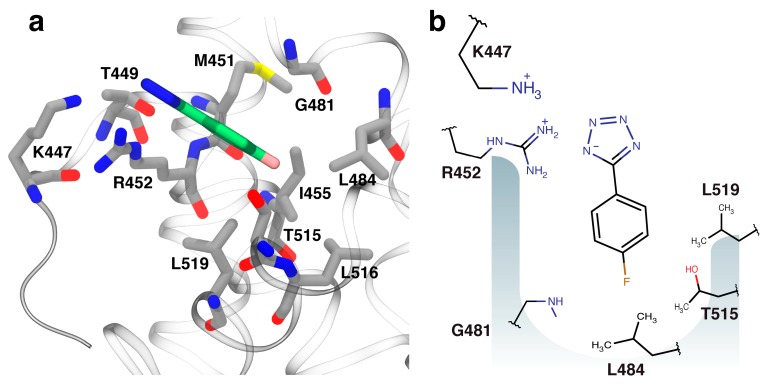
Modeled orientation of fragment 4. (a) The docked orientation of fragment 4 (green carbon atoms) in the single-stranded DNA-binding (SSB)-Ct binding pocket of DnaGC (gray carbon atoms). (b) A schematic representation of interactions between fragment 4 and its binding pocket. In all structural figures, the protein was visualized using visual molecular dynamics (VMD) [31].
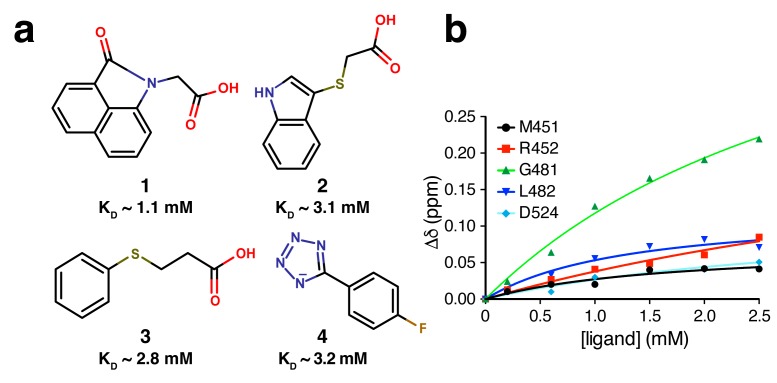
Comparison of the four hits revealed some similar features such as the presence of indole groups in fragments 1 and 2 or an aliphatic thioether-linked extension in 2 and 3. In addition, three (1–3) have a carboxylate attached (Figure 3a) and fragment 4 has a 1H-tetrazole group, which is a carboxylate bioisostere [32]. Tetrazoles have pKa values comparable to carboxylic acids (~5) and are good hydrogen bond acceptors. Modeling studies suggest that deprotonated tetrazoles form stronger hydrogen bonds than carboxylate groups [33].
Figure 3.
(a) Structure of hits with binding affinities for further optimization. (b) 15N–1H HSQC titration of fragment 4. Binding affinities (KD values) were derived from the change in chemical shift, Δδ, with incremental additions of ligand.
Affinity measurements of weakly binding ligands are challenging for most biophysical techniques [34,35]. Nevertheless, binding affinities of fragments 1–4 can be roughly estimated to be in the 1–3 mM range by NMR titration experiments that monitored the gradual change in chemical shift of a few well-resolved resonances in HSQC spectra (Figure 3b).
2.3. Orientation of Identified Hits Using Molecular Docking
The docked structure of fragment 4 bound into DnaGC (Figure 2a) is in agreement with the chemical shift perturbation (CSP) data. The tetrazole anion of 4 makes hydrogen bonds and/or a salt bridge with the side chains of K447, T449, and R452. The phenyl ring appears likely to mimic the last phenylalanine residue in the SSB-Ct peptide and makes hydrophobic interactions with P480, G481, T515 and L519 in the binding site. Fluorine is a strong hydrogen-bond acceptor [36], and the model suggests that there is a hydrogen bond to the para-fluoro group from the amide proton of G481. Methyl groups from the side chains of L448, L455, T515, L516, and L519 also interact with the para-fluoro group.
In the case of fragment 1, CSP-guided docking predicts that the carbonyl group interacts with the side-chain of R452. The fragment orientation enables favorable contact of the free carboxylate of the fragment and the positively charged side chain of K518 (Figure S4). Its improved binding affinity (~1.1 mM) is most likely explained by its hydrophobic skeleton occupying the shallow binding pocket. Comparison of fragment 1 with an analog where the carboxylate is substituted with a methyl group gave fewer peak shifts in the HSQC spectrum, confirming that the carboxylic acid group improves the binding (Figure S5).
Fragments 2 and 3 have similar aliphatic chains with thioether links to aromatic groups. As with other fragments, the carboxylate moieties are predicted to form electrostatic and hydrogen bond interaction, whereas the main chemical backbone mimics the phenylalanine residue in SSB-Ct. Thioethers may have a strong influence on the conformation of aliphatic chains [37]. In the proposed binding mode of fragment 3, the sulfur atom interacts with R452 while the carboxylate forms hydrogen bonds with I455 and G481 (Figure S6).
One of the fragments tested, N-acetylated L-Phe, is expected to mimic the C-terminal residue (F177) in the SSB-Ct peptide. STD and 15N–1H HSQC experiments confirmed the binding event (data not shown). However, binding affinities measured in titration experiments monitored by HSQC spectra showed that it has about two-fold lower affinity compared to other hits (3, 4). The modeled orientations of the fragment show indeed that the phenyl ring forms hydrophobic contacts in the binding site, and the carboxylate interacts with R452 (Figure S7). Substituting the carboxylate with a 1H-tetrazole ring, as found in fragment 4, increased the affinity 2-fold.
2.4. Fragment-to-Hit Optimization
With the knowledge that the tetrazole moiety might confer improved membrane-crossing properties compared with carboxylates owing to its higher lipophilicity, fragment 4 (Figure 3a) was chosen as a starting point for fragment-to-hit optimization. As docked into DnaGC, 4 has suitable vectors for fragment growth. The in silico analog screen identified 10 tetrazole analogs with favorable binding poses relative to the SSB-Ct peptide (not shown). Nevertheless, STD and 2D NMR experiments showed that most of these analogs did not bind to DnaGC. As an exception, 5-[2-fluoro-6-(4-fluorphenoxy)phenyl]tetrazol-1-ide (5) showed a STD signal and significant CSP in 2D NMR experiments (Figure S3e). Mapping of the CSP on the protein surface allowed a binding pose of compound 5 to be calculated (Figure 4).
Figure 4.
Visualization of binding of compound 5. (a) The lowest energy binding poses of 5 (green carbon atoms) bound to DnaGC (gray carbon atoms). (b) Schematic representation of residues involved in interaction with compound 5.
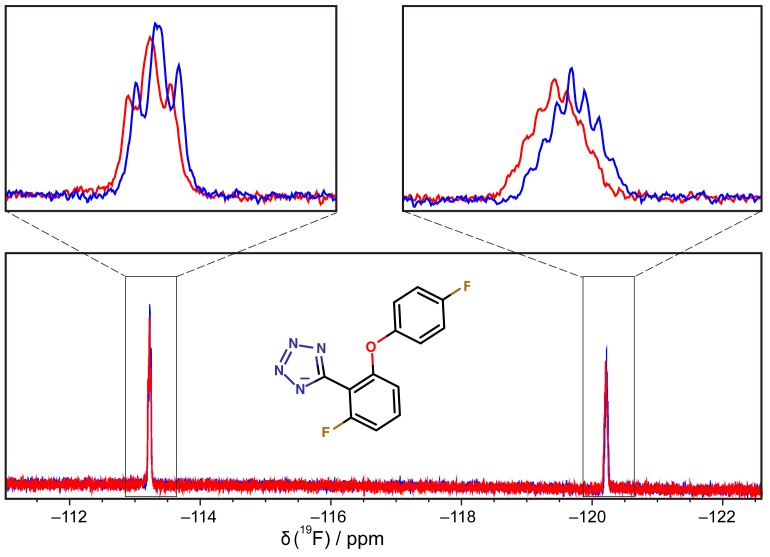
The observed NMR peak shifts were consistent with the docked orientation. The negatively charged tetrazole is predicted to form favorable electrostatic and hydrogen bond interactions with the side chains of K447 and R452. The methyl groups of L446, M451, L455, L484 T515 and L519 form a hydrophobic pocket accommodating two aromatic rings of the compound. One of the fluorine atoms is about 3 Å from the amide group of G481 while the second at the para-position of the phenoxy group points out of the pocket toward K518. In the docked conformation, these fluorine atoms are involved in hydrogen bond formation with these two residues (Figure 4). To test the docked orientation of 5, 1D 19F-NMR was carried out using a 20-fold excess of compound over protein. Fluorine signals were broadened and shifted slightly downfield, confirming binding of 5 to DnaGC (Figure 5). Nevertheless, the 3D NOESY-15N-HSQC spectrum failed to detect any protein–ligand NOEs.
Figure 5.
1D 19F-NMR spectra of compound 5 at 1 mM in the presence (red trace) and absence (blue trace) of 50 μM DnaGC.
Compound 5 was shown to bind to DnaGC with about a three-fold improved affinity compared to the starting fragment 4, as measured by titration experiments monitored by 15N–1H HSQC spectra (KD = 1.3 mM). Searching the ZINC database [38] for ligands structurally similar to 5 indicated that the only available analog was 6. It is missing the halogen atoms and has a meta-phenoxy group (Figure 6). The binding of 6 to DnaGC was also assessed by STD and 15N–1H HSQC experiments (Figure 6).
Figure 6.
(a) Saturation transfer difference (STD) spectrum of compound 6 using DnaG-RCD. In red is a 1D 1H-NMR reference spectrum, overlaid with a STD spectrum (blue). (b) Overlay of 15N–1H HSQC spectra of 15N-DnaGC (black) with 5 (blue) and 6 (red), each at 1 mM. The apo-protein spectrum is shown in black. Representative assignments of resonances that showed the highest weighted CSP (Figure S3e,f) are shown.
Compounds 5 and 6 showed similar patterns of CSPs (Figure S3e,f). Compounds that possess similar protein-binding modes are known to induce similar CSPs [39]. Changes in the position of the phenoxy group in 6 relative to 5 increased the magnitude of the CSP as a result of a slightly improved binding affinity, KD = 1.2 mM.
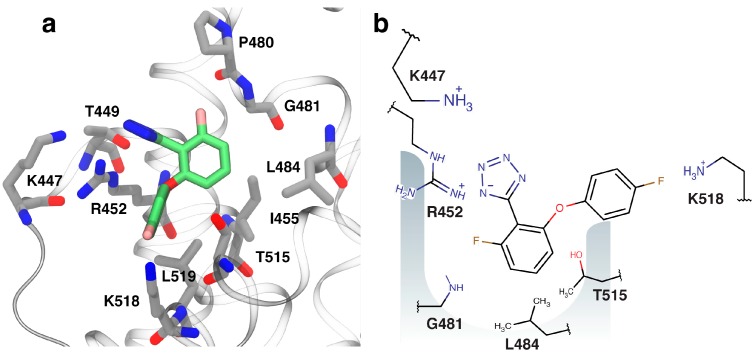
Compound 6 was in turn docked to DnaGC and is predicted to form electrostatic interactions with the side chains of K447, T449 ad R452 (Figure 7). The central aromatic ring sits in a hydrophobic groove formed by M451, I455 and L484, while the phenoxy oxygen atom forms a hydrogen bond with the side chain –OH of T515. In addition, the phenoxy ring forms a cation–π interaction with the guanidinium group of R452 and hydrophobic contacts with the L446 and W522 side chains.
Figure 7.
(a) Docked binding pose of 6 (green carbon atoms) bound to DnaGC (gray carbon atoms). (b) Schematic representation of interactions.
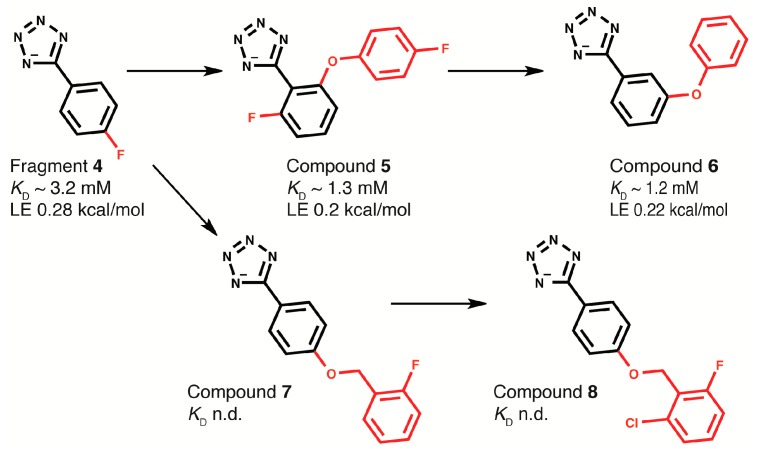
To sample the impact of substituents at the para-position of the phenyl group of 4, additional compounds were purchased (compounds 7 and 8; Figure 8). Molecular docking suggested that para-substitutions might dramatically change the orientation of the tetrazole moiety in the binding pocket, and STD and 15N–1H HSQC experiments showed that replacing the para-fluorine with a bulky substituent reduced the CSPs substantially.
Figure 8.
Schematic representation of optimization of fragment 4. The red labeled groups were added during fragment-to-hit optimization. LE: Ligand efficiency (∆G/[number of heavy atoms]), n.d.: not determined.
2.5. Binding of Compounds to Other SSB Partner Proteins
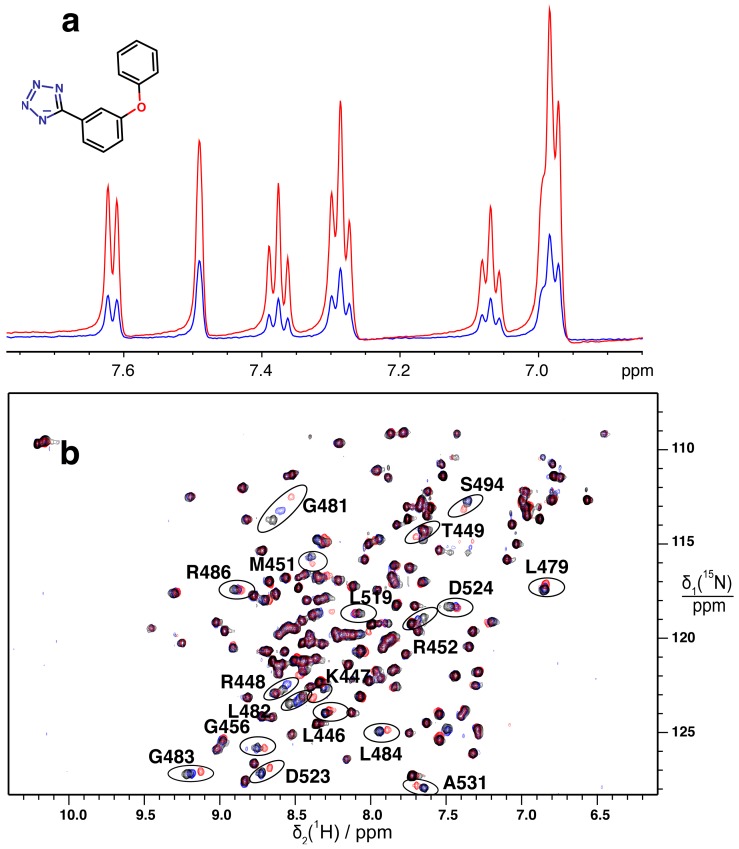
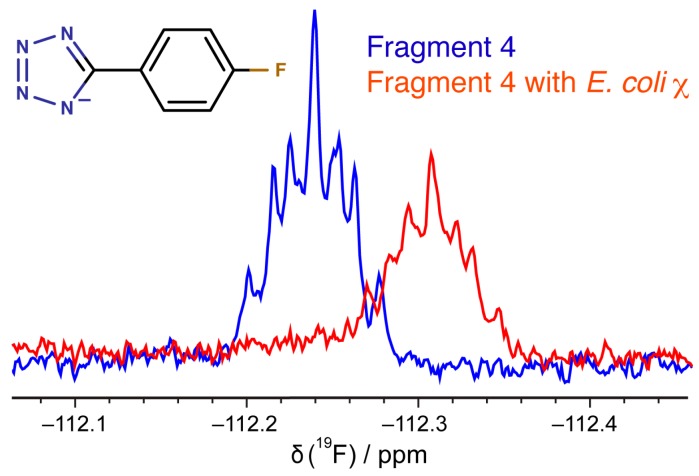
The fragments identified here and the first generation of optimized leads for DnaGC were tested against other SSB-Ct binding partners including E. coli PriA, E. coli RNAse HI, and the χ subunit of E. coli and Acinetobacter baumannii DNA polymerase III. STD-NMR was used to assess binding. All of the identified fragments showed STD signals, confirming binding (Figure S8). Moreover, the docked orientation of fragment 4 in the binding site of E. coli χ showed the possibility of hydrogen bonding with the fluorine atom. 1D 19F NMR (Figure 9) demonstrated binding.
Figure 9.
1D 19F-NMR spectra. The blue spectrum is of fragment 4 alone and its spectrum in the presence of E. coli χ is shown in red.
3. Materials and Methods
3.1. Protein Expression and Purification
A phage λ-promoter plasmid (pZX1404) that directs overexpression of a protein comprising the central and C-terminal domains of E. coli DnaG primase (residues 111–581, here called DnaG-RCD) was constructed by cloning a PCR fragment between the BamHI and EcoRI sites of vector pND706 [40]. PCR was performed using plasmid pPL195 [41] as template and the following primers (restriction sites in italics): dnaG_RCD_F, 5′-GCGGGATCCTAAGAAGGAGATATACATATG ACGCTTTATCAGTTGATG; dnaG_RCD_R, 5′-GCGGAATTCTTACTTTTTCGCCAGCTC C. The full sequence of the gene encoding RCD was then verified by nucleotide sequence determination. Another plasmid pZX1399 encoding amino acids 115–581 of E. coli DnaG was also constructed in a similar manner. However, the protein was expressed in insoluble form, and therefore was not used. Unlabeled DnaG-RCD and unlabeled and 15N-labeled DnaGC were expressed and purified as described previously for DnaGC [42].
3.2. Fragment Libraries
The “first pass screen” fragment library (Zenobia Therapeutics, San Diego, CA, USA) was used for the SPR competition assay. Each fragment (50 mM in DMSO) was diluted to 1 mM final concentration. Fragment library members were tested for chip surface binding to eliminate false positives.
The Monash Institute of Pharmaceutical Science (MIPS) library comprised of around 1140 fragments purchased from Maybridge was used for STD-NMR experiments. The individual fragments were diluted in 2H6-DMSO to give ~660 mM final stock concentrations [30]. The fragments were mixed in cocktails of up to 6 compounds with well-resolved resonances in their 1D 1H-NMR spectra.
3.3. SPR Competition Assay
SPR measurements utilized a Biacore T200 instrument (GE Healthcare, Little Chalfont, UK) at 20 °C to measure the competition of compounds for the DnaGC/SSB-Ct peptide interaction. The buffer contained 10 mM HEPES (pH 7.4), 3 mM EDTA, 100 mM NaCl, 2% DMSO, 1 mM dithiothreitol and 0.05% (v/v) surfactant P20 (GE Healthcare). An N-terminally biotinylated SSB-Ct peptide [Biotin-(Ahx)-GSAPS-NEPPMDFDDDIPF; where Ahx is an amino-hexanoate spacer, followed by the 16 C-terminal residues of SSB highlighted in bold] was immobilized onto a streptavidin (SA) chip surface. RCD at 30 μM and fragments at 1 mM concentrations were used in all SPR experiments. Each sample was mixed for approximately 15 min prior to measurements. Mixtures were injected separately onto two flow cells, one of which served as a reference.
Prior to measurements, each individual fragment was tested for solubility and non-specific binding to an unmodified surface at 1 mM concentration to eliminate false positive responses. Compounds that bound non-specifically to the chip surface were excluded from screening. A flow rate of 5 μL/min was used during the 60 s injection and 60 s dissociation phases for all experiments.
3.4. Saturation-Transfer Difference (STD) NMR Spectroscopy
STD-NMR experiments were carried out using 5 μM unlabeled DnaG-RCD and mixtures of 6 fragments in each sample, at ~250 μM for each fragment. The sample volume was 500 μL with 98–99% 2H2O buffer containing 50 mM phosphate (pH 7.8), 50 mM NaCl and 1 mM dithiothreitol. Spectra were recorded at 283 K using a Bruker Avance 600 MHz spectrometer (Bruker, Karlsruhe, Germany) equipped with a cryoprobe. Saturation of protein was achieved with a 4 s Gaussian pulse sequence train centered at −1 ppm. For reference spectra, a similar saturation pulse was applied 20 kHz off-resonance. A 20 ms spin-lock period was applied before acquisition to allow the residual protein signals to decay. The STD experiments were recorded over 64 scans. All NMR data were processed using TOPSPIN 3.1. Relative intensities were based on the most intense STD signal (Imax) identified across all STD spectra. A positive STD signal was categorized as “strong”, “moderate” or “weak” where the intensity was >50%, >25% or <25% of Imax, respectively [30].
3.5. 2D 15N–1H HSQC Spectra
Protein binding by compounds identified by SPR and STD screens was confirmed by recording 15N–1H HSQC spectra on uniformly 15N-labeled DnaGC (100 μM) in the presence of 3.3 mM compounds (from 2H6-DMSO stocks) with HSQC buffer (50 mM MES pH 6.0, 60 mM NaCl, 1 mM dithiothreitol) containing 3% 2H2O. The final volume of each sample was 150 μL. The recording time was 30 min for each 15N–1H HSQC experiment. A standard pulse sequence was used for data acquisition. Spectra were recorded at 298 K with a Bruker Avance 600 MHz NMR spectrometer equipped with cryoprobe and auto-sample changer. Compounds were regarded as hits if chemical shift perturbation was observed in the 15N–1H HSQC spectra. The spectra were processed with TOPSPIN 3.1 and analyzed using CCPN [43]. Weighted CSP values [44] were calculated as
| CSP = (ΔδH2 + 0.2 ΔδN2)0.5 |
Binding affinities were estimated by incremental titration of fragments into protein, with recording of a 15N–1H HSQC experiment at each concentration point. Compound solubilities were tested to determine the highest concentrations of ligands used in assays. Equilibrium dissociation constants from NMR titration data were derived using the “single site-specific binding with ligand depletion” model in GraphPad Prism v.6.0 (La Jolla, CA, USA).
3.6. 19F-NMR Spectroscopy
1D 19F-NMR spectra were recorded on a Bruker Avance III 400 MHz NMR spectrometer (Bruker, Karlsruhe, Germany) equipped with the two-channel BBO probe with z-gradient at 300 K. All 19F-NMR spectra were recorded with 256 scans for fragment and complex samples sequentially. Fragments dissolved in 2H6-DMSO were diluted in HSQC buffer to give final fragment and protein concentrations of 1 mM and 50 μM, respectively.
3.7. Molecular Docking
AutoDock Tools 1.5.6 [45] was used to prepare protein [46] and ligand structures for docking. The protonation state of the titratable groups in the protein were assigned at pH 7.0 using PROPKA 3.1 [47]. Polar hydrogen atoms and atom-based Gasteiger partial charges were added. Nonpolar hydrogen atoms were merged with the parent atom. The DnaGC structure was taken from the previously solved crystal structure (PDB ID: 1T3W) [13]. The protein was treated as a rigid body. The CSP docking calculations were performed using AutoDock Vina 1.1.2 [45]. The calculations utilized an exhaustiveness of 1024 with grid points separated by 1.0 Å and grid size large enough to include the SSB-Ct peptide binding site (16 × 16 × 14 Å). Ligand data were obtained from the ZINC database of commercially available compounds [38].
4. Conclusions
Fragment-based screening has successfully identified compounds targeting the DnaG primase and its SSB-Ct interaction. Compounds containing indole and 1H-tetrazole scaffolds were identified as first-generation hits. Based on CSP-guided molecular modeling studies, they are involved in formation of various electrostatic and hydrogen bond networks in the binding pockets, which makes them promising starting points for further optimization. Initial in silico fragment-to-lead optimization was carried out using the ZINC Database. Para-phenyl substituted tetrazoles were identified. The observation that compounds selected for binding to the C-terminal domain of DnaG primase also bind to other SSB-interacting proteins indicates that compounds may in future be derived that bind to similar binding pockets in multiple protein targets, which is a prerequisite for development of antibacterial compounds with a very low propensity for development of resistance.
Acknowledgments
This work was supported in part by an infrastructure grant from the Australian Research Council (LE160100047 to M.J.S., N.E.D., and A.J.O.), an ARC Future Fellowship (FT0990287 to A.J.O.), a project grant from the Australian National Health & Medical Research Council (APP1007947 to N.E.D., G.O., and A.J.O.), and a postgraduate scholarship from the University of Wollongong (to Z.C.).
Supplementary Materials
The following are available online at http://www.mdpi.com/2079-6382/7/1/14/s1, Figure S1. Surface plasmon resonance (SPR) competition sensorgram for one of the SPR hits, and saturation transfer difference (STD)-NMR spectrum of SPR hit D6; Figure S2. 15N–1H HSQC spectra of DnaGC protein with and without fragment hits 1–3; Figure S3. Residue-specific weighted chemical shift perturbations induced in 15N-DnaGC by binding to compounds 1–6; Figure S4. Modeled orientation of fragment 1; Figure S5. Comparison of chemical shift perturbation (CSP) induced by fragment 1 and compound L1C6; Figure S6. Modeled orientation of fragment 3; Figure S7. Modeled orientation of N-acetyl-l-Phe (green carbon atoms) in the SSB-Ct binding pocket of DnaGC (gray carbon atoms). Figure S8. STD NMR spectrum of fragment 4 with four SSB-Ct binding partners other than DnaG primase.
Author Contributions
Z.C., G.O., N.E.D., M.J.S. and A.J.O. conceived and designed the experiments; Z.C. and S.J.H performed the experiments; Z.C., S.J.H., G.O., M.J.S. and A.J.O. analyzed the data; A.T.Y.L. and Z.-Q.X. contributed reagents and new methods; Z.C., N.E.D. and A.J.O. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kobori J.A., Kornberg A. The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J. Biol. Chem. 1982;257:13763–13769. [PubMed] [Google Scholar]
- 2.Kobori J.A., Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB–dnaC protein complex. J. Biol. Chem. 1982;257:13770–13775. [PubMed] [Google Scholar]
- 3.Tougu K., Marians K.J. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 4.Kitani T., Yoda K., Ogawa T., Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J. Mol. Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- 5.Yoda K.-Y., Yasuda H., Jiang X.-W., Okazaki T. RNA-primed intitiation sites of DNA replication in the origin region of bacteriophage λ genome. Nucleic Acids Res. 1998;16:6531–6546. doi: 10.1093/nar/16.14.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouché J.P., Zechel K., Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J. Biol. Chem. 1975;250:5995–6001. [PubMed] [Google Scholar]
- 7.Pan H., Wigley D.B. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure. 2000;8:231–239. doi: 10.1016/S0969-2126(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 8.Keck J.L., Berger J.M. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 9.Podobnik M., McInerney P., O’Donnell M., Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 2000;300:353–362. doi: 10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- 10.Sun W., Godson G.N. Interaction of Escherichia coli primase with a phage G4oric-E. coli SSB complex. J. Bacteriol. 1996;178:6701–6705. doi: 10.1128/jb.178.23.6701-6705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tougu K., Marians K.J. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J. Biol. Chem. 1996;271:21391–21397. doi: 10.1074/jbc.271.35.21391. [DOI] [PubMed] [Google Scholar]
- 12.Yuzhakov A., Kelman Z., O’Donnell M. Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/S0092-8674(00)80968-X. [DOI] [PubMed] [Google Scholar]
- 13.Oakley A.J., Loscha K.V., Schaeffer P.M., Liepinsh E., Pintacuda G., Wilce M.C.J., Otting G., Dixon N.E. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J. Biol. Chem. 2005;280:11495–11504. doi: 10.1074/jbc.M412645200. [DOI] [PubMed] [Google Scholar]
- 14.Su X.-C., Schaeffer P.M., Loscha K.V., Gan P.H.P., Dixon N.E., Otting G. Monomeric solution structure of the helicase-binding domain of Escherichia coli DnaG primase. FEBS J. 2006;273:4997–5009. doi: 10.1111/j.1742-4658.2006.05495.x. [DOI] [PubMed] [Google Scholar]
- 15.Shereda R.D., Kozlov A.G., Lohman T.M., Cox M.M., Keck J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson A., Causer R.J., Dixon N.E. Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr. Drug Targets. 2012;13:352–372. doi: 10.2174/138945012799424598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glover B.P., McHenry C.S. The χψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 18.Kelman Z., Yuzhakov A., Andjelkovic J., O’Donnell M. Devoted to the lagging strand–the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witte G. DNA polymerase III χ subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedorov R., Witte G., Urbanke C., Manstein D.J., Curth U. 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 2006;34:6708–6717. doi: 10.1093/nar/gkl1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naue N., Fedorov R., Pich A., Manstein D.J., Curth U. Site-directed mutagenesis of the χ subunit of DNA polymerase III and single-stranded DNA-binding protein of E. coli reveals key residues for their interaction. Nucleic Acids Res. 2011;39:1398–1407. doi: 10.1093/nar/gkq988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marceau A.H., Bahng S., Massoni S.C., George N.P., Sandler S.J., Marians K.J., Keck J.L. Structure of the SSB–DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30:4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharyya B., George N.P., Thurmes T.M., Zhou R., Jani N., Wessel S.R., Sandler S.J., Ha T., Keck J.L. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl. Acad. Sci. USA. 2014;111:1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu D., Keck J.L. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl. Acad. Sci. USA. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu D., Windsor M.A., Gellman S.H., Keck J.L. Peptide inhibitors identify roles for SSB C-terminal residues in SSB/exonuclease I complex formation. Biochemistry. 2009;48:6764–6771. doi: 10.1021/bi900361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naue N., Beerbaum M., Bogutzki A., Schmieder P., Curth U. The helicase-binding domain of Escherichia coli DnaG primase interacts with the highly conserved C-terminal region of single-stranded DNA-binding protein. Nucleic Acids Res. 2013;41:4507–4517. doi: 10.1093/nar/gkt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryzhikov M., Koroleva O., Postnov D., Tran A., Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chilingaryan Z., Yin Z., Oakley A.J. Fragment-based screening by protein crystallography: Successes and pitfalls. Int. J. Mol. Sci. 2012;13:12857–12879. doi: 10.3390/ijms131012857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer M., Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 30.Doak B.C., Morton C.J., Simpson J.S., Scanlon M.J. Design and evaluation of the performance of an NMR screening fragment library. Aust. J. Chem. 2013;66:1465–1472. doi: 10.1071/CH13280. [DOI] [Google Scholar]
- 31.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 32.Malik M.A., Wani M.Y., Al-Thabaiti S.A., Shiekh R.A. Tetrazoles as carboxylic acid isosteres: Chemistry and biology. J. Incl. Phenom. Macrocycl. Chem. 2014;78:15–37. doi: 10.1007/s10847-013-0334-x. [DOI] [Google Scholar]
- 33.Allen F.H., Groom C.R., Liebeschuetz J.W., Bardwell D.A., Olsson T.S.G., Wood P.A. The hydrogen bond environments of 1H-tetrazole and tetrazolate rings: The structural basis for tetrazole–carboxylic acid bioisosterism. J. Chem. Inf. Model. 2012;52:857–866. doi: 10.1021/ci200521k. [DOI] [PubMed] [Google Scholar]
- 34.Fielding L. NMR methods for the determination of protein–ligand dissociation constants. Curr. Top. Med. Chem. 2007;3:39–53. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- 35.Lian L.-Y. NMR studies of weak protein–protein interactions. Prog. Nucl. Magn. Reson. Spectrosc. 2013;71:59–72. doi: 10.1016/j.pnmrs.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Dalvit C., Vulpetti A. Intermolecular and intramolecular hydrogen bonds involving fluorine atoms: Implications for recognition, selectivity, and chemical properties. ChemMedChem. 2012;7:262–272. doi: 10.1002/cmdc.201100483. [DOI] [PubMed] [Google Scholar]
- 37.Brameld K.A., Kuhn B., Reuter D.C., Stahl M. Small molecule conformational preferences derived from crystal structure data. A medicinal chemistry focused analysis. J. Chem. Inf. Model. 2008;48:1–24. doi: 10.1021/ci7002494. [DOI] [PubMed] [Google Scholar]
- 38.Irwin J.J., Shoichet B.K. ZINC−A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medek A., Hajduk P.J., Mack J., Fesik S.W. The use of differential chemical shifts for determining the binding site location and orientation of protein-bound ligands. J. Am. Chem. Soc. 2000;122:1241–1242. doi: 10.1021/ja993921m. [DOI] [Google Scholar]
- 40.Stamford N.P.J., Lilley P.E., Dixon N.E. Enriched sources of Escherichia coli replication proteins. The dnaG primase is a zinc metalloprotein. Biochim. Biophys. Acta. 1992;1132:17–25. doi: 10.1016/0167-4781(92)90047-4. [DOI] [PubMed] [Google Scholar]
- 41.Love C.A., Lilley P.E., Dixon N.E. Stable high-copy-number bacteriophage λ promoter vectors for overproduction of proteins in Escherichia coli. Gene. 1996;176:49–53. doi: 10.1016/0378-1119(96)00208-9. [DOI] [PubMed] [Google Scholar]
- 42.Loscha K., Oakley A.J., Bancia B., Schaeffer P.M., Prosselkov P., Otting G., Wilce M.C.J., Dixon N.E. Expression, purification, crystallization, and NMR studies of the helicase interaction domain of Escherichia coli DnaG primase. Protein Expr. Purif. 2004;33:304–310. doi: 10.1016/j.pep.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 44.Ziarek J.J., Peterson F.C., Lytle B.L., Volkman B.F. Binding site identification and structure determination of protein–ligand complexes by NMR: A semiautomated approach. Methods Enzymol. 2011;493:241–275. doi: 10.1016/B978-0-12-381274-2.00010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo A.T.Y., Oakley A.J., Dixon N.E. The DnaG primase interaction with single-stranded DNA-binding protein (SSB) Manuscript in preparation.
- 47.Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.