Abstract
Cells in the pluripotent state have the ability to self-renew indefinitely and to differentiate to all the cells of the embryo. These cells provide an in vitro window into development, including human development, as well as holding extraordinary promise for cell-based therapies in regenerative medicine. The recent demonstration that somatic cells can be reprogrammed to the pluripotent state has raised the possibility of patient and disease specific induced pluripotent cells. Here we review the molecular underpinning of pluripotency. We focus on the transcriptional and signaling networks that underlie the state of pluripotency and control differentiation. In general, the action of each of the molecular components and pathways is dose and context dependent highlighting the need for a systems approach to understanding pluripotency.
The three germ layers of the mammalian embryo all derive from the cells of the epiblast which is itself a derivative of the inner cell mass (ICM) (Figure 1). Mouse embryonic stem cell (mESC) lines were initially derived by plating cells from the ICM on a layer of embryonic feeder cells1, 2. The cells cultured from the ICM meet the defining criteria for pluripotency in that they: 1) self-renewal indefinitely and 2) give rise to all the cell types which comprise the embryo. More recently, pluripotent cells meeting these same criteria have been isolated from the early human embryo (Movies 1 and 2) 3.
Figure 1. Early Mammalian Embryonic Development.
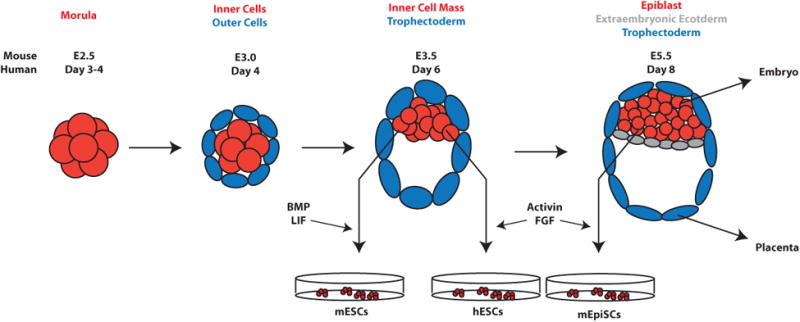
After morula stages, the first cell fate decisions are made, in which cells sort to outer and inner populations. Outer cells give rise to the extraembryonic trophectoderm (TE), while inner cells form the inner cell mass (ICM). The ICM is located asymmetrically at one side of the blastocoel cavity within the TE. Subsequently, the ICM further differentiates to the extraembryonic endoderm (ExEn) and the epiblast, which gives rise to the embryonic ectoderm, mesoderm and endoderm. Mouse and human embryonic stem cells are derived in vitro by explanting the ICM.
Functionally, pluripotency can be demonstrated by several experimental tests. These include differentiation to all three germ layers in vitro and in vivo (embroid body and teratoma formation, respectively), contribution to chimeric mice upon injection into blastocyst-stage embryos, and, most stringently, tetraploid complementation. In the latter technique, the pluripotent cells generate the entire mouse while the tetraploid cells contribute only to extraembryonic tissue4, 5. The pluripotency of mESCs has been demonstrating using all of the above techniques, while human embryonic stem cells (hESCs) have been used to generate embryoid bodies, teratomas and even mouse-human chimeric blastocyst-stage embryos6.
The study of ESCs holds significant promise for problems of both fundamental and clinical significance. ESCs provided a technical means to manipulate the mouse germline. Furthermore, while studies of mESCs in vitro can complement in vivo approaches, hESCs provide the only system for studying human development and its differences with other mammals. Finally, the ability to differentiate ESCs to specific cell types has the potential to lead to cell-based therapies for a wide range of disorders in regenerative medicine. The recent discovery that somatic cells can be reprogrammed into a pluripotent state7 (known as induced pluripotent stem cells or iPSCs) has raised the possibility of generating patient- and disease-specific stem cells through reprogramming. In this article, we review the molecular basis of pluripotency focusing in particular on the signaling and transcriptional networks that ESCs use to maintain pluripotency and to differentiate.
Signaling pathways in pluripotency and differentiation
During embryogenesis, signaling pathways provide the cues to establish positional information within the embryo and to instruct cells to differentiate. Pathways typically begin at the cell surface with ligand binding to a receptor complex and terminate in the cell nucleus with the activation of transcription thus allowing a transfer of information from outside the cell to inside the nucleus. Proper signaling cues are essential both for self-renewal in the state of pluripotency and for instructing cells to differentiate to particular lineages. In this section, we review several emerging themes in signaling in pluripotent cells with a focus on the developmentally essential LIF, BMP, Activin/Nodal, FGF, and Wnt pathways (Table 1).
Table 1.
Signaling pathways involved in the maintenance of pluripotency. Table summarizing properties of pathways that play a role in maintaining pluripotency either in mESCs or hESCs.
| Pathway | LIF | BMP | Activin/Nodal | FGF | Wnt |
|---|---|---|---|---|---|
| Receptor | gp130 | Alk2/3/6 | Alk4/5/7 | FGF-R | LRP5/6 |
| Signal transducer | Stat3 | Smad1/5/8 | Smad2/3 | MEK/ERK | β-catenin |
| mESCs? | + | + | - | - | + |
| hESCs? | - | - | + | + | + |
Proper signaling cues can maintain self-renewal by activating pluripotency and repressing differentiation-specific genes
Within the embryo, specific signals specify the ICM and allow its cells to remain pluripotent. Traditionally, in vitro, these signals have been replaced by culture on a feeder layer of mouse embryonic fibroblasts. These same cells are capable of maintaining the pluripotency of both mES and hESCs, however, elucidation of the signaling requirements to maintain each cell type without feeders has revealed large difference between mouse and human pluripotent cells.
Signaling pathways maintaining pluripotency in mESCs
Over twenty years ago, it was discovered that the feeder cells could be replaced by a combination of the signaling molecule LIF and serum8. LIF signals through the transcriptional activator STAT3 and activation of STAT3 alone is sufficient to replace the requirement for LIF in maintaining pluripotency9. The primary mechanism of action of LIF in maintaining pluripotency appears to be through STAT3 induction of Klf4, however, LIF also maintains Nanog expression by signaling through the PI3K pathway10. Thus, LIF appears to function mainly by directly inducing key pluripotency-associated genes.
More recently, it was discovered that under these culture conditions, the serum in the medium could be replaced with BMP ligands allowing the culture of mESCs in feeder-free, serum-free medium containing LIF and BMP11. In ESCs, BMP ligands signal through the Smad pathway to activate expression of Id genes that inhibit differentiation11. BMP also functions by inhibiting the ERK and p38 MAPK pathways which promote differentiation in mES (see below)12. Thus, LIF and BMP function synergistically in mES by promoting pluripotency and suppressing differentiation, respectively.
It is natural to ask whether activation of these pathways is necessary or merely sufficient for the maintenance of pluripotency. In fact, mESCs deficient in either LIF or its receptor gp130 can be propagated and mice generated from these cells develop nearly normally13. Further, the pluripotent state of mESCs can be maintained solely by inhibition of the FGF differentiation pathway using both an FGF receptor inhibitor and a MEK inhibitor, however, growth under these conditions is improved when GSK3β is inhibited as well14. Stat3-/- cells which are incapable of transducing LIF signals can be maintained in this formulation demonstrating that this pathway is not strictly required for pluripotency. The maintenance of pluripotency by these three inhibitors has been termed the “ground state” of ES cell self-renewal14.
Signaling pathways maintaining pluripotency in hESCs
Surprisingly, the LIF and BMP pathways do not play a role in self-renewal in hESCs (Table 2). Addition of LIF to hESC culture medium activates STAT3 but cannot substitute for the layer of feeder cells as is the case for mESC15. Additionally, BMP is a differentiation pathway in hESCs, and even relatively low doses cause differentiation to extraembryonic or mesodermal fates16-18. Instead, hESCs can be maintained in feeder-free, serum-free conditions through stimulation of the FGF19-21 and Activin/Nodal pathways22-24. Thus the signaling requirements of mouse and human pluripotent cells are significantly different. It has been suggested that hESCs may represent a later stage of development than mESCs and indeed stem cell populations derived from the E5.5 epiblast share many features with hESCs (Figure 2)25, 26. It has also been shown that it is possible to revert hESCs to an earlier developmental state that resembles mESCs in its signaling requirements27.
Table 2.
Comparison of Mouse ESCs, mouse EpiSCs with Human ESCs. Many features of mESCs, mEpiSCs and hESCs have been evaluated singly and in parallel. A summary of key characteristics is provided here. For additional information, see refs 25, 26, 113, 114.
| mESCs | hESCs | mEpiSCs | |
|---|---|---|---|
| Morphology | Rounded | Flattened | Flattened |
| Single cell survival | Good | Poor | Poor |
| Potency | All embryonic fates | All embryonic fates | All embryonic fates |
| Signaling inputs | BMP, LIF | Activin, FGF | Activin, FGF |
| Embryoid Body Formation | Yes | Yes | Yes |
| Teratoma Formation | Yes | Yes | Yes |
| Tetraploid Complementation | Yes | N/A | No |
| X inactivation | No | Yes | Yes |
Figure 2. Relationships between signaling pathways, pluripotency, and differentiation.
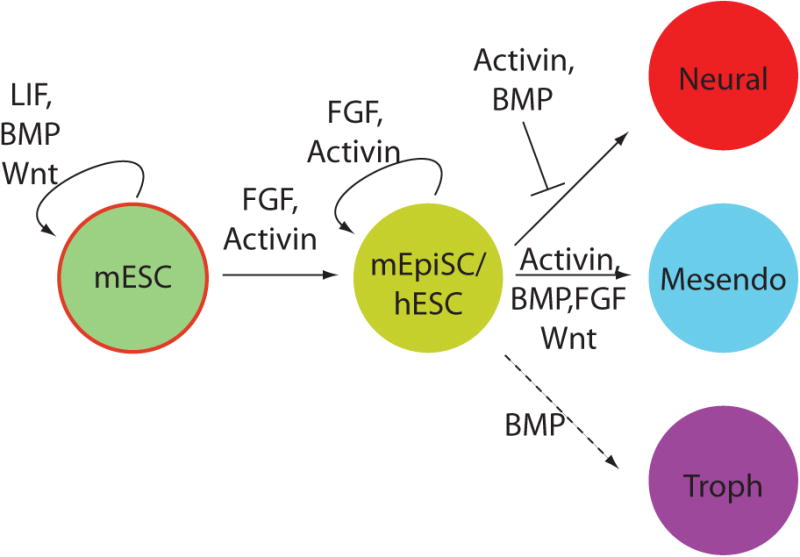
Schematic depicting the relationship between signaling pathways and the indicated cell states. The dashed line indicates a connection only present in human but not mouse ES cells while the red-circled state indicates a state only accessible to mouse but not human cells.
Similar to the roles LIF and BMP play in mESC, FGF and Actvin signaling both activate the expression of key pluripotency genes and suppress differentiation-related genes and pathways. Activin/Nodal signaling activates Nanog and the signal transducers Smad2/3 bind directly to the Nanog promoter28. This interaction has been suggested to occur in vivo in model organisms as well29. FGF signaling through the ERK pathway has been reported to sustain Nanog expression, however, this is likely an indirect effect acting through the Activin/Nodal pathway17. Independently, FGF activation of the PI3K pathway promotes pluripotency by directing Smad2/3 activity to pluripotency rather than differentiation genes (see below)30, 31. Further, both pathways play a role in suppressing the BMP differentiation pathway28, 32, although the molecular mechanisms of these interactions remain unclear. In other contexts, Activin/Nodal signaling has been suggested to suppress BMP signaling through competition for common pathway elements such as Smad433, while FGF signaling through ERK has been shown to induce inhibitory phosphorylations in the linker regions of the BMP signal transducers Smad1/5/834.
Despite the differences between mouse and human, Wnt signaling has emerged as a signaling pathway that plays a role in maintaining pluripotency in both mouse and human. In mESCs, Wnt signaling can maintain pluripotency under conditions that would otherwise promote differentiation15, 35 and may function both by upregulating LIF/Stat3 signaling36 and by suppressing the transition to the epiblast state37. Wnt signaling can also enhance the reprogramming of murine somatic cells to induced pluripotent cells38, 39. Wnt signaling appears to play a similar role in maintaining hESC pluripotency although the molecular mechanisms remain unclear15. As Wnt, FGF, and TGFβ all function as morphogens, it is very likely that particular concentrations are necessary for this activity as we now discuss.
The same signalling pathways recur in maintenance of pluripotency and induction of differentiation
Paradoxically, many of the pathways involved in maintaining pluripotency play a key role in differentiation as well. Decades of research in model organisms have delineated essential roles for the FGF, BMP, Activin/Nodal and Wnt signaling pathways in early developmental processes including mesoderm induction, dorsal-ventral patterning, and formation of Spemann's organizer40-44 and these pathways directly activate key differentiation genes such as Brachyury, Gooscoid, and Sox17. Furthermore, under differentiation conditions, these pathways play similar roles in ESCs. Taken together, these observations raise a central question: how do signaling pathways maintain pluripotency under some conditions while directing differentiation under others.
In mESCs, the key to answering this question may lie in the fact that mESCs represent an early stage of development and are not primed for differentiation. Recent studies argue that mESCs transition to a primed epiblast stem cell (EpiSC) state before differentiating to any of the three germ layer lineages. This ES to epiblast transition is induced by upregulation of FGF signaling, consistent with the expression of FGF5 in the epiblast in vivo. Subsequently, embryonic lineages are specified by particular activities and/or combinations of ligands such as BMP, Activin/Nodal or retinoic acid (RA)45, 46. Importantly, while some signals are instructive, others may potentiate differentiation directed by other pathways. Further, RA signaling also appears to initiate the upregulation of FGF signaling that leads to the EpiSC state45. Thus, complex signaling relationships mediate pluripotency versus differentiation toward specific germ layers, and differences in cell state may determine whether a signaling pathway promotes self-renewal or differentiation.
In human cells, the issue is more problematic as hESCs already represent a later stage of development and are primed for differentiation. Upon stimulation with growth factors such as BMP or Activin/Nodal, hESCs show both morphological and molecular signs of differentiation within 24 hours18, 47. Thus, how it is that Activin/Nodal or Wnt signaling can both promote pluripotency and direct differentiation in hESCs remains an important question. Indeed recent studies showing that activation of Wnt signaling in hESCs leads to mesendoderm differentiation have been used to suggest that Wnt is primarily a differentiation, not self-renewal, pathway in hESCs31, 48.
The answer to this issue may lie at least in part in the fact that nearly all of these pathways function as morphogens in vitro and in vivo, elucidating different outcomes depending on the concentration or duration of signaling40, 42, 49. In the case of Wnt signaling, recent evidence suggests that while low levels support pluripotency in hESCs, higher levels lead to differentiation50, 51. This may also provide part of the explanation for the differing effects of Activin/Nodal signaling in hESCs as the concentrations used to maintain self-renewal are typically significantly lower than those used in differentiation18, 47. Thus, in this view, the state of pluripotency can be considered one of many possible fate outcomes induced by Activin/Nodal or Wnt morphogens and is induced by low but not high concentrations.
Signaling pathways form a network that dictates the balance of self-renewal and differentiation
Signaling pathways do not function in isolation but the status of signaling through one pathway can dictate the outcome when another is activated. Thus, another part of the explanation for how the same signaling pathways guide both self-renewal and differentiation likely lies in considering the status of a network of pathways rather than evaluating each pathway separately (Figure 3). Similar principles are needed to understand differentiation as the result of adding identical concentrations of a differentiating ligand can be altered depending on the status of other pathways.
Figure 3. Network of signaling pathways governing pluripotency and differentiation.
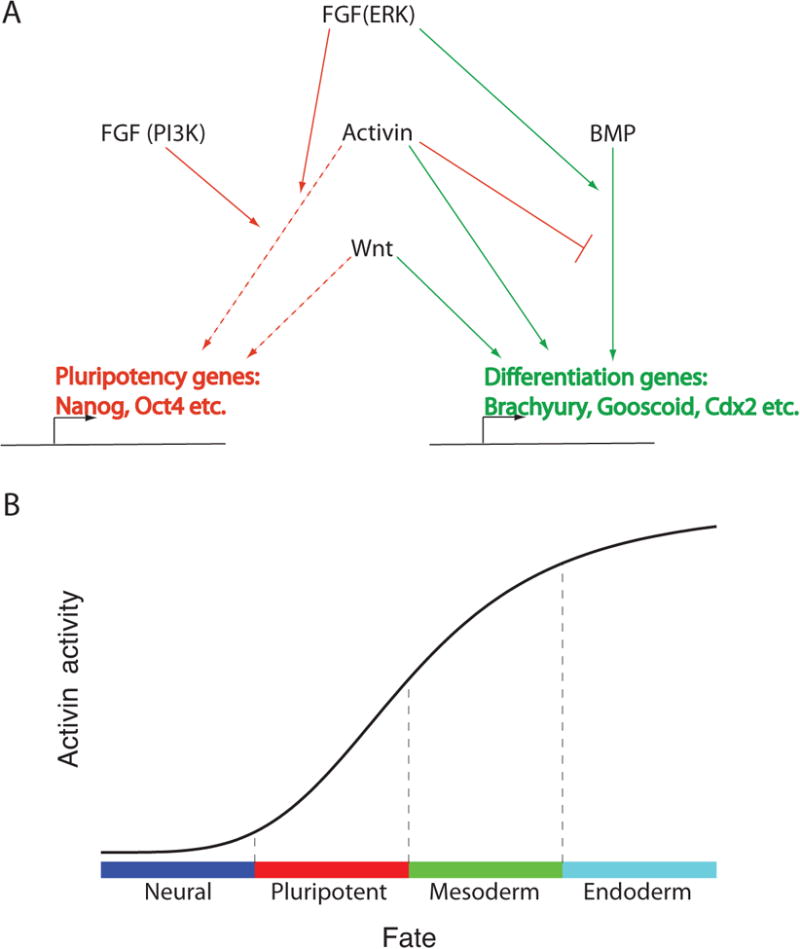
(A) Schematic depicting the relationships between signaling pathways and genes control cell fate. Red lines denote interactions promoting pluripotency and green lines denote interactions promoting differentiation. Dashed lines indicate interactions only operative at low to intermediate activity of the signaling pathway. (B) Pluripotency is one of a spectrum of possible fates that result from modulating the Activin/Nodal pathway. Lower or higher levels of pathway activity lead to neural or mesendodermal differentiation, respectively.
An illustration of these ideas has recently been uncovered in the interactions between FGF and Activin/Nodal signaling governing the balance between pluripotency and differentiation in hESCs31. As discussed above, Activin/Nodal signaling mediates both self-renewal and differentiation to mesendodermal lineages. These effects depend on the status of the ERK and PI3K pathways which function downstream of FGF. In pluripotency conditions, the PI3K pathway is active and directs Activin/Nodal signaling to pluripotency-promoting genes such as Nanog. Under differentiation conditions, PI3K is suppressed, leading to upregulation of the ERK pathway as well as activation of Wnt signaling. These combined changes redirect Activin/Nodal from maintaining pluripotency to inducing differentiation. Thus considering the status of an integrated signaling pathway elucidates how Activin/Nodal signaling can play context-specific roles.
A similar phenomenon occurs during BMP-mediated differentiation. Results from model systems in vivo has identified a role for BMP in inducing mesodermal lineages52. In hESCs, however, treatment with BMP ligands leads to induction of genes and cell morphology associated with trophoectedermal lineages16. These differing results can be explained by the status of the FGF pathway. When the FGF pathway is active, it cooperates with BMP signaling to induce mesoderm, while in its absence, BMP induces trophectoderm17. A recent study has challenged whether BMP-differentiated hESCs represent a true trophectodermal population18, however, whatever the outcome of that debate, it is clear that outcome of BMP-mediated differentiation depends on the status of the FGF pathway. These results highlight that the effects of signaling pathways on self-renewal and differentiation can only be unraveled by considering an integrated signaling network.
Epigenetic control of pluripotency and differentiation
The chromatin state of a cell provides the context in which the transcriptional changes that mediate self-renewal and differentiation must take place. Recently, modulators of chromatin have emerged as important players in the maintenance of pluripotency, in differentiation, and in reprogramming. A thorough discussion of this topic is beyond the scope of this review and the reader is directed to recent reviews devoted to this subject53, 54. Here we review basic concepts and highlight recent studies relevant to signaling and transcriptional networks in pluripotency.
ESCs are generally characterized by an open chromatin configuration54 and this is associated with a hyperactive transcriptome, including expression of low levels of many lineage specific genes55. This promiscuous transcription has been suggested to be associated with the plasticity of ESCs and differentiation is reflected in silencing genes from alternate lineages. In vivo studies have demonstrated a similar open chromatin configuration in cells from the E3.5 mouse blastocyst56.
Generally, genes active in the pluripotent state are associated with the chromatin mark H3K4me3 while those associated with differentiation have a repressive mark such as H3K27me353. Many differentiation genes have both active and repressive marks. This bivalent chromatin represents a state that is silent but poised for transcription57. These bivalent marks are recognized by the Polycomb complex that acts in a repressive role. Differentiation is accompanied by the loss of the repressive H3K27me3 mark and activation of transcription58. However, a simple role for Polycomb proteins in repressing differentiation-associated genes to maintain pluripotency is excluded by the observation that ESCs defective in a critical subunit of the Polycomb complex are pluripotent59. The maintenance of pluripotency in the absence of the Polycomb complex is likely due to the redundant role of several repressive complexes in silencing differentiation-related genes60. More recent studies have revealed a nuanced picture of Polycomb function with the composition and activity of the Polycomb complex changing during differentiation53.
A recent study has highlighted the role of poised chromatin in triggering differentiation61. This study revealed that complexes of the Activin/Nodal signal transducers Smad2/3 with TIF1γ/TRIM33 recognized the poised chromatin mark H3K9me3 on key mesendodermal regulators specifically in response to activation by Activin/Nodal. This recognition was necessary for activation of these genes by the canonical Activin/Nodal active signaling complex containing Smad2 and Smad4. Thus, Activin/Nodal/nodal signaling uses the poised chromatin marks to switch these genes from poised to active states. These finding remain controversial, however, as in other contexts TRIM33 has been shown to be antagonistic to Activin/Nodal signals62, 63 and the phenotype of the mouse knockout of TRIM33 is more consistent with overactive than repressed Activin/Nodal64.
There are extensive associations between the network of transcription factors governing pluripotency and chromatin modifications. In particular, c-Myc has emerged as a key factor involved in the core circuitry of ESCs and linked to multiple activities involving chromatin modifications65. This study shows that loss of Myc leads to widespread changes in chromatin modifications in both mESCs and the early mouse embryo.
Transcriptional networks controlling pluripotency
Core Transcriptional Circuitry
Through microarray analysis, a global view of gene expression associated with the state of stemness has begun to emerge66-69. Prominent results in each of these studies included genes that had previously been shown to play critical roles in embryogenesis and formation of ESCs. These include genes that have been utilized for the reprogramming of mouse and human somatic cells, such as Klf4 and c-Myc. In addition, focused studies in mESCs and hESCs have confirmed requirements for other factors like FoxD3 and Id proteins in maintenance of self-renewal or pluripotency70-72. Relative to differences in signaling requirements and epigenetic status between mESCs, mEpiSCs and hESCs, transcriptional profiles appear to be more conserved between these cell types25, 26. Expression levels of some differentiation-associated genes are elevated in mEpiSCs and hESCs compared to mESCs at the RNA level, consistent with the notion of “primed” versus “naive” states of pluripotency26, 27. On the other hand, evidence that these differences exist at the protein level is limited. Further, pluripotency transcription factors Oct4, Sox2, Nanog, and Myc levels are consistent between cell types26. In this section, we will focus on Oct4, Sox2 and Nanog, three transcription factors that have been shown repeatedly to be at the heart of the transcriptional network that supports pluripotency.
Oct4, Sox2 or Nanog loss-of-function results in failure of epiblast formation and embryonic lethality by implantation stages. Consequently, mESCs cannot be established from Oct4-/-, Sox2-/- or Nanog-/- blastocysts73-75. RNA interference-mediated knockdown of these genes in hESCs results in loss of pluripotency and self-renewal76-78, consistent with results in mice. Gene expression and knockout studies established indispensible roles for Oct4, Sox2 and Nanog in pre- and peri-implantation murine development. Oct4 is expressed from the 8 cell stage, transiently in the extraembryonic endoderm and later becomes restricted to the epiblast79, 80. Similarly, Sox2 is expressed in all cells at morula stages and in the epiblast, as well as trophectoderm81. Nanog expression is initiated slightly later, in post-compaction morulae, and persists in the ICM and epiblast74, 82. Analysis of gene expression in human blastocysts indicates that Oct4, Sox2 and Nanog are expressed in the ICM83, 84. Recently, numerous genome-wide studies have been undertaken to determine how these key factors regulate the transcriptional repertoire of ESCs. Although the picture remains incomplete, several evolutionarily conserved trends have emerged.
Coordinators of the Pluripotency Network
Cooperative regulation
Genome-scale analyses of Oct4, Sox2 and Nanog binding sites reveal that they frequently bind the same regulatory regions in undifferentiated mouse and human ESCs, and that these binding sites are often in close proximity to one another85-88. Oct/Sox composite binding sites have been identified in individual promoters, directly adjacent or separated by less than five base pairs89-91. These data point not only to coordinated regulation of targets, but in some cases physical interactions between the transcription factors themselves89, 90, 92. Further, it appears that combinatorial binding sites may be significantly more conserved between mouse and human than individual binding sites93, suggesting that this may be a dominant mode of transcriptional control in the state of pluripotency.
In addition to one another, Oct4, Sox2 and Nanog activities are modulated by downstream effectors of signaling pathways, such as Tcf3 and Smad3, and by Polycomb Repressor Complexes (PRCs). Tcf3 and Smad3 binding sites are enriched in promoters that are also bound by the Oct4/Sox2/Nanog combination94-96. In the presence of Wnt signaling levels that support pluripotency in mESCs, Tcf3 is likely to be associated with repression of transcription, whereas elevated levels of Wnt signaling overcome the repressive effect of Tcf3 and lead to differentiation of mESCs95, 97, 98. In the reciprocal experiment, knockdown of Tcf3 can substitute for the Wnt requirement in maintenance of pluripotency96. A critical component of TGFβ signalling, Smad3, appears to be recruited by Oct4 to specific promoters on a genome-wide scale94, providing another potential intersection between established signaling cues and the transcriptional network of ESCs. Finally, a subset of regulatory regions enriched for binding by each of these transcription factors is also enriched for binding by PRCs58, 85. Differential effects of Oct4, Nanog or Tcf3 knockdown on target gene expression (see below) may result from association of PRCs with some targets but not others95. Thus, a variety of inputs acting in concert, both cooperatively and antagonistically, are required to balance the transcriptional activity of the pluripotency network.
Targets
Oct4, Sox2 and Nanog have been shown to both activate and repress transcription in particular cases89, 90, 99. Indeed, global analyses have found Oct4, Sox2, and Nanog binding sites to be roughly equally distributed between genes that maintain pluripotency and those that promote differentiation85, 87. However, when Oct4 levels are experimentally manipulated, genes that are predicted to be activated by Oct4 are more likely to be affected than those predicted to be repressed88, 94. Suppression of Oct4 function in mESCs leads first to downregulation of targets that promote stemness, followed at later time points by upregulation of genes that promote differentiation85, 88. These observations suggest that Oct4 acts primarily through the activation of other pluripotency genes, which in turn repress genes associated with differentiation. In agreement with this hypothesis, overexpression of Oct4 coupled with cyclohexamide treatment to inhibit protein synthesis indicated that repressors of differentiation were far more likely to be direct targets than promoters of differentiation100. On the other hand, a slightly more even distribution between activated and repressed promoters was observed for Nanog binding sites88, and Nanog and Oct4 have both been associated with repressive protein complexes in mESCs101. Thus, the precise activity of Oct4, Nanog and Sox2 is likely to be context- and promoter-specific.
Roughly equal numbers of Oct4 and Nanog binding sites are found in intragenic regions compared to promoter regions86. Although it remains unclear whether functional binding is equivalent between these two categories, these findings point to the need for careful attention to the genomic regions examined in published studies and further analysis.
Stoichiometry and combinatorial regulation of cell fate
In addition to regulating other genes, Oct4, Sox2 and Nanog have been shown to cross- and auto-regulate75, 86, 100, 102-105. Such feed-forward mechanisms support robustness of transcriptional programs, and also serve to maintain balanced levels of key regulatory factors. Much like the case with signaling pathways, specific levels of transcription factor activity appear to be important for maintenance of pluripotency. Thus, even small perturbations of Oct4 or Sox2 levels lead to differentiation78, 106-108. Interestingly, upregulation of these transcription factors has different effects. For example, elevated levels of Oct4 expression lead to differentiation of ESCs to extraembryonic endoderm fates108, while overexpression of Sox2 generates a variety of fates that specifically exclude endoderm106. Similarly, in a growth factor-mediated differentiation paradigm, slightly elevated levels of Oct4 or Sox2 correlated with distinct differentiation outcomes109. Further, this study found that in pluripotent cells Oct4 and Sox2 bound promoters together in agreement with previous reports, but that they became enriched at opposing loci during differentiation. For example, Sox2 becomes enriched over Oct4 at its own enhancer and in the Brachyury regulatory region during neurectodermal differentiation, signaling positive and negative regulation, respectively. Together with the high degree of cooperative binding observed in ChIP experiments, these data support the notion that correct stoichiometry of key transcription factors is critical. Thus, concentration- and activity-specific mechanisms function at multiple levels in regulation of pluripotency, including extracellular signaling and intracellular transcription factors.
Reprogramming to the pluripotent state
Induced pluripotent stem cells (iPSCs) present an opportunity to further analyze and test our knowledge of pluripotency. Further, the ability to generate disease- or patient-specific iPSC lines for study and eventually for therapeutics holds great promise for both basic biology and medicine. Still in its infancy, the field of iPSC research began with the reprogramming of mouse fibroblasts7, followed closely by complementary experiments in human somatic cells110, 111. In these seminal studies, it was demonstrated that forced expression of a small number of transcription factors could alter the potency of highly derivative cells from embryos or adults, bringing them back to a status resembling embryonic stem cells. In the intervening five years, considerable progress has been made in our understanding of the reprogramming process. To thoroughly address this topic is the work of several reviews112-114. In this section, we focus on a few major developments in the field of reprogramming as they relate to our understanding of pluripotency.
The original study in mice found that four transcription factors, Oct4, Sox2, Klf4 and c-Myc (OSKM) were sufficient for reprogramming7. Although it was later postulated that these cells were only partially reprogrammed, the utility of OSKM has been upheld in numerous subsequent studies115-117. These requirements have further been dissected, revealing that different combinations of transcription factors are sufficient for the reprogramming of different somatic cell types118-124. In some cases, the function of individual transcription factors can be replaced by chemical or small molecule treatments125-127. Notably, the one requirement for which no substitute has been found is Oct4, underscoring the pivotal role that this transcription factor plays in pluripotency.
With very few exceptions, the efficiency of the reprogramming process continues to be low (< 1%)114. This suggests that numerous obstacles exist to the reacquisition of pluripotency by somatic cells114, 128. These include, but may not be limited to, deactivation of the somatic cell gene program, activation of endogenous pluripotency genes (both the factors being used to reprogram and additional loci), epigenetic remodeling, and a number of cell divisions. It has further been postulated that only a small number of cells in any given population have the capacity to be reprogrammed128. It is likely that low reprogramming efficiency is the result of a combination of these factors – many barriers exist, and thus only a small number of cells manage to clear all the hurdles to be fully reprogrammed.
In order to be considered fully reprogrammed, iPSCs must pass all the tests of pluripotency, including the abilities to self-renew and to form derivatives of each of the three embryonic germ layers in vitro and in vivo (teratomas). The tetraploid complementation test, in which mouse iPSCs are challenged to form an entire embryo, was recently passed129, 130 albeit with possible complications131. Global profiling methods for gene expression and DNA methylation have also been leveraged in characterization of iPSC lines132-134. However, our incomplete understanding of the state of pluripotency and of embryonic development in general leads to several key challenges in the effort to utilize iPSC technology114, 135.
First, a more precise definition of the state of pluripotency is necessary. Focused and global studies of gene expression, promoter activation and repression, DNA methylation and other characteristics continue to refine this picture. Second, due to variability in the precise nature of iPSC lines derived using different methods, any study of disease-specific iPSC lines must have an isogenic wild-type iPSC line generated in the same manner, preferably in parallel. This increases confidence that defects in differentiation of the disease-specific iPSC line result from the disease defect, rather than an artifact of the reprogramming process. This leads to a third requirement, the ability to genetically modify iPSC lines. Although homologous recombination has been a standard technique in mESCs for decades, efficiencies in human cells have until recently been very low. However, use of zinc finger (ZF) and transcription activator-like effector (TALE) nucleases has significantly improved efficiency of this process136, 137. Finally, methods for directed differentiation of ESCs and iPSCs to specific cell types affected in a given disease must be established. This issue has and continues to be a major focus of the field.
The discovery of the possibility for somatic cells to reacquire pluripotency was a tremendous leap forward for both basic biology and medical applications. The field continues to move forward at an impressive pace, bringing new perspectives and tools to bear on questions of the nature of pluripotency.
Contributor Information
Aryeh Warmflash, Laboratory of Molecular Vertebrate Embryology, The Rockefeller University, New York, NY.
Brigitte L Arduini, Laboratory of Molecular Vertebrate Embryology, The Rockefeller University, New York, NY.
Ali H Brivanlou, Laboratory of Molecular Vertebrate Embryology, The Rockefeller University, New York, NY.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Eggan K, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6209–14. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8424–8. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295:90–102. doi: 10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Smith AG, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–90. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 9.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–60. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–22. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 11.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–92. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 12.Qi X, et al. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6027–32. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart CL, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–9. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 14.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 16.Xu RH, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–4. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 17.Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–34. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernardo AS, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–55. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amit M, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–8. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–7. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 21.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 22.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–82. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 23.Beattie GM, et al. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489–95. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- 24.Xiao L, Yuan X, Sharkis SJ. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–86. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 25.Brons IGM, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 26.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 27.Hanna J, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9222–7. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu RH, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin M, Alev C, Wu Y, Nagai H, Sheng G. Activin/TGF-beta signaling regulates Nanog expression in the epiblast during gastrulation. Mech Dev. 2011;128:268–78. doi: 10.1016/j.mod.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 30.McLean AB, et al. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 31.Singh AM, et al. Signaling Network Crosstalk in Human Pluripotent Cells: A Smad2/3-Regulated Switch that Controls the Balance between Self-Renewal and Differentiation. Cell Stem Cell. 2012;10:312–26. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avery S, Zafarana G, Gokhale PJ, Andrews PW. The role of SMAD4 in human embryonic stem cell self-renewal and stem cell fate. Stem Cells. 2010;28:863–73. doi: 10.1002/stem.409. [DOI] [PubMed] [Google Scholar]
- 33.Candia AF, et al. Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–80. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 34.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389:618–22. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 35.Miyabayashi T, et al. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5668–73. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 37.ten Berge D, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–5. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marson A, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–5. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lluis F, Pedone E, Pepe S, Cosma MP. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Green JB, New HV, Smith JC. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–9. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 41.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–84. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- 43.Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–17. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- 44.Hemmati-Brivanlou A, Thomsen GH. Ventral mesodermal patterning in Xenopus embryos: expression patterns and activities of BMP-2 and BMP-4. Dev Genet. 1995;17:78–89. doi: 10.1002/dvg.1020170109. [DOI] [PubMed] [Google Scholar]
- 45.Stavridis MP, Collins BJ, Storey KG. Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development. 2010;137:881–90. doi: 10.1242/dev.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunath T, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 47.D'Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–41. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 48.Davidson KC, et al. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiecker C, Niehrs C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development. 2001;128:4189–201. doi: 10.1242/dev.128.21.4189. [DOI] [PubMed] [Google Scholar]
- 50.Tsutsui H, et al. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat Commun. 2011;2:167. doi: 10.1038/ncomms1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, Jiang K, Ding S. Concise review: A chemical approach to control cell fate and function. Stem Cells. 2012;30:61–8. doi: 10.1002/stem.768. [DOI] [PubMed] [Google Scholar]
- 52.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 53.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–50. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efroni S, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–47. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed K, et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 58.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 59.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chakravarthy H, Ormsbee BD, Mallanna SK, Rizzino A. Rapid activation of the bivalent gene Sox21 requires displacement of multiple layers of gene-silencing machinery. FASEB J. 2011;25:206–18. doi: 10.1096/fj.10-166926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xi Q, et al. A Poised Chromatin Platform for TGF-β Access to Master Regulators. Cell. 2011;147:1511–24. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dupont S, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–35. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 63.Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1γ to Chromatin via Its PHD Finger-Bromodomain Activates Its Ubiquitin Ligase and Transcriptional Repressor Activities. Mol Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 64.Morsut L, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–8. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 65.Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS ONE. 2009;4:e7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivanova NB, et al. A stem cell molecular signature. Science (New York, NY) 2002;298:601–4. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 67.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science (New York, NY) 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 68.Sato N, et al. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–13. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 69.Sperger JM, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13350–5. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hollnagel A, Oehlmann V, Heymer J, Rüther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. The Journal of biological chemistry. 1999;274:19838–45. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Labosky Pa. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem cells (Dayton, Ohio) 2008;26:2475–84. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanna La, Foreman RK, Tarasenko Ia, Kessler DS, Labosky Pa. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes & development. 2002;16:2650–61. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 74.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 75.Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nature cell biology. 2007;9:625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 76.Fong H, Hohenstein Ka, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem cells (Dayton, Ohio) 2008;26:1931–8. doi: 10.1634/stemcells.2007-1002. [DOI] [PubMed] [Google Scholar]
- 77.Hyslop L, et al. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem cells (Dayton, Ohio) 2005;23:1035–43. doi: 10.1634/stemcells.2005-0080. [DOI] [PubMed] [Google Scholar]
- 78.Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem cells (Dayton, Ohio) 2004;22:225–35. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- 79.Rosner MH, et al. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–92. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 80.Palmieri SL, Peter W, Hess H, Schöler HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Developmental biology. 1994;166:259–67. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 81.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & development. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 83.Hansis C, Grifo Ja, Krey LC. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Molecular human reproduction. 2000;6:999–1004. doi: 10.1093/molehr/6.11.999. [DOI] [PubMed] [Google Scholar]
- 84.Galán A, et al. Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PloS one. 2010;5:e13615. doi: 10.1371/journal.pone.0013615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sharov Aa, et al. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 87.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mathur D, et al. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome biology. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Molecular and cellular biology. 1999;19:5453–65. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tomioka M, et al. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic acids research. 2002;30:3202–13. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Molecular and cellular biology. 1997;17:6321–9. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei Z, et al. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem cells (Dayton, Ohio) 2009;27:2969–78. doi: 10.1002/stem.231. [DOI] [PubMed] [Google Scholar]
- 93.Göke J, et al. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS computational biology. 2011;7:e1002304. doi: 10.1371/journal.pcbi.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mullen AC, et al. Master Transcription Factors Determine Cell-Type-Specific Responses to TGF-β Signaling. Cell. 2011;147:565–76. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–55. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yi F, et al. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nature cell biology. 2011;13:762–70. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wray J, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nature cell biology. 2011;13:838–45. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yi F, Pereira L, Merrill BJ. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem cells (Dayton, Ohio) 2008;26:1951–60. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pan G, Thomson Ja. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell research. 2007;17:42–9. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 100.Onichtchouk D, et al. Zebrafish Pou5f1-dependent transcriptional networks in temporal control of early development. Molecular systems biology. 2010;6:354. doi: 10.1038/msb.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature cell biology. 2008;10:731–9. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 102.Chew Jl, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Molecular and cellular biology. 2005;25:6031–46. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuroda T, et al. Octamer and Sox Elements Are Required for Transcriptional cis Regulation of Nanog Gene Expression Octamer and Sox Elements Are Required for Transcriptional cis Regulation of Nanog Gene Expression. Society. 2005 doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20:1730–2. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 105.Rodda DJ, et al. Transcriptional regulation of nanog by OCT4 and SOX2. The Journal of biological chemistry. 2005;280:24731–7. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 106.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem cells (Dayton, Ohio) 2008;26:903–11. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 107.Rodriguez RT, et al. Manipulation of OCT4 levels in human embryonic stem cells results in induction of differential cell types. Experimental biology and medicine (Maywood, NJ) 2007;232:1368–80. doi: 10.3181/0703-RM-63. [DOI] [PubMed] [Google Scholar]
- 108.Niwa H, Miyazaki Ji, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature genetics. 2000;24:372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 109.Thomson M, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–89. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 111.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 112.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–25. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes & Development. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sugii S, et al. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3558–63. doi: 10.1073/pnas.0910172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Loh YH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–9. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nature genetics. 2009;41:968–76. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Giorgetti A, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell stem cell. 2009;5:353–7. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–50. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 120.Tsai SY, et al. Oct4 and klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem cells (Dayton, Ohio) 2010;28:221–8. doi: 10.1002/stem.281. [DOI] [PubMed] [Google Scholar]
- 121.Feng B, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nature cell biology. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 122.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell stem cell. 2008;2:10–2. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 123.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 124.Zhao Hx, et al. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation; research in biological diversity. 2010;80:123–9. doi: 10.1016/j.diff.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 125.Lyssiotis CA, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8912–7. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li Y, et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell research. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feng B, Ng JH, Heng JCD, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell stem cell. 2009;4:301–12. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 128.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 129.Boland MJ, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–4. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 130.Zhao Xy, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 131.Tong M, et al. Mice generated from tetraploid complementation competent iPS cells show similar developmental features as those from ES cells but are prone to tumorigenesis. Cell research. 2011;21:1634–7. doi: 10.1038/cr.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carey BW, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell stem cell. 2011;9:588–98. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 133.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li W, et al. iPS cells generated without c-Myc have active Dlk1-Dio3 region and are capable of producing full-term mice through tetraploid complementation. Cell research. 2011;21:550–3. doi: 10.1038/cr.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell stem cell. 2009;5:584–95. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zou J, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]


