Abstract
Genome integrity is monitored by a checkpoint that delays mitosis in response to DNA damage. This checkpoint is enforced by Chk1, a protein kinase that inhibits the mitotic inducer Cdc25. In fission yeast, Chk1 is regulated by a group of proteins that includes Rad3, a protein kinase related to human ATM and ATR. These kinases phosphorylate serine or threonine followed by glutamine (SQ/TQ). Fission yeast and human Chk1 proteins share two conserved SQ motifs at serine-345 and serine-367. Serine-345 of human Chk1 is phosphorylated in response to DNA damage. Here we report that Rad3 and ATM phosphorylate serine-345 of fission yeast Chk1. Mutation of serine-345 (chk1-S345A) abrogates Rad3-dependent phosphorylation of Chk1 in vivo. The chk1-S345A cells are sensitive to DNA damage and are checkpoint defective. In contrast, mutations of serine-367 and other SQ/TQ sites do not substantially impair the checkpoint or cause damage sensitivity. These findings attest to the importance of serine-345 phosphorylation for Chk1 function and strengthen evidence that transduction of the DNA damage checkpoint signal requires direct phosphorylation of Chk1 by Rad3.
Genome integrity is under constant assault from cellular processes and genotoxic agents that damage DNA. Several strategies exist for coping with this problem—foremost among them are a varied array of DNA repair mechanisms. Acting in concert with repair systems are checkpoint mechanisms that arrest cell division in response to DNA damage (1). These checkpoints provide time to repair damaged DNA before the initiation of mitosis. Determining how checkpoints sense DNA damage and arrest the cell cycle is of central importance in understanding how genome stability is maintained.
Most of the known checkpoint genes were discovered in studies of the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe (2). Fission yeast has been particularly important in analysis of the G2-M DNA damage checkpoint that prevents mitosis (M) when damaged DNA is sensed during the G2 phase. The serine-threonine protein kinase Chk1 is the effector of this checkpoint (3, 4). Chk1 accomplishes this task by indirectly inhibiting the cyclin-dependent kinase Cdc2 (5). Chk1 phosphorylates and thereby inhibits Cdc25, the mitotic inducer phosphatase that activates Cdc2 by dephosphorylating tyrosine-15 (6–9). Chk1 also induces accumulation of Mik1, one of two tyrosine kinases that phosphorylates Cdc2 on tyrosine-15 (10).
Transmission of the checkpoint signal to Chk1 requires six “checkpoint Rad” proteins (Rad1, Rad3, Rad9, Rad 17, Rad26, and Hus1), as well as Cut5/Rad4, Crb2/Rhp9, and Rfc3 (2). Biochemical functions of these proteins are poorly understood, although it is known that subgroups of these proteins form multisubunit complexes (11, 12). Rad3 may be the most interesting protein in the context of signal transduction. Rad3 is a large protein that shares sequence similarity to phosphatidylinositol kinases but is thought to function as a protein kinase in vivo (13). Phosphorylation of Chk1 that occurs in response to DNA damage requires Rad3 and the other checkpoint proteins mentioned above (14). The possibility that Rad3 directly phosphorylates Chk1 is supported by coprecipitation of overproduced Rad3 and Chk1, as well as evidence of in vitro phosphorylation of Chk1 by Rad3 (15).
Rad3 is related to ATM and ATR, two kinases involved in G2-M checkpoints in metazoan species (16–21). ATR appears to have a direct role in regulating Chk1 phosphorylation (22–24). Serine-345 of human Chk1 and the conserved site serine-344 of Xenopus Chk1 are phosphorylated when checkpoints are activated. Serine-345 is part of a SQ/TQ motif that is preferred by ATM and ATR (25). The physiological significance of serine-345 phosphorylation is uncertain, although a mutant form of Xenopus Chk1 that lacks serine-344 and three additional SQ/TQ sites is defective in an oocyte extract checkpoint assay involving unreplicated DNA, and its activation in this in vitro assay is impaired (23). These findings suggest that ATR regulates Chk1 by direct phosphorylation.
In this report we investigate how Rad3 regulates Chk1 in fission yeast. We test the hypothesis that Chk1 function requires phosphorylation that is controlled by Rad3. These studies identified serine-345 as a site that is required for Chk1 phosphorylation in vivo and is essential for activation of the G2-M checkpoint arrest. No other SQ/TQ motifs are required for Chk1 function. These findings provide evidence that direct phosphorylation of Chk1 by Rad3 is required for cell cycle arrest in response to DNA damage in fission yeast.
Materials and Methods
Fission Yeast Strains, Plasmid Construction, and in Vitro Mutagenesis.
The following yeast strains were used in this study: BF2471, chk1+; AL2768, chk1-S345A; AL2769, chk1-S367A; AL2805, chk1-T186A; AL2806, chk1-S314A; AL2807, chk1-T323A; AL2808, chk1-S494A; AL2770, nmt1:GST:chk1+; AL2771, nmt1:GST:chk1-S345A; BF2503, nmt1:GST:chk1:K38A; and NR1592, chk1∷ura4+. All strains are leu1–32 ura4-D18, and all chk1 genes contained a C-terminal 9myc:2HA:6HIS:ura4+ tag. BF2471, AL2768, AL2769, and AL2805-AL2808 all express Chk1 from the endogenous chk1 promoter at single copy. AL2770 and AL2771 were generated by PCR-based gene targeting (26).
Phosphorylation of Glutathione S-Transferase (GST)-Chk1 (330) by ATM and Rad3.
GST-Chk1 (273) and GST-Chk1 (330) proteins were expressed in BL21-DE3 and purified with glutathione-Sepharose (Amersham Pharmacia). GST-Rad3, GST-Rad3KD, GST, and ATM were purified and used in kinase assays as described (27, 28). GST and GST-HsCds1 (1–91) were used as negative and positive controls, respectively (28).
Immunolocalization and Immunoblotting.
Indirect immunofluorescence studies were done with formaldehyde-fixed cells processed as described (29). Anti-Myc (9E10; Santa Cruz Biotechnology) and Cy3-conjugated anti-mouse IgG (The Jackson Laboratory) were used as primary and secondary antibodies, respectively. The DNA stain 4′,6-diamidino-2-phenylindole was used to visualized the nuclei. Cells were photographed with a Nikon Eclipse E800 microscope equipped with a Photometrix Quantix charge-coupled device camera. Images were acquired and analyzed with iplab spectrum software (Signal Analytics, Vienna, VA). Immunoblot analysis was performed with 15 μg of total lysate. Samples were electrophoresed on 8% SDS/PAGE gel and wet-transferred to Immobilon (Millipore). Blots were probed with mouse anti-Myc antibody (9E10; Santa Cruz Biotechnology). Primary antibody was detected with the use of an horseradish peroxidase-conjugated anti-mouse IgG antibody (Promega) and Luminol reagents (Pierce).
Checkpoint Arrest and UV Survival.
Checkpoint studies were done with synchronous cultures made by centrifugal elutriation at 30°C with a Beckman Coulter JE-5.0 elutriation rotor. DNA damage was inflicted from a 137C source at 3 Gy min−1 for 33 min to a total of 100 Gy or by addition of 5 milliunits ml−1 bleomycin sulfate (Calbiochem). Cells were scored for progression through mitosis by microscopic observation (5). UV irradiation was performed with a Stratalinker UV 1800 (Stratagene).
Results
Serine-345 Is Required for the Chk1 Electrophoretic Mobility Shift Associated with Rad3-Dependent Phosphorylation.
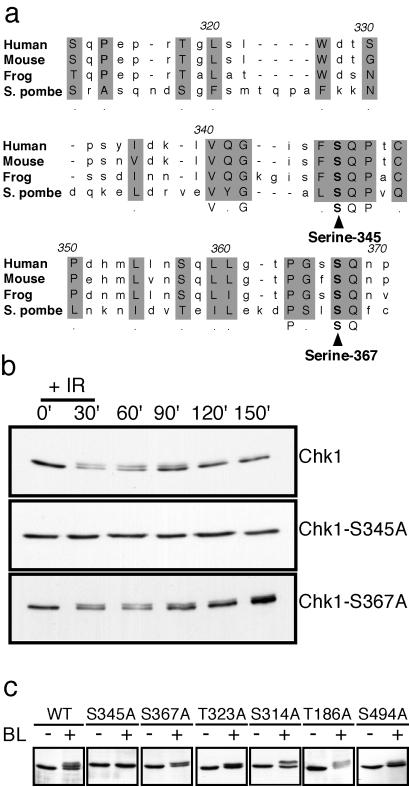
ATM, ATR, and Rad3 phosphorylate serine or threonine residues in the motif SQ or TQ (25, 30). Fission yeast Chk1 contains six serine or threonine residues followed by glutamine: T186 in the N-terminal kinase domain; S314, T323, S345, and S367 clustered in the C-terminal regulatory domain; and S494 near the extreme C terminus. Serine-345 and serine-367 appear to align with SQ motifs in human, mouse, and frog (Xenopus laevis) Chk1 homologs (Fig. 1a). In response to DNA damage, Chk1 is phosphorylated by a Rad3-dependent mechanism (14). Chk1 phosphorylation is readily monitored by a reduction of the electrophoretic mobility of Chk1. We carried out experiments to determine whether any of the SQ/TQ motifs were required for this effect. The relevant serine or threonine codons were changed to alanine, and the mutant forms of chk1 were integrated and expressed from the endogenous chk1 promoter in a chk1Δ background (see Materials and Methods). In the initial experiments, chk1+, chk1-S345A, and chk1-S367A cells were exposed to ionizing radiation. Immunoblot analysis showed that ionizing radiation caused wild-type Chk1 to migrate with reduced mobility (Fig. 1b). Ionizing radiation caused an electrophoretic mobility of Chk1S367A but not Chk1S345A (Fig. 1b). These findings indicated that serine-345 is required for the Chk1 electrophoretic mobility shift induced by ionizing radiation. These studies were expanded to examine all of the SQ/TQ motif mutants in experiments that used bleomycin to inflict DNA damage. Bleomycin is a radiomimetic drug that causes double-strand breaks in DNA. Immunoblot analysis showed that bleomycin treatment caused wild-type Chk1 to migrate with reduced mobility (Fig. 1c). Bleomycin caused an electrophoretic mobility of all mutant forms of Chk1, with the exception of Chk1S345A (Fig. 1c). These findings showed that serine-345 is required for the Chk1 electrophoretic mobility shift that is caused by Rad3-dependent phosphorylation and thus suggested that serine-345 is phosphorylated by Rad3.
Figure 1.
Serine-345 is required for the electrophoretic mobility shift of Chk1 that is caused by phosphorylation. (a) Alignment of the C-terminal SQ/TQ cluster domain of human (amino acids 310–369), mouse (amino acids 310–369), frog (amino acids 305–366), and S. pombe (amino acids 304–370) Chk1. Alignment was performed with the clustal w program. Conserved SQ motifs at serine-345 and serine-367 are indicated. (b) Cultures of chk1+, chk1-S345A, and chk1-S367A cells that expressed 9-myc-tagged Chk1 were mock-treated or treated with ionizing radiation (100 Gy). After irradiation, cells were collected every 30 min, and lysates were prepared for immunoblotting with anti-myc antibodies. DNA damage induced the appearance of a slower mobility form of Chk1 in chk1+ and chk1-S367A cells. (c) Wild-type, chk1-S345A, chk1-S367A, chk1-T323A, chk1-S314A, chk1-T186A, and chk1-S494A cultures that expressed 9-myc-tagged Chk1 were mock-treated or treated with bleomycin (5 milliunits/ml) for 45 min. Immunoblotting analysis showed that all forms of Chk1 except Chk1S345A underwent a mobility shift in response to bleomycin treatment.
Rad3 and ATM Phosphorylate Serine-345 of Fission Yeast Chk1 in Vitro.
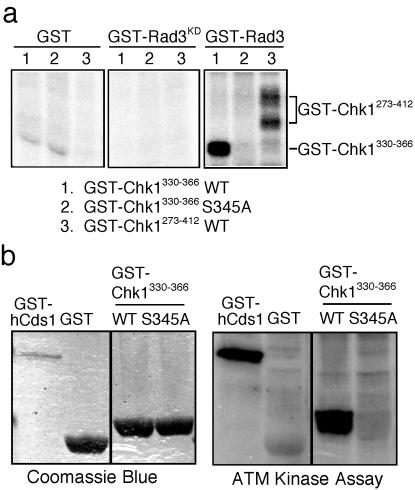
We performed in vitro kinase assays to determine whether Rad3 phosphorylates serine-345 of Chk1. A GST fusion protein that contains amino acids 273–412 of fission yeast Chk1 was expressed and purified from bacteria. GST-Rad3 isolated from fission yeast phosphorylated GST-Chk1 (273) (Fig. 2a). Unfused GST purified from fission yeast failed to phosphorylate GST-Chk1 (273). Likewise, a kinase-inactive form of Rad3 (GST-Rad3KD) was unable to phosphorylate GST-Chk1 (273) (Fig. 2a). Expression of GST-Chk1 (273) was very weak in bacteria; therefore, we also tested GST-Chk1 (330) as a substrate of GST-Rad3. GST-Rad3 readily phosphorylated GST-Chk1 (330), whereas no phosphorylation of the S345A mutant form of GST-Chk1 (330) was detected (Fig. 2a). These results indicated that GST-Rad3 is able to phosphorylate serine-345 of Chk1 in vitro. In view of the relationship between fission yeast Rad3 and human ATM, we explored the possibility that ATM shared the ability to phosphorylate GST-Chk1 (330). ATM isolated by immunoprecipitation from HeLa cells phosphorylated GST-Chk1 (330) but not unfused GST (Fig. 2b). ATM failed to phosphorylate the S345A mutant form of GST-Chk1 (330) (Fig. 2b). These results showed that Rad3 and ATM share an ability to phosphorylate serine-345.
Figure 2.
Rad3 and ATM phosphorylate a peptide derived from Chk1 at serine-345. (a) Rad3 phosphorylates serine-345 of Chk1. Unfused GST, a kinase dead form of GST-Rad3 (GST-Rad3KD) and active GST-Rad3 were expressed and purified from fission yeast and tested with the substrates GST-Chk1330–366 (wild type and S345A mutant) and GST-Chk1273–412 expressed and purified from bacteria. GST-Rad3 phosphorylated GST-Chk1273–412 and GST-Chk1330–366, but failed to phosphorylate GST-Chk1330–366 that contained the S345A mutation. (b) ATM phosphorylates serine-345 of Chk1. ATM was purified from HeLa cells and incubated with [Δ-32P]ATP and wild type (WT) or S345A mutant GST-Chk1330–366. GST and GST-HsCds11–91 were use as negative and positive controls, respectively.
DNA Damage Sensitivity Caused by chk1-S345A Mutation.
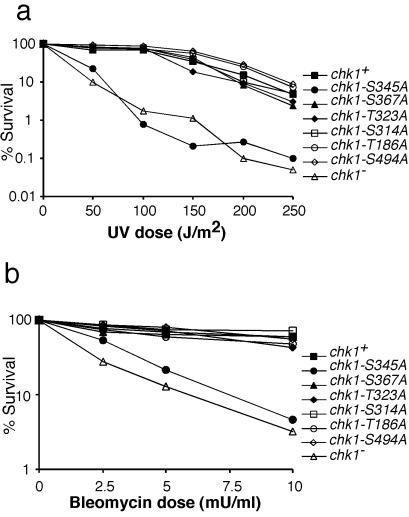
To evaluate the physiological importance of serine-345 in the function of Chk1, we measured the ability of wild-type and chk1 mutants to survive exposure to UV light. As seen (3), the chk1Δ mutation substantially decreased survival after exposure to UV (Fig. 3a). The chk1Δ cells were ≈100-fold more sensitive than wild type to UV radiation. The chk1-S345A mutation caused UV sensitivity to a degree that was equivalent to that of chk1Δ (Fig. 3a). In contrast, mutations of other SQ/TQ motifs had no effect on UV survival (Fig. 3a). Damage survival studies also were performed with bleomycin. As expected, chk1Δ cells were quite sensitive to a range of bleomycin doses (Fig. 3b). With the exception of the chk1-S345A strain, all mutants that harbored mutations at Chk1 SQ/TQ motifs were indistinguishable from wild type (Fig. 3b). On the other hand, chk1-S345A cells displayed bleomycin sensitivity at a level approximately equivalent to that of chk1Δ cells (Fig. 3b). Thus, in both UV and bleomycin survival assays, the chk1-S345A and chk1Δ alleles caused nearly identical phenotypes. These findings provided an indication that phosphorylation of Chk1 at serine-345 is a physiologically important part of the DNA damage response in fission yeast. As a corollary to these findings, we can conclude that any phosphorylation of other SQ/TQ motifs in Chk1, if it occurs, has no obvious role in survival after DNA damage.
Figure 3.
UV and bleomycin survival are impaired in chk1-S345A cells. (a) chk1-S345A cells are UV sensitive. Wild type (chk1+), chk1Δ, and mutants of all of the SQ/TQ motifs were tested in a UV survival assay. With the exception of chk1-S345A cells, all SQ/TQ motif mutants displayed UV resistance equivalent to that of wild type. The chk1-S345A and chk1Δ cells were equally sensitive to UV. (b) A similar damage survival assay was performed with bleomycin. Cells were exposed to the indicated doses of bleomycin for 1 h. The chk1-S345A and chk1Δ cells were sensitive to bleomycin, whereas all other strains were indistinguishable from wild type.
DNA Damage Checkpoint Defect of the chk1-S345A Mutant.
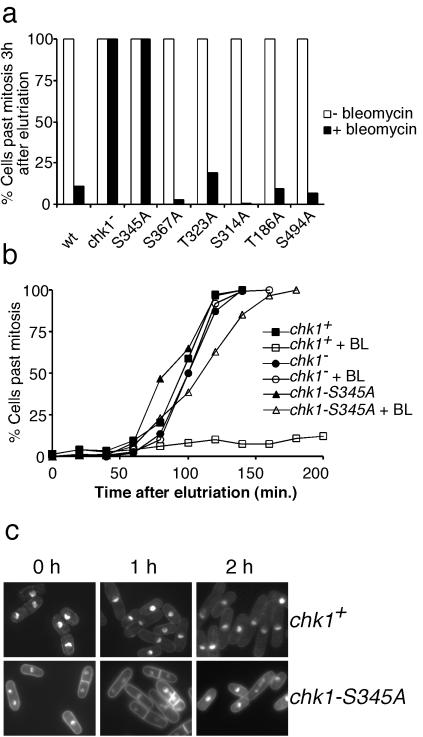
Chk1 is required for the G2-M DNA damage checkpoint in fission yeast (3). Therefore, we compared the DNA damage checkpoint response of wild type, chk1Δ, and the strains that had mutations in the SQ/TQ motifs. These strains were synchronized in early G2 phase by centrifugal elutriation and then treated continuously with bleomycin or mock-treated. Continuous treatment with bleomycin causes prolonged checkpoint arrest in wild-type cells (7, 10, 29). Progression through mitosis was monitored for 3 h. At the completion of the experiment, bleomycin treatment had effectively blocked division in chk1+ cells and all of the SQ/TQ motif mutants, with the exception of chk1-S345A (Fig. 4a). All of the chk1-S345A cells divided during the experiment, as did the chk1Δ cells (Fig. 4a). A more detailed kinetic analysis of the experiment showed that there was a very slight delay of mitosis (≈10 min) in chk1-S345A cells that was not observed in chk1Δ cells (Fig. 4b). These findings demonstrated that chk1-S345A cells were profoundly defective for the G2-M checkpoint, although perhaps not quite as defective as chk1Δ cells. As expected, microscopic observation of chk1-S345A cells exposed to bleomycin revealed septated cells that frequently had unequally divided nuclei, indicative of checkpoint failure (Fig. 4c). Collectively, these data demonstrated that serine-345 of Chk1 is required for a DNA damage checkpoint arrest.
Figure 4.
DNA damage checkpoint delay is abrogated in chk1-S345A cells. (a) Wild type (chk1+), chk1Δ, and mutants of all of the SQ/TQ motifs were synchronized in G2 phase by centrifugal elutriation and exposed to bleomycin (5 milliunits/ml). Passage through mitosis was scored by microscopic observation. Data for the 3-h time are presented. Checkpoint arrest failed in chk1-S345A and chk1Δ cells. Checkpoint arrest was proficient in wild type and all SQ/TQ mutants except chk1-S345A. All mock-treated cultures underwent mitosis with similar kinetics. (b) Detailed kinetic analysis of chk1+, chk1Δ, and chk1-S345A cells in experiment described above. (c) Photographs of chk1+ and chk1-S345A cells after the addition of bleomycin to asynchronous cultures.
Nuclear Localization of Chk1S345A.
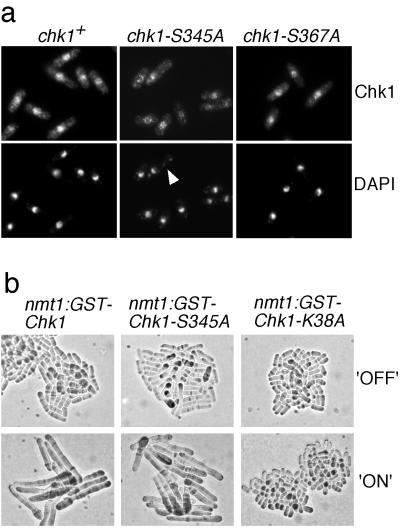
The checkpoint Rad proteins localize in the nucleus of fission yeast (12), where they presumably sense damaged DNA and transmit the checkpoint signal to Chk1. Chk1 was detected as a nuclear protein when it was expressed at low levels as a GST fusion protein (29). In view of the effect of the chk1-S345A mutation, we wondered whether phosphorylation of S345A might control the localization of Chk1. We were also concerned by the fact that some mutations in the C-terminal domain of Chk1 abolish Chk1 function but also unexpectedly diminish Chk1 nuclear localization (A.L.-G. and P.R., unpublished data). Wild-type amounts of the myc-tagged Chk1, Chk1S345A, and Chk1S367A were detected by indirect immunofluorescence in cells treated with bleomycin. The wild-type and mutant Chk1 proteins yielded identical patterns of strong nuclear fluorescence and weaker cytoplasmic signals (Fig. 5a). As anticipated, DNA staining with 4′,6-diamidino-2-phenylindole revealed that chk1-S345A underwent an abnormal mitosis (“cut” phenotype) in the presence of bleomycin. More than 50% of the mitosis observed in chk1-S345A showed a “cut” phenotype (Fig. 5a). This phenotype is typical of cells that undergo mitosis in the presence of damaged DNA. These findings confirmed the checkpoint defect of chk1-S345A cells and showed that this phenotype could not be attributed to defective localization of Chk1S345A.
Figure 5.
Chk1S345A is localized in the nucleus and retains the ability to arrest mitosis when overproduced. (a) Serine-345 mutation does not affect Chk1 nuclear localization. Immunolocalization of 9-myc-tagged Chk1, Chk1S345A, and Chk1S367A in cells treated with 5 milliunits/ml bleomycin for 45 min. Nuclei were visualized with 4′,6-diamidino-2-phenylindole. The arrowhead indicates a “cut” cell in which DNA is unequally partitioned at mitosis. (b) Chk1S345A arrests division when overexpressed. Wild-type cells that had nmt1-GST-chk1+, nmt1-GST-chk1S355A, or nmt1-GST-chk1-K38A genomic constructs were grown in nmt1-repressing (+B1) or -inducing (−B1) medium. Cells expressing Chk1 and Chk1S345A became highly elongated, which is indicative of cell cycle arrest, whereas cells expressing Chk1K38A, a kinase-inactive form of Chk1, did not arrest division.
Chk1S345A Overexpression Arrests Division.
Overexpression of Chk1 protein causes elongation in wild-type cells and prevents entry into mitosis (5, 31). This effect is independent of Rad3 and the other checkpoint Rad proteins. If the S345A mutation specifically eliminated regulation of Chk1 by Rad3, then Chk1S345A should retain the ability to arrest division when overexpressed. On the other hand, if the S345A mutant disrupted the intrinsic kinase activity of Chk1, or Chk1 activity required basal phosphorylation of serine-345 that was carried out by a kinase other than Rad3, then Chk1S345A should be unable to arrest division when overexpressed. These possibilities were tested by the construction of strains that expressed GST-Chk1 proteins under the control of the thiamine-repressible nmt1 promoter. As shown (5), GST-Chk1 overproduction caused a cell cycle arrest (Fig. 5b). We observed that GST-Chk1S345A caused an identical phenotype with kinetics that were similar to that observed with GST-Chk1 overproduction (Fig. 5b). To ascertain whether these effects require Chk1 kinase activity, a strain was constructed that expressed a kinase-inactive form of Chk1 (chk1-K38A). Overproduction of GST-Chk1K38A had no effect on cell division (Fig. 5b). These findings strongly suggest that Chk1S345A retains a basal activity that cannot be regulated in response to DNA damage.
Discussion
A central aim of checkpoint studies in fission yeast is to understand how the DNA damage signal is transmitted from the point of damage, through Rad3 and other checkpoint proteins, to the checkpoint effector kinase Chk1. Evidence has been building that Chk1 phosphorylation is regulated by Rad3 in fission yeast and ATR in metazoan organisms (14, 15, 22, 23), but the physiological importance of this phosphorylation was uncertain. In this article, we have shown that serine-345 of fission yeast Chk1, which is phosphorylated in vitro by Rad3, is required for Chk1 phosphorylation, DNA damage survival, and checkpoint enforcement in vivo. These data provide persuasive evidence that phosphorylation of Chk1 at serine-345 is required to engage the G2-M DNA damage checkpoint.
These findings confirm and extend several recent investigations that have linked Chk1 regulation to ATR in metazoan species. Studies with human tissue culture cells showed that serine-345 of Chk1 is phosphorylated in response to DNA damage (22). Serine-345 phosphorylation was modulated by high expression of ATR. This work established a link between ATR and serine-345 phosphorylation. A separate study showed that ATR controls Chk1 phosphorylation in Xenopus egg extracts (23). Xenopus ATR phosphorylated Chk1 at multiple SQ/TQ sites in vitro. One of these sites, serine-344, appears to align with serine-345 in human and fission yeast Chk1 (Fig. 1a). Mutant Chk1 that lacked serine-345 and three additional SQ/TQ sites was unable to restore a mitotic checkpoint when added to Chk1-depleted Xenopus egg extracts that were supplemented with sperm nuclei and a replication inhibitor (23). More recently, another study with human tissue culture cells showed that ATR controls phosphorylation of serine-317 and serine-345 of human Chk1 (24). A mutant form of Chk1 that had alanine substitutions at these two positions exhibited impaired activation. These studies showed that one or possibly a combination of SQ/TQ sites is important for Chk1 function in these assay systems.
This work extends the understanding of DNA damage checkpoints in several ways. Most importantly, by finding that chk1-S345A cells are highly sensitive to killing by UV radiation, at a level equivalent to that of chk1Δ cells, we have established the physiological importance of a conserved phosphorylation site in Chk1. DNA damage checkpoints serve to enhance genome stability and survival of DNA damage; thus it is important to demonstrate that elimination of conserved phosphorylation site in Chk1 diminishes the ability of cells to survive exposure to a DNA-damaging agent. Furthermore, we have refined the analysis of individual SQ/TQ sites in Chk1 by showing that serine-345 of Chk1 is essential for the DNA damage checkpoint arrest and important for UV survival in vivo. Previous work left unresolved whether any of the individual SQ/TQ sites in the C-terminal domain of Chk1 were important for checkpoint arrest (22, 23). We further demonstrated the physiological importance of serine-345 by showing that the UV survival profile of the chk1-S345A mutant was equivalent to a chk1Δ mutant. These studies establish the functional relevance of a conserved phosphorylation site in Chk1 in the context of a living organism.
In contrast to the chk1-S345A mutation, none of the other mutations of SQ/TQ motifs enhanced sensitivity to DNA damage. Furthermore, all of the mutant strains (except chk1-S345A) arrested division in response to DNA damage. These findings argue that serine-345 is the crucially important site of phosphorylation on Chk1, and that if phosphorylation occurs at other SQ/TQ motifs, these phosphorylations have little measurable effect on Chk1 function. In carefully performed studies with synchronous cultures, we noticed that chk1-S345A cells exhibit a very abbreviated checkpoint delay in response to bleomycin treatment (Fig. 4b). Therefore, it is formally possible that Chk1 is very weakly regulated by phosphorylation at another position. In fact, we have noted that chk1-T323A cells slowly resume division after arresting for 3 h in bleomcyin, whereas wild-type cells and other SQ/TQ mutants remain arrested for at least 4 h (unpublished data). These findings suggest that minor regulation of Chk1 might be mediated through phosphorylation of threonine-323, although ablation of threonine-323 has no measurable effect on survival of ionizing radiation, UV, or bleomycin when these agents are applied as a pulse of DNA damage.
These studies also provide some hints as to how Chk1 might be regulated by phosphorylation of serine-345. The simplest possibility was that serine-345 phosphorylation was required for the kinase activity of Chk1. We found that wild-type GST-Chk1 and mutant GST-Chk1S345A were equally potent in their ability to arrest division when overexpressed in fission yeast. In contrast, a kinase-inactive form of Chk1 was unable to arrest division when overexpressed. These findings strongly suggest that Chk1S345A is active as a protein kinase and argue against the possibility that the chk1-S345A mutation severely disrupts Chk1 structure. Thus, it appears that Chk1 is active without serine-345 phosphorylation, but serine-345 phosphorylation must in some way further enhance Chk1 activity in vivo. We have been unable to measure an increase in the kinase activity of immunoprecipitated Chk1 in response to irradiation (Beth Baber-Furnari and P.R., unpublished data). However, recent studies measured an increase in kinase activity of hyperphosphorylated Chk1 isolated from checkpoint-activated Xenopus egg extracts treated with a phosphatase inhibitor and from checkpoint-arrested human tissue culture cells (24, 32). Thus far, there have been no reports of direct in vitro activation of Chk1 with an ATM-related kinase. It is possible that other proteins are required for effective phosphorylation of Chk1 by Rad3. One such protein might be Crb2/Rhp9, a protein that is required for Chk1 phosphorylation and associates with Chk1 in a yeast two-hybrid assay (33, 34). In this regard it is notable that Chk1 phosphorylation in Xenopus oocyte extracts correlates with a physical association with Claspin, a novel checkpoint protein (32). It is possible that Crb2 and Claspin perform similar functions in their respective organisms. Interestingly, certain deletions or point mutations in the C terminus of Chk1 activate Chk1 when assayed with in vitro kinase assays or the Xenopus extract mitotic induction system (35–37). These findings suggest that the C terminus of Chk1 might inhibit Chk1 activity.
One limitation of our studies is that they do not provide direct evidence that serine-345 is phosphorylated in vivo. Metabolic labeling experiments of Chk1 with radioactive phosphate have been unsuccessful, and phosphospecific antisera for serine-345 are not yet available. Serine-345 is required for mobility shift of Chk1 that is caused by Rad3-dependent phosphorylation, but it is formally possible that serine-345 is not an actual site of phosphorylation. However, the simplest interpretation of the data is that serine-345 is phosphorylated by Rad3 in vivo. This explanation is consistent with studies of mammalian cells demonstrating that serine-345 is phosphorylated in vivo (22).
Our findings provide evidence that Rad3 controls Chk1 by direct phosphorylation. This phosphorylation is required for Chk1 function and, as a consequence, the G2-M DNA damage checkpoint. Chk1 phosphorylation also requires Rad1, Rad9, Rad17, Rad26, Hus1 (14), Rfc3 (and presumably other small subunits of RFC) (38), and Crb2/Rhp9 (33). It appears that Rad3 activation is independent of all of these proteins, with the exception of Rad26 (11). The requirement for Rad26 is readily understandable, as Rad3 associates with Rad26 in vivo, and it is likely that the two proteins function as a complex. It is yet to be understood why so many proteins are required for Rad3 to phosphorylate Chk1 in vivo.
Acknowledgments
We thank the Scripps Cell Cycle Groups for support and encouragement. K.T. was supported by the Naito Foundation. This work was funded by National Institutes of Health grants awarded to C.H.M. and P.R.
Abbreviations
- GST
glutathione S-transferase
- M
mitosis
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Rhind N, Russell P. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walworth N, Davey S, Beach D. Nature (London) 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 4.Rhind N, Russell P. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhind N, Furnari B, Russell P. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 6.Furnari B, Rhind N, Russell P. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 7.Furnari B, Blasina A, Boddy M N, McGowan C H, Russell P. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Girona A, Kanoh J, Russell P. Curr Biol. 2001;11:50–54. doi: 10.1016/s0960-9822(00)00026-9. [DOI] [PubMed] [Google Scholar]
- 10.Baber-Furnari B A, Rhind N, Boddy M N, Shanahan P, Lopez-Girona A, Russell P. Mol Biol Cell. 2000;11:1–11. doi: 10.1091/mbc.11.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards R J, Bentley N J, Carr A M. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 12.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay H D, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr A M. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 14.Walworth N C, Bernards R. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 15.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N J. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 17.Keegan K, Holtzman D, Plug A, Christenson E, Brainerd E, Flaggs G, Bentley N, Taylor E, Meyn M, Moss S, et al. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 18.Cimprich K A, Shin T B, Keith C T, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 20.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright J A, Keegan K S, Herendeen D R, Bentley N J, Carr A M, Hoekstra M F, Concannon P. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Guntuku S, Cui X S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Z, Kumagai A, Wang S X, Dunphy W G. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Piwnica-Worms H. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S T, Lim D S, Canman C E, Kastan M B. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 26.Bahler J, Wu J, Longtine M S, Shah N G, McKenzie A, Steever A B, Wach A, Phileppsen P, Pringle J R. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 27.Blasina A, Price B D, Turenne G A, McGowan C H. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 28.Melchionna R, Chen X B, Blasina A, McGowan C H. Nat Cell Biol. 2000;2:762–765. doi: 10.1038/35036406. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nature (London) 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka K, Boddy M N, Chen X B, McGowan C, Russell P. Mol Cell Biol. 2001;21:3398–3404. doi: 10.1128/MCB.21.10.3398-3404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ford J C, al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Carr A M. Science. 1994;265:533–535. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- 32.Kumagai A, Dunphy W G. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 33.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willson J, Wilson S, Warr N, Watts F Z. Nucleic Acids Res. 1997;25:2138–2145. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, Tempczyk-Russell A, Nguyen B, Myers P, Lundgren K, et al. Cell. 2000;100:681–692. doi: 10.1016/s0092-8674(00)80704-7. [DOI] [PubMed] [Google Scholar]
- 36.Oe T, Nakajo N, Katsuragi Y, Okazaki K, Sagata N. Dev Biol. 2001;229:250–261. doi: 10.1006/dbio.2000.9968. [DOI] [PubMed] [Google Scholar]
- 37.Wang S X, Dunphy W G. FEBS Lett. 2000;487:277–281. doi: 10.1016/s0014-5793(00)02370-x. [DOI] [PubMed] [Google Scholar]
- 38.Shimida M, Okuzaki D, Tanaka S, Tougan T, Tamai K, Shimoda C, Nojima H. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]