Abstract
Background
Weight loss success is determined by genetic factors, which may differ according to treatment strategy.
Methods
From a multidisciplinary obesity treatment program involving dietary advice, psychological counseling, and increased physical activity, 587 subjects (68% female; 46.1 ± 12.4 years; BMI 39.9 ± 6.3) were recruited. At baseline, a blood sample was drawn for DNA isolation. Genotypes were determined for 30 polymorphisms in 25 candidate genes. The association between genotypes and weight loss was assessed after 3 months (short-term) and after 12 months of treatment (long-term). Weight loss was categorized as ≥5% or <5% of initial weight.
Results
The G/G genotype of PLIN1 (rs2289487) and PLIN1 (rs2304795), the T/T genotype of PLIN1 (rs1052700), and the C/C genotype of MMP2 predicted ≥5% weight loss in the first 3 months. The C/G-G/G genotype of PPARγ (rs1801282) and the T/C genotype of TIMP4 (rs3755724) predicted ≥5% weight loss after 12 months. Subjects with the combination of PPARγ (rs1801282) C/G-G/G and TIMP4 (rs3755724) T/C lost even more weight.
Conclusion
Polymorphisms in genes related to regulation of fat storage and structural adaptation of the adipocytes are predictors for weight loss success with different genes being relevant for short-term and long-term weight loss success.
Keywords: Obesity, Lifestyle intervention, Polymorphisms, Predictors, Weight loss maintenance, Fat storage, Adipocyte adaptation, Extracellular matrix
Introduction
Obesity is a multifactorial trait influenced by various environmental factors such as lifestyle and diet, but also by genetic background. Treating obese people with lifestyle advice is a challenge, because responses to treatment vary strongly. Weight loss maintenance and continuation of a modified lifestyle are the greatest challenges, where some turn out to be more successful than others. Stubbs et al. [1] have reviewed the different types of pre-treatment predictors and correlates of weight loss and maintenance. Many potential predictors and correlates were identified, which were from genetic, physiological, psychosocial and behavioral origin [1]. In 2015, a genome-wide association study in >300,000 individuals showed that 97 DNA loci, enriched for a role in hypothalamic control of energy balance and with overrepresentation of pathways involved in both food intake and physical activity, were associated with BMI, explaining ∼2.7% of the variation [2]. In addition, 49 loci associated with body fat distribution (waist-to-hip ratio adjusted for BMI) were identified [3]. The first evidence for a role of genetic predisposition in weight loss and weight regain came from a study by Delahanty et al. [4], which showed that several gene variants were associated with short- and long-term weight loss and weight regain. Based on a meta-analysis of available studies, Xiang et al. [5] concluded that individuals carrying the homozygous fat mass and obesity-associated (FTO) obesity-predisposing allele lose more weight by means of diet or lifestyle interventions than noncarriers. Knowledge of such predictors might help to explain interindividual variations in weight loss success and lead to more personalized treatments for weight loss and weight loss maintenance.
The focus of our study was on the role of variants in genes involved in the regulation of adipocyte structure and function in short- or longer-term weight loss success. Obesity is associated with hypertrophy of adipocytes demanding adaptation of the extracellular matrix (ECM), the outer protective layer of the cells [6]. Conversely, weight loss requires ECM adaptations, and inability of the adipocyte to respond adequately may lead to adipocyte cell stress, providing a risk factor for increased fat storage and weight regain [7, 8]. Thus, variation in genes coding for components and modulators of the ECM and for fat storage capacity may play an important role in the weight loss response to a weight loss intervention.
We therefore selected candidate genes and gene loci involved in the storage of fat in adipocytes (PLIN1 [perilipin-1], PPARγ [peroxisome proliferator-activated receptor-γ] [4, 9, 10, 11]), in lipolysis (ADRB2 [adrenergic β2-receptor] [12]), and in the formation of the ECM (COL4A1, COL4A2, COL6A1, COL6A2, COL6A3, MMP2 [matrix metalloproteinase 2], TIMP4 [tissue inhibitor of metalloproteinase 4] [6]). For comparison, we added genes that had previously been shown to play a role in other body weight regulation processes: hypothalamic activity/eating behavior (BDNF [brain-derived neurotrophic factor], CNTF [ciliary neurotrophic factor], FTO, GNDPA2, LEP [leptin], MC4R [melanocortin 4 receptor], near TMEM18, SEC16B, SH2B1 [13, 14]), whole-body energy homeostasis (ADIPOQ [adiponectin] [15]), weight regain (ACE [angiotensin I converting enzyme] [16]), and obesity risk in the knockout mouse (GPRC5B [17]). In total, 25 genes with 30 single nucleotide polymorphisms (SNPs) were selected (full list in suppl. Table 1; see www.karger.com/doi/10.1159/000469662 for all supplementary material), most of which have previously been associated with body mass index based on genome-wide association studies [18, 19, 20, 21]. Variation in these genes was used for analysis of the genetic association with 3- and 12-month weight loss success in severely obese subjects enrolled in a commercial lifestyle modification program. Our results indicate that some of the gene variants are predictors of weight loss in this lifestyle-treated cohort.
Methods
Subjects
For this study, 587 consecutive subjects from a local commercial obesity treatment center (CO-EUR, Heerlen, The Netherlands, co-eur.com) were recruited. Subjects were at least 18 years old and had a BMI >30. The study protocol and informed consent document were approved by the Medical Ethical Committee of Maastricht University Medical Centre+. All subjects gave written informed consent before being enrolled into the study.
Study Protocol
All participants of this study followed the lifestyle program by CO-EUR, a commercial enterprise that offers an intensive lifestyle program to obese individuals with the aim to improve their lifestyle and attain long-term weight loss. The CO-EUR obesity treatment program consisted of an 18-month multidisciplinary program targeting lifestyle modification. It included a physical activity program, psychological counseling based on cognitive behavioral therapy (CBT), and nutritional advice to promote a healthy lifestyle. A detailed description of the treatment program and its effect has been published previously [22]. The goal of the program was to achieve lifestyle modification resulting in long-term weight loss.
At baseline, before the start of the treatment program, a blood sample was drawn for isolation of DNA. In addition, body weight and height were measured. After entering the treatment program, body weight was measured every 3 months.
Measurements
Body weight was measured on a digital scale (Omron HBF-500E, Omron Healthcare Europe) to the nearest 0.1 kg. Height was measured with a stadiometer.
DNA Isolation and Genotyping
Blood was drawn from a forearm vein into an EDTA-containing tube (BD Vacutainer, 10 mL). The buffy coat was obtained by centrifugation (5°C, 3,000 rpm, 10 min) and stored at −80°C until analysis. Genomic DNA was isolated from peripheral blood leukocytes in the buffy coat using a QIAamp kit (QIAgen, Amsterdam, The Netherlands). Genotypes of 25 genes were determined using TaqMan allelic discrimination (Applied Biosystems, Foster City, CA, USA) and competitive allele-specific PCR (KASP; LGC, Teddington, UK) by Kbioscience (Hoddeston, UK). The genotyping success rate was >95%. The full list of the 25 genes with the 30 variation identifiers as well as the allele frequencies and Hardy-Weinberg values (χ2 test) can be found in supplementary Table 1. For PLIN1 rs2289487, the results from the genotyping assay were obtained as the forward “A” and “G” alleles, but in the text we have used the reverse annotation “T” and “C” in line with previous scientific publications.
Data Handling and Statistical Analysis
Allelic and genotype frequencies were calculated. All polymorphisms were in Hardy-Weinberg equilibrium (online suppl. Table 1). For each variant, the genetic inheritance model that best described the weight loss effect was determined by logistic regression analysis using SNPStats [23]. SNPStats is publicly available at http://bioinfo.iconcologia.net/snpstats. The association between genotypes, grouped according to the best inheritance model, and weight loss was assessed after 3 months (n = 558) and after 12 months (n = 275) of treatment. For these analyses, weight loss was categorized as ≥5% of initial weight (high weight loss) or <5% of initial weight (low weight loss). This cutoff level was chosen because a ≥5% weight loss is associated with a meaningful improvement of health in obese individuals [24, 25]. Binary logistic regression analysis was used to determine the odds for ≥5% weight loss of the genotypes. For SNPs of the 4 genes showing association with ≥5% weight loss, weight loss trajectories over 12 months were calculated and compared by mixed model analysis of variance. We also analyzed the weight loss trajectories of pairwise combinations of these genes, where we compared the combination of genotypes of the 2 genes showing the most beneficial effect on weight loss with all other combinations of the genotypes of the 2 genes.
For finding relevant associated SNPs, an odds ratio (OR) with a 95% confidence interval (CI) deviating from unity and a p value <0.05 was considered as significant. Our final results were checked against a corrected p value of 0.0017 (0.05/30). The reported p values are the uncorrected p values. Statistical analyses were performed using the SPSS v20.0 statistical software (IBM, Armonk, NY, USA).
Results
Predictors of Weight Loss at 3 Months
Out of 587 subjects, 558 had a body weight measurement after 3 months of treatment, 336 (60%) had a weight loss <5%, and 222 (40%) a weight loss ≥5%. The high weight loss group was significantly older and had a lower BMI at baseline than the low weight loss group (Table 1). ORs for ≥5% weight loss with a 95% CI deviating from unity were found for 4 SNPs in 2 of the 25 genes (PLIN1 [3 SNPs] and MMP2). Subjects with the most beneficial genotypes of the PLIN1 SNPs (rs2289487, rs2304795, and rs1052700) were 1.7–1.9 times more likely to be in the high weight loss group than subjects with the less favorable PLIN1 genotypes (Table 2). Furthermore, subjects with the C/C genotype of MMP2 rs1132896 were 1.8 times more likely to be in the high weight loss group than subjects with the G/G-G/C genotype (Table 2). Adjustment for age and BMI as covariates did not change these results (Table 2). Results for all SNPs can be found in online supplementary Table 1.
Table 1.
Baseline characteristics of subjects with ≥5% or <5% weight loss at 3 or 12 months
| <5% weight loss | ≥5% weight loss | p value1 | |
|---|---|---|---|
| 3 months | |||
| n (% male) | 336 (32.4) | 222 (30.2) | 0.575 |
| Age, years | 45.0±12.7 | 48.0±11.7 | 0.004 |
| BMI | 38.9±6.4 | 36.9±5.7 | <0.001 |
| 12 months | |||
| Sex (% male) | 97 (28.9) | 178 (38.2) | 0.122 |
| Age, years | 44.5±11.6 | 48.4±11.7 | 0.008 |
| BMI | 39.9±6.5 | 34.4±5.6 | <0.001 |
Data presented as mean ± SD
p value for the difference between the <5% and ≥5% weight loss groups, independent samples t test.
Table 2.
Results of binary logistic regression analysis with 3- and 12-month weight loss as dependent variable (<5% weight loss is reference) and the genotypes as independent variables
| Gene | rs number | Genotype/model | OR (95% CI) | p value1 | OR2 (95% CI) | p value1, 2 |
|---|---|---|---|---|---|---|
| 3 months | ||||||
| PLIN1 | rs2289487 | C/C vs. T/T-T/C3 | 1.92 (1.17–3.13) | 0.009 | 1.94 (1.18–3.18) | 0.009 |
| PLIN1 | rs2304795 | G/G vs. A/A-G/A3 | 1.75 (1.10–2.79) | 0.018 | 1.75 (1.09–2.80) | 0.020 |
| PLIN1 | rs1052700 | T/T vs. A/A-T/A3 | 1.72 (1.04–2.82) | 0.033 | 1.74 (1.05–2.88) | 0.031 |
| MMP2 | rs1132896 | C/C vs. G/G-G/C3 | 1.83 (1.13–2.96) | 0.014 | 1.84 (1.13–2.99) | 0.014 |
| 12 months | ||||||
| PPARγ | rs1801282 | C/C vs. C/G-G/G4 | 2.03 (1.07–3.87) | 0.025 | 2.25 (1.17–4.34) | 0.012 |
| TIMP4 | rs3755724 | T/C vs. C/C-T/T5 | 2.03 (1.21–3.41) | <0.001 | 2.03 (1.19–3.45) | 0.008 |
OR, odds ratio; 95% CI, 95% confidence interval.
p value not corrected for multiple SNPs.
Adjusted for BMI and age as covariates.
Recessive.
Dominant.
Overdominant.
Predictors of Weight Loss at 12 Months
Out of 587 subjects, 275 had a valid 12-month measurement. Of these subjects, 97 (35%) had low (<5%) weight loss, and 178 (65%) had high (≥5%) weight loss. The high weight loss group was significantly older and had a lower BMI than the low weight loss group (Table 1). ORs for ≥5% weight loss with a 95% CI deviating from unity were found for 2 SNPs in 2 of the 25 genes (PPARγ and TIMP4). Subjects with the C/C genotype of PPARγ rs1801281 were 2.0 times more likely to be in the high weight loss group than subjects with the C/G-G/G genotype (Table 2). For TIMP4 rs3755724, subjects with the T/C genotype were also 2.0 times more likely to be in the high weight loss group than subjects with the C/C-T/T genotype (Table 2). Adjustment for age and BMI as covariates did not change these results (Table 2). We also analyzed the association of a genetic risk score based on the SNPs that were associated with weight loss success at 12 months, but there was no association between the risk score and weight loss success (p = 0.177). Results for all SNPs can be found in online supplementary Table 2.
Mixed Model Analysis for Weight Changes
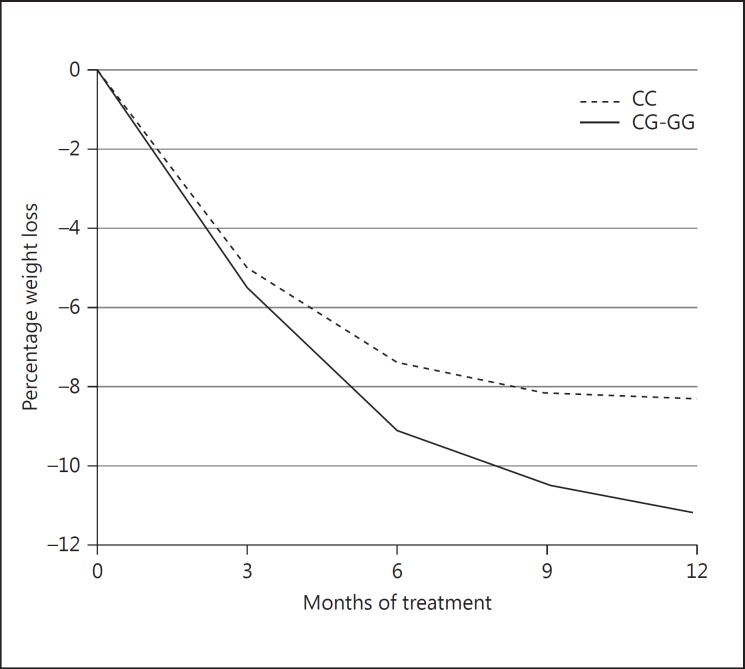
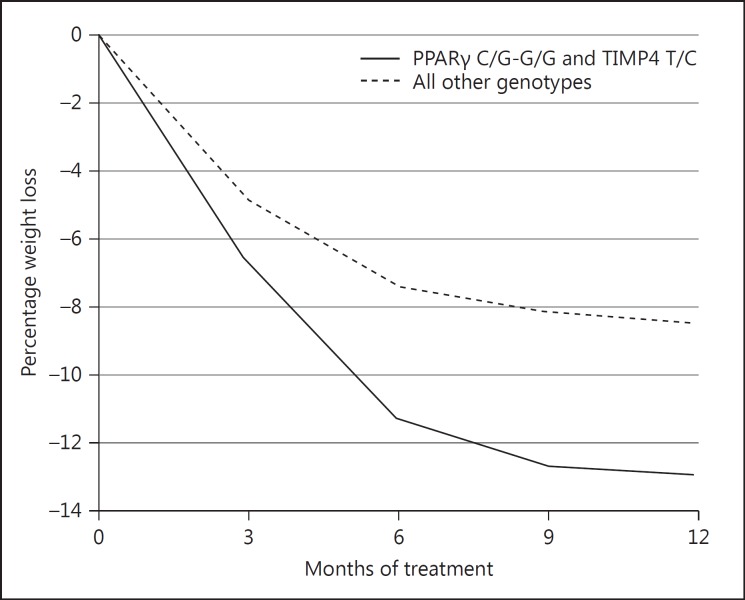
In the 275 subjects with a weight measurement after 12 months of treatment, we analyzed the weight change trajectories for the genotypes that were associated with differences in weight loss at 3 or 12 months. In Table 3, the outcomes of the mixed model analysis for the weight loss (as % of initial body weight) over 12 months of treatment are shown for the various genotypes. Only subjects with the C/G-G/G genotype compared to the C/C genotype of PPARγ rs1801282 showed significantly more weight loss during the treatment program (p = 0.027; Fig. 1, Table 3). We also paired the genotypes associated with better odds of ≥5% weight loss from PLIN1 rs2289487, MMP2 rs1132896, PPARγ rs1801282, and TIMP4 rs3755724 and compared them to all other genotype combinations. Only subjects with the combination of PPARγ rs1801282 C/G-G/G and TIMP4 rs3755724 T/C showed significantly higher weight loss than subjects with all other genotype combinations (p < 0.001) (Fig. 2).
Table 3.
Weight change (as % of initial body weight; mean, 95% confidence interval) per genotype at 3 and 12 months of treatment in individuals that completed 12 months of treatment (n = 275)
| Gene | rs number | Genotype | Weight change at 3 months | Genotype difference | p value1 | Weight change at 12 months | Genotype difference | p value1 |
|---|---|---|---|---|---|---|---|---|
| PLIN1 | rs2289487 | C/C (n = 39) | −5.3 (−5.9, −4.7) | −0.2 (−1.2, 0.8) | 0.722 | −9.3 (−112, −7.5) | −1.9 (−3.9, 0.1) | 0.059 |
| T/T-T/C (n = 234) | −5.1 (−6.0, −4.2) | −7.4 (−8.2, −6.7) | ||||||
| PLIN1 | rs2304795 | G/G (n = 38) | −4.8 (−6.0, −3.7) | 0.5 (−0.8, 1.8) | 0.473 | −6.7 (−8.8, −5.1) | 0.9 (−11, 2.9) | 0.378 |
| A/A-G/A (n = 230) | −5.3 (−5.9, −4.8) | −7.9 (−8.1, −6.6) | ||||||
| PLIN1 | rs1052700 | T/T (n = 42) | −6.4 (−8.0, −4.8) | −1.3 (−2.7, 0.1) | 0.058 | −9.1 (−10.9, −7.4) | −1.7 (−3.7, 0.2) | 0.077 |
| A/A-T/A (n = 232) | −5.1 (−5.6, −4.6) | −7.4 (−8.1, −6.6) | ||||||
| MMP2 | rs1132896 | C/C (n = 43) | −5.6 (−6.6, −4.5) | −0.4 (−1.7, 0.9) | 0.552 | −8.2 (−9.9, −6.4) | −0.6 (−2.5, 1.3) | 0.578 |
| C/G-G/G (n = 227) | −5.2 (−5.7, −4.7) | −7.6 (−8.3, −6.8) | ||||||
| PPARγ | rs1801282 | C/C (n = 211) | −3.9 (−6.4, −1.5) | 1.3 (−2.2, 4.8) | 0.454 | −7.2 (−8.0, −6.4) | 1.9 (0.2, 3.5) | 0.027 |
| C/G-G/G (n = 63) | −5.3 (−5.8, −4.8) | −9.1 (−10.5, −7.6) | ||||||
| TIMP4 | rs3755724 | T/C (n = 122) | −5.0 (−5.8, −4.3) | 0.4 (−0.6, 1.4) | 0.422 | −8.2 (−9.2, −7.1) | −0.9 (−2.3, 0.5) | 0.191 |
| C/C-T/T (n = 148) | −5.4 (−6.0, −4.8) | −7.2 (−8.2, −6.3) |
p value from a mixed model ANOVA analysis comparing weight loss at 3- and 12-month follow-up among genotypes (not corrected for multiple SNPs).
Fig. 1.
Weight loss (as % of initial weight) during 12 months of treatment for carriers of the C/C genotype vs. the C/G-G/G genotypes of PPARγ rs1801282.
Fig. 2.
Weight loss (as % of initial weight) during the 12 months of treatment for carriers of the combination of PPARγ rs1801282 C/G-GG and TIMP4 rs3755724 T/C, in comparison with carriers of all other genotype combinations of these PPARγ and TIMP4 SNPs.
Discussion
In this study, we have investigated the genetic predisposition to a ≥5% weight loss after 3 and 12 months in subjects participating in a multidisciplinary obesity treatment program. The genetic factors for 3-month weight loss (PLIN1, MMP2) and 12-month weight loss (PPARγ, TIMP4) were identified. The genetic variation in PPARγ and TIMP4 had a synergistic effect. Thus, genetic factors related to adipose tissue structure and function rather than hypothalamic regulation of food intake appear to be important for weight loss through a multidisciplinary lifestyle intervention. This is, to our knowledge, the first study to report genetic and weight loss data on such a large number of severely obese subjects undergoing a commercial multidisciplinary treatment program.
We acknowledge that our study has some limitations. There was a high dropout rate of 54% over the 12-month treatment period. Dropout rates are known to be a major problem when analyzing data from obesity treatments in a real-life setting, such as in this study, compared to well-controlled scientific studies [26]. Therefore, the high dropout may have reduced the power of our study. In order to check if the dropout might have influenced the allele frequencies of those who were still in the program at 12 months, we compared the allele frequencies of the 4 polymorphisms in the different genes between the total group, the group at 12 months and the dropouts, but we found no significant difference. Instead of using multiple test correction, we looked for associations by scoring if the 95% CI was different from unity with a value of p < 0.05. We have done this in order not to dismiss potential clues in this rather small population. However, when the more strict p value of 0.0017 (0.05/30) was applied, only the association of weight loss success at 12 months with TIMP4 (p < 0.001) remained. For the SNPs not reaching this level of statistical significance, results from previous studies can confirm their relevance.
PLIN1 is a well-characterized effector of energy and lipid metabolism and as a lipid droplet-coating protein it controls access to the adipocyte triglyceride stores that supply most tissues with fuel under a negative energy balance [27]. It has been reported to play a role in body weight regulation. In a weight loss/maintenance study of obese/overweight men and women, Soenen et al. [9] found that the haplotype of PLIN1 rs2289487 (C-allele) and PLIN1 rs894160 (A-allele) was related to a lower body mass, fat mass and fat-free mass in men at baseline and throughout the intervention. This haplotype was also related to a better weight reduction after 12-month follow-up in women [9]. In addition, PLIN1 rs2289487 T>C has previously been associated with a reduced risk of obesity in Spanish women [28]. In the present study, we found that for the total group the C/C genotype of PLIN1 rs2289487 predicted high weight loss in the first 3 months, and C/C-genotype carriers showed a trend for a higher weight loss over 12 months of treatment. Since from other studies it is clear that there is a sex difference in the association between weight loss and PLIN1 SNPs, we decided to test for association with PLIN1 rs2289487 separately among females and males. At 3 months, the C/C genotype of this SNP of the PLIN1 gene showed significance as a predictor of high weight loss in females (p = 0.002; OR 2.61 [1.41 to 4.85]). At 12 months, there was a trend for high weight loss (p = 0.07; OR 2.33 [−0.89 to 6.14]). As such, our findings are in line with studies by others. For PLIN1 rs1052700, the T/T genotype predicted high weight loss after 3 months, and T/T genotype carriers showed a trend for higher weight loss over 12 months of treatment. Soenen et al. [9] reported that men with the T/T genotype compared to the A/A-T/A genotype had a significantly lower weight, fat mass and fat-free mass at baseline and throughout the intervention, whereas women with the T/T genotype had a significantly lower plasma leptin level. In other studies, the analysis of weight loss and obesity risk in T-allele carriers of PLIN1 rs1052700 showed a lower obesity risk in women [29] and a higher weight loss after a weight-loss intervention in children and adults [30, 31], although one study reported a higher obesity risk in women [32]. Our results are in accordance with the data from most of these studies.
For yet another polymorphism in the PLIN1 gene, rs2304795, the G/G genotype was found to be associated with higher odds for high weight loss at 3 months, but no relation with weight loss over a period of 12-month treatment was observed. In the study by Soenen et al. [9], it was observed that the G-allele in a haplotype with the A-allele of PLIN1 rs2304796 was associated with a significantly larger reduction of fat mass and fat percentage, but only in female subjects. In 2 studies performed by Qi et al. [29, 32] regarding PLIN1 gene polymorphisms, PLIN1 rs2304795 was associated with higher obesity risk in women. In our study, there was only a trend at 3 months for women with the G/G genotype of this SNP of the PLIN1 gene to have lower odds for high weight loss (p = 0.07; OR 0.35 [0.11 to −1.16]). Lower odds for high weight loss could be interpreted as a higher obesity risk, which would comply with the studies by Qi et al. [28, 32].
MMPs are essential for ECM remodeling. The remodeling of the adipocyte ECM occurs during the development and growth of the fat depot [8], but probably also when adipocytes shrink under conditions of calorie restriction [7]. The results from our study indicate that MMP2 rs1132896 (C/C genotype) is a predictor for high weight loss at 3 months. In a recent study on long-term weight development, the C-allele was found to be associated with risk for weight gain in women [33]. In a study among Koreans, the C-allele of MMP2 rs1132896 was found to increase the risk for the development of obesity [34]. It suggests that the C-allele is representing a form of MMP2 that provides higher flexibility to the adipocyte ECM both under requirements of cell shrinkage or cell growth.
Two polymorphisms had odds for better weight loss at 12 months of treatment: TIMP4 rs3755724 (T/C genotype) and PPARγ (C/C genotype). Like MMPs, TIMPs are also active during adipose tissue remodeling [8]. To the best of our knowledge, this is the first report of a relation between TIMP genetic variation and weight loss. The significant OR for weight loss success for the TIMP4 rs3755724 heterozygotes compared to both homozygotes may confer a survival advantage in the heterozygotes. Functional analysis of this polymorphism is relatively difficult. Associations have been described with mental disorders like focal epilepsy and schizophrenia [35, 36]. A complicating fact is that the SNP is located at −55 in the promoter region of TIMP4 but also in an intron of the gene for synapsin II, which is transcribed from the opposite strand. New and larger studies are required to confirm this finding and address the underlying mechanism. No association with mean weight loss over 12 months was observed (Table 3). PPARγ is known to have a role in adipocyte differentiation, lipid metabolism, and glucose homeostasis. It also controls the expression of adiponectin, resistin, leptin, and tumor necrosis factor-α secreted from adipose tissue [37]. For rs1801282, a C/G SNP also known concerning the protein as Pro12Ala [38], we found that at 12 months subjects with the C/C genotype are at higher odds for high weight loss than subjects with the C/G-G/G genotype, and the G-allele carriers showed a higher loss of relative body weight over the 12-month treatment period. G-allele carriers have slightly higher baseline body weight (+0.7 kg) compared to subjects with the C/C genotype. These findings are in accordance with a meta-analysis showing that carriers of the Ala12 allele had a higher average BMI [39], and also with the study of Delahanty et al. [4], who found that 6-month and 2-year weight loss was associated with the minor Ala12 allele of the PPARγ gene. Bozina et al. [40] found that carriers of the Pro12Ala (C/G) or Ala12Ala (G/G) genotype had greater odds for obesity, but this was in a specific subsample with the ACE DD genotype.
Apart from PPARγ, we included other SNPs in our analysis that were also studied by Delahanty et al. [4] (BDNF, FTO, nearGNPDA2, SEC16B, SH2B1). In agreement with Delahanty et al. [4], we did not find associations between these SNPs and weight loss, although the FTO SNP showed a treatment-specific effect. A recent meta-analysis [5] suggested that the AA genotype of the FTO SNP, but not the TA genotype, was associated with significantly more weight loss than the TT genotype in diet and lifestyle interventions (difference −0.72 kg [95% CI: −1.21, +0.23 kg]; p = 0.004).
Interestingly, both the 3-month and 12-month analyses provide evidence on the involvement of the same processes in the genetic predisposition to weight loss by dietary and lifestyle advice, i.e. fat storage in adipocytes (PLIN1 and PPARγ) and adipose tissue remodeling (MMP2, TIMP4). Variation in genes involved in other processes like the sympathetic nervous system (ADRB2), hypothalamic regulation of eating behavior (CNTF, BDNF, FTO, LEP), or peripheral energy metabolism (ACE, ADIPOQ) did not show associations. It indicates that the capacity to lose weight in the investigated cohort depends largely on the (genetically determined) performance to preserve the stored fat and on the structural flexibility of the adipocytes. Those characteristics may also be reflected in a lower baseline BMI for the high weight loss group at 3 and 12 months. In vivo, regulation of fat storage and structural adaptation of the adipocytes are expected to be linked processes. Here, we found with the mixed model analysis for mean weight change over the 12-month period that subjects with the combination of the genotypes associated with higher weight loss of PPARγ and TIMP4 (PPARγ rs1801282 C/G-G/G and TIMP4 rs3755724 T/C) showed significantly higher weight loss than all other genotype combinations (p < 0.001) and a trend for combining SNPs of PLIN1 and MMP2 (p = 0.09). This synergistic effect seems to confirm that both processes are interacting and determine the success of weight loss.
In conclusion, weight loss of severely obese subjects by dietary advice, psychological counseling, and increased physical activity in real-life conditions depends on the genetic background. Relevant genetic variation relates to genes involved in adipocyte fat storage and structural adaptation of the adipocytes during fat reduction. These findings can be of importance when searching for explanations regarding treatment success or failure.
Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Stubbs J, Whybrow S, Teixeira P, Blundell J, Lawton C, Westenhoefer J, Engel D, Shepherd R, McConnon A, Gilbert P, Raats M. Problems in identifying predictors and correlates of weight loss and maintenance: implications for weight control therapies based on behaviour change. Obesity Rev. 2011;12:688–708. doi: 10.1111/j.1467-789X.2011.00883.x. [DOI] [PubMed] [Google Scholar]
- 2.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delahanty LM, Pan Q, Jablonski KA, Watson KE, McCaffery JM, Shuldiner A, Kahn SE, Knowler WC, Florez JC, Franks PW. Genetic predictors of weight loss and weight regain after intensive lifestyle modification, metformin treatment, or standard care in the Diabetes Prevention Program. Diabetes Care. 2012;35:363–366. doi: 10.2337/dc11-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang L, Wu H, Pan A, Patel B, Xiang G, Qi L, Kaplan RC, Hu F, Wylie-Rosett J, Qi Q. FTO genotype and weight loss in diet and lifestyle interventions: a systematic review and meta-analysis. Am J Clin Nutr. 2016;103:1162–1170. doi: 10.3945/ajcn.115.123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariman EC, Wang P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell Mol Life Sci. 2010;67:1277–1292. doi: 10.1007/s00018-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariman EC. Human biology of weight maintenance after weight loss. J Nutrigenet Nutrigenomics. 2012;5:13–25. doi: 10.1159/000337081. [DOI] [PubMed] [Google Scholar]
- 8.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J Biol Chem. 2003;278:11888–11896. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 9.Soenen S, Mariman EC, Vogels N, Bouwman FG, den Hoed M, Brown L, Westerterp-Plantenga MS. Relationship between perilipin gene polymorphisms and body weight and body composition during weight loss and weight maintenance. Physiol Behav. 2009;96:723–728. doi: 10.1016/j.physbeh.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Vogels N, Mariman EC, Bouwman FG, Kester AD, Diepvens K, Westerterp-Plantenga MS. Relation of weight maintenance and dietary restraint to peroxisome proliferator-activated receptor gamma2, glucocorticoid receptor, and ciliary neurotrophic factor polymorphisms. Am J Clin Nutr. 2005;82:740–746. doi: 10.1093/ajcn/82.4.740. [DOI] [PubMed] [Google Scholar]
- 11.Erez G, Tirosh A, Rudich A, Meiner V, Schwarzfuchs D, Sharon N, Shpitzen S, Bluher M, Stumvoll M, Thiery J, et al. Phenotypic and genetic variation in leptin as determinants of weight regain. Int J Obes (Lond) 2011;35:785–792. doi: 10.1038/ijo.2010.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Wu J, Yu L. Association of Gln27Glu and Arg16Gly polymorphisms in Beta2-adrenergic receptor gene with obesity susceptibility: a meta-analysis. PLoS One. 2014;9:e100489. doi: 10.1371/journal.pone.0100489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariman EC. Future nutrigenetics: in search of the missing genetic variation. J Nutrigenet Nutrigenomics. 2009;2:257–262. doi: 10.1159/000297212. [DOI] [PubMed] [Google Scholar]
- 14.Mariman EC, Bouwman FG, Aller EE, van Baak MA, Wang P. Extreme obesity is associated with variation in genes related to the circadian rhythm of food intake and hypothalamic signaling. Physiol Genomics. 2015;47:225–231. doi: 10.1152/physiolgenomics.00006.2015. [DOI] [PubMed] [Google Scholar]
- 15.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Holst C, Wodzig WK, Andersen MR, Astrup A, van Baak MA, Larsen TM, Jebb SA, Kafatos A, Pfeiffer AF, et al. Circulating ACE is a predictor of weight loss maintenance not only in overweight and obese women, but also in men. Int J Obes (Lond) 2012;36:1545–1551. doi: 10.1038/ijo.2011.278. [DOI] [PubMed] [Google Scholar]
- 17.Kim YJ, Sano T, Nabetani T, Asano Y, Hirabayashi Y. GPRC5B activates obesity-associated inflammatory signaling in adipocytes. Sci Signal. 2012;5:ra85. doi: 10.1126/scisignal.2003149. [DOI] [PubMed] [Google Scholar]
- 18.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, Henning E, Blackburn H, Loos RJ, Wareham NJ, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45:513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Bradfield JP, Zhang H, Sleiman PM, Kim CE, Glessner JT, Deliard S, Thomas KA, Frackelton EC, Li M, et al. Role of BMI-associated loci identified in GWAS meta-analyses in the context of common childhood obesity in European Americans. Obesity. 2011;19:2436–2439. doi: 10.1038/oby.2011.237. [DOI] [PubMed] [Google Scholar]
- 21.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A, Volzke H, Wallaschofski H, Abecasis GR, Edward L, et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22:3597–3607. doi: 10.1093/hmg/ddt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aller EE, van Baak MA. Evaluation of an 18-month commercial multidisciplinary obesity treatment programme. Clin Obesity. 2016;6:33–41. doi: 10.1111/cob.12122. [DOI] [PubMed] [Google Scholar]
- 23.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 24.Franz MJ, VanWormer JJ, Crain AL, Boucher JL, Histon T, Caplan W, Bowman JD, Pronk NP. Weight-loss outcomes: a systematic review and meta-analysis of weight-loss clinical trials with a minimum 1-year follow-up. J Am Diet Assoc. 2007;107:1755–1767. doi: 10.1016/j.jada.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Klein S, Burke LE, Bray GA, Blair S, Allison DB, Pi-Sunyer X, Hong Y, Eckel RH. Clinical implications of obesity with specific focus on cardiovascular disease: a statement for professionals from the American Heart Association Council on Nutrition, Physical Activity, and Metabolism: endorsed by the American College of Cardiology Foundation. Circulation. 2004;110:2952–2967. doi: 10.1161/01.CIR.0000145546.97738.1E. [DOI] [PubMed] [Google Scholar]
- 26.Dalle Grave R, Calugi S, Molinari E, Petroni ML, Bondi M, Compare A, Marchesini G, Group QS. Weight loss expectations in obese patients and treatment attrition: an observational multicenter study. Obesity Res. 2005;13:1961–1969. doi: 10.1038/oby.2005.241. [DOI] [PubMed] [Google Scholar]
- 27.Smith CE, Ordovas JM. Update on perilipin polymorphisms and obesity. Nutr Rev. 2012;70:611–621. doi: 10.1111/j.1753-4887.2012.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi L, Corella D, Sorli JV, Portoles O, Shen H, Coltell O, Godoy D, Greenberg AS, Ordovas JM. Genetic variation at the perilipin (PLIN) locus is associated with obesity-related phenotypes in White women. Clin Genet. 2004;66:299–310. doi: 10.1111/j.1399-0004.2004.00309.x. [DOI] [PubMed] [Google Scholar]
- 29.Qi L, Tai ES, Tan CE, Shen H, Chew SK, Greenberg AS, Corella D, Ordovas JM. Intragenic linkage disequilibrium structure of the human perilipin gene (PLIN) and haplotype association with increased obesity risk in a multiethnic Asian population. J Mol Med (Berl) 2005;83:448–456. doi: 10.1007/s00109-004-0630-4. [DOI] [PubMed] [Google Scholar]
- 30.Deram S, Nicolau CY, Perez-Martinez P, Guazzelli I, Halpern A, Wajchenberg BL, Ordovas JM, Villares SM. Effects of perilipin (PLIN) gene variation on metabolic syndrome risk and weight loss in obese children and adolescents. J Clin Endocrinol Metab. 2008;93:4933–4940. doi: 10.1210/jc.2008-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang Y, Kim OY, Lee JH, Koh SJ, Chae JS, Kim JY, Park S, Cho H, Lee JE, Ordovas JM. Genetic variation at the perilipin locus is associated with changes in serum free fatty acids and abdominal fat following mild weight loss. Int J Obes (Lond) 2006;30:1601–1608. doi: 10.1038/sj.ijo.0803312. [DOI] [PubMed] [Google Scholar]
- 32.Qi L, Shen H, Larson I, Schaefer EJ, Greenberg AS, Tregouet DA, Corella D, Ordovas JM. Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obesity Res. 2004;12:1758–1765. doi: 10.1038/oby.2004.218. [DOI] [PubMed] [Google Scholar]
- 33.Bouwman FG, Boer JM, Imholz S, Wang P, Verschuren WM, Dolle ME, Mariman EC. Gender-specific genetic associations of polymorphisms in ACE, AKR1C2, FTO and MMP2 with weight gain over a 10-year period. Genes Nutr. 2014;9:434. doi: 10.1007/s12263-014-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han DH, Kim SK, Kang S, Choe BK, Kim KS, Chung JH. Matrix metallopeptidase 2 gene polymorphism is associated with obesity in Korean population. Korean J Physiol Pharmacol. 2008;12:125–129. doi: 10.4196/kjpp.2008.12.3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haerian BS, Sha'ari HM, Fong CY, Tan HJ, Wong SW, Ong LC, Raymond AA, Tan CT, Mohamed Z. Contribution of TIMP4 rs3755724 polymorphism to susceptibility to focal epilepsy in Malaysian Chinese. J Neuroimmunol. 2015;278:137–143. doi: 10.1016/j.jneuroim.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Yim SV, Kim SK, Park HJ, Jeon HS, Jo BC, Kang WS, Lee SM, Kim JW, Chung JH. Assessment of the correlation between TIMP4 SNPs and schizophrenia and autism spectrum disorders. Mol Med Rep. 2013;7:489–494. doi: 10.3892/mmr.2012.1221. [DOI] [PubMed] [Google Scholar]
- 37.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsiao TJ, Lin E. The Pro12Ala polymorphism in the peroxisome proliferator-activated receptor gamma (PPARG) gene in relation to obesity and metabolic phenotypes in a Taiwanese population. Endocrine. 2015;48:786–793. doi: 10.1007/s12020-014-0407-7. [DOI] [PubMed] [Google Scholar]
- 39.Galbete C, Toledo E, Martinez-Gonzalez MA, Martinez JA, Guillen-Grima F, Marti A. Pro12Ala variant of the PPARG2 gene increases body mass index: an updated meta-analysis encompassing 49,092 subjects. Obesity. 2013;21:1486–1495. doi: 10.1002/oby.20150. [DOI] [PubMed] [Google Scholar]
- 40.Bozina T, Sertic J, Lovric J, Jelakovic B, Simic I, Reiner Z. Interaction of genetic risk factors confers increased risk for metabolic syndrome: the role of peroxisome proliferator-activated receptor gamma. Genet Test Mol Biomarkers. 2014;18:32–40. doi: 10.1089/gtmb.2013.0344. [DOI] [PubMed] [Google Scholar]