Abstract
Objective
Adipose tissue stem cells (ADSCs) present a promising therapeutic method to alleviate liver failure (LF). The purpose of this prospective study was to evaluate the efficacy of undifferentiated ADSC transplantation on liver regeneration and on the expression of liver regeneration- and liver-specific genes, following 60% partial hepatectomy (PHx).
Methods
Sixty female rats were subjected to PHx and were transplanted with 106 or 2 × 106 ADSCs, either into the portal vein (PV) or into the hepatic parenchyma. Animals of the control group were not transplanted and served as controls. Animals were sacrificed on the 4th, the 7th, or the 15th postoperative day (POD).
Results
The transplanted ADSCs were successfully engrafted into the liver parenchyma and ameliorated the histopathologic damage on the 7th and 15th POD. All transplanted animals demonstrated a significantly higher liver regeneration rate on the 4th and 7th POD, compared with the control group. The expression of hepatocyte growth factor, α-fetoprotein, tyrosine aminotransferase, hepatocyte nuclear factor 4a, and cytochrome P450 1A2 was significantly upregulated, compared with the control group.
Conclusions
Although undifferentiated, ADSC transplantation significantly enhanced the liver regeneration process. These findings may be proven clinically valuable, especially in cases of acute LF.
1. Introduction
Liver failure (LF) is one of the leading causes of morbidity and mortality worldwide [1]. The only effective treatment so far for acute and chronic LF is liver transplantation [2], with its associated limitations, including the shortage of liver donors and the need for continuous immunosuppression. These facts have prompted the efforts for an alternative treatment of end-stage liver disease (ESLD).
Mesenchymal stem cells (MSCs) present a promising therapeutic method to alleviate ESLD. According to a growing body of evidence in recent years, adipose tissue stem cells (ADSCs), a certain type of MSCs, represent the most promising candidate progenitor cells for transplantation, as they show a stronger commitment to hepatic lineage, as well as higher rates of proliferation, compared to bone marrow mesenchymal stem cells (BM-MSCs) [3, 4].
Despite the encouraging experimental outcomes, many questions still remain, such as type and quantity of transplanted ADSCs, optimal route of administration, and pretreatment or not with growth factors.
In the present prospective study, we aimed to investigate the effect of undifferentiated ADSC transplantation on liver regeneration, as well as on the expression of liver-specific genes, in a rat model of partial hepatectomy (PHx), in relation to the number and their route of administration.
2. Materials and Methods
2.1. Animals
One hundred Wistar rats of conventional microbiological status (n = 90 female, n = 10 male), weighing 190–260 g, were purchased from the same breeder (Democritus, Agia Paraskevi, Greece). All rats were grouped and housed in type IV cages with 400 cm2 floor area per rat, with a controlled environment of 12 h : 12 h light-dark cycle. All animals had ad libitum access to food and water. They were allowed to acclimate to the laboratory conditions for at least one week prior to the experiment. All studies carried out at the Experimental Research Center, ELPEN conform to the Presidential Decree 56/2013 for the Protection of Animals used for Scientific Purposes. Male Wistar rats were used as ADSC donors, while the female rats as the recipients of ADSCs.
2.2. Experimental Design
Female rats were randomly allocated to one of six different experimental groups. Control group (n = 15) underwent 60% PHx, without transplantation. Sham-operated group (n = 15) underwent a midline laparotomy with incision of the liver ligaments, followed by abdominal closure. Groups A and B (n = 15/group) underwent 60% PHx with subsequent administration of 106 and 2 × 106 ADSCs into the portal vein (PV), respectively. Groups C and D (n = 15/group) underwent 60% PHx with subsequent administration of 106 and 2 × 106ADSCs into the hepatic parenchyma, respectively. Group N (n = 90), although not belonging to experimental groups, represents the preoperative values of all animals. Each experimental group was subdivided into three subgroups (n = 5/subgroup), depending on the postoperative day (POD) of sacrifice (Table 1).
Table 1.
Experimental design.
| Experimental group | Subgroup | Number of animals/subgroup | Subgroup identification number | POD1 of sacrifice | Route of ADSC2 transplantation | Number of transplanted ADSCs |
|---|---|---|---|---|---|---|
| Control group | 4 days | 5 | CN1 | 4th | — | — |
| 7 days | 5 | CN2 | 7th | — | — | |
| 15 days | 5 | CN3 | 15th | — | — | |
|
| ||||||
| Sham group | 4 days | 5 | S1 | 4th | — | — |
| 7 days | 5 | S2 | 7th | — | — | |
| 15 days | 5 | S3 | 15th | — | — | |
|
| ||||||
| Group A | 4 days | 5 | A1 | 4th | PV3 | 106 |
| 7 days | 5 | A2 | 7th | PV | 106 | |
| 15 days | 5 | A3 | 15th | PV | 106 | |
|
| ||||||
| Group B | 4 days | 5 | B1 | 4th | PV | 2 × 106 |
| 7 days | 5 | B2 | 7th | PV | 2 × 106 | |
| 15 days | 5 | B3 | 15th | PV | 2 × 106 | |
|
| ||||||
| Group C | 4 days | 5 | C1 | 4th | HP4 | 106 |
| 7 days | 5 | C2 | 7th | HP | 106 | |
| 15 days | 5 | C3 | 15th | HP | 106 | |
|
| ||||||
| Group D | 4 days | 5 | D1 | 4th | HP | 2 × 106 |
| 7 days | 5 | D2 | 7th | HP | 2 × 106 | |
| 15 days | 5 | D3 | 15th | HP | 2 × 106 | |
Indicating the six different experimental groups as well as the three subgroups in each experimental group, according to the number and route of transplantation of ADSCs and the postoperative day of euthanasia. 1POD: postoperative day; 2ADSCs: adipose tissue stem cells; 3PV: portal vein; 4HP: hepatic parenchyma.
2.3. Isolation and Culture of Rat ADSCs
White adipose tissue was collected from rats and was immediately transferred to the laboratory at 4°C. The tissue was washed with phosphate-buffered saline (PBS), minced using two scalpels, and then was digested in crude collagenase (1 mg/ml DMEM) for 30 min at 37°C. Subsequently, the digest was centrifuged (200g for 5 min) to discard the supernatant and the pellet was resuspended in DMEM/10% FBS/1% penicillin/streptomycin and transferred to a culture flask. After an overnight incubation, the medium was changed as to remove the nonadherent cells and the attached cells were further cultured in the same medium.
2.4. Surgical Procedure and Euthanasia
All rats had no access to food and water for the last 4 hours before the surgical procedure. General gas anesthesia was induced and maintained by a mixture of O2 and N2O and isoflurane (Forenium®, 4% for induction and 2% for maintenance). All interventions were performed under sterile conditions. A midline laparotomy was performed, followed by incision of liver ligaments. The intestinal loops were shifted towards the left side, while the left lateral lobe (LLL) and the median lobe (ML) of the liver were shifted cranially. Rats were subjected to 60% PHx, by dividing the LLL and ML near the origin of their vasculature using electrocautery, followed by suture ligation and resection (Figures 1 and 2). The resected liver specimen was immediately weighed to estimate the resected liver mass, as well as the percentage of PHx performed, as the liver mass represents approximately 5% of the rat's total body weight [5]. Following PHx, gentle dissection was carried out, and the PV was exposed posteriorly and laterally to the hepatic artery (HA) and common bile duct (CBD). ADSCs were administered either into the PV or into the remnants of the resected liver lobes, with the use of a 30-gauge needle, at a dialysis of 106 ADSCs in 0.2 ml of saline (Figure 3). Finally, 1 ml NaCl 0.9% was administered intraperitoneally and the abdomen was closed in a continuous one-layer fashion. Next, rats were placed under heat-producing lamps to recover from anesthesia.
Figure 1.
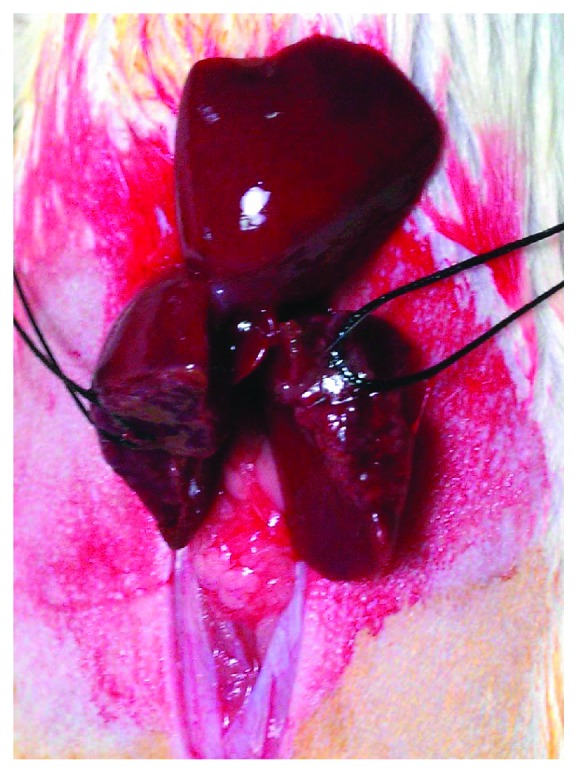
60% partial hepatectomy (PHx) was performed, by dividing the left lateral lobe (LLL), and median lobe (ML) of the liver near the origin of their vasculature by using electrocautery, followed by suture ligation and resection.
Figure 2.
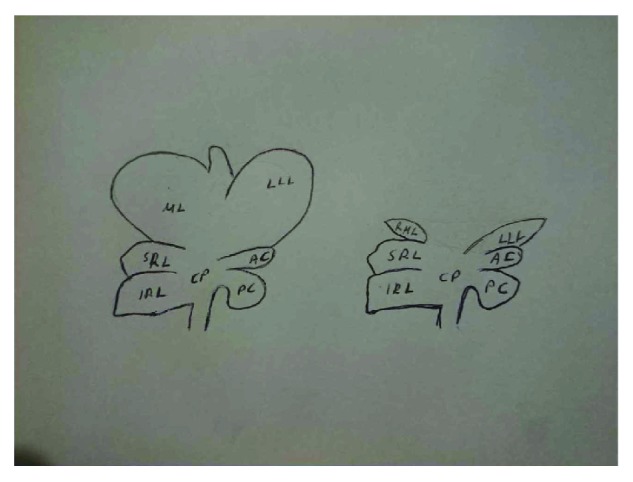
Indicating the normal liver anatomy of the rat (on the left) and the liver anatomy after our surgical technique of nearly totally resecting the left lateral lobe (LLL) and median lobe (ML) of the liver (on the right), thus achieving a 60% partial hepatectomy.
Figure 3.
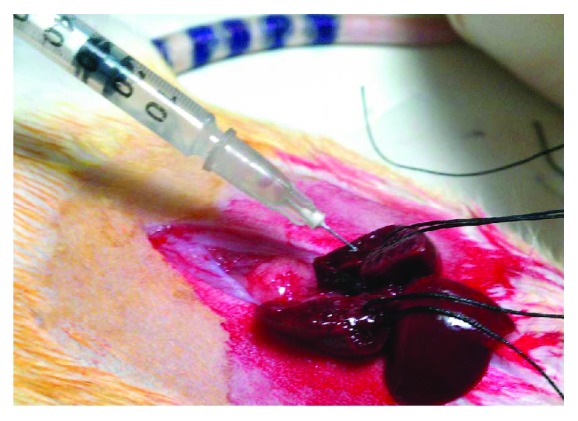
106 ADSC transplantation into the remnants of the resected liver lobes (LLL and ML), with the use of a 30-gauge needle, at a dialysis of 106 ADSCs in 0.2 ml of saline.
Rats of each group were randomly allocated to be sacrificed either on the 4th or on the 7th day or on the 15th POD. Rats were anesthetized before euthanasia, followed by animal weighing. All rats had no access to food and water for the last 4 hours before euthanasia. A midline laparotomy was performed and blood samples were taken from the inferior vena cava (IVC), followed by harvesting of the liver, which was also weighed (Figure 4). Four representative tissue samples from the nearly totally resected liver lobes as well as four tissue samples from the nonresected lobes were harvested. Half of the tissue was fixed in 4% buffered formaldehyde, embedded in paraffin, and routinely stained with haematoxylin and eosin (H&E staining), while the other half was immediately transferred in liquid nitrogen and then stored at −80°C for future RNA extraction.
Figure 4.
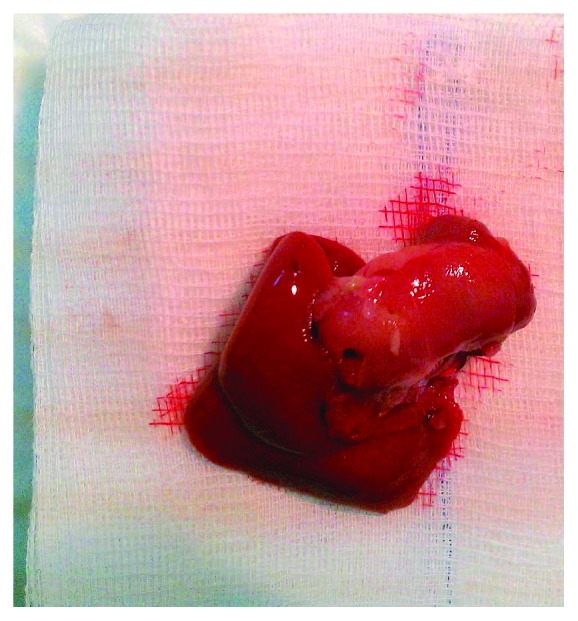
The harvested regenerated liver at the time of euthanasia.
2.5. Peripheral Blood Sample Analysis
The levels of platelets (PLT), aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyltransferase (GGT), alkaline phosphatase (ALP), albumin (ALB), prothrombin time (PT), (INR), total proteins (PR), total bilirubin (TBIL), direct bilirubin (DBIL), indirect bilirubin (IBIL), and phosphorus were measured in peripheral blood sample, in a single center, by standard laboratory methods.
2.6. Liver Regeneration Rate
Liver regeneration rate (%) was calculated on the day of sacrifice, using the following equation: 100 × {C–(A − B)}/A, where A is the estimated total liver weight at the time of PHx, which represents approximately 5% of the rat's total body weight [5], B is the weight of the excised liver, and C is the weight of the harvested regenerating liver at the time of sacrifice [6, 7].
2.7. Body Weight Assessment
Total body weight was measured prior to the surgical procedure (initial weight (IW)) as well as at the time of sacrifice, with the rats under general gas anesthesia and still alive (preeuthanasia weight (PEW)). Preeuthanasia weight (PEW%) was calculated, using the following equation: PEW = 100 × (PEW − IW)/IW.
2.8. Fluorescence In Situ Hybridization
10 μl of ZytoLight Rat Y/12 Fluorescence in Situ Hybridization (FISH) Y- chromosome probe (ZytoVision GmbH) was applied onto each individual deparaffinized liver section of 4 μm thickness, according to the guidelines of the manufacturer. Sample material was evaluated by fluorescence microscopy, with filter sets for the wavelength ranges applied.
2.9. Histological Analysis
Multiple 4 μm sections were scored blindly, by two independent observers, for the following parameters: sinusoidal congestion, vacuolization of hepatocyte cytoplasm, parenchymal necrosis, and inflammation, with the total score representing the sum of all parameters for each individual animal. Each parameter was graded numerically as follows: congestion, vacuolization, and inflammation: 0 = none, 1 = minimal, 2 = mild, 3 = moderate, and 4 = severe. The numerical graduation for necrosis was as follows: 0 = nonnecrotic cells, 1 = single-cell necrosis, 2 ≤ 30% necrosis, 3 ≤ 60% necrosis, and 4 ≥ 60% necrosis.
2.10. RNA Extraction and Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
Total RNA was extracted using NucleoSpin® RNA Plus (Macherey-Nagel GmbH & Co. KG, Germany). RNA concentration and quality were determined using the NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, DE, USA). 250 ng of total RNA was converted to cDNA using Superscript II RT-PCR kit (Invitrogen Life Technologies, CA, USA). Liver regeneration- as well as liver-specific genes (hepatocyte growth factor (HGF), α-fetoprotein (AFP), albumin (ALB), glypican 3 (GPC3), tyrosine aminotransferase (TAT), hepatocyte nuclear factor 4A (HNF-4a), and cytochrome P450 1A2 (CYP1A2)) relative mRNA expression levels were determined by reverse transcription quantitative real-time PCR (RT-qPCR), by using the 2−ΔΔCT method, on LightCycler® 480 System (Roche Diagnostics GmbH, Germany), using Maxima® SYBR Green/ROX kit (Thermo Fisher Scientific, DE, USA). The samples were run at least in duplicates, and for each sample, the mean Cp value was calculated. As an appropriate endogenous control, the GAPDH gene was selected according to the literature [8, 9]. Three pool samples (control group) were prepared in total, respective to the day of sacrifice, containing all individuals of each subset, and each sample was analyzed with the time-matching pool sample as a calibrator. Relative expression was then assessed by LightCycler 480 Software, Version 1.5 (Roche Diagnostics GmbH, Germany). Sequences of gene- and rat-specific primers used are depicted in Table 2.
Table 2.
Sequences of primers used for reverse transcription quantitative real-time PCR (RT-qPCR).
| Gene | Forward | Reverse |
|---|---|---|
| AFP1 | AGAAAACAGGGCGATGTCCA | TGCCTTGTCATACTGAGCGG |
| GPC32 | TAAAAGTCGCCCGTGTCGAA | ATGTAGCCTGGCAAAGCACT |
| HGF3 | CCCTATTTCCCGTTGTGAAGGA | ACCATCCACCCTACTGTTGTTT |
| TAT4 | GATTTTGGCAGTGGCTGAAAGG | GAACATTGGTGCTGAGGTTGG |
| ALB5 | AAGAGAAAGCACTGGTCGCA | GGGGAATCGCTGGCTCA-TAC |
| HNF-4a6 | AGGATGAAGAAGTTGCCCCC | GATGTGTCTGGTGGGTCCTG |
| CYP1A27 | CATCCTTTGTCCCCTTCACCA | GGTCTTTCCACTGCTTCTCATC |
| GAPDH8 | CTCTCTGCTCCTCCCTGTTC | TACGGCCAAATCCGTTCACA |
The sequences of primers (forward and reverse) used for reverse transcription quantitative real-time PCR (RT-qPCR). 1AFP: α-fetoprotein; 2GPC3: glypican 3; 3HGF: hepatocyte growth factor; 4TAT: tyrosine aminotransferase; 5ALB: albumin; 6HNF-4a: hepatocyte nuclear factor 4a; 7CYP1A2: cytochrome P450 1A2; 8GAPDH: glyceraldehyde 3-phosphate dehydrogenase (GAPDH was used as an endogenous control).
2.11. Statistical Analysis
Statistical analysis was done with IBM SPSS Statistics ver. 20 (SPSS Inc., Chicago, IL, USA), by a statistician specializing in medical statistics. The level of statistical significance was set at 5% (α = 0.05).
Analysis of variance (with pairwise post hoc Bonferroni tests) was used to compare the mean values of all parameters by time period (4 days, 7 days, and 15 days) for each group (CN, S, A, Β, C, and D), respectively. Each time the assumption of homogeneity of variances, which is crucial for ANOVA, was checked with Levene's test.
3. Results
3.1. Tracing of Transplanted ADSCs
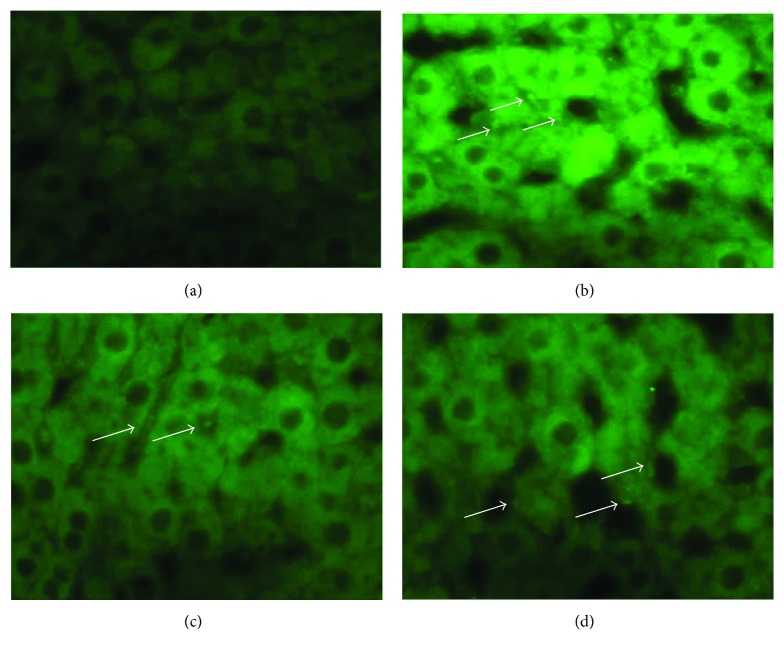
The expression of the rat Y chromosome was observed in the liver parenchyma of all transplanted rats of all POD of sacrifice, irrespective of the number or the route of transplantation, whereas it was not observed in nontransplanted animals, thus demonstrating the successful transplantation and localization of ADSCs (Figure 5).
Figure 5.
(a–d) Fluorescent in situ hybridization (FISH) of the Y chromosome, with fluorescein stain used for nuclear staining. (a) Animal of the control group, in which no signals are present. (b) Localization of the intraportally transplanted ADSCs in the host liver on the 4th POD (as depicted by arrows). (c and d) Localization of the intraparenchymaly transplanted ADSCs in the host liver on the 7th and 15th POD, respectively (as depicted by arrows).
3.2. Analysis of Histopathologic Damage
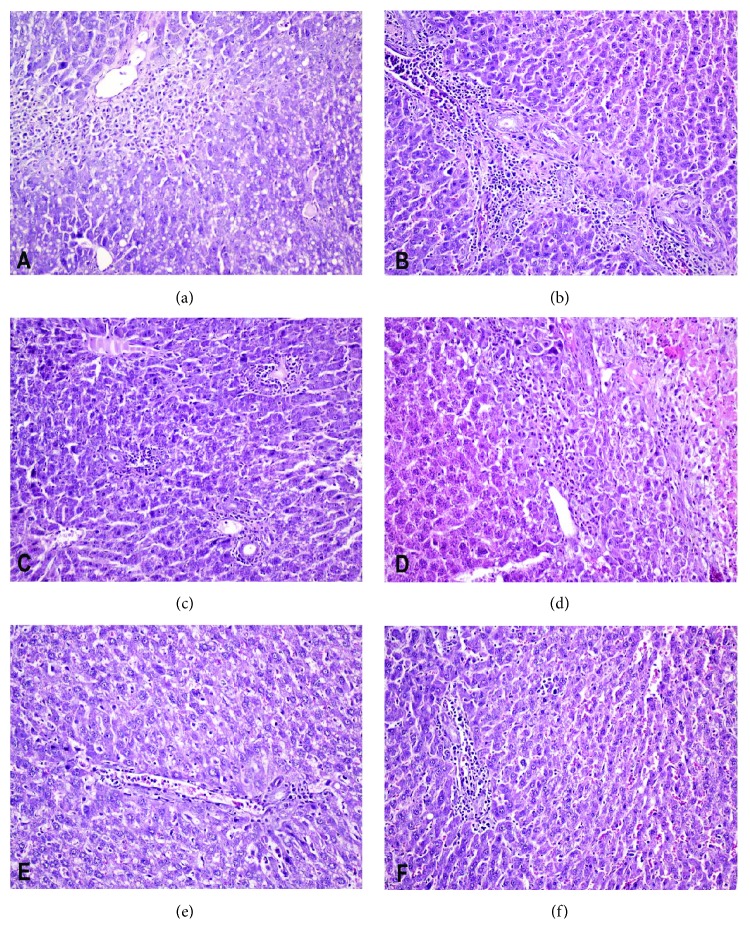
Regarding the animals that were sacrificed on the 4th POD, no statistically significant differences in the total score as well as in each individual parameter were identified between the transplanted as well as between the transplanted and the nontransplanted animals. On the 7th and 15th POD, however, a significantly improved total score was observed in the subgroups B2 (p = 0.044) and C2 (p = 0.044), as well as in the subgroups A3 (p = 0.047) and B3 (p = 0.047), compared with the respective control animals, with no significant differences between the transplanted groups (Table 3) (Figure 6).
Table 3.
Liver histological analysis scoring.
| Animal | Subgroup ID1 | Congestion | Vacuolization | Necrosis | Inflammation | Total score2 |
|---|---|---|---|---|---|---|
| CN1-1 | CN1 | 1 | 0 | 0 | 0 | 1 |
| CN1-2 | CN1 | 1 | 0 | 1 | 1 | 3 |
| CN1-3 | CN1 | 1 | 1 | 1 | 1 | 4 |
| CN1-4 | CN1 | 1 | 0 | 0 | 0 | 1 |
| CN1-5 | CN1 | 1 | 1 | 1 | 3 | 6 |
| CN2-6 | CN2 | 1 | 1 | 0 | 0 | 2 |
| CN2-7 | CN2 | 1 | 1 | 1 | 0 | 3 |
| CN2-8 | CN2 | 1 | 1 | 0 | 1 | 3 |
| CN2-9 | CN2 | 0 | 0 | 0 | 0 | 0 |
| CN2-10 | CN2 | 2 | 2 | 3 | 3 | 10 |
| CN3-11 | CN3 | 1 | 1 | 1 | 1 | 4 |
| CN3-12 | CN3 | 1 | 0 | 1 | 0 | 2 |
| CN3-13 | CN3 | 0 | 1 | 0 | 0 | 1 |
| CN3-14 | CN3 | 0 | 0 | 1 | 0 | 1 |
| CN3-15 | CN3 | 1 | 0 | 1 | 0 | 2 |
| S1-1 | S1 | 0 | 0 | 0 | 0 | 0 |
| S1-2 | S1 | 0 | 0 | 0 | 0 | 0 |
| S1-3 | S1 | 0 | 0 | 0 | 0 | 0 |
| S1-4 | S1 | 0 | 0 | 0 | 0 | 0 |
| S1-5 | S1 | 0 | 0 | 0 | 0 | 0 |
| S2-6 | S2 | 0 | 0 | 0 | 0 | 0 |
| S2-7 | S2 | 0 | 0 | 0 | 0 | 0 |
| S2-8 | S2 | 0 | 0 | 0 | 0 | 0 |
| S2-9 | S2 | 0 | 0 | 0 | 0 | 0 |
| S2-10 | S2 | 0 | 0 | 0 | 0 | 0 |
| S3-11 | S3 | 0 | 0 | 0 | 0 | 0 |
| S3-12 | S3 | 0 | 0 | 0 | 0 | 0 |
| S3-13 | S3 | 0 | 0 | 0 | 0 | 0 |
| S3-14 | S3 | 0 | 0 | 0 | 0 | 0 |
| S3-15 | S3 | 0 | 0 | 0 | 0 | 0 |
| Α1-1 | A1 | 1 | 0 | 2 | 2 | 5 |
| Α1-2 | A1 | 0 | 3 | 1 | 1 | 5 |
| Α1-3 | A1 | 0 | 3 | 1 | 1 | 5 |
| Α1-4 | A1 | 0 | 1 | 0 | 0 | 1 |
| Α1-5 | A1 | 1 | 3 | 1 | 1 | 6 |
| Α2-6 | A2 | 0 | 1 | 0 | 0 | 1 |
| Α2-7 | A2 | 0 | 0 | 0 | 0 | 0 |
| Α2-8 | A2 | 0 | 0 | 0 | 0 | 0 |
| Α2-9 | A2 | 1 | 0 | 1 | 2 | 4 |
| Α2-10 | A2 | 0 | 0 | 0 | 0 | 0 |
| Α3-11 | A3 | 0 | 0 | 0 | 0 | 0 |
| Α3-12 | A3 | 0 | 0 | 0 | 0 | 0 |
| Α3-13 | A3 | 0 | 0 | 0 | 0 | 0 |
| Α3-14 | A3 | 0 | 0 | 0 | 0 | 0 |
| Α3-15 | A3 | 0 | 0 | 0 | 0 | 0 |
| B1-1 | B1 | 0 | 0 | 0 | 0 | 0 |
| B1-2 | B1 | 0 | 1 | 0 | 0 | 1 |
| B1-3 | B1 | 0 | 1 | 1 | 1 | 3 |
| B1-4 | B1 | 0 | 0 | 0 | 0 | 0 |
| B1-5 | B1 | 0 | 1 | 0 | 0 | 1 |
| B2-6 | B2 | 0 | 0 | 0 | 0 | 0 |
| B2-7 | B2 | 0 | 0 | 0 | 0 | 0 |
| B2-8 | B2 | 0 | 0 | 0 | 0 | 0 |
| B2-9 | B2 | 0 | 0 | 0 | 0 | 0 |
| B2-10 | B2 | 0 | 0 | 0 | 0 | 0 |
| B3-11 | B3 | 0 | 0 | 0 | 0 | 0 |
| B3-12 | B3 | 0 | 0 | 0 | 0 | 0 |
| B3-13 | B3 | 0 | 0 | 0 | 0 | 0 |
| B3-14 | B3 | 0 | 0 | 0 | 0 | 0 |
| B3-15 | B3 | 0 | 0 | 0 | 0 | 0 |
| C1-1 | C1 | 0 | 0 | 0 | 0 | 0 |
| C1-2 | C1 | 0 | 1 | 0 | 0 | 1 |
| C1-3 | C1 | 1 | 0 | 1 | 1 | 3 |
| C1-4 | C1 | 1 | 2 | 2 | 1 | 6 |
| C1-5 | C1 | 0 | 2 | 2 | 1 | 5 |
| C2-6 | C2 | 0 | 0 | 0 | 0 | 0 |
| C2-7 | C2 | 0 | 0 | 0 | 0 | 0 |
| C2-8 | C2 | 0 | 0 | 0 | 0 | 0 |
| C2-9 | C2 | 0 | 0 | 0 | 0 | 0 |
| C2-10 | C2 | 0 | 0 | 0 | 0 | 0 |
| C3-11 | C3 | 1 | 0 | 0 | 1 | 2 |
| C3-12 | C3 | 0 | 1 | 0 | 0 | 1 |
| C3-13 | C3 | 0 | 0 | 0 | 0 | 0 |
| C3-14 | C3 | 1 | 0 | 0 | 1 | 2 |
| C3-15 | C3 | 0 | 0 | 0 | 0 | 0 |
| D1-1 | D1 | 0 | 0 | 0 | 0 | 0 |
| D1-2 | D1 | 0 | 1 | 0 | 0 | 1 |
| D1-3 | D1 | 1 | 1 | 1 | 1 | 4 |
| D1-4 | D1 | 1 | 0 | 1 | 1 | 3 |
| D1-5 | D1 | 0 | 0 | 0 | 0 | 0 |
| D2-6 | D2 | 0 | 1 | 0 | 0 | 1 |
| D2-7 | D2 | 0 | 0 | 1 | 0 | 1 |
| D2-8 | D2 | 0 | 1 | 0 | 0 | 1 |
| D2-9 | D2 | 0 | 0 | 0 | 0 | 0 |
| D2-10 | D2 | 0 | 0 | 0 | 0 | 0 |
| D3-11 | D3 | 0 | 0 | 0 | 0 | 0 |
| D3-12 | D3 | 0 | 1 | 0 | 0 | 1 |
| D3-13 | D3 | 0 | 0 | 0 | 0 | 0 |
| D3-14 | D3 | 0 | 0 | 0 | 0 | 0 |
| D3-15 | D3 | 0 | 0 | 0 | 0 | 0 |
Liver histological analysis for the following parameters: sinusoidal congestion, vacuolization of hepatocyte cytoplasm, parenchymal necrosis, and inflammation, with the total score representing the sum of all parameters for each individual animal. Each parameter was graded numerically as follows: congestion, vacuolization, and inflammation: 0 = none, 1 = minimal, 2 = mild, 3 = moderate, and 4 = severe. The numerical graduation for necrosis was as follows: 0 = nonnecrotic cells, 1 = single-cell necrosis, 2 ≤ 30% necrosis, 3 ≤ 60% necrosis, and 4 ≥ 60% necrosis. 1The subgroup ID is depicted in Table 1 (experimental design). 2Total score represents the sum of all parameters for each individual animal.
Figure 6.
Histological analysis of the liver. (a, c, and e) Indicating the presence of severe congestion, vacuolization, inflammation, and necrosis in an animal of the CN2 subgroup at the 7th postoperative day (a), which is statistically significantly improved in a transplanted animal of the B2 subgroup (c) and resembles normal liver histology in a transplanted animal of the C2 subgroup (e). (b, d, and f) Indicating the presence of moderate congestion, vacuolization, and inflammation in an animal of the CN3 subgroup at the 15th postoperative day (b), which is statistically significantly improved in a transplanted animal of the B3 subgroup (d) and resembles normal liver histology in a transplanted animal of the D3 subgroup (f).
3.3. ADSC Transplantation Promotes Hepatic Regeneration
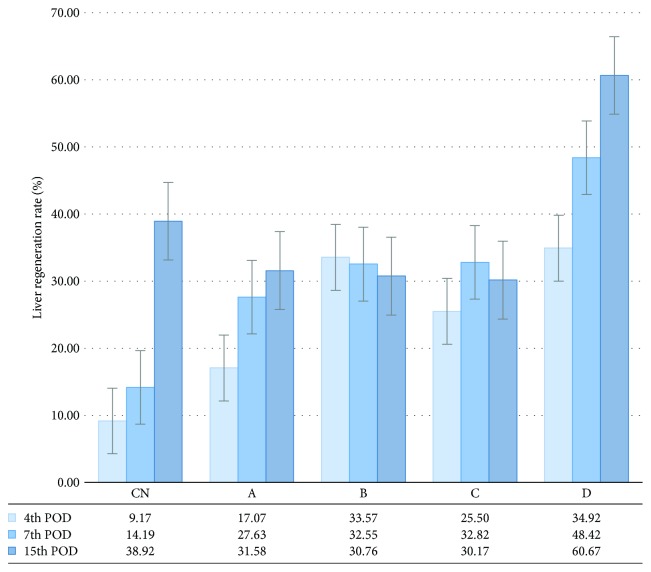
The mean percentage of PHx that all rats were subjected to was 59.7%, without any significant difference between the transplanted and the nontransplanted groups. A significantly greater liver regeneration rate was observed in the subgroups B1 (p = 0.022) and D1 (p = 0.014), as well as in the subgroup D2 (p = 0.021), compared with the respective control subgroups, without any significant differences between the transplanted animals of the same POD. On the 15th POD, no significant differences in the regeneration rate were observed between the transplanted animals, as well as between the transplanted and the respective control animals (Figure 7).
Figure 7.
Liver regeneration rate (%), as calculated on the POD1 of sacrifice, using the equation: 100 × {C − (A − B)}/A, where A is the estimated total liver weight at the time of PHx2, B is the weight of the excised liver, and C is the weight of the harvested regenerating liver at the time of sacrifice. The vertical lines indicate the standard error of the mean (SEM). Liver regeneration rate was not calculated for the sham group, as these animals did not undergo PHx. The experimental groups are depicted in Table 1 (experimental design). 1POD: postoperative day; 2PHx: partial hepatectomy.
3.4. Body Weight as a Nutritional Status Parameter
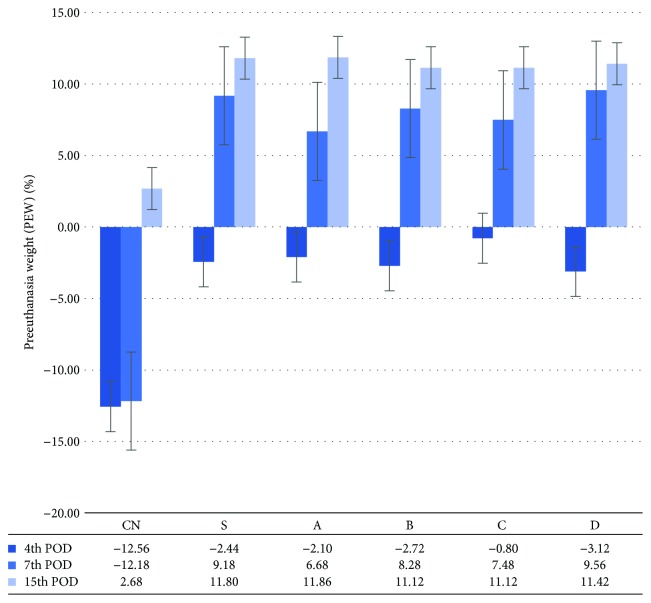
A significantly greater PEW was observed in all the transplanted subgroups of the 4th and 7th POD, compared with the respective control animals, whereas no significant differences were observed between the transplanted animals. On the 15th POD, no significant differences in PEW were observed between the transplanted, as well as between the transplanted and the nontransplanted animals (Figure 8).
Figure 8.
Preeuthanasia weight (%) (PEW (%)), as calculated on the POD1 of sacrifice, using the equation: preeuthanasia weight (PEW) = 100 × (PEW − IW)/IW, where PEW is the weight at the time of sacrifice and IW is the initial weight prior to PHx2. The vertical lines indicate the standard error of the mean (SEM). The experimental groups are depicted in Table 1 (experimental design). 1POD: postoperative day. 2PHx: partial hepatectomy.
3.5. Peripheral Blood Sample Markers of Liver Function
The levels of AST and ALT in the transplanted animals decreased, compared with their control counterparts, without though any significant difference on the 4th and 7th POD. On the 15th POD, a nonsignificant increase in the levels of AST and ALT was recorded, compared with the levels on the 4th and 7th POD (Tables 4 and 5).
Table 4.
Serum values of aspartate aminotransferase, alanine aminotransferase, and number of platelets.
| Aspartate aminotransferase (AST) | Alanine aminotransferase (ALT) | Platelet number (PLT) (x1000) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subgroup ID1 | Number of animals | Mean | Standard deviation | 95% confidence interval for mean | Subgroup ID | Number of animals | Mean | Standard deviation | 95% confidence interval for mean | Subgroup ID | Number of animals | Mean | Standard deviation | 95% confidence interval for mean | |||
| Lower bound | Upper bound | Lower bound | Upper bound | Lower bound | Upper bound | ||||||||||||
| 4th postoperative day of sacrifice | |||||||||||||||||
| CN1 | 5 | 164.80 | 27.004 | 131.27 | 198.33 | CN1 | 5 | 65.00 | 24.197 | 34.96 | 95.04 | CN1 | 5 | 939.40 | 123.253 | 786.36 | 1092.44 |
| S1 | 5 | 106.00 | 31.401 | 67.01 | 144.99 | S1 | 5 | 41.00 | 6.892 | 32.44 | 49.56 | S1 | 5 | 1022.60 | 132.657 | 857.88 | 1187.32 |
| A1 | 5 | 1646.60 | 3457.812 | −2646.84 | 5940.04 | A1 | 5 | 884.80 | 1911.953 | −1489.20 | 3258.80 | A1 | 5 | 808.20 | 221.719 | 532.90 | 1083.50 |
| B1 | 5 | 123.60 | 44.909 | 67.84 | 179.36 | B1 | 5 | 47.80 | 18.404 | 24.95 | 70.65 | B1 | 5 | 1044.20 | 86.757 | 936.48 | 1151.92 |
| C1 | 5 | 125.60 | 24.785 | 94.83 | 156.37 | C1 | 5 | 57.20 | 16.022 | 37.31 | 77.09 | C1 | 5 | 839.00 | 134.030 | 672.58 | 1005.42 |
| D1 | 5 | 154.80 | 70.183 | 67.66 | 241.94 | D1 | 5 | 43.60 | 10.502 | 30.56 | 56.64 | D1 | 5 | 1016.20 | 132.315 | 851.91 | 1180.49 |
| 7th postoperative day of sacrifice | |||||||||||||||||
| CN2 | 5 | 619.80 | 1069.440 | −708.08 | 1947.68 | CN2 | 5 | 83.20 | 54.642 | 15.35 | 151.05 | CN2 | 5 | 948.20 | 141.111 | 772.99 | 1123.41 |
| S2 | 5 | 107.80 | 27.725 | 73.37 | 142.23 | S2 | 5 | 53.00 | 16.263 | 32.81 | 73.19 | S2 | 5 | 1028.20 | 78.004 | 931.34 | 1125.06 |
| A2 | 5 | 146.60 | 40.692 | 96.07 | 197.13 | A2 | 5 | 41.80 | 13.737 | 24.74 | 58.86 | A2 | 5 | 1006.00 | 96.946 | 885.63 | 1126.37 |
| B2 | 5 | 107.60 | 48.588 | 47.27 | 167.93 | B2 | 5 | 53.00 | 22.869 | 24.60 | 81.40 | B2 | 5 | 1044.80 | 226.942 | 763.01 | 1326.59 |
| C2 | 5 | 164.00 | 142.367 | −12.77 | 340.77 | C2 | 5 | 70.60 | 37.038 | 24.61 | 116.59 | C2 | 5 | 854.80 | 237.579 | 559.81 | 1149.79 |
| D2 | 5 | 116.80 | 12.050 | 101.84 | 131.76 | D2 | 5 | 52.80 | 7.396 | 43.62 | 61.98 | D2 | 5 | 1059.40 | 178.106 | 838.25 | 1280.55 |
| 15th postoperative day of sacrifice | |||||||||||||||||
| CN3 | 5 | 127.40 | 49.772 | 65.60 | 189.20 | CN3 | 5 | 45.80 | 14.618 | 27.65 | 63.95 | CN3 | 5 | 675.00 | 249.933 | 364.67 | 985.33 |
| S3 | 5 | 99.00 | 38.216 | 51.55 | 146.45 | S3 | 5 | 42.60 | 7.765 | 32.96 | 52.24 | S3 | 5 | 772.60 | 83.395 | 669.05 | 876.15 |
| A3 | 5 | 99.00 | 24.197 | 68.96 | 129.04 | A3 | 5 | 45.40 | 20.428 | 20.04 | 70.76 | A3 | 5 | 625.60 | 91.172 | 512.40 | 738.80 |
| B3 | 5 | 150.40 | 48.418 | 90.28 | 210.52 | B3 | 5 | 63.20 | 10.569 | 50.08 | 76.32 | B3 | 5 | 909.40 | 47.221 | 850.77 | 968.03 |
| C3 | 5 | 148.38 | 52.407 | 83.31 | 213.45 | C3 | 5 | 44.38 | 6.485 | 36.33 | 52.43 | C3 | 5 | 813.40 | 278.302 | 467.84 | 1158.96 |
| D3 | 5 | 242.60 | 99.354 | 119.24 | 365.96 | D3 | 5 | 60.80 | 20.499 | 35.35 | 86.25 | D3 | 5 | 914.40 | 56.801 | 843.87 | 984.93 |
Demonstrating the mean serum values of aspartate aminotransferase, alanine aminotransferase, and the number of platelets, as well as the standard deviation and the lower and upper bound of the 95% confidence interval for the mean value, in each experimental subgroup. 1The subgroup ID is depicted in Table 1 (experimental design).
Table 5.
AST, ALB, PT, and INR statistical significant differences.
| Aspartate aminotransferase (AST) | Albumin (ALB) | Prothrombin time (PT) | International normalized ratio (INR) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subgroup ID number (I)1 | Subgroup ID number (J)1 | Mean difference (I–J) | Statistical significance (p value2) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) |
| D3 | N | −134.600 | 0.010 | N | B1 | 1.9600 | 0.000 | N | CN1 | −1.79600 | 0.0001 | N | CN1 | −.14600 | 0.0001 |
| S3 | 143.600 | 0.008 | C1 | 1.2000 | 0.020 | A1 | −1.01600 | 0.0001 | A1 | −0.08600 | 0.0001 | ||||
| A3 | 143.600 | 0.008 | CN1 | B1 | 1.3800 | 0.005 | B1 | −0.95600 | 0.0001 | B1 | −0.07600 | 0.0001 | |||
| S1 | B1 | 1.7600 | 0.000 | C1 | −0.97400 | 0.0001 | C1 | −0.07600 | 0.0001 | ||||||
| B1 | D1 | −1.2200 | 0.017 | D1 | −0.91600 | 0.0001 | D1 | −0.07600 | 0.0001 | ||||||
| N | CN2 | 0.5200 | 0.004 | CN1 | S1 | 1.92000 | 0.0001 | CN1 | S1 | 0.15600 | 0.0001 | ||||
| A2 | 0.5000 | 0.007 | A1 | 0.78000 | 0.0001 | A1 | 0.06000 | 0.0001 | |||||||
| C2 | 0.9400 | 0.0001 | B1 | 0.84000 | 0.0001 | B1 | 0.07000 | 0.0001 | |||||||
| D2 | 0.5800 | 0.001 | C1 | 0.82200 | 0.0001 | C1 | 0.07000 | 0.0001 | |||||||
| CN2 | S2 | −0.4400 | 0.028 | D1 | 0.88000 | 0.0001 | D1 | 0.07000 | 0.0001 | ||||||
| C2 | 0.4200 | 0.042 | S1 | A1 | −1.14000 | 0.0001 | S1 | A1 | −0.09600 | 0.0001 | |||||
| S2 | A2 | 0.4200 | 0.042 | B1 | −1.08000 | 0.0001 | B1 | −0.08600 | 0.0001 | ||||||
| C2 | 0.8600 | 0.000 | C1 | −1.09800 | 0.0001 | C1 | −0.08600 | 0.0001 | |||||||
| D2 | 0.5000 | 0.009 | D1 | −1.04000 | 0.0001 | D1 | −0.08600 | 0.0001 | |||||||
| A2 | C2 | 0.4400 | 0.028 | CN2 | N | −1.03400 | 0.0001 | CN2 | N | −0.08400 | 0.0001 | ||||
| B2 | C2 | 0.5800 | 0.002 | S2 | 1.08200 | 0.0001 | S2 | 0.08800 | 0.000 | ||||||
| N | A3 | 1.5400 | 0.016 | A2 | 0.73800 | 0.001 | A2 | 0.06000 | 0.001 | ||||||
| B3 | 3.1400 | 0.0001 | B2 | 0.77800 | 0.001 | B2 | 0.06000 | 0.001 | |||||||
| B3 | CN3 | −2.8400 | 0.000 | C2 | 0.75600 | 0.001 | C2 | 0.06000 | 0.001 | ||||||
| S3 | −3.0200 | 0.000 | D2 | 0.80800 | 0.000 | D2 | 0.07000 | 0.000 | |||||||
| A3 | −1.6000 | 0.019 | CN3 | N | −0.76200 | 0.001 | CN3 | N | −0.06200 | 0.001 | |||||
| C3 | −2.8400 | 0.000 | S3 | 0.81200 | 0.000 | S3 | 0.06600 | 0.000 | |||||||
| D3 | −2.4600 | 0.000 | A3 | 0.71800 | 0.000 | A3 | 0.06000 | 0.000 | |||||||
| B3 | 0.76200 | 0.000 | B3 | 0.06200 | 0.000 | ||||||||||
| C3 | 0.74800 | 0.000 | C3 | 0.06000 | 0.000 | ||||||||||
| D3 | 0.79200 | 0.000 | D3 | 0.06800 | 0.000 | ||||||||||
Statistically significant differences between different experimental subgroups, regarding the serum levels of aspartate aminotransferase, albumin, prothrombin time, and international normalized ratio. 1The subgroup ID is depicted in Table 1 (experimental design). 2The level of statistical significance was set at 5% (α = 0.05).
Significantly improved PT and INR values were demonstrated in the transplanted animals of all POD of sacrifice, compared with their control counterparts, with no significant differences between the transplanted ones (Table 5). The PLT number increased in all animals of the 4th and the 7th POD, compared with the preoperative values, with significant differences specifically identified, without, however, any significant difference between the transplanted and the nontransplanted animals (Table 4). Although the serum levels of ALB were slightly higher in the control group, compared with the majority of the transplanted groups of all POD, significant differences were specifically identified on each POD (Table 5).
The levels of GGT, ALP, TBIL, DBIL, IBIL, and PR were evaluated in blood serum, as markers of function of hepatocytes and cholangiocytes, and the significant differences are depicted in Tables 6 and 7. Although the serum levels of phosphorus in all the transplanted animals of all POD were greater than the preoperative ones, they were still decreased on the 4th and 7th POD, compared with their control counterparts, without though any significant difference.
Table 6.
GGT, ALP, and TBIL statistical significant differences.
| Gamma-glutamyltransferase (GGT) | Alkaline phosphatase (ALP) | Total bilirubin (TBIL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subgroup ID number (I)1 | Subgroup ID number (J)1 | Mean difference (I–J) | Statistical significance (p value2) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) |
| D1 | B1 | 3.720 | 0.007 | B1 | S1 | 140.000 | 0.048 | CN1 | A1 | −0.15800 | 0.012 |
| C1 | 3.200 | 0.032 | D2 | N | 153.800 | 0.014 | A1 | S1 | 0.15800 | 0.012 | |
| D3 | N | 4.800 | 0.003 | C1 | 0.14800 | 0.023 | |||||
| S3 | 4.200 | 0.014 | CN2 | N | 0.10600 | 0.006 | |||||
| A3 | 5.520 | 0.001 | S2 | 0.09200 | 0.024 | ||||||
| B3 | 5.760 | 0.000 | A2 | 0.10000 | 0.010 | ||||||
| C3 | 5.180 | 0.001 | B2 | 0.10800 | 0.004 | ||||||
| C2 | 0.11800 | 0.002 | |||||||||
| D2 | 0.10800 | 0.004 | |||||||||
| CN3 | N | 0.15600 | 0.001 | ||||||||
| S3 | 0.16800 | 0.000 | |||||||||
| A3 | 0.12000 | 0.024 | |||||||||
| C3 | S3 | 0.12200 | 0.021 | ||||||||
Statistically significant differences between different experimental subgroups, regarding the serum levels of Gamma-glutamyltransferase, alkaline phosphatase, and total bilirubin. The level of statistical significance was set at 5% (α = 0.05). 1The subgroup ID is depicted in Table 1 (experimental design).
Table 7.
DBIL, IBIL, and PR statistical significant differences.
| Direct bilirubin (DBIL) | Indirect bilirubin (IBIL) | Total proteins (PR) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) | Subgroup ID number (I) | Subgroup ID number (J) | Mean difference (I–J) | Statistical significance (p value) |
| A1 | N | 0.09400 | 0.014 | C3 | N | 0.10000 | 0.001 | N | A1 | 1.1400 | 0.003 |
| CN1 | 0.09800 | 0.009 | S3 | 0.10200 | 0.000 | B1 | 1.3000 | 0.001 | |||
| S1 | 0.09800 | 0.009 | A3 | 0.09000 | 0.002 | C1 | 1.2800 | 0.001 | |||
| C1 | 0.09800 | 0.009 | B3 | 0.07800 | 0.011 | D1 | 1.0600 | 0.007 | |||
| CN2 | N | 0.07600 | 0.009 | D3 | N | 0.08600 | 0.004 | S1 | B1 | 0.8800 | 0.042 |
| S2 | 0.07000 | 0.021 | S3 | 0.08800 | 0.003 | N | CN2 | 0.9200 | 0.006 | ||
| A2 | 0.06400 | 0.048 | A3 | 0.07600 | 0.014 | A2 | 0.8200 | 0.019 | |||
| B2 | 0.07400 | 0.012 | B2 | 1.0800 | 0.001 | ||||||
| C2 | 0.08400 | 0.003 | C2 | 0.9600 | 0.004 | ||||||
| D2 | 0.08200 | 0.004 | D2 | 1.2000 | 0.0001 | ||||||
| CN3 | N | 0.10800 | 0.0001 | N | CN3 | 1.0000 | 0.004 | ||||
| S3 | 0.11800 | 0.000 | A3 | 1.0400 | 0.002 | ||||||
| A3 | 0.08200 | 0.016 | B3 | CN3 | 1.0600 | 0.002 | |||||
| C3 | 0.09800 | 0.002 | A3 | 1.1000 | 0.001 | ||||||
| D3 | 0.11000 | 0.000 | |||||||||
Statistically significant differences between different experimental subgroups, regarding the serum levels of direct bilirubin, indirect bilirubin, and total proteins. The level of statistical significance was set at 5% (α = 0.05). The subgroup ID is depicted in Table 1 (experimental design).
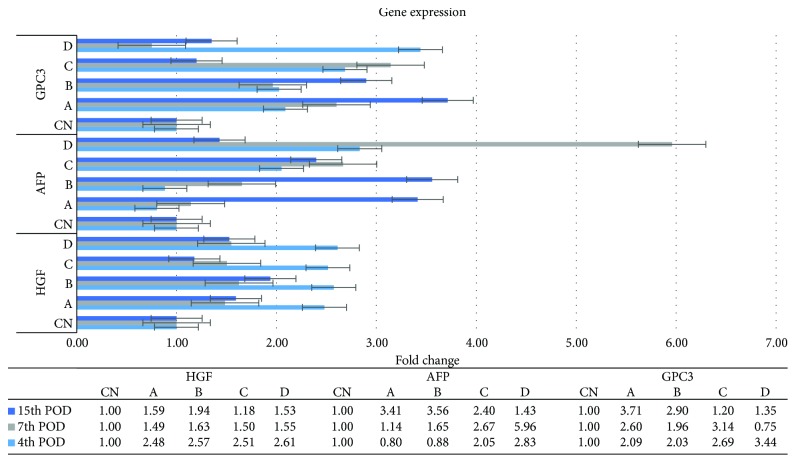
3.6. Expression Levels of Liver Regeneration- and Liver-Specific Genes
All the transplanted animals showed a significantly higher expression ratio of the HGF gene on the 4th and 7th POD of sacrifice, compared with the respective control animals. On the 15th POD, the expression of the HGF gene was nearly identical to that of the 7th POD, without, though, any significant difference (Figure 9).
Figure 9.
HGF, AFP, and GPC3 mRNA expression levels, as determined by reverse transcription quantitative real-time PCR (RT-qPCR), by using the 2−ΔΔCT method. The horizontal lines indicate the standard error of the mean (SEM). The experimental groups are depicted in Table 1 (experimental design).
The expression ratio of the AFP gene was significantly higher in the D1 subgroup, compared with the CN1 (p = 0.04), A1 (p = 0.02), and B1 (p = 0.03) subgroups, as well as in the D2 subgroup, compared with the CN2 subgroup (p = 0.04). A completely different pattern was observed on the 15th POD, with the intraportally transplanted animals demonstrating a significantly higher ratio, compared with the control animals, without other significant differences (Figure 9).
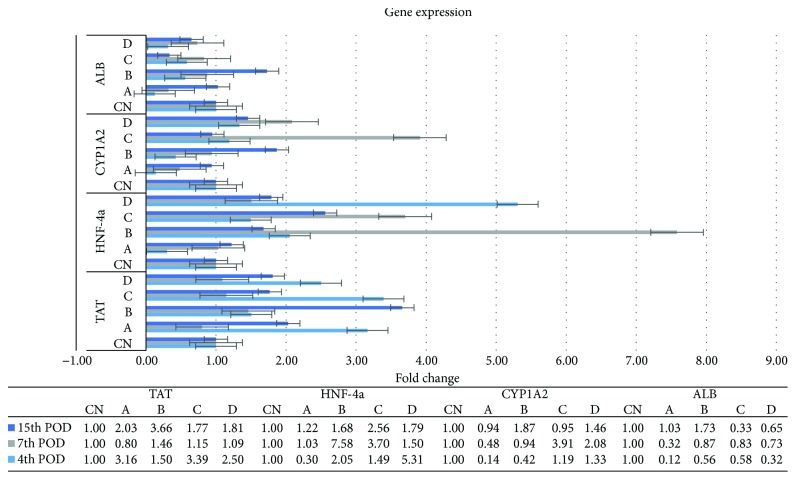
The expression profile of TAT showed two peaks, on the 4th and on the 15th POD. Except for the B1 subgroup, all the transplanted animals of the 4th and 15th POD demonstrated an increased expression of the TAT gene, compared with the respective control subgroups, with significant differences recorded only on the 15th POD (Figure 10).
Figure 10.
TAT, HNF-4a, CYP1A2, and ALB mRNA expression levels, as determined by reverse transcription quantitative real-time PCR (RT-qPCR), by using the 2−ΔΔCT method. The horizontal lines indicate the standard error of the mean (SEM). The experimental groups are depicted in Table 1 (experimental design).
On the 4th POD, the expression of the HNF-4a gene was higher in nearly all the transplanted animals, compared with their control counterparts, with significant differences identified between the D1 and the CN1 (p = 0.04) and A1 (p = 0.02) subgroups. A different pattern was recorded on the 7th and the 15th POD, with the B2, C2, and C3 subgroups showing a higher but not significant ratio, compared with their control counterparts (Figure 10).
Despite the differences in the expression ratio of the CYP1A2 gene in all animals of all POD of sacrifice, no statistically significant differences were demonstrated (Figure 10).
Although the GPC3 gene was highly expressed in nearly all the transplanted animals of all POD, compared with their control counterparts, a significant difference was solely recorded between the A3 and the CN3 subgroup (p = 0.04) (Figure 9).
A significantly lower expression ratio of the ALB gene was demonstrated in all the transplanted animals of the 4th and 7th POD, compared with their control counterparts. On the 15th POD, however, the B3 subgroup showed an increased, but not significant, expression, compared with the control counterparts, with significant differences identified between the B3, and C3 (p = 0.001) and D3 (p = 0.009) subgroups (Figure 10).
4. Discussion
ADSCs are in abundance and may be harvested with the use of minimally invasive procedures [10]. They secrete growth factors and cytokines associated with liver regeneration, such as HGF, vascular endothelial growth factor (VEGF), and interleukin-6 (IL-6) [10–13].
It was initially thought that MSCs' therapeutic potential originated from their pretransplantation hepatic differentiation. On the other hand, undifferentiated MSCs are less receptive to oxidative stress and thus more likely to survive the initial hypoxic phase following transplantation [14]. Moreover, stem cells could act either in a paracrine or in an endocrine fashion, thus affecting adjacent cells by secreting growth factors and cytokines [11, 13]. This attenuation of the promoted liver regeneration process in the transplanted animals of the present study after the 7th POD could be explained by the fact that the liver regeneration process is a well-orchestrated phenomenon, which in rats is completed within 7 days following PHx; thus, no difference in the liver regeneration rate should be expected from the 7th POD and onwards. Our findings are also in accordance with several other reports, which have demonstrated that circulating stem cells mobilized to the injured liver and then they began to proliferate and restored liver histology and function [7, 15–24].
Despite the lack of statistical significance, possibly due to the small number of animals in each experimental subgroup, the decreased levels of AST and ALT in the transplanted animals of the 4th and 7th POD, compared with the respective control subgroups, are further supported in the literature [25, 26]. On the other hand, several factors may be responsible for the increase in the levels of AST and ALT on the 15th POD, including the neovascularization phenomenon, which may lead to an increased outflow of accumulated proteins of hepatocellular damage, such as AST and ALT [26]. This phenomenon may be further triggered by the enhancement of the liver regeneration process in the transplanted animals and necessitates further investigation.
HGF is present in liver matrix and is mainly produced by the stellate cells but also by endothelial cells of the liver [27, 28]. Given its properties as a direct mitogen for hepatocytes as well as its activation early in the liver regeneration process, it is considered as the initiator of the liver regeneration and the most irreplaceable factor of this process [29]. Considering that HGF levels increase as a response to PHx, as well as that active HGF is consumed from the intrahepatic stores in the first 3 hours after PHx, followed by de novo HGF synthesis, our results demonstrate that the transplanted ADSCs promoted the expression of the HGF gene, which in turn lead to an upregulated liver regeneration rate, as also described in other studies [7].
AFP is expressed in hepatic oval cells, as well as in cells differentiating towards the hepatic lineage [30]. In maturing hepatocytes, the expression of AFP gradually declines; thus, AFP is consequently used as a marker of early hepatic differentiation [31]. The significantly increased expression of the AFP gene in the majority of the transplanted animals of all POD of sacrifice in the present study is indicative of an upregulated differentiation process of the regenerating cells, including the ADSCs, towards the hepatic lineage. One step further, differentiated cells still expressed genes of their former early differentiation state, even on the 15th POD, as also described by other studies [8, 32].
Previous reports have demonstrated that an increase in TAT expression levels precedes its increase in activity, manifested as a peak at 8 to 18 hours following PHx [33, 34]. There is evidence that the increase in the enzyme's activity is due to de novo enzyme synthesis [33]. Our results indicate a positive effect of ADSC transplantation on the expression of the TAT gene, throughout the entire follow-up period, which due to the lack of further studies with a follow-up period of more than 7 days following PHx requires further investigation.
HNF-4a was initially thought of as stably expressed in hepatocytes, without any significant changes during liver regeneration; therefore, it was used as a marker of mature hepatocytes [35]. Contrary to that, the results of the present study are in agreement with the data published by other studies [36], demonstrating that HNF-4a expression is determined by the liver regeneration and differentiation process and is significantly upregulated during the intermediate phase of liver regeneration [34].
Cytochrome 1A2 is responsible for the metabolism of drugs and toxic compounds. It has been demonstrated that hepatic progenitor cells do not express many of the P450 isoforms, whereas the increase of their expression coincides with the differentiation process [37, 38]. Our findings are indicative of an upregulated differentiation process of the regenerating cells, including the ADSCs, towards the hepatic lineage in the intraparenchymal transplanted animals, which attenuates after the 7th POD, possibly due to the already complete or near-complete liver regeneration process, as well as restoration of the synthetic and metabolic activity of the regenerated liver.
Several reports have demonstrated that MSCs, either pretreated or not with growth factors, transplanted in animals with liver injury, expressed the ALB gene, which is a gene expressed in mature hepatocytes [39]. In the present study, ALB followed a time-dependent expression pattern, as also demonstrated in other studies [39], in which the lower expression levels in the transplanted animals on the 4th and 7th POD were followed by a slight overexpression of the ALB gene on the 15th POD in the intraportally transplanted animals, compared with the respective control animals. Considering, however, the expression pattern of the ALB gene and the serum levels of ALB, it seems that the higher expression ratio of the gene in the intraportally transplanted animals of the 15th POD is not translated into higher serum levels of ALB. Several changes in the posttranscription level may be responsible for this phenomenon and need further investigation.
Multiple studies have focused on the role of GPC3 in liver regeneration. It has been implicated that GPC3 may be a negative regulator of liver regeneration and hepatocyte proliferation [40]. However, our results indicate that GPC3 expression is upregulated throughout the entire postoperative follow-up period, possibly controlling the liver regeneration process, through its negative regulatory action. However, more light has to be shed on its exact mechanism of action during liver regeneration, which still remains elusive.
The optimal route of transplantation as well as the optimal number of transplanted MSCs is still a matter of debate. Some studies support the systemic transplantation of MSCs, via a peripheral vein, as a better route of transplantation, compared with the PV or the intrahepatic administration [17], while other studies support the PV as the optimal route [15, 41]. In the present study, although the IH administration of ADSCs rendered the best results in promoting the liver regeneration process, compared with the respective control group on the 4th and 7th POD, no other specific significant differences were identified among the transplanted animals, as for the number and route of administration of ADSCs, a finding also supported by several other studies [42, 43].
In the context of the limited time in treating and saving a patient suffering from ALF, the results of the present study are of particular value, when applied to a clinical setting. The cornerstone of our study is the successful transplantation of ADSCs, without a previous in vitro differentiation towards the hepatic lineage, which needs a considerable amount of time to complete, ranging from 2 weeks to several months [15, 18, 21, 44–46]. This results in a fast and definitive enhancement of the liver regeneration process, as well as in an upregulation of the synthetic ability of the liver, without sacrificing any time in the differentiation process, which may be proven clinically valuable.
Although the PHx model in rats is a well-studied area [29], the present study demonstrated a new surgical approach for PHx, by dividing the LLL and ML near the origin of their vasculature using electrocautery, followed by suture ligation and resection, instead of totally resecting these two lobes. This approach allowed us to administer the ADSCs into the liver parenchyma of the nearly totally resected lobes, instead of administering them into the parenchyma of the intact remaining lobes. Moreover, this approach allowed us, at the time of euthanasia, to harvest representative tissue samples from both the nearly totally resected liver lobes, where the histological and regeneration changes are more likely to be more evident, and from the intact lobes. On the other hand, the major limitation of the present study is the fact that it compared the outcomes between experimental groups that were transplanted via specific routes and with specific number of ADSCs in each case, instead of comparing the outcomes between all the available transplantation routes as well as numbers of transplanted ADSCs, which would shed more light on the optimal route and number of transplanted ADSCs. However, this fact might have challenged a complex study, with a huge number of animals and statistical comparisons, as well as abundant human and financial resources required.
5. Conclusion
An in vitro differentiation of the ADSCs towards the hepatic lineage is not a prerequisite for a successful outcome, as the transplanted undifferentiated ADSCs managed to successfully engraft into the liver parenchyma and promote the liver regeneration process. Moreover, they ameliorated the histopathologic damage of the liver and at the same time upregulated the expression of liver regeneration- and liver-specific genes, irrespective of the number and route of transplantation. This promotion of the liver regeneration process, without sacrificing any time in a pretransplantation differentiation process, may be proven valuable in the clinical context, especially in cases of acute LF, and opens new horizons in the treatment of ESLD.
Acknowledgments
This study was funded as a Research Scholarship by the Experimental Research Center, ELPEN, which does not lead to any conflict of interest regarding the publication of this manuscript. The authors would also like to express their sincere thanks to Mrs. A. Zacharioudaki, DVM, Μ. Karamperi, E. Karampela, N. Psychalakis, K. Tsarea, E. Gerakis, and S. Gerakis for their assistance in the implementation of the experiments.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
George C. Zografos and Apostolos E. Papalois contributed equally to this work. Themistoklis Feretis and Ioannis G. Papanikolaou contributed equally to the preparation and editing of the present manuscript.
References
- 1.Bellentani S., Tiribelli C. The spectrum of liver disease in the general population: lesson from the Dionysos study. Journal of Hepatology. 2001;35(4):531–537. doi: 10.1016/S0168-8278(01)00151-9. [DOI] [PubMed] [Google Scholar]
- 2.Mehrabi A., Fonouni H., Müller S. A., Schmidt J. Current concepts in transplant surgery: liver transplantation today. Langenbeck's Archives of Surgery. 2008;393(3):245–260. doi: 10.1007/s00423-007-0262-6. [DOI] [PubMed] [Google Scholar]
- 3.Banas A. Purification of adipose tissue mesenchymal stem cells and differentiation toward hepatic-like cells. Methods in Molecular Biology. 2012;826:61–72. doi: 10.1007/978-1-61779-468-1_6. [DOI] [PubMed] [Google Scholar]
- 4.Saulnier N., Lattanzi W., Puglisi M. A., et al. Mesenchymal stromal cells multipotency and plasticity: induction toward the hepatic lineage. European Review for Medical and Pharmacological Sciences. 2009;13(Supplement 1):71–78. [PubMed] [Google Scholar]
- 5.Martins P. N. A., Neuhaus P. Surgical anatomy of the liver, hepatic vasculature and bile ducts in the rat. Liver International. 2007;27(3):384–392. doi: 10.1111/j.1478-3231.2006.01414.x. [DOI] [PubMed] [Google Scholar]
- 6.Selzner M., Clavien P. A. Failure of regeneration of the steatotic rat liver: disruption at two different levels in the regeneration pathway. Hepatology. 2000;31(1):35–42. doi: 10.1002/hep.510310108. [DOI] [PubMed] [Google Scholar]
- 7.Seki T., Yokoyama Y., Nagasaki H., Kokuryo T., Nagino M. Adipose tissue-derived mesenchymal stem cell transplantation promotes hepatic regeneration after hepatic ischemia-reperfusion and subsequent hepatectomy in rats. The Journal of Surgical Research. 2012;178(1):63–70. doi: 10.1016/j.jss.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Bae S. H., Choi J. Y., Yoon S. K., et al. Thy1-positive bone marrow stem cells express liver-specific genes in vitro and can mature into hepatocytes in vivo. Hepatology International. 2008;2(1):63–71. doi: 10.1007/s12072-007-9031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grozdanov P. N., Yovchev M. I., Dabeva M. D. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Laboratory Investigation. 2006;86(12):1272–1284. doi: 10.1038/labinvest.3700479. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa T., Banas A., Hagiwara K., Iwaguro H., Ochiya T. Stem cells for hepatic regeneration: the role of adipose tissue derived mesenchymal stem cells. Current Stem Cell Research & Therapy. 2010;5(2):182–189. doi: 10.2174/157488810791268636. [DOI] [PubMed] [Google Scholar]
- 11.Banas A., Teratani T., Yamamoto Y., et al. IFATS collection: in vivo therapeutic potential of human adipose tissue mesenchymal stem cells after transplantation into mice with liver injury. Stem Cells. 2008;26(10):2705–2712. doi: 10.1634/stemcells.2008-0034. [DOI] [PubMed] [Google Scholar]
- 12.Kondo K., Shintani S., Shibata R., et al. Implantation of adipose-derived regenerative cells enhances ischemia-induced angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2009;29(1):61–66. doi: 10.1161/ATVBAHA.108.166496. [DOI] [PubMed] [Google Scholar]
- 13.Rehman J., Traktuev D., Li J., et al. Secretion of angiogenic and antiapoptotic factors by human adipose stromal cells. Circulation. 2004;109(10):1292–1298. doi: 10.1161/01.CIR.0000121425.42966.F1. [DOI] [PubMed] [Google Scholar]
- 14.Kuo T. K., Hung S. P., Chuang C. H., et al. Stem cell therapy for liver disease: parameters governing the success of using bone marrow mesenchymal stem cells. Gastroenterology. 2008;134(7):2111–2121.e3. doi: 10.1053/j.gastro.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang L., Ma T., Chen W., et al. Therapeutic potential and related signal pathway of adipose-derived stem cell transplantation for rat liver injury. Hepatology Research. 2009;39(8):822–832. doi: 10.1111/j.1872-034X.2009.00506.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhu X., He B., Zhou X., Ren J. Effects of transplanted bone-marrow-derived mesenchymal stem cells in animal models of acute hepatitis. Cell and Tissue Research. 2013;351(3):477–486. doi: 10.1007/s00441-012-1524-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim S. J., Park K. C., Lee J. U., Kim K. J., Kim D. G. Therapeutic potential of adipose tissue-derived stem cells for liver failure according to the transplantation routes. Journal of the Korean Surgical Society. 2011;81(3):176–186. doi: 10.4174/jkss.2011.81.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banas A., Teratani T., Yamamoto Y., et al. Rapid hepatic fate specification of adipose-derived stem cells and their therapeutic potential for liver failure. Journal of Gastroenterology and Hepatology. 2009;24(1):70–77. doi: 10.1111/j.1440-1746.2008.05496.x. [DOI] [PubMed] [Google Scholar]
- 19.Yin L., Zhu Y., Yang J., et al. Adipose tissue-derived mesenchymal stem cells differentiated into hepatocyte-like cells in vivo and in vitro. Molecular Medicine Reports. 2015;11(3):1722–1732. doi: 10.3892/mmr.2014.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pascual-Miguelañez I., Salinas-Gomez J., Fernandez-Luengas D., et al. Systemic treatment of acute liver failure with adipose derived stem cells. Journal of Investigative Surgery. 2015;28(2):120–126. doi: 10.3109/08941939.2014.987407. [DOI] [PubMed] [Google Scholar]
- 21.Sgodda M., Aurich H., Kleist S., et al. Hepatocyte differentiation of mesenchymal stem cells from rat peritoneal adipose tissue in vitro and in vivo. Experimental Cell Research. 2007;313(13):2875–2886. doi: 10.1016/j.yexcr.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Sun J., Yuan Y., Qin H., et al. Serum from hepatectomized rats induces the differentiation of adipose tissue mesenchymal stem cells into hepatocyte-like cells and upregulates the expression of hepatocyte growth factor and interleukin-6 in vitro. International Journal of Molecular Medicine. 2013;31(3):667–675. doi: 10.3892/ijmm.2013.1257. [DOI] [PubMed] [Google Scholar]
- 23.Liu T., Mu H., Shen Z., Song Z., Chen X., Wang Y. Autologous adipose tissue-derived mesenchymal stem cells are involved in rat liver regeneration following repeat partial hepatectomy. Molecular Medicine Reports. 2016;13(3):2053–2059. doi: 10.3892/mmr.2016.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saidi R. F., Rajeshkumar B., Shariftabrizi A., et al. Human adipose-derived mesenchymal stem cells attenuate liver ischemia-reperfusion injury and promote liver regeneration. Surgery. 2014;156(5):1225–1231. doi: 10.1016/j.surg.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Zhao S., Wang Y., Gao C., et al. Superparamagnetic iron oxide magnetic nanomaterial-labeled bone marrow mesenchymal stem cells for rat liver repair after hepatectomy. The Journal of Surgical Research. 2014;191(2):290–301. doi: 10.1016/j.jss.2014.03.064. [DOI] [PubMed] [Google Scholar]
- 26.Koellensperger E., Niesen W., Kolbenschlag J., Gramley F., Germann G., Leimer U. Human adipose tissue derived stem cells promote liver regeneration in a rat model of toxic injury. Stem Cells International. 2013;2013:10. doi: 10.1155/2013/534263.534263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeCouter J., Moritz D. R., Li B., et al. Angiogenesis-independent endothelial protection of liver: role of VEGFR-1. Science. 2003;299(5608):890–893. doi: 10.1126/science.1079562. [DOI] [PubMed] [Google Scholar]
- 28.Schirmacher P., Geerts A., Jung W., Pietrangelo A., Rogler C. E., Dienes H. P. The role of Ito cells in the biosynthesis of HGF-SF in the liver. EXS. 1993;65:285–299. [PubMed] [Google Scholar]
- 29.Michalopoulos G. K. Liver regeneration. Journal of Cellular Physiology. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kania G., Blyszczuk P., Jochheim A., Ott M., Wobus A. M. Generation of glycogen- and albumin-producing hepatocyte-like cells from embryonic stem cells. Biological Chemistry. 2004;385(10):943–953. doi: 10.1515/BC.2004.123. [DOI] [PubMed] [Google Scholar]
- 31.Fiegel H. C., Kluth J., Lioznov M. V., et al. Hepatic lineages isolated from developing rat liver show different ways of maturation. Biochemical and Biophysical Research Communications. 2003;305(1):46–53. doi: 10.1016/S0006-291X(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 32.Deng X., Chen Y. X., Zhang X., et al. Hepatic stellate cells modulate the differentiation of bone marrow mesenchymal stem cells into hepatocyte-like cells. Journal of Cellular Physiology. 2008;217(1):138–144. doi: 10.1002/jcp.21481. [DOI] [PubMed] [Google Scholar]
- 33.Della Fazia M. A., Servillo G., Viola-Magni M. Different expression of tyrosine aminotransferase and serine deydratase in rat livers after partial hepatectomy. Biochemical and Biophysical Research Communications. 1992;182(2):753–759. doi: 10.1016/0006-291X(92)91796-S. [DOI] [PubMed] [Google Scholar]
- 34.Fukuhara Y., Hirasawa A., Li X. K., et al. Gene expression profile in the regenerating rat liver after partial hepatectomy. Journal of Hepatology. 2003;38(6):784–792. doi: 10.1016/S0168-8278(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 35.Flodby P., Antonson P., Barlow C., Blanck A., Porsch-Hällström I., Xanthopoulos K. G. Differential patterns of expression of three C/EBP isoforms, HNF-1, and HNF-4 after partial hepatectomy in rats. Experimental Cell Research. 1993;208(1):248–256. doi: 10.1006/excr.1993.1244. [DOI] [PubMed] [Google Scholar]
- 36.Xu M., Alwahsh S. M., Ramadori G., Kollmar O., Slotta J. E. Upregulation of hepatic melanocortin 4 receptor during rat liver regeneration. The Journal of Surgical Research. 2016;203(1):222–230. doi: 10.1016/j.jss.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 37.Czekaj P., Bryzek A., Czekaj T. M., et al. Cytochrome P450 mRNA expressions along with in vitro differentiation of hepatocyte precursor cells from fetal, young and old rats. Folia Histochemica et Cytobiologica. 2010;48(1):46–57. doi: 10.2478/v10042-008-0085-5. [DOI] [PubMed] [Google Scholar]
- 38.Ichinohe N., Tanimizu N., Ooe H., et al. Differentiation capacity of hepatic stem/progenitor cells isolated from D-galactosamine-treated rat livers. Hepatology. 2013;57(3):1192–1202. doi: 10.1002/hep.26084. [DOI] [PubMed] [Google Scholar]
- 39.Cho K. A., Ju S. Y., Cho S. J., et al. Mesenchymal stem cells showed the highest potential for the regeneration of injured liver tissue compared with other subpopulations of the bone marrow. Cell Biology International. 2009;33(7):772–7. doi: 10.1016/j.cellbi.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 40.Liu B., Bell A. W., Paranjpe S., et al. Suppression of liver regeneration and hepatocyte proliferation in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2010;52(3):1060–1067. doi: 10.1002/hep.23794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanazawa H., Fujimoto Y., Teratani T., et al. Bone marrow-derived mesenchymal stem cells ameliorate hepatic ischemia reperfusion injury in a rat model. PLoS One. 2011;6(4, article e19195) doi: 10.1371/journal.pone.0019195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng S., Yang J., Yang J., et al. Transplantation of umbilical cord mesenchymal stem cells via different routes in rats with acute liver failure. International Journal of Clinical and Experimental Pathology. 2015;8(12):15854–15862. [PMC free article] [PubMed] [Google Scholar]
- 43.Li D. L., He X. H., Zhang S. A., Fang J., Chen F. S., Fan J. J. Bone marrow-derived mesenchymal stem cells promote hepatic regeneration after partial hepatectomy in rats. Pathobiology. 2013;80(5):228–234. doi: 10.1159/000346796. [DOI] [PubMed] [Google Scholar]
- 44.Banas A., Teratani T., Yamamoto Y., et al. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology. 2007;46(1):219–228. doi: 10.1002/hep.21704. [DOI] [PubMed] [Google Scholar]
- 45.Seo M. J., Suh S. Y., Bae Y. C., Jung J. S. Differentiation of human adipose stromal cells into hepatic lineage in vitro and in vivo. Biochemical and Biophysical Research Communications. 2005;328(1):258–264. doi: 10.1016/j.bbrc.2004.12.158. [DOI] [PubMed] [Google Scholar]
- 46.Sato Y., Araki H., Kato J., et al. Human mesenchymal stem cells xenografted directly to rat liver are differentiated into human hepatocytes without fusion. Blood. 2005;106(2):756–763. doi: 10.1182/blood-2005-02-0572. [DOI] [PubMed] [Google Scholar]