Abstract
GIGGLE is a genomics search engine that identifies and ranks the significance of genomic loci shared between query features and thousands of genome interval files. GIGGLE (https://github.com/ryanlayer/giggle) scales to billions of intervals and is over three orders of magnitude faster than existing methods. Its speed extends the accessibility and utility of resources such as ENCODE, Roadmap Epigenomics, and GTEx by facilitating data integration and hypothesis generation.
The results from genome-wide assays such as ChIP-seq, RNA-seq, and variant calling are often interpreted by comparing experimentally identified genomic loci to other known genomic features such as open chromatin, enhancers, and transcribed regions. Large-scale functional genomics projects have greatly empowered this type of analysis by characterizing the genomic regions associated with a wide range of genomic processes. However, interpretation is complicated by the size of these data set collections, which consist of thousands of results that span hundreds of different tissue types, assays, and biological conditions. Effectively integrating these large, complex, and heterogeneous resources requires the ability to rapidly search the full data set and identify the most statistically relevant features. While existing software such as BEDTOOLS1 and TABIX2 identify regions that are common to genome interval files, these methods were designed to investigate a limited number of files. More recent methods3,4 describe improved statistical measures, yet they do not scale to the vast amount of data that is now available.
We introduce GIGGLE, a fast and highly scalable genomic interval searching strategy that, much like web search engines did for the Internet, provides users with the ability to conduct large-scale comparisons of their results with thousands of reference data sets and genome annotations in seconds. GIGGLE enables the identification of novel and unexpected relationships among local data sets as well as the vast amount of publicly available genomics data. It works through command line and web interfaces, as well as APIs in the C, Go, and Python programming languages.
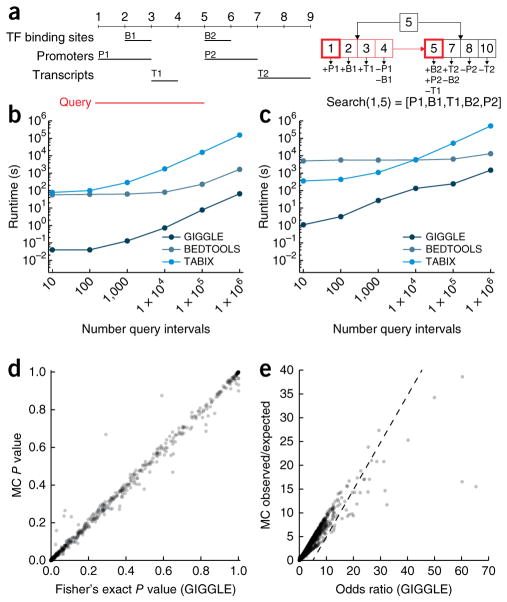
GIGGLE is based on a temporal indexing scheme5 that uses a B+ tree to create a single index of the genome intervals from thousands of annotations and genomic data files (Fig. 1a). Each interval in an indexed file is represented by two keys in the tree that correspond to the interval’s bounds (start and end + 1). Each key in a leaf node contains a list of intervals that either start at a chromosomal position (indicated by a “+”) or have ended (indicated by a “−”) just before that position. We give an example (Fig. 1a) in which position 7 corresponds to a key in the second leaf node with the list [+T2, −B2]. This indicates that at chromosomal position 7, the second interval in the “Transcripts” file (T2) has started, and the second interval in the “TF binding sites” file (B2) has ended. To find the intervals in the index that intersect a query interval (e.g., [1,5] in Fig. 1a), the tree is searched for the query start and end, the keys within that range are scanned, and intervals in the lists of those keys are identified as intersecting the query interval (see Supplementary Fig. 1 and Online Methods for complete algorithmic details).
Figure 1.
Indexing, searching, performance, and score calibration. (a) A set of three genomic intervals files (transcription factor (TF) binding sites, promoters, and transcripts) (left, black) is indexed using a single (simplified) B+ tree (right). Intervals among the annotations overlapping a query interval (left, red) are found by searching the tree for the query start and end (right, boxed red) and scanning the keys between these positions (right, boxed red). (b) Runtimes for GIGGLE, BEDTOOLS, and TABIX considering random query sets with between 10 and 1 million random 100-base-pair intervals against the ChromHMM processing of Roadmap Epigenomics (1,905 files and over 55 million intervals). (c) Runtimes for the same method and queries against UCSC genome browser annotations (5,603 files and over 6.9 billion intervals). While GIGGLE and BEDTOOLS runtimes converge for query sizes exceeding hundreds of millions of intervals, this scenario far exceeds typical query set sizes. (d,e) A comparison between GIGGLE’s relationship estimates using a contingency table and Monte-Carlo-based methods for (d) significance (Fisher’s Exact two-tailed test) and (e) enrichment considering a search of MyoD ChIP-seq peaks (631 intervals) against ChromHMM predictions from Roadmap.
GIGGLE’s potential for high scalability is based on two factors. First, identifying the number of overlaps between a query and any given annotation file is determined entirely within the unified index, thus eliminating the inefficiencies of existing methods, which must instead open and inspect the underlying data files. Second, the B+ tree structure minimizes disk reads; this is vital to performance since databases of this scale will grow beyond the capacity of main memory and must be stored on disk. To measure GIGGLE’s query performance (Supplementary Software), we created an index of the ChromHMM6 annotations curated by the Roadmap Epigenomics Project (Roadmap) from 127 tissues and cell lines. Each genome was segmented into 15 genomic states, yielding over 55 million intervals in the resulting GIGGLE index (2.2 GB index, indexed in 80 s). When testing query performance with a range of 10 to 1,000,000 query intervals, GIGGLE was 2,336× faster than TABIX and 25× faster than BEDTOOLS (Fig. 1b; see Supplementary Data 1 for the data used to create Fig. 1) for the largest comparison. Similarly, using an index of 5,603 annotation files for the human genome (GRCh37, a total of 6.9 billion intervals) from the UCSC Genome browser (554 GB index, indexed in 269 min), GIGGLE was up to 345× faster than TABIX and 8× faster than BEDTOOLS (Fig. 1c).
Speed is essential for searching data of this scale, but, as with internet searches, it is arguably more important to rank results by their relevance to the set of query intervals. Ranking requires a metric that quantifies the degree of similarity between the query intervals and each interval file in the GIGGLE index. Monte Carlo (MC) simulations are commonly used in genomics analyses7,8 to compare the observed number of intersections to a null distribution of intersections obtained by randomly shuffling intervals thousands of times and testing the number of intersections in each trial. While MC simulations are an effective method for pairs of interval sets, they are computationally intractable for large-scale data sets since thousands of permutations are required for each interval file.
GIGGLE eliminates this complexity by estimating the significance and enrichment between the query intervals and each indexed interval file with a Fisher’s Exact two-tailed test and the odds ratio of a 2 × 2 contingency table containing the number of intervals that are in (i) both the query and indexed file, (ii) solely the query file, (iii) solely the indexed file, and (iv) neither the query file nor the indexed file. The first three values are directly computed with a GIGGLE search, and the last value is estimated by the difference between the union of the two sets and the quotient of the mean interval size of both sets and the genome size. These estimates are well correlated with the MC results (Fig. 1d,e) and have the favorable property of near-instant computation.
GIGGLE ranks query results by a composite of the product of −log10(P value) and log2(odds ratio). This ‘GIGGLE score’ avoids some of the issues that arise when using only P values to select top hits9. In MC simulations, the proportion of values that are more extreme than the observation (i.e., the P value) is highly dependent on the variance of the trials. When the variance of the MC distribution is low, observations that are only marginally larger than the expected value may be significant, yet not interesting biologically. For example, one result from a search of MyoD (a muscle differentiation transcription factor) ChIP-seq peaks against Roadmap had a low enrichment (1.7×), but the variance of the MC simulations was also low, making the observation significant (P < 0.001). Similarly, when the MC distribution variance is high, large enrichments may not reach significance. These effects are mitigated by combining significance and enrichment into the GIGGLE score.
While the GIGGLE score can be used to rank results, it is also insightful to use all scores to visualize the full spectrum of relationships between a query set and all indexed interval sets (Fig. 2; see Supplementary Data 2 for the data used to create Fig. 2). For example, a heatmap of GIGGLE scores from a search of MyoD ChIP-seq peaks against the GIGGLE index of Roadmap (Fig. 2a) illustrates MyoD’s important and specific role in muscle tissues10. Similarly, a search of GWAS SNPs associated with Crohn’s disease11 (Fig. 2b) shows that variants cluster in immune cell enhancers. While these results illuminate the dynamics of individual features, the speed of GIGGLE (<0.3 s for Crohn’s SNPs) allows researchers to conduct exploratory research on a massive scale. For example, GIGGLE can quickly (3.5 s) query sets of GWAS SNPs for 39 different traits11 against Roadmap to not only confirm that the enrichment of SNPs in immune cell enhancers is present in other autoimmune diseases and absent in nonautoimmune traits11 (Fig. 2c, left), but also to show that there is no cell-specific pattern in transcribed regions for either set of traits (Fig. 2c, right).
Figure 2.
Visualization of GIGGLE scores from various searches. (a,b) The relationships between 15 genomic states across 127 different cell types and tissues predicted by ChromHMM for Roadmap and (a) MyoD ChIP-seq peaks and (b) genome-wide significant SNPs for Crohn’s disease. Black boxes within panels highlight (a) muscle and (b) immune tissues and cell types. (c) Results from the enhancer and strong transcription tracks from ChromHMM for Roadmap data when considering GWAS SNPs for 21 autoimmune disorder and 18 nonautoimmune traits. The black boxes highlight immune tissues and cell types. (d) The relationships between ESR1 ChiP-seq binding sites from 53 different experiments and the binding sites from 105 other ChIP-seq experiments (38 different unique factors) in MCF-7 cells. Higher GIGGLE scores indicate more enrichment. Black boxes highlight the relationships between ESR1 and FOXA1, GATA3, PR, EP300, and NCAPG. Color lookup tables indicate GIGGLE scores.
We emphasize that GIGGLE is a completely general framework that enables researchers to efficiently explore any collection of interval sets for any species. For example, using a GIGGLE index of all ChIP-seq data sets available from Cistrome12 (5,992 files; 8,716,024 intervals; 521 MB index; indexed in 17 s), we quickly (<3 min) performed a full pair-wise comparison of the 270 factors (734,249 intervals) available for the MCF-7 breast cancer cell line. From this comparison, distinct subsets become clear, including coordinated genomic binding of CTCF, RAD21, and STAG1, indicative of regions involved in long-range interactions13–15, and estrogen receptor α (ER) co-occurrence with other transcription factors (Group 1 and Group 2 in Supplementary Fig. 2, respectively). Focusing specifically on ER (Fig. 2d) uncovers sequence-specific transcription factors known to play important roles in ER genomic binding (FOXA116, GATA317 and PR18) and cofactors that are involved in estrogen-induced gene regulation (EP30019 and NCAPG20). One unexpected finding from this large-scale analysis of MCF-7 ChIP-seq data is the strong co-occurrence of histone variant H2AFX20,21 and ER cofactor GREB122 (Group 3 in Supplementary Fig. 2), suggesting a potential physical interaction between these factors.
GIGGLE also provides the infrastructure to integrate data sources. For example, we developed a web interface that allows users to further investigate interesting results from Roadmap (e.g., MyoD ChIP-seq and Myoblast enhancers) by a querying a GIGGLE index of the UCSC genome browser data (Supplementary Fig. 3). Those results are visualized in the genome browser as a dynamic ‘smartview’, where only the tracks with at least one overlap are visible. Other GIGGLE indices can also be used to verify results. For example, we recapitulated the top hits from the GIGGLE search of both MyoD ChIP-seq peaks and Crohn’s disease GWAS SNPs against Roadmap with similar searches against an index of the FANTOM5 data23 (1,825 files; 11,284,790 intervals) (Supplementary Tables 1–4). This is especially promising since FANTOM5 and Roadmap are based on fundamentally different assays and therefore provide orthogonal corroboration of these biological relationships. These examples illustrate GIGGLE’s ability to confirm previously characterized associations and demonstrate the discovery potential afforded by GIGGLE’s rapid, prioritized searches.
The exploratory power of a single interface from which many data sets can be searched has the possibility to dramatically advance large-scale, integrative analyses. GIGGLE is capable of powering a single access point that will inform researchers and clinicians of all known experiments and curated annotations that are associated with a particular genomic region. In summary, GIGGLE provides a new engine with which to conduct large-scale, in silico ‘screens’ of multidimensional genomics data sets in search of insights into genome biology in diverse experimental contexts.
ONLINE METHODS
The GIGGLE index
The GIGGLE index is based on a previously described5 temporal indexing method and consists of a set of B+ Trees, one for each chromosome, represented among the database interval files. A B+ Tree is a generalization of a binary tree where each node can have multiple keys, internal nodes contain only keys and facilitate tree searching, leaf nodes contain the key-value pairs, and adjacent leaf nodes are linked. Each key in an internal node is linked to two child nodes. The ‘left’ link points to a node with keys that are less than the current key, and the ‘right’ link points to a node with keys that are greater than or equal to the current key. In the GIGGLE index, keys represent chromosomal positions, and the values associated with keys are lists of intervals that either start at that position (indicated by a “+” in Fig. 1a and Supplementary Fig. 1) or have ended just before that position (indicated by a “−” in Fig. 1a and Supplementary Fig. 1). Leaf nodes also contain a ‘leading’ key (“L” in Supplementary Fig. 1, omitted from Fig. 1a) that stores intervals that start before the first key in the leaf, but have not ended by the last key in the prior leaf node (e.g., interval T1 in Supplementary Fig. 1). While the leading values contain redundant information and require extra storage, they improve performance by preventing queries from having to load and scan other leaf nodes for intervals that start in some earlier leaf node (e.g., interval T1).
Bulk indexing
To improve indexing efficiency, GIGGLE performs ‘bulk’ indexing across many presorted interval files (see Supplementary Fig. 1a). In bulk indexing, a priority queue is used to select the interval with the next lowest start position among the full set of files. The queue is loaded with one (the first) interval from each file; and, as intervals are removed from the queue, the next interval from the corresponding file is added to the queue. For example, in Supplementary Figure 1b, step 1 considers interval P1 from the Promoters file, and step 2 considers interval B1 from the TF binding sites file. After P1 is considered, the next interval in the Promoters file (P2) is added to the queue. Similarly, in step 2, B2 from the TF binding sites file is added after B1 is considered.
Each interval is inserted into both the B+ Tree based on its start position and an auxiliary priority queue that is keyed by the end coordinate (plus one). This priority queue is used to add intervals to the leading values and insert end positions into the B+ Tree. If the start position of the current interval has been previously observed, then the interval start is added to the list of the existing value (intervals B2, P2, and T2 in steps 4, 5, and 6 in Supplementary Fig. 1b). Otherwise, a new key is added to the current leaf for the start position (assuming the current leaf has not reached its maximum number of keys, which is set to 100 by default), and the interval start (“+”) is added to the new list that is associated with that key (intervals P1, B1, and T1 in steps 1, 2, and 3 in Supplementary Fig. 1b). Before a key is added for a new start position, all intervals with end values less than or equal to the start value are removed from the priority queue, and the interval ends (“−”) are either added to the lists of existing keys (interval B1 in step 4 in Supplementary Fig. 1b), or new keys are created (intervals P1, T1, B2, P2, and T2 in steps 4 and 6 in Supplementary Fig. 1b). If at any point the current node becomes full, then a new leaf node is created, all intervals in the priority queue are added to the leading key value of the new node, and the new key is added to the new node (step 4 in in Supplementary Fig. 1b). Once all files have been processed and leaf node construction is complete, internal nodes are added by promoting the first key in each leaf node (other than the leftmost node) to a parent node (step 7 in Supplementary Fig. 1b). This process continues one level at a time until there is only one parent node.
Searching the GIGGLE index
A B+ Tree search starts at the root node, proceeds down internal nodes, and terminates at a key in a leaf node. At each node in the search path, an internal search is performed among the keys in the node. While the current node is not a leaf node, the result of that search determines the next node in the path. If the matching key is less than or equal to the query, the path will follow the key’s ‘right’ link, otherwise it will follow the key’s ‘left’ link. When the keys of a leaf node have been searched, the leaf and matching key are returned.
For a given query interval, GIGGLE performs a specialized range query to find overlapping intervals (the intersecting set) across all indexed files. First, the B+ Tree is searched for the query interval’s start and end values, which gives the start leaf node and start key and the end leaf node and end key, respectively (step 1 in Supplementary Fig. 1c). Next, the intervals in the leading value of the start leaf node are added to the intersecting set (step 2 in Supplementary Fig. 1c). Then the keys in the leaf node are scanned from the first value up to and including the start key. At each key, starting intervals (“+”) are added to the intersecting set, and ending intervals (“−”) are removed (steps 3 and 4 in Supplementary Fig. 1c). Last, the remaining keys up to and including the end key are scanned, and the starting intervals are added at each key (steps 5 and 6 in Supplementary Fig. 1c). If the start key does not equal the end key, then the search will use the links between leaf nodes.
The main advantage of a GIGGLE index is in minimizing disk accesses. This benefit is most apparent when the database contains thousands of files, and query intervals overlap only a small fraction of the database. Both BEDTOOLS and TABIX are efficient algorithms that may be faster than GIGGLE when considering small databases. BEDTOOLS does not take advantage of an index and must, in general, perform a full scan of both the query file and the database file. In cases where the query intervals intersect only a small proportion of the database intervals (the most common use case), BEDTOOLS must read and parse most of the database. In contrast, GIGGLE only considers the intervals that either intersect or are immediately adjacent to the query intervals. In the unlikely case where nearly every database interval intersects a query interval, BEDTOOLS may be more efficient than GIGGLE because it does not have the overhead of the index. TABIX uses an index that is based on an R-Tree but has fixed bin sizes. Like GIGGLE, TABIX uses the index only to consider the database intervals near queries. However, TABIX is less efficient for two reasons. First, TABIX is optimized for small index files, and most queries require opening, decompressing, and parsing the source data files. Second, TABIX creates one index for each source data file. When taken together, TABIX is less efficient than GIGGLE because it must perform disk accesses on both the index and the source data file.
Several search options are available to increase GIGGLE’s usability. First, a ‘verbose’ mode (−v) prints all overlapping intervals along with the source file so that users can filter results. Second, a ‘per query’ mode (−o) lists each query interval followed the reference hits so that users can recover specific hits. Third, searches can consider only a subset of reference files (−f) by providing a comma-separated list of regular expressions. Results are giving for only those files with names that match one of those expressions. Specific examples of each of these options are given at https://github.com/ryanlayer/giggle/blob/master/README.md#example.
Data format and sorting requirements
For indexing, GIGGLE supports VCF files (https://samtools.github.io/hts-specs/VCFv4.3.pdf) and BED files (https://genome.ucsc.edu/FAQ/FAQformat.html#format1) that have been sorted and bgzipped (https://github.com/samtools/htslib). Sorting is ascending lexicographical by chromosome, then ascending numerically by start and then by end. For searching, GIGGLE supports bgzipped VCF and BED files. These need not be sorted, but sorted files are likely to perform better because of cache performance. We provide a script in the GIGGLE repository (https://github.com/ryanlayer/giggle/blob/master/scripts/sort_bed) that can sort and bgzip full directories using multiple processors. Otherwise, the following command can be used on individual, uncompressed BED files:
LC_ALL = C sort–buffer-size 2G -k1,1 -k2,2n -k3,3n track.bed | bgzip -c > track.bed.gz
Data sources
CHROMHMM, roadmap epigenomics data source
Tissue-based annotations were downloaded from:
These files were subsequently split and renamed into tissue/ state-based files (e.g., Spleen/Enhancers). Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/examples/rme/README.md
UCSC Genome Browser data source
The full set of hg19 annotations was downloaded from: http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database. The files with identifiable chromosome, start, and end values are converted to BED files. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/examples/ucsc/README.md
MyoD ChIP-seq data source
ChIP-seq peaks from GSM1218850 are downloaded from:
ftp://ftp.ncbi.nlm.nih.gov/geo/samples/GSM1218nnn/GSM1218850/suppl/GSM1218850_MB135DMMD.peak.txt.gz
Peaks with a q-value greater than or equal to 100 are retained; detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/examples/myod/README.md
GWAS variants for 39 autoimmune and non-autoimmune traits data source
A spreadsheet with a list of traits, chromosome, start, end, and other fields was downloaded from:
https://www.nature.com/nature/journal/v518/n7539/extref/nature13835-s1.xls
Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/examples/gwas/README.md
Fantom5 data source
The enhance expression matrix and associated metadata was downloaded from:
http://fantom.gsc.riken.jp/5/datafiles/latest/extra/Enhancers/Human.sample_name2library_id.txt
Values were extracted from the matrix and placed in tissue-specific files. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/examples/fantom/README.md
Cistrome data source
Reanalyzed ChIP-seq narrow peaks from raw GEO data was downloaded from:
http://cistrome.org/db/interface.html
With the following fields selected: Human_TF, Human_histone, Human_chromatin_accessibility, Human_other. Only peaks with a q-value greater than 100 were retained. For Figure 2d, all files had to pass two Cistrome CQ metrics: fraction of reads in peaks, and at last 500 peaks had to have ten-fold enrichment. We also removed files with less than 100 peaks with a q-value greater than or equal to 100. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/examples/cis-trome/README.md
Experiments
Speed tests
Runtimes were for counting the number of intersections between a query set and a database set for GIGGLE (https://github.com/ryanlayer/giggle), BEDTOOLS (https://github.com/arq5x/bedtools2), and TABIX (https://github.com/samtools/htslib). The query sets had between 10 and 1 million 100 base pair intervals, and the databases were the ChromHMM predictions from Roadmap Epigenomics and the hg19 annotations from the UCSC genome browser. All tests were performed using a single core on the 2. 4 GHz Intel Xeon processor (E5-2680 v4) with 25 MB of cache and a 510 MB/s read 485 MB/s write SSD drive (SM863a). Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/experiments/speed_test/README.md
Relationship comparison
Two pairs of methods for quantifying the relationship between a query interval set and a database interval set were compared: the Fisher’s Exact two-tail test of a 2 × 2 contingency table versus a Monte Carlo base P value and the odds ratio of a 2 × 2 contingency table versus a Monte Carlo base enrichment. The query set was the GWAS variants associated with Crohn’s disease, and the database was the ChromHMM predictions from Roadmap Epigenomics. Monte Carlo simulations were performed using BITS (https://github.com/arq5x/bits), a simulation was performed for the intersection of the GWAS variants and each tissue/genomic state interval set, and each simulation consisted of 1,000 rounds. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/experiments/mc_vs_table/README.md
MyoD heat map
The GIGGLE scores for MyoD ChIP-seq peaks searched against the ChromHMM predictions from Roadmap Epigenomics. Only the peaks with a q-value greater than 100 were used. The cell line and tissue names are in Supplementary File 1. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/experiments/chipseq/README.md
Crohn’s disease heat map and autoimmune/nonautoimmune heat map
The GIGGLE scores for the sets of GWAS variants associated with Crohn’s disease and other traits were searched against the ChromHMM predictions from Roadmap Epigenomics. For Figure 2c, the left two columns correspond to the scores from the Enhancer state from ChromHMM for each tissue, and the right two columns correspond to the scores from the Strong Transcription state. Within these major columns, the left minor column corresponds to the autoimmune disorders, and the right minor column to the nonautoimmune traits. The categorization of these traits was retained from Farh et al.11. The cell line and tissue names are in Supplementary Data 3. The autoimmune disorders and the non-autoimmune traits are listed in Supplementary Data 3. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/experiments/gwas/README.md
Cistrome ER
The GIGGLE scores for the ChIP-seq peak files of ERS1 from the MCF-7 cells line that passed quality control (described above) were searched against all other MCF-7 cell line results that also passed quality control. Only the peaks with a q-value greater than 100 were used. The full set of accession numbers is Supplementary Data 3. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/experiments/cistrome/README.md
Cistrome MCF-7
The GIGGLE scores for all ChIP-seq peak files from the MCF-7 cell line that passed quality control were searched against themselves. Only the peaks with a q-value greater than 100 were used. The full set of accession numbers is Supplementary Data 3. Detailed methods underlying this process can be found at:
https://github.com/ryanlayer/giggle/blob/master/experiments/cistrome/README.md
GIGGLE command line and programming interfaces
Command line interface
Indexing:
giggle index \ -i “intervals/*.bed.gz” \ -o interval_index -s
Searching:
giggle search \ -i interval_index -r chr1:1000000-2000000 giggle search \ -i interval_index -q query.bed.gz
C interface
Indexing:
#include “giggle_index.h”
int main(int argc, char **argv) {
uint64_t num_intervals =
giggle_bulk_insert(
“intervals/*.bed.gz”,
“interval_index”,
1);
return 0;
}
Searching:
#include”giggle_index.h”
int main(int argc, char **argv) {
struct giggle_index *gi =
giggle_load(
“interval_index”,
block_store_giggle_set_data_handler);
struct giggle_query_result *gqr =
giggle_query(
gi,”chr1”,1000000,2000000,NULL);
uint32_t i;
for(i = 0; i < gqr->num_files; i++) {
struct file_data *fd =
file_index_get(gi->file_idx, i);
if (giggle_get_query_len(gqr, i) > 0)) {
char *result;
struct giggle_query_iter *gqi =
giggle_get_query_itr(gqr, i);
while (giggle_query_next(gqi,
&result) == 0)
printf(“%s\t%s\n”,
result,
fd->file_name);
giggle_iter_destroy(&gqi);
}
}
giggle_query_result_destroy(&gqr);
giggle_index_destroy(&gi);
return 0;
}
Python interface: https://github.com/brentp/python-giggle
Indexing:
from giggle import Giggle
index = Giggle.create(‘interval_index’,
‘intervals/*.bed.gz’)
Searching:
from giggle import Giggle index = Giggle(‘interval_index’) print(index.files) result = index.query(‘chr1’, 9999, 20000) print(result.n_files) print(result.n_total_hits) print(result.n_hits(0)) for hit in result[0]: print(hit) # hit is a string
Go interface: https://github.com/brentp/go-giggle
Indexing:
import (
giggle “github.com/brentp/go-giggle”
“fmt”
)
func main() {
index := giggle.New(“interval_indexr”,
“intervals/*.bed.gz”)
}
Searching:
import (
giggle “github.com/brentp/go-giggle”
“fmt”
)
func main() {
index:= giggle.Open(“interval_index”)
res:= index.Query(“1”, 565657, 567999)
// all files in the index
index.Files()
// int showing total count
res.TotalHits()
// []uint32 giving number of hits for each file
res.Hits()
var lines []string
# access results by index of file.
lines = res.Of(0)
fmt.Println(strings.Join(lines, “\n”))
lines = res.Of(1)
}
Code availability
All source code is available at https://github.com/ryanlayer/giggle.
Life Sciences Reporting Summary
Further information regarding the experimental design may be found in the Life Sciences Reporting Summary.
Data availability
URLs for Roadmap Epigenomics, the UCSC Genome browser, and Fantom5 indices and a hosted interactive heatmap are available at https://github.com/ryanlayer/giggle/blob/master/README.md#hosted-data-and-services.
Supplementary Material
Supplementary Figure 1. GIGGLE indexing process.
(a) Three example annotation sets shown graphically (left) and encoded in files (right) by start position, end position, and ID. (b) GIGGLE’s bulk indexing process. (c) The GIGGLE interval search process.
Supplementary Figure 2. The GIGGLE scores for all pairwise combinations of the ChIP-seq datasets for the MCF-7 cell line.
Group 1 highlights the relationship between CTCF, RAD21, and STAG1. Group 2 highlights ERS1, FOXA1, GATA3, and EPS300. Group 3 shows an unexpected relationship between H2AFX and GREB1.
Supplementary Figure 3. A web interface that integrates data from of Roadmap and the UCSC genome browser.
(a) Users specify either a single interval or file to upload as the query, and the server responds with the GIGGLE results from an index in a heatmap. In this case the index is of CHROMHMM prediction from Roadmap. The color of each cell indicates the GIGGLE score, and users can click on a cell (e.g., Myoblast enhancers, marked in red) for more information. (b) When a cell is selected by the user, a window opens that contains the list of intervals in that particular Roadmap cell type/genome state annotation that overlap the query. Each interval is a link that can be followed (e.g., chr1:33642000-33642800, marked in red) for more information. (c) When an interval is selected, that interval becomes a query to a GIGGLE index of the UCSC genome browser tracks. The result gives the set of tracks that contain an interval that overlaps the query, and the web interface opens a window with a “smartview” where only those tracks with overlaps are displayed.
Supplementary Table 1. Top GIGGLE results for MyoD ChIP-seq peaks (631 intervals) against the CHROMHMM Roadmap data set (1,905 files and 55,605,005 intervals)
Supplementary Table 2. Top GIGGLE results for MyoD ChIP-seq peaks (631 intervals) against the FANTOM5 data set (1,825 files and 11,284,790 intervals)
Supplementary Table 3. Top GIGGLE results for Crohn’s disease GWAS SNPs (84) against the CHROMHMM Roadmap data set (1,905 files and 55,605,005 intervals)
Supplementary Table 4. Top GIGGLE results for Crohn’s disease GWAS SNPs (84) against the FANTOM5 data set (1,825 files and 11,284,790 intervals)
Acknowledgments
We are grateful to the anonymous reviewers for their suggestions and comments. This research was funded by US National Institutes of Health awards to R.M.L. (K99HG009532) and A.R.Q. (R01HG006693, R01GM124355, U24CA209999).
Footnotes
AUTHOR CONTRIBUTIONS
R.M.L. conceived and designed the study, developed GIGGLE, and wrote the manuscript. B.S.P. developed the GIGGLE score and the PYTHON and GO APIs. T.D. developed the web interface. G.T.M. provided input in the development of the web interface. J.G. conceived and designed the ChIP-seq experiment. A.R.Q. conceived and designed the study and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheffield NC, Bock C. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Favorov A, et al. PLOS Comput Biol. 2012;8:e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elmasri R, Wuu GTJ, Kim Y-J. The time index: an access structure for temporal data. In: McLeod D, Sacks-Davis R, Schek H-J, editors. Proceedings of the 16th International Conference on Very Large Data Bases (VLDB ‘90) Morgan Kaufmann; San Francisco, California, USA: 1990. pp. 1–12. [Google Scholar]
- 6.Ernst J, Kellis M. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Layer RM, Skadron K, Robins G, Hall IM, Quinlan AR. Bioinformatics. 2013;29:1–7. doi: 10.1093/bioinformatics/bts652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De S, Pedersen BS, Kechris K. Brief Bioinform. 2014;15:919–928. doi: 10.1093/bib/bbt053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Y, et al. Bioinformatics. 2014;30:801–807. doi: 10.1093/bioinformatics/btr671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacQuarrie KL, et al. Mol Cell Biol. 2013;33:773–784. doi: 10.1128/MCB.00916-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farh KKH, et al. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mei S, et al. Nucleic Acids Res. 2017;45:D658–D662. doi: 10.1093/nar/gkw983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Splinter E, et al. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nativio R, et al. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, et al. PLoS Genet. 2016;12:e1005992. doi: 10.1371/journal.pgen.1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll JS, et al. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Theodorou V, Stark R, Menon S, Carroll JS. Genome Res. 2013;23:12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammed H, et al. Nature. 2015;523:313–317. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanstein B, et al. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, et al. Mol Cell. 2015;59:188–202. doi: 10.1016/j.molcel.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Periyasamy M, et al. Cell Rep. 2015;13:108–121. doi: 10.1016/j.celrep.2015.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed H, et al. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lizio M, et al. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. GIGGLE indexing process.
(a) Three example annotation sets shown graphically (left) and encoded in files (right) by start position, end position, and ID. (b) GIGGLE’s bulk indexing process. (c) The GIGGLE interval search process.
Supplementary Figure 2. The GIGGLE scores for all pairwise combinations of the ChIP-seq datasets for the MCF-7 cell line.
Group 1 highlights the relationship between CTCF, RAD21, and STAG1. Group 2 highlights ERS1, FOXA1, GATA3, and EPS300. Group 3 shows an unexpected relationship between H2AFX and GREB1.
Supplementary Figure 3. A web interface that integrates data from of Roadmap and the UCSC genome browser.
(a) Users specify either a single interval or file to upload as the query, and the server responds with the GIGGLE results from an index in a heatmap. In this case the index is of CHROMHMM prediction from Roadmap. The color of each cell indicates the GIGGLE score, and users can click on a cell (e.g., Myoblast enhancers, marked in red) for more information. (b) When a cell is selected by the user, a window opens that contains the list of intervals in that particular Roadmap cell type/genome state annotation that overlap the query. Each interval is a link that can be followed (e.g., chr1:33642000-33642800, marked in red) for more information. (c) When an interval is selected, that interval becomes a query to a GIGGLE index of the UCSC genome browser tracks. The result gives the set of tracks that contain an interval that overlaps the query, and the web interface opens a window with a “smartview” where only those tracks with overlaps are displayed.
Supplementary Table 1. Top GIGGLE results for MyoD ChIP-seq peaks (631 intervals) against the CHROMHMM Roadmap data set (1,905 files and 55,605,005 intervals)
Supplementary Table 2. Top GIGGLE results for MyoD ChIP-seq peaks (631 intervals) against the FANTOM5 data set (1,825 files and 11,284,790 intervals)
Supplementary Table 3. Top GIGGLE results for Crohn’s disease GWAS SNPs (84) against the CHROMHMM Roadmap data set (1,905 files and 55,605,005 intervals)
Supplementary Table 4. Top GIGGLE results for Crohn’s disease GWAS SNPs (84) against the FANTOM5 data set (1,825 files and 11,284,790 intervals)
Data Availability Statement
URLs for Roadmap Epigenomics, the UCSC Genome browser, and Fantom5 indices and a hosted interactive heatmap are available at https://github.com/ryanlayer/giggle/blob/master/README.md#hosted-data-and-services.