Summary
Efficient retrograde access to projection neurons for the delivery of sensors and effectors constitutes an important and enabling capability for neural circuit dissection. Such an approach would also be useful for gene therapy, including the treatment of neurodegenerative disorders characterized by pathological spread through functionally connected and highly distributed networks. Viral vectors, in particular, are powerful gene delivery vehicles for the nervous system, but all available tools suffer from inefficient retrograde transport or limited clinical potential. To address this need, we applied in vivo directed evolution to engineer potent retrograde functionality into the capsid of adeno-associated virus (AAV) — a vector that has shown promise in neuroscience research and the clinic. A newly evolved variant, rAAV2-retro, permits robust retrograde access to projection neurons with efficiency comparable to classical synthetic retrograde tracers, and enables sufficient sensor/effector expression for functional circuit interrogation and in vivo genome editing in targeted neuronal populations.
Introduction
Brain functions such as perception, cognition, and the control of movement depend on the coordinated action of large-scale neuronal networks, local circuit modules linked together by long-range connections (Miller and Vogt, 1984; Tomioka et al., 2005; Ramón y Cajol, 1911). Such long-range connections are formed by projection neurons that often comprise several intermingled classes, each projecting to a distinct constellation of downstream targets. Projection neurons have also been implicated in the spread of several neurodegenerative diseases from spatially localized sites of onset (Guo and Lee, 2014; Seeley et al., 2009). Thus, the ability to selectively target specific classes of projection neurons for transgene delivery (e.g. for activity monitoring or manipulation, or genome editing for targeted gene knock-outs or repair of pathological mutations) will be important both for gaining insights into how large-scale networks contribute to brain function and, in the long-run, for therapeutic intervention in neurodegenerative diseases.
Viral vectors constitute an important class of tools for introducing transgenes into specific neuronal populations and are by far the best option for genetic access to target projection neurons through entry at axonal terminals and retrograde transport of their payload to the cell nuclei. A number of naturally evolved neurotropic viruses exhibit retrograde spread as part of their lifecycle, including rabies (Baer et al., 1965; Callaway and Luo, 2015; Wickersham et al., 2007), poliovirus (Ohka et al., 1998) and herpes simplex virus (HSV) (Ugolini et al., 1987) among others. Of these, rabies virus is particularly neuro-invasive (Coulon et al., 1989) and quickly propagates through the nervous system through trans-cellular transfer (Kelly and Strick, 2000). However, its potential for both biological investigation and gene therapy is hampered by excessive virulence (Schnell et al., 2010), though progress is being made to reduce its toxicity (Reardon et al., 2016). In addition to naturally neurotropic strains, many other viruses can infect neurons when administered directly to the nervous system, with “pseudorabies” (SuHV1, actually a herpesvirus), adenoviruses and lentiviruses used most commonly in animal research. Canine adenovirus-2 (CAV-2) displays the best infectivity and retrograde transport of this class of viruses (Soudais et al., 2001) and has increasingly become the reagent of choice for accessing projection neurons (Junyent and Kremer, 2015). However, CAV-2 permits only modest levels of transgene expression, displays some toxicity (Piersanti et al., 2013; Simão et al., 2015) and is currently not easily compatible with scalable, robust production for the generation of clinical-grade or even large animal studies (Kremer et al., 2000). Thus, the development of a non-toxic, readily manufactured viral vector that affords flexible packaging of different transgenes, is robustly internalized and retrogradely transported by axons, and supports long-term, high-level payload expression remains a pressing need.
Recombinant adeno-associated viruses (rAAVs) have emerged as an effective platform for in vivo gene therapy, as they mediate high-level transgene expression, are non-toxic, and evoke minimal immune responses (Kaplitt et al., 2007). These properties were at the core of the decision to grant the first full regulatory approval of any gene therapy treatment to rAAV-mediated restoration of lipoprotein lipase deficiency (Gaudet et al., 2010). rAAVs hold great promise in clinical trials for a range of neurological disorders (Ojala et al., 2014), and they constitute some of the most widespread vectors in neuroscience research (Murlidharan et al., 2014). Since the original discovery that AAV can undergo retrograde transport (Kaspar et al., 2002), rAAVs have afforded some degree of retrograde access to projection neurons in select circuits (Castle et al., 2014; Hollis Ii et al., 2008; Kaspar et al., 2002; Kaspar et al., 2003; Rothermel et al., 2013; Taymans et al., 2007; Towne et al., 2010), but their natural propensity for retrograde transport is low, hampering efforts to address the role of projection neurons in circuit computations or disease progression. Here we describe the in vivo directed evolution (Kotterman and Schaffer, 2014) of a new rAAV variant (rAAV2-retro) that – in addition to its regular ability to infect neuronal cell bodies at the site of exposure – is robustly internalized by axons and mediates retrograde access to projection neurons with efficiency comparable to classical retrograde labeling reagents such as synthetic dyes. The rAAV2-retro gene delivery system can be used on its own or in conjunction with Cre recombinase driver lines to achieve long-term, high-level transgene expression that is sufficient for effective functional interrogation of neural circuit function, as well as for genome editing in targeted neuronal populations.
Results
Directed evolution of rAAV2-retro
To engineer novel rAAVs with enhanced retrograde transport, we designed an in vivo directed-evolution approach to enrich for rAAV capsid variants that were efficiently transported to cell bodies of neurons sending long-range projections to the site of virus injection in the mouse brain (Figure 1A and Experimental Procedures). To maximize the likelihood of recovering a variant with the desired properties, we started with a diverse mixture of previously described libraries of rAAV cap variants (Koerber et al., 2008; Koerber et al., 2009; Koerber et al., 2006; Müller et al., 2003). Viral particles were packaged in a way that linked each capsid variant to the AAV genome containing the corresponding cap gene, and the final pool of capsid variants included point mutants, variants with a random 7-mer (plus 3 constant flanking residues) peptide insertion into the region AAV2 utilizes to bind its co-receptor heparin (Kern et al., 2003), and random chimeras of capsid gene sequences from seven parental serotypes (Figure 1A and Experimental Procedures). To identify variants with broad retrograde tropism, we targeted two independent populations of projection neurons: striatal GABAergic neurons projecting to substantia nigra pars reticulata (SNr) and glutamatergic hindbrain neurons projecting to the cerebellar cortex. Three weeks following injection of the entire pool of rAAV variants into the SNr or cerebellum (one injection per animal), striatal or hindbrain tissue, respectively, was removed, the cap sequences were recovered by PCR, and virus was repackaged (Figure 1B). After two more selection steps, error-prone PCR was performed to further diversify the library, followed by two final in vivo selection steps.
Figure 1. Directed evolution of rAAV2-retro.
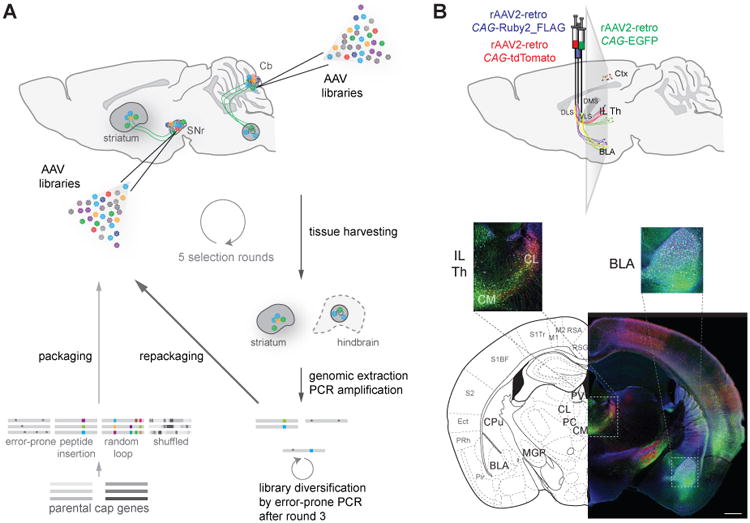
(A) Schematic of the directed evolution procedure. Plasmid libraries containing variant AAV cap genes previously generated by error prone PCR, peptide insertion, randomization of loop regions, and DNA shuffling were packaged, and the resulting viral vector libraries were injected into substantia nigra or cerebellar cortex. Three weeks later, striatal or hindbrain, respectively, tissues were removed, viral genomes isolated, and selected cap genes amplified and packaged for the next round of selection.(B) Example retrograde labeling of projection neurons with the final chosen variant, rAAV2-retro. Retrograde access to intermingled sub-populations of neurons in various brain regions projecting to three striatal compartments was assessed two weeks after delivery of rAAV2-retro carrying different fluorescent proteins into the corresponding axonal fields. rAAV2-retro-tdTomato was injected into dorsolateral striatum (DLS), rAAV2-retro Ruby2_FLAG into dorsomedial striatum (DMS), and rAAV2-retro EGFP into ventrolateral striatum (VLS). Unlike in subsequent experiments (see Figures 2-6), the three probes were visualized by immunohistochemistry to ensure robust detection of retrograde transport at this early time point. Ctx: cortex; BLA: basolateral amygdala; IL TH: intralaminar thalamic nuclei; CL: centrolateral thalamic nucleus; CM: centromedial thalamic nucleus. Scale bar: 1mm. See also Figures S1 and S4, and Movie S1.
We sequenced 30 cap variants after the fourth round of evolution, and the majority originated from the insertion library and contained exogenous 10-mer peptides between N587 and R588 of the wild-type AAV2 VP1 capsid gene (Müller et al., 2003) (see Experimental Procedures). Intriguingly, all of the recovered inserts were of the form LAxxDxTKxA or LAxDxTKxxA; mutations elsewhere in the sequence were also enriched (Table S1). We sequenced a further 22 clones after the fifth round of evolution, and with this additional round of evolution, all sequences were AAV2 mutants with LAxxDxTKxA/LAxDxTKxxA insertions (Table S1), demonstrating a marked degree of further convergence. Such convergence suggested that specific peptide insertions into the heparin-binding loop were largely responsible for the retrograde functionality, with potential secondary contributions from the other sites (see Discussion).
We next focused a secondary screen (not shown) on seventeen isolated variants possessing different combinations of peptide insertions and point mutations in the capsid sequence. To apply a high level of stringency, the chosen variants were packaged with enhanced green fluorescent protein (EGFP) transgene driven by the CMV promoter, which is typically weak in neurons. For each capsid variant, we evaluated its ability to deliver sufficient payload to cell bodies in key afferent regions to permit the detection of non-antibody-amplified EGFP signal three weeks after injection. The clone (insert LADQDYTKTA+ V708I + N382D) that displayed the strongest retrograde transport in two independent circuits in this secondary screen (cortex to globus pallidus and inferior olive/basal pontine nuclei (BPN) to cerebellum) was chosen for further analysis and named rAAV2-retro. When we assessed two additional promoters more commonly used in rodent in vivo studies (CAG – Figure 1B, or human Synapsin-1, data not shown), we observed remarkable retrograde labeling efficiency with this rAAV variant in a range of different circuits in mice and rats (Figure 1B, Figure S1, Movie S1 and see below). Swapping the 7-mer insertion to one of the other recovered sequences or adding additional point mutations identified in the screen did not lead to further increases in retrograde transport (data not shown).
Efficient retrograde access to projection neurons
The corticopontine tract is a remarkably convergent descending motor pathway, contributing more than95% of the afferents to the basal pontine nuclei (BPN) (Brodal and Bjaalie, 1992). This pathway is therefore particularly advantageous for quantifying the efficiency of viral uptake and retrograde transport by axonal terminals of projection neurons. Indeed, injection of rAAV2-retro into the BPN resulted in dense labeling of layer V neurons (Figure 2A), consistent with past tracing studies (Legg et al., 1989).
Figure 2. Quantification of retrograde transport efficiency.
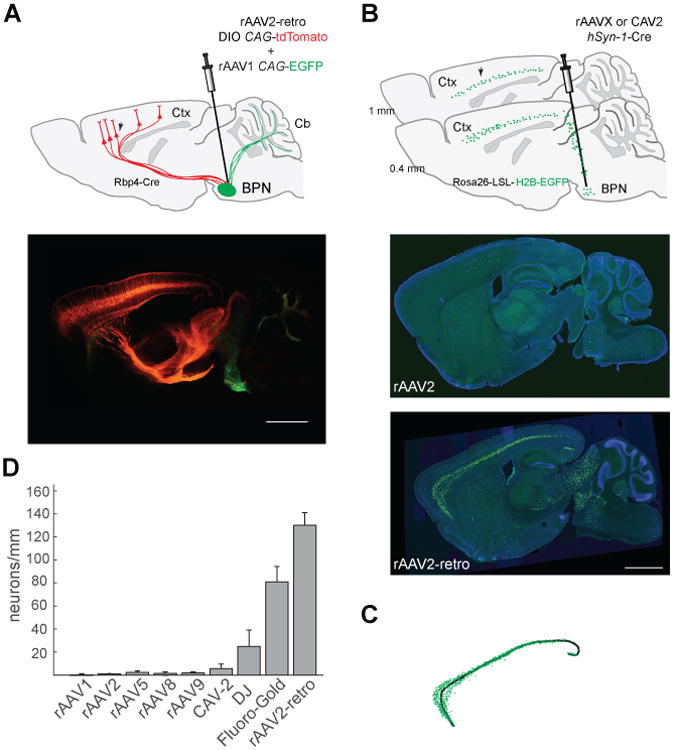
(A) Efficient labeling of the corticopontine tract throughout the rostro-caudal axis via basal pontine injection of rAAV2-retro-DIO-CAG-tdTomato in a layer V-specific Cre mouse line (Rbp4_KL100 Cre). Top panel: schematic of the experiment. Injection site was marked by co-injecting rAAV1-CAG-EGFP. BPN: Basal pontine nuclei. Bottom panel: unamplified tdTomato and EGFP expression three weeks after injection. Scale bar: 1 mm. (B) Quantification assay design. Top panel: schematic of the experiment. Corticopontine labeling was assessed in sagittal sections lateral to the injection tract (∼1 mm lateral with respect to midline) taken from Rosa26-Lox-STOP-Lox-H2B-EGFP animals injected in the BPN with various AAV serotypes carrying Cre recombinase transgene. Arrow indicates expected nuclear GFP labeling in cortical neurons of the corticopontine tract. Middle panel: representative image of an rAAV2-injected brain. Bottom panel: representative image of rAAV2-retro-injected brain. Scale bar for both the panels: 1 mm. (C) Schematic of the semi-automated quantification procedure. Fluorescent nuclei (green) were automatically detected and counted along a manually drawn line that traced the length of cortical layer V (black). (D) Retrograde transport efficiency for different AAV serotypes and for canine adenovirus type 2 (CAV-2) (See Experimental Procedures). Error bars represent the S.E.M. See also Figures S2 and S3.
We therefore next compared the efficacy of retrograde transport in the corticopontine circuit for rAAV2-retrovs. several commonly used AAV serotypes, under identical infection and processing conditions (Figure 2B–D, Experimental Procedures). To ensure quantification accuracy and to eliminate the possible confound of cell-to-cell variability in transgene expression level, we used the AAV to deliver Cre recombinase to Rosa26-Lox-STOP-Lox-H2B-EGFP (histone-fused EGFP) transgenic mice (He et al., 2012). Even a low concentration of the Cre enzyme is sufficient to turn on the expression of such a Cre-dependent cassette (Nagy, 2000), and the stringent nuclear localization of the histone-fused reporter affords unambiguous identification of infected cells, without confounding neuropil signal.
We used a semi-automated analysis procedure to calculate the linear density of infected cortical projection neurons in imaged sagittal sections from mouse brains harvested three weeks after local virus injection in the BPN (Figure 2C, Experimental Procedures). For animals infected with rAAV2, we observed minimal cortical GFP expression (linear density 0.98 ± 0.20 neurons/mm, mean ± S.E.M., n=5; Figures 2B, middle panel). In contrast, and in agreement with the earlier observation (Figure 2A), a dense layer of GFP-positive layer V projection neurons could be observed throughout the rostro-caudal axis of the cortex in rAAV2-retro-injected animals (linear density 130.11±11.08 neurons/mm, n=4; Figure 2B, lower panel). None of the other commonly used AAV serotypes, nor the canine adenovirus-2,matched the retrograde efficiency of the engineered rAAV2-retro variant (linear density AAV1: 0.05 ± 0.04, AAV2: 0.98 ± 0.2, AAV5: 2.38 ± 1.24, AAV8: 1.43 ± 1.43, AAV9: 1.98 ± 0.86, DJ: 24.82 ± 14.32, CAV-2: 5.56 ± 4.13, n=3 to 5 each; Figure 2D, Figure S2). The poor performance by the common AAV serotypes could not have arisen simply as a result of differences in infectivity since even the serotypes displaying similar labeling efficiency at the injection site to that of rAAV2-retro (rAAV2, rAAV9) did not permit efficient retrograde access to corticopontine neurons (Figure S3). Strikingly, the density of cortical projection neurons labeled by rAAV2-retro was comparable to that achieved with Fluoro-Gold (Schmued and Fallon, 1986), a robust synthetic retrograde tracer (linear density 81.03 ± 11.08 neurons/mm, n=3). Thus, rAAV2-retro exhibits up to two orders of magnitude enhancement over existing serotypes in retrograde access to corticopontine projection neurons, and rivals the efficacy of synthetic retrograde tracers.
Generality of retrograde functionality
We next examined whether the retrograde functionality of rAAV2-retro extended to other circuits, specifically by characterizing the extent to which it labeled various afferents to the dorsomedial striatum (DMS) – a part of the basal ganglia that receives long-range inputs from a variety of cortical and subcortical areas (Pan et al., 2010). We found that the efficacy of rAAV2-retro-mediated retrograde access to the strongest afferent inputs into DMS—cortex, thalamus and amygdala—was comparable to that of fluorescent beads classically used for retrograde tracing (Figure 3A). To provide an unbiased estimate for the number of retrogradely labeled neurons in all brain areas known to provide significant long-range input to the DMS, we next assigned any detected fluorescent label in an imaged section of the mouse brain to specific brain areas by aligning the section to the annotated Allen Brain Atlas (Figure 3B–C). Quantitative analysis (Figure 3B-C) revealed that strong retrograde labeling was found in the vast majority of regions that have been previously reported to send prominent projections to the striatum (Pan et al., 2010). In one noticeable exception, only modest labeling was observed in the substantia nigra pars compacta (SNc), despite it being the source of a strong dopaminergic input to the DMS (Figure 3C, arrow at cell count for SNc). A small subset of projection neuron classes was found to be similarly refractory to retrograde access by rAAV2-retro in some of the other circuits tested (Table S2; note that all other AAV serotypes tested also failed to label these projections). Despite these exceptions, rAAV2-retro is broadly applicable for retrograde access to projection neurons within the central nervous system. We note that no rAAV2-retro-mediated labeling of unexpected pathways has been reported to date (Table S2), however additional studies will be necessary to fully rule out its ability to infect axons of passage.
Figure 3. Generality of retrograde transport afforded by rAAV2-retro.
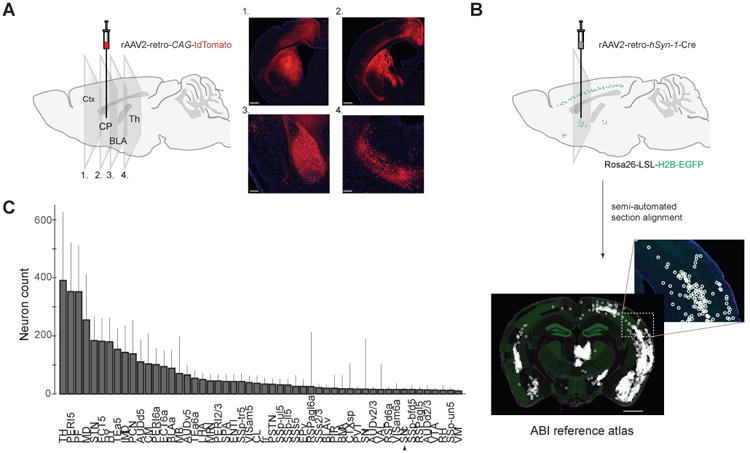
(A) Representative images showing extensive labeling in the main input structures to the dorsal striatum including cortex (panel 1), amygdala (panel 3), and thalamus (panel 4). Scale bars: 800 micrometers for panels 1 and 2,200 micrometers for panels 3 and 4.Ctx: cortex; CP: caudate/putamen; BLA: basolateral amygdala; Th: thalamus; (B) Schematic of automated whole-brain quantification of retrograde labeling. Brains of Rosa26-LSL-H2B-EGFP injected with rAAV2-retro-hSyn1-Cre were imaged to visualize DAPI stained nuclei and green fluorescence from H2B-GFP expressing nuclei. The green channel was used to detect labeled neurons; the blue channel was aligned to the Nissl images from the Allen Brain Institute's standard mouse brain (see Experimental Procedures). The alignment permitted detected neurons to be assigned to different regions using the annotation provided by the brain atlas. Scale bar: 1.25 mm. (C) Whole-brain quantification of retrograde labeling out of a small region of the dorsomedial striatum. Abbreviations for the different brain areas are given according to the Allen Brain Atlas. Arrow highlights the SNc. Error bars represent the S.E.M.
Retrograde access to genetically defined neuronal populations
We also determined whether the retrograde functionality of rAAV2-retro could be combined with the specificity of Cre transgenic lines to enable the interrogation of specific classes of projection neurons. Specifically, we asked whether we could combine rAAV2-retro and Cre transgenic lines to segregate two functionally distinct long-range connections that run in parallel between two brain areas. Projections from cerebral cortex to the striatum arise primarily from neurons in layer V but some neurons in layers II and III also provide striatal inputs. It has been suggested that inputs to the striatum from different cortical layers constitute separate pathways, with neurons in layer V projecting to the patch compartment of the striatum, and neurons in layers II and III to the matrix (Gerfen, 1989). The interdigitated nature of patch and matrix micro-compartments makes it difficult to selectively target these pathways for functional interrogation with the rAAV2-retro system alone. We therefore explored whether we could segregate the two inputs by combining rAAV2-retro with a transgenic line that expresses Cre recombinase in most layer V neurons (Gerfen et al., 2013), but not in neurons in layers II and III (Figure 4A).
Figure 4. Combining rAAV2-retro system with Cre driver lines for selective access to parallel corticostriatal pathways.
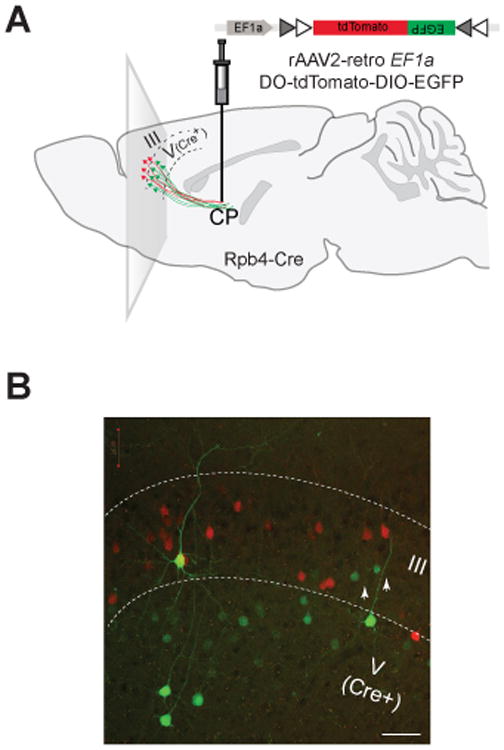
(A) Schematic of the experiment. rAAV2-retro carrying a Cre-dependent color flipping fluorescent reporter was injected into the striatum of a cortical layer V-specific Cre line. (B) Two corticostriatal pathways differentially labeled through a Cre-dependent inversion of the reporter in one (layer V), but not the other (layer II/III), pathway. A presumed Cre-independent inversion— typical for all AAV vectors—was observed in a small fraction of corticostriatal neurons in layer II/III (white arrows). Scale bar: 50 micrometers.
To highlight both pathways in the same experiment, we chose a Cre-dependent color-flipping payload, which expresses tdTomato in the absence of Cre, but inverts to drive the expression of EGFP in Cre-positive cells (Saunders et al., 2012). When rAAV2-retro carrying this payload was injected into the dorsomedial striatum of a layer V-specific Cre driver line, Rbp4_KL100 Cre (Gerfen et al., 2013), layer V, but not layer II/III, neurons projecting to the striatum were robustly labeled with EGFP. Furthermore, the corticostriatal pathway originating in layer II/III was clearly distinguishable by the expression of tdTomato (Figure 4B). In accordance with the topographic nature of the corticostriatal projection, highly localized injections of the virus into the dorsomedial striatum led to a labeling of correspondingly small sections of the cortex. Cortical regions with selective layer II/III labeling were not observed, however, suggesting that the two pathways from the same cortical area traverse through the neighboring matrix and patch micro-compartments within striatum.
While this experiment highlighted both pathways, choosing a “Cre-on” or a “Cre-off” payload will permit selective interrogation of one at the exclusion of the other. This example, therefore, highlights the added specificity of circuit interrogation that can be achieved by combining a highly efficient retrograde virus with available Cre (or Flp) driver lines.
UsingrAAV2-retro for circuit interrogation and gene manipulation
The utility of rAAV2-retro for circuit interrogation will depend on its ability to mediate high levels of expression of genetically encoded indicators and effectors. We first assessed the ability to monitor neural activity in defined classes of projection neurons, through rAAV2-retro-mediated expression of GCaMP6f (Chen et al., 2013) (Figure 5A-C). Using in vivo two-photon Ca2+ imaging, we detected dendritic and somatic Ca2+ transients in the primary motor cortex as early as 7 days after viral delivery to the BPN (Figure 5D). The temporal profile of Ca2+ signals reflected the structure of a cued reaching task, with the signal in many corticopontine neurons being tightly linked to the “go” cue (Figure 5E–F) (Li et al., 2015). Repeated recordings from identified neurons were possible both across trials within a session early in the expression time course (Figure 5E, F), and across many behavioral sessions for over two months post-infection. Thus, rAAV2-retro confers the ability to express sensors in projection neurons at levels sufficient for imaging, creating many new opportunities for deciphering the contributions of specific projections to neural circuit computations.
Figure 5. rAAV2-retro supports sufficient transgene expression for functional circuit interrogation.
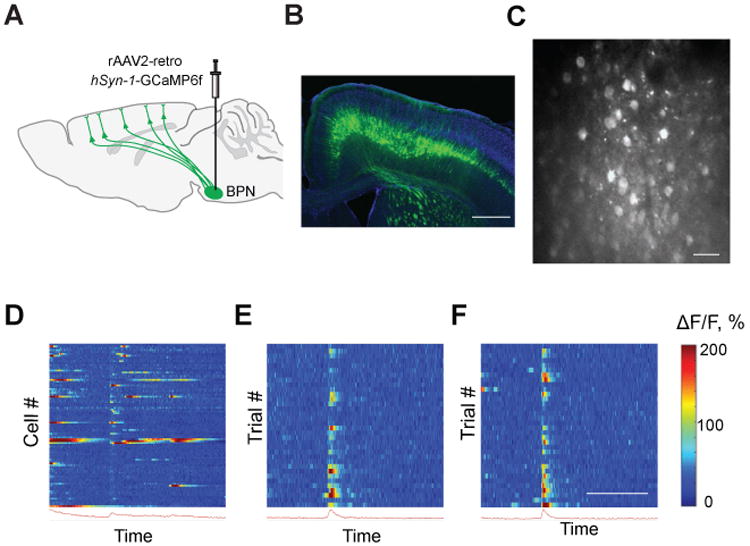
(A) Schematic of the experiment. Expression of calcium indicator GCaMP6f is restricted to corticopontine neurons using localized injection of rAAV2-retro into the BPN. (B) Cross section of the mouse brain showing GCaMP6f expression throughout the corticopontine tract. Scale bar: 50 micrometers. (C) Maximum projection of an in vivo two-photon calcium image taken during a single reach showing layer V-pyramidal tract somas and apical dendrites. Scale bar: 50 micrometers. (D) Activity of 89 ROIs during a single paw reach repetition (broken line denotes the tone “go” signal). (E-F) Two examples of single corticopontine neurons during 40 consecutive trials (same animal as in B-D). Scale bar: 4 seconds.
Finally, we evaluated the utility of rAAV2-retro for delivery of effectors, such as the CRISPR/Cas9 gene editing system, to projection neurons (Figure 6). Specifically, we packaged into rAAV2-retro the Staphylococcus aureus Cas9 (SaCas9, (Slaymaker et al., 2016)) and a single guide RNA designed to ablate the expression of tdTomato (Experimental Procedures). Delivery of rAAV2-retro-SaCas9-anti-tdTomato to the BPN of animals expressing tdTomato in cortical layer V excitatory neurons resulted in suppression of tdTomato expression in 88.6 ± 0.7% of SaCas9-expressing layer V neurons (Figure 6B, bottom panel and Figure 6C, n=3). In contrast, delivery of a non-targeted SaCas9 did not lead to any discernible changes, with only 4.4 ± 3.2% of cells displaying potential reduction in tdTomato expression (Figure 6B, top panel, and Figure 6C, n=3). Furthermore, tdTomato expression remained unaffected in layer V neurons that were inaccessible via pontine injection. The rAAV2-retro system thus permits efficient gene modification selectively in neurons projecting to specific areas of interest.
Figure 6. rAAV2-retro system enables in vivo genome editing using CRISPR/Cas9.
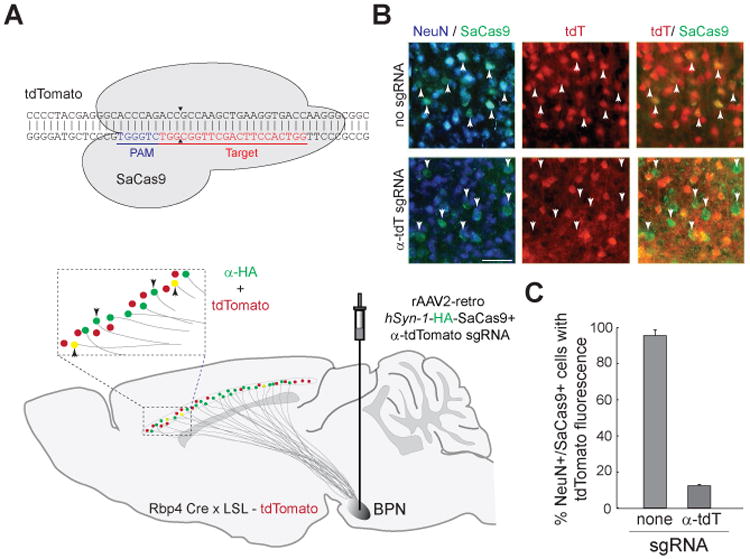
(A) Schematic of the experiment. Top panel: The rAAV2-retro system was used to deliver Staphylococcus aureus Cas9 (SaCas9) single guide RNA combination engineered to ablate expression of tdTomato. Bottom panel: rAAV2-retro carrying the SaCas9-anti-tdTomato payload was injected into the BPN of mice expressing tdTomato from a single genomic locus in layer V neurons. SaCas9 was epitope tagged, permitting identification of retrogradely labeled neurons (green channel). Upward arrows: expected labeling following successful ablation of tdTomato. Downward arrows: expected labeling if tdTomato expression is unaffected. (B) Representative images from brain sections of animals that received the CRISPR/Cas9 system targeted against tdTomato or carrying a non-targeted guide. tdT: tdTomato. Scale bar: 50 micrometers. (C) Efficiency of ablation. Error bars represent the S.E.M.
Collectively, these observations establish rAAV2-retro as an effective reagent to genetically access projection neurons for functional interrogation of neural circuits and, in the long run, for possible therapeutics.
Discussion
Recombinant adeno-associated viruses (rAAVs) can greatly facilitate the functional dissection of mammalian neural circuits, and hold promise for therapeutic intervention in disorders of the nervous system. We have used directed evolution to endow the AAV capsid with the additional capacity for efficient retrograde access to projection neurons in many neural circuits. The newly engineered rAAV2-retro offers up to two orders of magnitude enhancement in retrograde transport compared to commonly used AAV serotypes, matching the efficacy of synthetic retrograde tracers in many circuits. The level of transgene expression achieved with rAAV2-retro via retrograde access is ample for interrogating neural circuit function, as well as for targeted manipulations of the neuronal genome. Thus, by enabling selective monitoring and manipulation of projection neurons connecting different brain areas, rAAV2-retro based tools are poised to provide insights into how large-scale networks enable brain function, and may form the basis for future therapeutic intervention in diseases characterized by progressive large-scale network dysfunction.
The markedly increased efficacy of retrograde access afforded by rAAV2-retro compared to its parental serotype AAV2 might have been enabled through an insertion-mediated disruption of the native binding site for heparin and/or through the creation of a new binding surface that incorporates the inserted peptide. This variant does have reduced heparin affinity (Figure S4), which could decrease virus sequestration in the extracellular matrix of the synaptic cleft and enhance local vector spread, as has been observed with rAAV1 and rAAV6 (Arnett et al., 2013). However, the resulting increase in vector spread alone cannot explain the efficacy of retrograde transport, as other inserted 7-mer sequences disrupt heparin binding similarly but do not affect retrograde transport (Dalkara et al., 2013). Furthermore, rAAV5 and rAAV9 do not bind heparin, yet their retrograde transport efficiency is similar to that of rAAV2. In support of the alternative explanation, the peptide insertions selected in the original selection (LAxxDxTKxA/LAxDxTKxxA) share the same overall composition, differing simply in the register of the conserved motif. The engineered peptide insertion might support enhanced binding to an existing cellular co-factor in the AAV pathway (e.g. the recently identified common AAV receptor (Pillay et al., 2016)), or it might create a novel interaction with the cellular machinery such as a cell surface receptor and/or a component of the vesicular trafficking or nuclear entry pathway. Further experiments will be required to elucidate the mechanism of the profound enhancement in the efficacy of retrograde transport.
Despite the multiple orders of magnitude improvement in retrograde transport that rAAV2-retro offers over existing serotypes, a small set of projection neuron classes appears refractory to efficient retrograde infection by this newly evolved rAAV variant (Table S2). It should be noted, however, that other AAV serotypes fail to label these projections as well. Whether the expression level of the critical cellular factor that interfaces with this novel AAV variant in these neurons is just extremely low—a conclusion supported for the corticothalamic and corticocollicular projections, for instance, by the observation that rAAV2-retro could still deliver sufficient Cre recombinase to cell bodies to direct high-level of expression for locally delivered Cre-dependent payloads (Table S2, highlighted entries)—or whether the factor is missing entirely, remains to be determined in each case. Other viral reagents with retrograde functionality (such as HSV1 (Enquist et al., 1998) or Hi-Ret lentivirus (Kato et al., 2010)) might prove useful for targeting some of these projection neuron classes. Alternatively, additional AAV capsid engineering could extend retrograde transduction to all neurons.
Since gaining access to individual classes of projection neurons will be a critical enabling step in elucidating how local circuit dynamics and large-scale network function are coordinated, the rAAV2-retro vector system provides an important addition to the genetic toolkit for dissecting neural circuit function. Local circuit computations are increasingly thought to depend on the dynamics of the entire neuronal population within a particular local circuit module (Fusi et al., 2016). How these dynamics map onto the different classes of projection neurons—and thus what information is passed on to the different downstream targets—remains unresolved for most circuits. rAAV2-retro based vectors, alone or in combination with specific Cre transgenic lines, permit genetic access to specific populations of projections neurons. In turn, rAAV2-retro carrying rabies G glycoprotein can be used to trans-complement conditional rabies vectors (e.g. the newly developed, less toxic version described by Reardon et al., 2016) for access to presynaptic microcircuits impinging on particular classes of projection neurons (Brown and Hestrin, 2009). The resulting ability to selectively monitor and manipulate activity of individual projection neuron classes and their local microcircuits should provide insight into how projection neurons translate local circuit dynamics for their respective large-scale networks.
rAAV2-retro also holds promise for therapeutic intervention, with several possible applications. For example, in situations where pathology impacts large volumes of neural tissue—such as Alzheimer's or lysosomal storage diseases—multiple injections pose a safety risk and may be insufficient to achieve the needed levels of transduction. However, a small number of injections in strategic locations can enable vector dispersal over large volumes (e.g. corticopontine tract from the point of convergence in the BPN), or difficult-to-access tissue (e.g. spinal motor neurons from the muscle (Kaspar et al., 2003)). Furthermore, large-scale functional networks have been implicated in the spread of many neurodegenerative disorders from their spatially localized onset. A prominent emerging view posits that deposition of abnormal protein assemblies in vulnerable neuronal populations triggers a pathological cascade of aberrant neuronal activity within, and disintegration of, large-scale functional networks, ultimately leading to failure of neurological functions (for a recent review see (Brettschneider et al., 2015)). Intriguingly, the affected neural networks appear capable of transiently overcoming the aberrant dynamics early in the disease, as patients with many neurodegenerative diseases often display periods of dramatic improvement. Thus, early intervention aimed at slowing down the spread of aggregates from the cortical origin of pathology might be sufficient to stabilize, or even restore, cognitive function. From this perspective, subcortically-projecting neurons in the mesocortical regions where pathological protein aggregates first appear, in both Alzheimer's (Braak and Braak, 1996) and Parkinson's (Braak et al., 2003) disorders, constitute an attractive intervention target. Accessing those projection neurons with rAAV2-retro-based tools designed, for instance, to introduce a mutation that is capable of arresting further aggregation in cis (Gregoire et al., 2014) or to deliver chaperones capable of dissembling the aggregates (Jackrel et al., 2014) thus has the potential to slow down the progression of the most debilitating, cognitive, symptoms. Evaluating the efficiency and long-term safety of rAAV2-retro reagents in non-human primates will pave the path to their eventual consideration for these, and other, gene therapy approaches.
Experimental Procedures
All procedures were in accordance with protocols approved by the Janelia Research Campus and the University of California Berkeley Institutional Animal Care and Use Committees.
Library generation and viral production
Four previously generated virus libraries were used at the start of the directed evolution procedure: 1) a random mutagenesis library, generated by subjecting the AAV2 cap gene (encoding virus proteins VP1-3 and Assembly-Activating Protein, AAP) to error prone PCR (Maheshri et al., 2006); 2) library of AAV2 cap gene variants containing 7-mer randomized peptide within a 10-mer insert of the form LAXXXXXXXA between N587 and R588 (Müller et al., 2003); 3) library of AAV2 cap gene variants containing randomized loop regions (Koerber et al., 2009); and 4) a DNA shuffling library generated from wild-type AAV1, AAV2, AAV4, AAV5, AAV6, AAV8 and AAV9 cap gene sequences (Koerber et al., 2008). Each pool of mutant DNA had been originally sub-cloned into the replication-competent AAV packaging plasmid to create a viral plasmid library that, when packaged into AAV virions, can be selected for any new property or function. The replication-competent AAV system incorporates the mutant cap gene into the viral payload, and thus the genotype of each variant is linked to its phenotype. Capsid sequences of the desired property can then be recovered by DNA sequence analysis of the encapsulated AAV genome.
The four replication-competent AAV libraries were packaged by calcium phosphate transient transfection of HEK293-T cells followed by virus harvest, iodixanol gradient centrifugation, and Amicon filtration (Maheshri et al., 2006).
In vivo virus or tracer injections
For virusinjections, ∼50-100 nl (mice) or ∼250-500nl (rats) of virus-containing solution was slowly injected at each depth into the tissue. For tracer injections, 50 nl of 5% Fluoro-Gold (Fluorochrome, Denver, CO) in 0.9% NaCl, or 100 nl of retrobeads (LumaFluor, Durham, NC) diluted 1:1 in 0.9% NaCl were injected at the same set of sites for each injection target.
Library selection and evolution
The four mutant virus libraries were pooled and injected either into the SNr or into the cerebellum of adult (6-8 week old) wild-type C57/Bl6J mice (either sex; Charles River). Three weeks following injections, striatal or hindbrain tissue was accordingly removed, DNA was extracted, and virions that had successfully reached the remote retrograde target tissue were PCR-amplified and recloned into an rcAAV packaging plasmid to create a new replication competent AAV library for the next round of selection. After three selection steps, the rescued cap genes were randomly mutated by error prone PCR using 5′-ACGCGGAAGCTTCGATCAACTACGCAG-3′ and 5′-AGACCAAAGTTCAACTGAAACGAATTAAACGG-3′ as forward and reverse primers, respectively. Two additional in vivo selection rounds were then performed. Individual isolates were sequenced after rounds 4 and 5 to evaluate the degree of library enrichment (Table S1). Seventeen variants were chosen for secondary screening at the end of round five, and the corresponding cap gene sequences were re-cloned into an rAAV helper plasmid. Individual high-titer preps of the parental wild type AAV2 and of each of the seventeen chosen mutant variants carrying a CMV-EGFP payload were then performed by Vector BioLabs, Inc (Philadelphia, PA). The CMV promoter is typically weak in neurons, and thus this secondary screen provided a stringent test of the efficiency of retrograde transport. The individual AAV variants were injected either into the cerebellum or into the globus pallidus. After three weeks, endogenous, unamplified EGFP fluorescence was visualized in the regions expected to be labeled if retrograde transport was efficient. Mutant r5H6 (insert LADQDYTKTA + V708I + N382D) displayed the strongest retrograde transport in both circuits, and was thus chosen for further analysis and named rAAV2-retro (see Supplemental Information for the complete gene sequence).
Payloads and promoters used in the study
For all subsequent experiments, the CMV promoter was replaced with a promoter known to be more robust in adult neurons. Cre recombinase and the GCaMP6f calcium sensor were driven by the human Synapsin-1 (hSyn1) promoter; all of the fluorophores were driven by the CAG promoter; and the color-flipping construct was driven by the EF1α promoter.
Virus production for the quantification of retrograde efficiency
hSyn-Cre payload was packaged using rAAV1, rAAV2, rAAV5, rAAV8, rAAV9, DJ and rAAV2-retro capsids at the Janelia Virus Shared Resource. All seven virus preparations were processed in parallel, and were titer matched before in vivo injections. All lots were diluted to the lowest measured titer (1.3 E12 GC/ml), and each virus was injected into the right pontine nucleus of three to five adult Rosa26-Lox-STOP-Lox-H2B-EGFP mice (He et al., 2012).
Histology
Animals were sacrificed two-three weeks following virus injections, at which point the brains were harvested and the right hemisphere was sagittally sectioned at a thickness of 50 μm. In the triple labeling experiment in Figure 1B and Movie S1, the fluorphores were antibody-amplified; otherwise, unstained sections were used. The sections were mounted in VECTASHIELD Antifade Mounting Medium containing DAPI (Vector Laboratories), and imaged using a P-E Pannoramic slide scanner (3D Histech) using a 20× objective and FITC and DAPI filters.
Retrograde transport quantification
To quantify retrograde transport, the extent of corticopontine labeling was assessed in sagittal sections lateral to the injection tract (∼1 mm lateral with respect to midline) taken from Rosa26-Lox-STOP-Lox-H2B-EGFP animals injected in the basal pons with various AAV serotypes carrying a Cre recombinase transgene. Images obtained with the Pannoramic slide scanner were stitched together and then analyzed using custom software written in Matlab (Mathworks) to detect the GFP-labeled nuclei across the cortex.
For each chosen sagittal section, a region of interest (ROI) was manually drawn around the cortex to isolate the area in the image for automated cell counting. To enhance the detection of nuclei, the image was then convolved with a “Mexican Hat” kernel comprising the difference of two Gaussians (26.00 μm variance and 3.25 μm variance). Image noise was reduced using a median filter, and peak detection was then performed. Linear density was computed as the number of labeled nuclei per mm of the total length of a line drawn manually to trace the outline of cortical layer V.
Quantification of infection efficiency at the injection site
For rAAV2, rAAV9 and rAAV2-retro, additional analysis of infection efficiency at the injection site was performed. A single sagittal section spanning the middle of the injection site—as judged by the presence of extensive labeling along the injection tract—was chosen for each of the injected brains. The green channel was convolved with the “Mexican Hat” kernel (see above), and peaks were then detected as local maxima on these threshold images using custom functions written in Matlab. An ROI was drawn over the midbrain area containing the basal pontine nuclei but excluding the injection tract, and the total number of labeled nuclei within the ROI was tabulated.
Analysis of the generality of retrograde transport
Rosa26-Lox-STOP-Lox-H2B-EGFP mice were injected with 25 nl of rAAV2-retro hSyn1-Cre (1.3E12 GC/ml) in the dorsal striatum. Three weeks after injection, coronally-sectioned brains were imaged using a Pannoramic Scanner to visualize DAPI stained nuclei and green fluorescence from H2B-GFP expressing nuclei. The green channel was similarly used for the detection of labeled nuclei (see above). The blue channel of each section was aligned to the Nissl images from the Allen Brain Institute's standardized mouse brain atlas using custom analysis routines written with the help of the Matlab Image Processing Toolbox. The annotated regions from the Allen Brain Institute's mouse brain atlas were then used to assign detected neurons in aligned sections to specific brain regions (Kuan et al., 2015).
We note that finite precision of the reference atlas together with anatomical variability of individual brains limit the robustness of this semi-automated process to prominent afferent inputs.
Imaging of neuronal population activity in vivo following retrograde delivery of GCaMP6f
Seven adult mice were implanted with custom-made headposts as previously described (Osborne and Dudman, 2014). rAAV2-retro carrying a hSyn1-GCaMP6f (1.0E13 GC/ml) payload was injected into the BPN using a Nanoliter 2010 injector (World Precision Instruments). A cranial window was placed over the primary motor cortex. All animals had visually identifiable GCaMP6f expressing cells in layer V of M1 one-week post injection. Then the animals were habituated to head fixation in a custom-built apparatus, trained to retrieve a food pellet as previously described (Guo et al., 2015). Calcium transients during behavior were imaged by exciting GCaMP6f at 920 nm. Emission light passed through a 565 DCXR dichroic filter (Chroma Technology) and an ET525/70m-2p filter (Chroma Technology) and was detected by a GaAsP photomultiplier tube (10770PB-40, Hamamatsu). Images (512 × 512 pixels) were acquired at ∼30 Hz with resonating scanners using ScanImage software.
CRISPR/Cas9 genome editing
The CMV promoter in pAAV-CMV-SaCas9-empty (Slaymaker et al., 2016) was replaced with hSyn1 to generate pAAV-hSyn1-SaCas9-empty. Oligonucleotides encoding sgRNA protospacer sequences were custom ordered, phosphorylated, hybridized and ligated into the BsaI restriction sites of pAAV-hSyn1-SaCas9-emtpy to generate pAAV-hSyn1-SaCas9-tdTomato-1 to -10 (see Supplemental Information for oligonucleotide sequences tested and details of the in vitro selection procedure). sgRNA 7—one of two that appeared to direct two cleavage events within the tdTomato sequence in vitro—was packaged into AAV2-retro and used for in vivo genome editing.
Approximately 100 nl of rAAV2-retro-hSyn1-SaCas9-tdTomato (5.0 E13 GC/ml) or rAAV2-retro-hSyn1-SaCas9-empty was then injected into the BPN of Rbp4_KL100 Cre × tdTomato mice as described above. Six weeks after injections, brains were harvested and 40 μm-thick coronal sections were cut, stained against the HA-tagged Cas9 and imaged using a Zeiss Axio Observer A1 inverted microscope. Quantification of immunostaining was performed using ImageJ analysis software (NIH).
Supplementary Material
Acknowledgments
We are indebted to K. Svoboda for suggesting the use of Rosa26-lox-STOP-lox-H2B-EGFP mice. We thank H. Adesnik for providing the Rbp4_KL 100Cre × tdTomato mouse line, J. Brousseau for invaluable assistance with the pilot experiments, M. Proskurin for help with the rat experiments, and J. Santiago-Ortiz for help with the CRISPR/Cas9 experiments. We are grateful to the members of the Dudman, Hantman, Jessell, Ji, Karpova, Petreanu, Sabatini, Spruston, Svoboda and Zuker labs for beta-testing rAAV2-retro and contributing their findings to the knowledge base summarized in Table S2.
T. Jessell, N. Spruston and K. Svoboda offered useful discussions and comments on the manuscript. D.V.S. is supported by NIH R01EY022975, T.G. by F32GM113446. This work was supported by the Howard Hughes Medical Institute.
Footnotes
Author Contributions: L.L.L, D.V.S, and A.Y.K. conceived the project; D.G.R.T., B-Y.H., J.T.D., A.W.H., L.L.L., D.V.S. and A.Y.K. designed and oversaw the molecular evolution process; B-Y.H. performed library packaging, amplifications, and sequencing; K.D.R. designed and oversaw viral production and quality control process; D.G.R.T., S. V., T. G., M.L., S. L., S.M., E.K., D.O. and C-C.H. performed experiments; D.G.R.T. wrote custom analysis code, D.G.R.T., S.V., T.G., and D.O. analyzed the data; D.G.R.T., L.L.L., D.V.S, and A.Y.K. wrote the manuscript. C.R.G. and J.S. provided essential reagents, expertise, and input to the manuscript.
References
- Arnett AL, Beutler LR, Quintana A, Allen J, Finn E, Palmiter RD, Chamberlain JS. Heparin-binding correlates with increased efficiency of AAV1-and AAV6-mediated transduction of striated muscle, but negatively impacts CNS transduction. Gene therapy. 2013;20:497–503. doi: 10.1038/gt.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer GM, Shanthaveerappa TR, Bourne GH. Studies on the pathogenesis of fixed rabies virus in rats. Bull World Health Organ. 1965;33:783–794. [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Development of Alzheimer-related neurofibrillary changes in the neocortex inversely recapitulates cortical myelogenesis. Acta neuropathologica. 1996;92:197–201. doi: 10.1007/s004010050508. [DOI] [PubMed] [Google Scholar]
- Braak H, Rüb U, Gai W, Del Tredici K. Idiopathic Parkinson's disease: possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. Journal of neural transmission. 2003;110:517–536. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Lee VMY, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nature Reviews Neuroscience. 2015;16:109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodal P, Bjaalie JG. Organization of the pontine nuclei. Neurosci Res. 1992;13:83–118. doi: 10.1016/0168-0102(92)90092-q. [DOI] [PubMed] [Google Scholar]
- Brown SP, Hestrin S. Intracortical circuits of pyramidal neurons reflect their long-range axonal targets. Nature. 2009;457:1133–1136. doi: 10.1038/nature07658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM, Luo L. Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Gershenson ZT, Giles AR, Holzbaur EL, Wolfe JH. Adeno-associated virus serotypes 1, 8, and 9 share conserved mechanisms for anterograde and retrograde axonal transport. Human gene therapy. 2014;25:705–720. doi: 10.1089/hum.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon P, Derbin C, Kucera P, Lafay F, Prehaud C, Flamand A. Invasion of the peripheral nervous systems of adult mice by the CVS strain of rabies virus and its avirulent derivative AvO1. Journal of virology. 1989;63:3550–3554. doi: 10.1128/jvi.63.8.3550-3554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D, Byrne LC, Klimczak RR, Visel M, Yin L, Merigan WH, Flannery JG, Schaffer DV. In vivo–directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Science translational medicine. 2013;5:189ra176–189ra176. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- Enquist L, Husak PJ, Banfield BW, Smith GA. Infection and spread of alphaherpesviruses in the nervous system. Advances in virus research. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- Fusi S, Miller EK, Rigotti M. Why neurons mix: high dimensionality for higher cognition. Current Opinion in Neurobiology. 2016;37:66–74. doi: 10.1016/j.conb.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Gaudet D, de Wal J, Tremblay K, Déry S, van Deventer S, Freidig A, Brisson D, Méthot J. Review of the clinical development of alipogene tiparvovec gene therapy for lipoprotein lipase deficiency. Atherosclerosis Supplements. 2010;11:55–60. doi: 10.1016/j.atherosclerosissup.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: striatal patch-matrix organization is related to cortical lamination. Science. 1989;246:385–388. doi: 10.1126/science.2799392. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Zhang S, Costanzo J, Wilson K, Fernandez EJ, Kwon I. Cis-suppression to arrest protein aggregation in mammalian cells. Biotechnology and bioengineering. 2014;111:462–474. doi: 10.1002/bit.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Lee VM. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nature medicine. 2014;20:130–138. doi: 10.1038/nm.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis ER, Ii, Kadoya K, Hirsch M, Samulski RJ, Tuszynski MH. Efficient retrograde neuronal transduction utilizing self-complementary AAV1. Molecular Therapy. 2008;16:296–301. doi: 10.1038/sj.mt.6300367. [DOI] [PubMed] [Google Scholar]
- Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JH, Koerber JT, Kim JS, Asuri P, Vazin T, Bartel M, Keung A, Kwon I, Park KI, Schaffer DV. An evolved adeno-associated viral variant enhances gene delivery and gene targeting in neural stem cells. Molecular Therapy. 2011;19:667–675. doi: 10.1038/mt.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junyent F, Kremer EJ. CAV-2—why a canine virus is a neurobiologist's best friend. Current opinion in pharmacology. 2015;24:86–93. doi: 10.1016/j.coph.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, Bland RJ, Young D, Strybing K, Eidelberg D, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Erickson D, Schaffer D, Hinh L, Gage FH, Peterson DA. Targeted retrograde gene delivery for neuronal protection. Molecular Therapy. 2002;5:50–56. doi: 10.1006/mthe.2001.0520. [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Llado J, Sherkat N, Rothstein JD, Gage FH. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- Kato S, Kobayashi K, Inoue Ki, Kuramochi M, Okada T, Yaginuma H, Morimoto K, Shimada T, Takada M, Kobayashi K. A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Human gene therapy. 2010;22:197–206. doi: 10.1089/hum.2009.179. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. Journal of neuroscience methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- Kern A, Schmidt K, Leder C, Müller O, Wobus C, Bettinger K, Von der Lieth C, King J, Kleinschmidt J. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. Journal of virology. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Jang JH, Schaffer DV. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Molecular Therapy. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Klimczak R, Jang JH, Dalkara D, Flannery JG, Schaffer DV. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Molecular Therapy. 2009;17:2088–2095. doi: 10.1038/mt.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Maheshri N, Kaspar BK, Schaffer DV. Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles. Nature protocols. 2006;1:701–706. doi: 10.1038/nprot.2006.93. [DOI] [PubMed] [Google Scholar]
- Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer EJ, Boutin S, Chillon M, Danos O. Canine adenovirus vectors: an alternative for adenovirus-mediated gene transfer. J Virol. 2000;74:505–512. doi: 10.1128/jvi.74.1.505-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan L, Li Y, Lau C, Feng D, Bernard A, Sunkin SM, Zeng H, Dang C, Hawrylycz M, Ng L. Neuroinformatics of the allen mouse brain connectivity atlas. Methods. 2015;73:4–17. doi: 10.1016/j.ymeth.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Legg C, Mercier B, Glickstein M. Corticopontine projection in the rat: the distribution of labelled cortical cells after large injections of horseradish peroxidase in the pontine nuclei. Journal of Comparative Neurology. 1989;286:427–441. doi: 10.1002/cne.902860403. [DOI] [PubMed] [Google Scholar]
- Li N, Chen TW, Guo ZV, Gerfen CR, Svoboda K. A motor cortex circuit for motor planning and movement. Nature. 2015;519:51–56. doi: 10.1038/nature14178. [DOI] [PubMed] [Google Scholar]
- Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nature biotechnology. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Miller MW, Vogt BA. Direct connections of rat visual cortex with sensory, motor, and association cortices. J Comp Neurol. 1984;226:184–202. doi: 10.1002/cne.902260204. [DOI] [PubMed] [Google Scholar]
- Müller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA, Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nature biotechnology. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- Murlidharan G, Samulski RJ, Asokan A. Biology of adeno-associated viral vectors in the central nervous system. Frontiers in molecular neuroscience. 2014;7 doi: 10.3389/fnmol.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Ohka S, Yang WX, Terada E, Iwasaki K, Nomoto A. Retrograde transport of intact poliovirus through the axon via the fast transport system. Virology. 1998;250:67–75. doi: 10.1006/viro.1998.9360. [DOI] [PubMed] [Google Scholar]
- Ojala DS, Amara DP, Schaffer DV. Adeno-associated virus vectors and neurological gene therapy. The Neuroscientist. 2014:1073858414521870. doi: 10.1177/1073858414521870. [DOI] [PubMed] [Google Scholar]
- Pan WX, Mao T, Dudman JT. Inputs to the dorsal striatum of the mouse reflect the parallel circuit architecture of the forebrain. Front Neuroanat. 2010;4:147. doi: 10.3389/fnana.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersanti S, Astrologo L, Licursi V, Costa R, Roncaglia E, Gennetier A, Ibanes S, Chillon M, Negri R, Tagliafico E. Differentiated neuroprogenitor cells incubated with human or canine adenovirus, or lentiviral vectors have distinct transcriptome profiles. PloS one. 2013;8:e69808. doi: 10.1371/journal.pone.0069808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S, Meyer N, Puschnik A, Davulcu O, Diep J, Ishikawa Y, Jae L, Wosen J, Nagamine C, Chapman M. An essential receptor for adeno-associated virus infection. Nature. 2016 doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajol S. Histologie du système nerveux de l'homme et des vertébrés. Vol. 2 Paris: Maloine; 1911. [Google Scholar]
- Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, Losonczy A. Rabies Virus CVS-N2c ΔG Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron. 2016 doi: 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel M, Brunert D, Zabawa C, Díaz-Quesada M, Wachowiak M. Transgene expression in target-defined neuron populations mediated by retrograde infection with adeno-associated viral vectors. The Journal of neuroscience. 2013;33:15195–15206. doi: 10.1523/JNEUROSCI.1618-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AB, Johnson C, Sabatini BL. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. 2012 doi: 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmued LC, Fallon JH. Fluoro-Gold: a new fluorescent retrograde axonal tracer with numerous unique properties. Brain Res. 1986;377:147–154. doi: 10.1016/0006-8993(86)91199-6. [DOI] [PubMed] [Google Scholar]
- Schnell MJ, McGettigan JP, Wirblich C, Papaneri A. The cell biology of rabies virus: using stealth to reach the brain. Nature Reviews Microbiology. 2010;8:51–61. doi: 10.1038/nrmicro2260. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão D, Pinto C, Fernandes P, Peddie C, Piersanti S, Collinson L, Salinas S, Saggio I, Schiavo G, Kremer E. Evaluation of helper-dependent canine adenovirus vectors in a 3D human CNS model. Gene therapy. 2015 doi: 10.1038/gt.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. The FASEB Journal. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- Taymans JM, Vandenberghe LH, Haute CVD, Thiry I, Deroose CM, Mortelmans L, Wilson JM, Debyser Z, Baekelandt V. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Human gene therapy. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- Tomioka R, Okamoto K, Furuta T, Fujiyama F, Iwasato T, Yanagawa Y, Obata K, Kaneko T, Tamamaki N. Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. European Journal of Neuroscience. 2005;21:1587–1600. doi: 10.1111/j.1460-9568.2005.03989.x. [DOI] [PubMed] [Google Scholar]
- Towne C, Schneider B, Kieran D, Redmond D, Aebischer P. Efficient transduction of non-human primate motor neurons after intramuscular delivery of recombinant AAV serotype 6. Gene therapy. 2010;17:141–146. doi: 10.1038/gt.2009.119. [DOI] [PubMed] [Google Scholar]
- Ugolini G, Kuypers HG, Simmons A. Retrograde transneuronal transfer of herpes simplex virus type 1 (HSV 1) from motoneurones. Brain Res. 1987;422:242–256. doi: 10.1016/0006-8993(87)90931-0. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


