Abstract
Coronary artery disease (or coronary heart disease), is the leading cause of mortality in many of the developing as well as the developed countries of the world. Cholesterol-enriched plaques in the heart’s blood vessels combined with inflammation lead to the lesion expansion, narrowing of blood vessels, reduced blood flow, and may subsequently cause lesion rupture and a heart attack. Even though several environmental risk factors have been established, such as high LDL-cholesterol, diabetes, and high blood pressure, the underlying genetic composition may substantially modify the disease risk; hence, genome composition and gene-environment interactions may be critical for disease progression. Ongoing scientific efforts have seen substantial advancements related to the fields of genetics and genomics, with the major breakthroughs yet to come. As genomics is the most rapidly advancing field in the life sciences, it is important to present a comprehensive overview of current efforts. Here, we present a summary of various genetic and genomics assays and approaches applied to coronary artery disease research.
Keywords: Coronary artery disease, Genomics, Genetics, Transcriptomics, Epigenetics, GWAS
Introduction
The term genomics was reportedly coined by Dr. Thomas H. Roderick and a group of scientists in a pub after a scientific meeting in Bethesda in 1986 [1] and, generally, is taken to mean the simultaneous study of a large number of genes in the context of a biological process or disease. A transforming sequence of events, beginning with the development and rapid improvement in microarray technology in the 1990s, followed by sequencing of the first draft of human genome in 2000, and especially development of next-generation sequencing technologies in 2004–2005 and genome-wide association studies (GWAS) in 2005, have led to an exponential growth of genome-wide methodologies and the applications of genomics across diverse fields of study.
This review focuses on the current methods and overall advances in genomics of cardiovascular diseases and provides an overview of how genomics may help illuminate some of the mechanisms of this complex disease. We discuss recent progress with regards to the genetic basis of coronary artery disease (CAD) in the context of GWAS, transcriptomic profiling, probing of in vivo protein-DNA interactions, as well as regulatory and higher order chromatin structure. We highlight a few examples of investigations at CAD GWAS loci, 9p21.3 and TCF21, which have applied functional genomics and mouse models to link these genetic associations to vascular wall disease processes. Finally, we discuss the critical advantages of applying the CRISPR/Cas9 system towards functional studies at CAD loci.
GWAS as Initial Discovery Tool for CAD Mechanisms
GWAS aims to discover novel susceptibility loci for complex diseases through hypothesis-free case-control studies and subsequent validation in a different or expanded cohort. The GWAS era started from the first successful GWAS publication for macular degeneration in 2005 and continues to expand to date with a total number of publications reaching above 2300. GWAS databases are getting larger, as exemplified with a Catalog of Published Genome-Wide Association Studies from National Human Genome Research Institute (NHGRI) and European Bioinformatics Institute (EMBL-EBI) that contains in total 15020 SNPs and 16831 SNP-trait associations from 2334 studies (data release on 2015-11-21).
The most recent large scale meta-analysis of GWAS in CAD identified a total of 2213 CAD-associated variants at 48 loci (that had previously already been reported to be associated with CAD at genome-wide significance, P < 5 × 10−8) as well as an additional 10 novel loci [2••]. This study included 60,801 cases and 123,504 controls from 48 GWA studies of CAD, out of which 57.5 % of the cases and 40.1 % of the controls had been included in a previous Metabochip-based CAD meta-analysis [3], indicating that massive data aggregation has the potential to resolve many previously unreplicated loci. A unique aspect of this study was the examination of a very large number of imputed genotypes based on the 1000 Genomes Project. This effort has identified 38 million variants and after selecting variants with minor allele frequency (MAF) > 0.005 that were detectable in over 60 % of the 48 GWA studies, 8.6 million SNPs and 836,000 indels were included in the meta-analysis, which represents to date the largest collection of variants tested for association with CAD. One of the limitations of the study is that the majority (77 %) of the participants were of European ancestry, suggesting that new CAD loci functional in other racial/ethnic groups remain to be discovered [4].
With the ever-increasing numbers of tested subjects, GWAS will likely continue to identify novel CAD-associated variants. However, these discovered variants contribute a fraction of the total estimated heritability of common disease risk and may have limited clinical and predictive value on their own. A recent study demonstrated that a CAD genetic risk score (GRS) of nearly 50,000 common variants was highly predictive of major adverse events, suggesting these variants collectively capture a lifetime of risk within individuals (Abraham G et al. bioRxiv 2016). The missing heritability may be accounted for by less common or rare variants that were missed on the genotyping array or inaccurately imputed due to allelic heterogeneity or population-specific signals. Thus, it is important to perform meta-analyses and fine-mapping in different racial/ethnic groups as well as apply robust statistical frameworks to correct for these [5] potential confounders and uncover new biology [6]. For instance, in the latest 1000G-based imputation meta-analysis of CAD, 10 novel loci were discovered (including SMAD3, SWAP70, ABHD2, ADORA2A, MFGE8, BCAS3, and NOS3), proposing novel genes that have a role in vessel wall and SMC-related processes as critical determinants of CAD risk. In addition, gene-environment interactions and epistatic interactions of multiple alleles may confer non-additive effects on disease susceptibility and broad-sense heritability, suggesting that missing CAD heritability estimates may be accounted for with as-yet unknown risk factors/interactions [2••, 7].
Several statistical methods have been applied to fine-mapping GWAS loci by calculating the posterior probabilities of causality for candidate variants [5, 8, 9]. By integrating disease-specific functional genomics data as prior knowledge, the credible set of causal variants at a susceptibility locus and overall complexity may be reduced [9, 10]. Similarly, Bayesian network analysis has been used to detect interaction between loci in order to elucidate underpinning biological pathways [11]. Bayesian networks could be input to key driver analysis (KDA), in order to define disease driver genes [12], an approach that has been used in CAD [13]. In addition, Bayesian networks have been coupled with protein–protein interaction (PPI) networks and differential modules (DM) from co-expression networks and integrated into KDA, an approach that has identified TNFRSF13C and EBF1 as key drivers for CAD [14].
Application of Transcriptomics to CAD Mechanisms
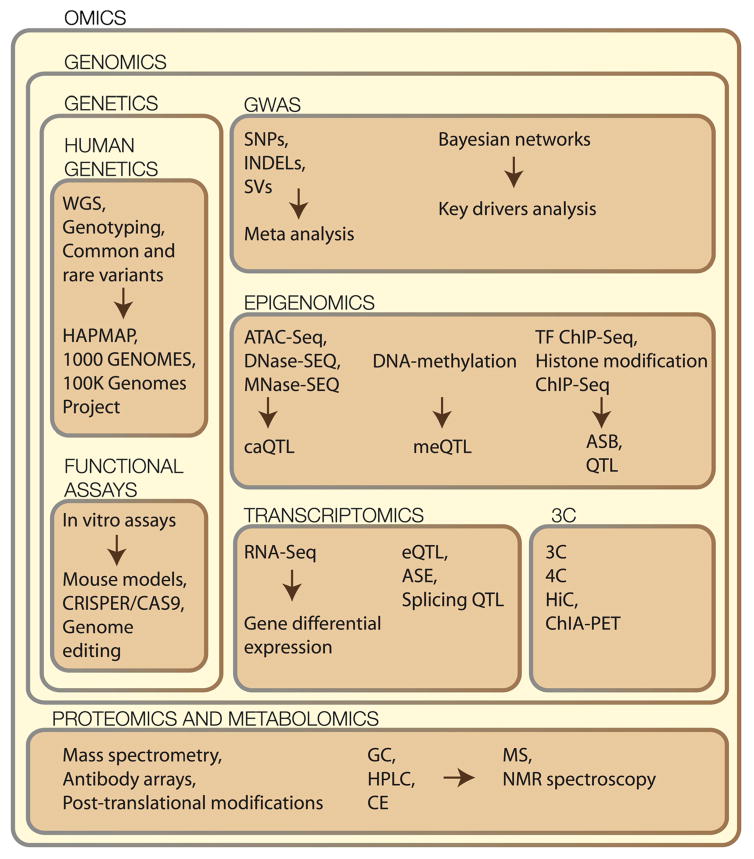
Contemporary genomics often involves integrative and simultaneous analysis of multiple datasets from diverse experiments [15•]. Our ultimate understanding of complex systems depends on successful integration of various GWAS, epigenomics, transcriptomics, and proteomics datasets (Fig. 1) [16, 17].
Fig. 1.
Integrative omics for application in vascular wall disease. SNP single nucleotide polymorphism, INDEL insertion or deletion, SV structural variant, TF transcription factor, caQTL chromatin accessibility quantitative trait locus, meQTL methylation quantitative trait locus, ASB allele-specific binding, QTL quantitative trait locus, eQTL expression quantitative trait locus, ASE allele-specific expression, GC gas chromatography, HPLC high performance liquid chromatography, CE capillary electrophoresis, MS mass spectrometry
A transcriptome is a complete collection of RNA molecules transcribed in a given cell or organism. RNA-sequencing (RNA-Seq) represents an evolution of microarray-based technology, which has now reached mainstream adoption [18, 19] and offers several advantages in transcriptome profiling [18, 20, 21]. For instance, RNA-Seq provides single-base level resolution, and thus enables detection of transcribed genetic variation, such as single nucleotide polymorphisms (SNPs), indels, gene fusions, and structural variation, and may resolve alternative transcripts of multi-transcript genes from the sequence data alone [22]. In addition, it has a greater dynamic range to quantitatively detect expression changes from weakly expressed genes encoding transcription factors to the highly expressed housekeeping genes.
RNA-Seq has been used in combination with genotyping to determine how genetic variation influences gene expression changes through expression quantitative trait loci (eQTL). These molecular traits may function locally, in the same genomic locus (cis) or distally, in a different genomic locus (trans). In addition, RNA-Seq may detect allele-specific expression (ASE), which involves the differential expression between two alleles at heterozygous sites. [23, 24]. Both eQTL and ASE have been instrumental tools to prioritize functional variants among thousands of candidate GWAS variants, and may point to the underlying causal mechanism(s) of the disease/trait association [25]. Given that ASE measures transcript changes within individuals it may pinpoint functional cis-acting variants that alter gene expression at GWAS loci, thus making the combination of GWAS and RNA-Seq a powerful approach for dissecting the mechanisms of complex phenotypes.
Large-scale RNA-Seq-based transcriptomic profiling has been used in lymphoblastoid cell lines (LCLs) to uncover the genetic architecture of complex traits such as autoimmune diseases; however, to define the disease mechanisms for CAD, there remains a need for large studies in primary cardiovascular cells/tissues. For example, in vivo studies of specific cardiac and vascular cells/ tissues will benefit from improvements in RNA-Seq methodology that will enable extraction and sequencing of RNA from limited numbers of cells/tissues that are difficult to process [26]. RNA-Seq in left ventricular myocardial tissue has been used to systematically identify the disease status of heart failure patients using a limited training dataset of six individuals, including three controls, one ischemic heart disease (ISCH), and two dilated cardiomyopathy (DCM) patients and has been successfully applied to a much larger set of 313 individuals [27]. In addition, RNA-Seq has been used in human cardiac fibroblasts [28], mouse embryonic epicardium that undergoes epithelial-to-mesenchymal transition [29], but only recently in smooth muscle cells of the coronary artery vessel wall [30••], and has not been well studied in coronary artery endothelial cells. RNA-sequencing in cultured human coronary artery smooth muscle cells (HCASMC) revealed a complex network of interconnected genes, including genes for structural proteins such as matrix metalloproteinases, collagens, and actin, as well as genes for signaling molecules, e.g., prostaglandins, cytokines, and interleukins [30••]. In addition, RNA-sequencing in HCASMC revealed 31 unannotated long non-coding RNAs (lncRNAs), some of which were associated with HCASMC migration and differentiation, e.g., SENCR [31].
Application of Epigenomics to CAD Mechanisms: DNA Methylation and Histone Modifications
Epigenetics comprises heritable changes in gene function in cells or organisms that are not attributable to changes in DNA. In particular, epigenetic changes such as DNA methylation and histone modification can switch on or off genes without altering the DNA base sequence itself. Given the dynamic nature of these changes in response to the environment [32], genome-wide epigenomics may reveal greater insights into complex disease mechanisms [33].
DNA methylation at CpG sites has been widely studied in the context of cancer, as a mode of tumor suppressor gene inactivation [34], and likely represents a common phenomenon accumulating in all age-related diseases including CAD [35]. Global DNA methylation profiling by sodium bisulfite conversion demonstrated a positive correlation of peripheral blood leukocyte methylation with the prevalence of cardiovascular disease and obesity [36]. Similarly, methylation-sensitive restriction enzymes detected increased global methylation in peripheral lymphocytes in CAD patients [37]. Further, an epigenetic-wide association study (EWAS) identified increased methylation at the hypoxia inducible factor, HIF3A locus in blood and adipose to be associated with greater body mass index (BMI) [38], a proposed risk factor for CAD [39]. There are also reports of methylation quantitative trait loci (meQTLs, genotype dependent patterns of DNA methylation) at the T-cadherin encoding locus CDH13 associated with cardiometabolic traits [40]. A report in LCLs revealed meQTLs to be coordinated with other epigenetic marks such as histone modification and chromatin accessibility and proximal gene expression, implicating cis-acting changes in TF occupancy [41]. Similar genome-wide studies in primary vascular cell types are needed to fully interpret the effects of CAD GWAS variants on methylation-mediated gene regulation.
Histones are protein molecules that package the DNA into nucleosomal units and, as such, are closely linked to the function of DNA molecule as genetic material. Post-translational covalent modifications (e.g., methylation, acetylation, or phosphorylation) to the N-terminal histone tail of nucleosomes contribute to conformational changes in chromatin structure, have a profound effect on gene expression, and often occur in specific combinations or signatures. The collaborative efforts of the ENCODE Project [42] and Roadmap Epigenomics [43] consortia have already revealed a compendia of genome-wide histone modification signatures for various regulatory features in multiple primary tissues and cell lines. These datasets have been applied to global mapping studies and databases to prioritize functional regulatory variants [44, 45]. While these assays have been employed extensively in LCLs, and tumor cell lines to follow-up autoimmune disease [8, 46] and cancer loci [47, 48], respectively, only a few studies have surveyed CAD loci [49, 50]. Specific histone modifications have been linked to different activation states and lineage commitment. For instance, H3K27ac has been shown to distinguish active from poised enhancers of gene expression and predict developmental state [51], whereas H3K27me3 labels stably repressed chromatin often associated with polycomb complex [52]. In coronary arteries, the active/ poised H3K4me2 mark of the MYH11 gene represents a highly specific epigenetic marker of smooth muscle cell (SMC) lineage that persists even with the occurrence of SMC synthetic phenotype within atherosclerotic lesions [53].
Chromatin Accessibility, Higher Order Structures, and Protein-DNA Interactions
DNA in the interphase nucleus is organized in two basic chromatin states: accessible/open and inaccessible/closed. Chromatin accessibility thus represents another layer of genomic regulation, which has been scaled genome-wide through assays such as DNase-seq [54], MNase-seq [55], FAIRE-seq [56], and more recently ATAC-seq [57••]. DNaseI enzyme was classically used to cleave specific sites along the genome that have less condensed chromatin [58, 59]. Like histone modifications, DNaseI hypersensitive sites mark various regulatory elements including enhancers, promoters, insulators, repressors, and locus-control regions [60]. Regions of accessible chromatin often coincide with transcription factor occupancy and depend on specific transacting factors or chromatin-remodeling factors [60, 61]. DNase-seq profiling in LCLs previously detected up to 55 % of disease-associated variants as chromatin accessibility QTLs (caQTLs), accounting for the gene regulatory effects in these cells [62]. While most of these effects were proximal (<10 kb) to the lead-associated variant or linkage disequilibrium (LD) variants, it is still unclear how multiple independent variants influence gene networks through changes in chromatin states. The Assay for Transpose Accessible Chromatin (ATAC-seq) was recently developed to address the need for sensitive assays requiring less starting material, which also has the ability to simultaneously profile open chromatin, transcription factor-binding footprints, as well as nucleosome positioning in a single assay [57••]. Given the limited availability of primary vascular tissues, this approach has been valuable to investigate the genetic mechanisms of CAD loci [63].
Chromatin accessibility assays have been useful to detect local nucleosome occupancy and positioning [55, 64, 65]; however, other assays are required to investigate higher order chromatin state, such as long-range interactions and three-dimensional structure of topologically associating domains (TAD). For instance, chromosome conformation capture (3C) [66] was utilized to investigate pairwise interactions within the CAD-associated 9p21 locus between CDKN2A/B and MTAP, and IFN2A, located ~1 Mb away, and it was shown that these chromosomal interactions were modulated by IFNγ [67]. Similarly, 3C was used to identify physical interactions between enhancer and promoter elements for an unexpected autoimmune disease gene, DEXI [68]. The 3C assay has been adapted to detect interactions between one locus and all other genomic loci through chromosome conformation capture-on-chip (4C) techniques. A 4C-seq-based approach was used to map the functional interactions between variants at the FTO obesity locus and the hypothalamus-expressed homeobox gene, IRX3 [69]. Further modifications of this approach have enabled high-resolution mapping of genome-wide interactions (Hi-C) [70] or involve coupling antibody-mediated pulldown with paired-end tag sequencing (ChIA-PET) [71], which has been instrumental in detecting unbiased and de novo higher order chromatin interactions. It should be noted that local chromatin accessibility assay ATAC-seq has been applied to single cells and has been shown to capture a higher order chromatin structure resembling the profiles generated by Hi-C [72].
Additionally, for CAD candidate genes that are transcription factors (TF), such as TCF21 and STAT3, protein-DNA interactions could be studied on a genome-wide scale using chromatin immunoprecipitation sequencing (ChIP-Seq). Recently, ChIP-Seq performed against TCF21 in human coronary artery smooth muscle cells (HCASMC) revealed downstream genes enriched for pathways relevant to CAD, including growth factor binding, TF interaction, and smooth muscle contraction [63]. The global binding pattern of TCF21 also identified likely co-regulatory mechanisms involving the AP-1 binding motifs, and subsequent ChIP-Seq against AP-1 family members JUN and JUND confirmed that there was indeed a significant overlap between TCF21 and AP-1 binding sites, demonstrating the significance of their cooperative action in the pathophysiology of CAD.
In Vivo Mouse Models to Study CAD Targets—9p21 Locus
Currently, mice are the most widely used animal model for studying the genetics of cardiovascular disease due to the ease of manipulating their genome and their ability to develop atherosclerotic plaques [73]. Indeed, genetic knockout models are straightforward if a disease-associated SNP is found within the coding sequence of a gene or is located within the proximal promoter region. More often, determining which gene is affected by these SNPs is difficult to ascertain as has been observed in one of the strongest CAD associations in the GWAS era, the 9p21 susceptibility locus for CAD. This locus was discovered in 2007 when four independent studies identified SNPs within a 58-kb block of the 9p21 chromosomal locus that were associated with an increased risk for CAD and myocardial infarction (MI) for homozygous risk allele carriers [74–77]. Subsequent studies and meta-analyses confirmed these associations and identified risk in additional cardiovascular disease states such as stroke, abdominal aortic aneurysm (AAA), saccular aneurysms, and peripheral artery disease (PAD) [78]. This discovery generated great enthusiasm to identify the causal genes influenced by this locus and provided an ideal opportunity to model the risk allele using genetically engineered mice. However, fine mapping studies were unable to significantly narrow the disease-associated region to a smaller set of SNP [79]. The nearest protein coding genes lie over 90 kb away from the core region of association and consist of the tumor suppressor genes CDKN2B encoding p15INK4B and CDKN2A encoding p16INK4a and p14ARF [80]. Additionally, the disease-associated SNPs are located within and proximal to a lncRNA, ANRIL, which has multiple alternatively processed transcripts [81], thereby adding further complexity to the situation.
Genomic studies have been employed to investigate this association, with limited success. Allelic imbalance and eQTL studies demonstrated that the 9p21 locus contains enhancer elements, which regulate expression of the nearby gene products and suggest that all nearby genes may contribute to the enhanced CAD risk [82–84]. However, these findings are confounded by additional studies, which are unable to replicate these gene expression changes leading to contention within the field [79, 85–87]. Interestingly, several studies suggest that ANRIL may regulate cell proliferation and behavior by inhibiting expression of CDKN2B, placing this gene product as a main regulator of the INK4/ARF locus [82, 86]. However, despite the strong sequence conservation of ANRIL in primates, an orthologous transcript cannot be found in mice, thereby impeding progress in identifying potential mechanisms using genetic mouse models [86]. Fortunately, the mouse 9p21 locus shares enough homology to allow the development of an inducible mouse knockout model that deleted the ortholog to the human linkage disequilibrium block [88]. This mouse showed decreased expression of both Cdkn2a and Cdkn2b, supporting the role of an enhancer region at 9p21 in both mice and humans [88]. Recently, several mouse genetic knockout studies have been performed to identify the role of these genes in CAD development. Whole body knockout of p14ARF led to increased atherosclerosis development [89], whereas p16INK4a deletion increased cell proliferation following carotid injury [90], but bone marrow specific loss or overexpression had no effect on atherogenesis [91, 92]. Notably, bone marrow-specific heterozygous deficiency of both p14ARF and p16INK4a had been shown to accelerate atherosclerosis [93]. Recently, loss of Cdkn2b in mice was shown to enhance aneurysm and atherosclerotic plaque formation primarily through altering smooth muscle cell (SMC) physiology [94, 95]. In these studies, SMCs of Cdkn2b-null mice exhibited increased apoptosis during aneurysm formation and an accumulation of apoptotic bodies in atherosclerotic plaques, which were resistant to cell clearance, leading to increased foam cell formation and inflammatory cytokine secretion. Taken together, these findings support a role for both Cdkn2a and Cdkn2b in regulating cellular behavior during vascular disease development but the exact role of each gene in CAD development has yet to be fully elucidated [96]. Collectively, these studies illustrate the inherent complexity of unraveling the biological mechanisms of GWAS hits in disease, even with the use of large-scale genomic efforts.
In Vivo Mouse Models to Study CAD Targets—TCF21 Locus
In 2011, a meta-analysis of 14 CAD GWA studies expanded the number of genome-wide significant loci to 46 [97], including a locus at 6q23.2. The lead SNP at this locus, rs12190287, is located in the 3′ untranslated region (UTR) of the basic helix-loop-helix transcription factor TCF21, a gene for which it is also a strong eQTL [97]. Subsequent investigation revealed that the minor protective G allele of rs12190287 alters both an activator protein 1 (AP-1) enhancer element and a repressive WT1 binding site, resulting in altered transcription factor binding in response to PDGF-β signaling and decreased TCF21 expression in vitro [49]. Interestingly, the minor protective G allele was also found to alter a seed binding sequence for miR-224, resulting in reduced miR-224 binding and thus increased levels of the G allele-containing TCF21 mRNA transcript [98]. Due to this variant’s existence at the intersection of multiple mechanisms of gene regulation, its influence on TCF21 expression in vivo during the disease process is expected to be complex.
TCF21 is a particularly compelling CAD-associated gene because of its role in coronary vascular development. In the context of cardiac development, Tcf21-expressing cells are initially observed in the proepicardium around E9.5 [99] and then migrate across the surface of the developing heart where they contribute to the multipotent epicardium [100]. Lineage tracing studies in mouse embryos have shown that these Tcf21-expressing epicardial cells undergo epithelial to mesenchymal transition (EMT), migrate into the heart and differentiate into both cardiac fibroblasts and coronary artery smooth muscle cells (caSMCs) [101]. In Tcf21-null mouse embryos, caSMCs are formed but show premature differentiation [102] and adventitial fibroblasts are almost completely absent [101], indicating that Tcf21 plays key roles in the differentiation of both cell types from a common progenitor. Indeed, Tcf21 was subsequently found to directly bind to and suppress expression of alpha smooth muscle actin (ACTA2) in cultured human caSMCs, and siRNA-mediated TCF21 knockdown in these cells resulted in a more differentiated gene expression program including upregulation of ACTA2, TAGLN, and MYH11 [30••].
Strikingly, Tcf21 lineage tracing studies, in which cells expressing Tcf21 are permanently labeled with a fluorescent marker at a specific time point, revealed that these cells actively participate in the evolving atherosclerotic lesion. In the LDLR−/− model of atherosclerosis, Tcf21 linage-traced cells in the adventitia are observed to proliferate, migrate through the media into the neointima, and ultimately align at the fibrous cap of the lesion where they express markers of mature smooth muscle cells including Acta2, Tagln, and Tgfbrb [30••]. The expression of this CAD-associated gene in cells that appear to be playing an important role in the formation of the lesion’s fibrous cap further implicates TCF21 in the vascular wall pathobiology of atherosclerosis. However, to ultimately confirm a causal role for TCF21 and to determine the effect of TCF21 on plaque stability, it will be necessary to perturb Tcf21 expression and determine the effect on plaque size, architecture, and vulnerability.
Application of CRISPR/Cas9 Genome Editing to Study CAD Loci
We are now in an unprecedented era of genome technology development, including a repertoire of nuclease-based tools to quickly and precisely edit specific genomic regions in various cells and organisms. The applications of zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and recently developed clustered regularly interspaced short palindromic repeats (CRISPR) coupled to CRISPR-associated Cas9 system each have their selective advantages and limitations with regard to specificity, efficiency, and off-target effects [103–105].
The highly efficient and versatile CRISPR/Cas9 systems require only a 20-bp sequence near a 5′-NGG-3′ protospacer-adjacent motif (PAM) for the guide RNA to hybridize and allow Cas9 to generate an adjacent double strand break (DSB) or single-strand nick, depending on the application. CRISPR/Cas9 was recently used to disrupt the CAD-associated TRIB1 locus in HepG2 cells to measure the effects on lipoprotein metabolism [106]. Interestingly, another study generated a deletion of a MEF2 binding site harboring a SNP rs9349379, located in the intronic region of the CAD-associated PHACTR1 locus [107]. Upon deletion, endothelial cells heterozygous for this SNP had reduced PHACTR1 levels, which established a missing functional link between this variant and CAD risk in the vessel wall. It remains to be determined precisely how this gene alters vascular injury responses, as PHACTR1 levels were unchanged with VEGF, TNFa, or shear stress perturbations [107]. CRISPR/Cas9 was further applied in vivo using adenovirus to induce loss-of-function mutations in the PCSK9 locus [108]. These studies revealed decreased plasma PCSK9 levels, consistent with a reduction in plasma cholesterol levels, together supporting an attractive therapeutic approach to control hypercholesterolemia in humans. The major advantage is that this adenoviral intervention would represent a one-time injection to effectively modify and shut down a critical LDL-cholesterol regulatory pathway in the circulation.
Recently, TALEN genome-editing strategies have targeted a GWAS variant for fetal hemoglobin levels located in an intronic enhancer of the BCL11A gene. These experiments have shown that biallelic excision of the intronic segment carrying the variant profoundly reduced BCL11A transcripts levels and almost completely abolished BCL11A protein synthesis [109]. More recently, CRISPR/Cas9 in situ saturating mutagenesis was used to dissect the 12-kb composite enhancer in BCL11A and showed that enhancer deletion abolished the expression of BCL11A and induced beta-globin and fetal hemoglobin (HbF) levels, thus providing a potential targeting strategy in restoring hemoglobin levels [110]. These high-throughput applications of Cas9 have already been applied to dissect the function of regulatory variants in large haplotype blocks (up to several kb), and as they are designed to probe regulatory enhancer function within their in vivo chromatin environment will likely continue to advance our understanding of the impact of common variants on CAD phenotypes. Whether CRISPR/Cas9 will be applicable to therapeutic targeting pathways operating in the vessel wall remains to be established.
Conclusion
In brief, this review summarizes current progress in the genomics field related to CAD. We emphasize that high-throughput genomics studies in vascular cells/tissues are needed to identify causal variants and mechanisms contributing to maladaptive or adaptive vessel wall responses during disease. We highlight some of the more widely adopted genomics approaches critical to prioritize candidate variants and genes for downstream follow-up analysis, ideally using established injury and disease models in vivo. Finally, forthcoming studies using CRISPR/Cas9 technology, inducible and cell-specific gene deletions will be of great importance to unravel the complex web of gene and disease interactions.
There are many future challenges and pitfalls in defining the exact causal CAD variants, genes, and mechanisms, and ultimately, the combination of big data from diverse experiments and tissues will lead to a more comprehensive and explanatory view on CAD [35]. Currently, the high cost of genomics experiments and required computational expertise represent drawbacks that may divert many groups away from genomics. As the price of next-generation sequencing continues to decline and biomedical groups become increasingly more multidisciplinary (wet-lab and bio-computational), one would expect future scientific endeavors to harness significant biological insights from genomics findings alone.
Acknowledgments
Robert Wirka receives grant support from the National Institute of Health (NIH) (F32HL129670-01). Clint L. Miller receives grant support from NIH (HL125912). Thomas Quertermous receives grant support from NIH (U01HL107388, HL109512, R21HL120757) and from the LeDucq Foundation.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Milos Pjanic, Clint L. Miller, Robert Wirka, Juyong B. Kim, Daniel M. DiRenzo, and Thomas Quertermous declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Kuska B. Beer, Bethesda, and biology: how “genomics” came into being. J Natl Cancer Inst. 1998;90(2):93. doi: 10.1093/jnci/90.2.93. [DOI] [PubMed] [Google Scholar]
- 2••.Consortium C A.D. A comprehensive 1000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–30. doi: 10.1038/ng.3396. This study provides a largest GWAS meta-analysis for the coronary artery disease including 60,801 cases and 123,504 controls from 48 individual GWAS studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium CAD et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park DS, et al. Adapt-Mix: learning local genetic correlation structure improves summary statistics-based analyses. Bioinformatics. 2015;31(12):i181–9. doi: 10.1093/bioinformatics/btv230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagliano SA, et al. A Bayesian method to incorporate hundreds of functional characteristics with association evidence to improve variant prioritization. PLoS One. 2014;9(5):e98122. doi: 10.1371/journal.pone.0098122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuk O, et al. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109(4):1193–8. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kichaev G, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10(10):e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trynka G, et al. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am J Hum Genet. 2015;97(1):139–52. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang IM, et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol Syst Biol. 2012;8:594. doi: 10.1038/msb.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makinen VP, et al. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10(7):e1004502. doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huan T, et al. A systems biology framework identifies molecular underpinnings of coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33(6):1427–34. doi: 10.1161/ATVBAHA.112.300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Miller CL, Pjanic M, Quertermous T. From locus association to mechanism of gene causality: the devil is in the details. Arterioscler Thromb Vasc Biol. 2015;35(10):2079–80. doi: 10.1161/ATVBAHA.115.306366. This editorial provides a good overview of methods to identify causal variation and causal genes, and reviews a recent paper in the field. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon SM, et al. Perspectives of integrative cancer genomics in next generation sequencing era. Genome Inform. 2012;10(2):69–73. doi: 10.5808/GI.2012.10.2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11(7):476–86. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S, et al. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32(9):926–32. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPherson A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7(5):e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Geijn B, et al. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods. 2015;12(11):1061–3. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayba O, et al. MBASED: allele-specific expression detection in cancer tissues and cell lines. Genome Biol. 2014;15(8):405. doi: 10.1186/s13059-014-0405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirinen M, et al. Assessing allele-specific expression across multiple tissues from RNA-seq read data. Bioinformatics. 2015;31(15):2497–504. doi: 10.1093/bioinformatics/btv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen SK, et al. Integrative DNA, RNA, and protein evidence connects TREML4 to coronary artery calcification. Am J Hum Genet. 2014;95(1):66–76. doi: 10.1016/j.ajhg.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105(2):83–9. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali SR, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res. 2014;115(7):625–35. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- 29.Chu M, et al. A novel role of CDX1 in embryonic epicardial development. PLoS One. 2014;9(7):e103271. doi: 10.1371/journal.pone.0103271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Nurnberg ST, et al. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet. 2015;11(5):e1005155. doi: 10.1371/journal.pgen.1005155. This study using multiple functional and in vivo assays demonstrates that the TCF21 gene, one of the lead CAD GWAS hits, is indeed causal for CAD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell RD, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34(6):1249–59. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez-Arcelus M, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feinberg AP. Epigenomics reveals a functional genome anatomy and a new approach to common disease. Nat Biotechnol. 2010;28(10):1049–52. doi: 10.1038/nbt1010-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–40. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 35.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim M, et al. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5(3):e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma P, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27(7):357–65. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 38.Dick KJ, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–8. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 39.Lamon-Fava S, Wilson PW, Schaefer EJ. Impact of body mass index on coronary heart disease risk factors in men and women. The Framingham Offspring Study. Arterioscler Thromb Vasc Biol. 1996;16(12):1509–15. doi: 10.1161/01.atv.16.12.1509. [DOI] [PubMed] [Google Scholar]
- 40.Putku M, et al. CDH13 promoter SNPs with pleiotropic effect on cardiometabolic parameters represent methylation QTLs. Hum Genet. 2015;134(3):291–303. doi: 10.1007/s00439-014-1521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banovich NE, et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10(9):e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30(11):1095–106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.del Rosario RC, et al. Sensitive detection of chromatin-altering polymorphisms reveals autoimmune disease mechanisms. Nat Methods. 2015;12(5):458–64. doi: 10.1038/nmeth.3326. [DOI] [PubMed] [Google Scholar]
- 47.Hazelett DJ, et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 2014;10(1):e1004102. doi: 10.1371/journal.pgen.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao L, et al. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller CL, et al. Disease-related growth factor and embryonic signaling pathways modulate an enhancer of TCF21 expression at the 6q23.2 coronary heart disease locus. PLoS Genet. 2013;9(7):e1003652. doi: 10.1371/journal.pgen.1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reschen ME, et al. Lipid-induced epigenomic changes in human macrophages identify a coronary artery disease-associated variant that regulates PPAP2B Expression through Altered C/EBP-beta binding. PLoS Genet. 2015;11(4):e1005061. doi: 10.1371/journal.pgen.1005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez D, et al. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods. 2013;10(2):171–7. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crawford GE, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16(1):123–31. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2(9):e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Buenrostro JD, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. doi: 10.1038/nmeth.2688. This study demonstrates the application of ATAC-Seq, a method for probing open chomatin regions, that dramatically reduces the number of cells needed for the experiment while preserving the resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193(4256):848–56. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 59.Enver T, Brewer AC, Patient RK. Simian virus 40-mediated cis induction of the Xenopus beta-globin DNase I hypersensitive site. Nature. 1985;318(6047):680–3. doi: 10.1038/318680a0. [DOI] [PubMed] [Google Scholar]
- 60.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bell O, et al. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12(8):554–64. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 62.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482(7385):390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sazonova O, et al. Characterization of TCF21 downstream target regions identifies a transcriptional network linking multiple independent coronary artery disease loci. PLoS Genet. 2015;11(5):e1005202. doi: 10.1371/journal.pgen.1005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winter DR, et al. DNase-seq predicts regions of rotational nucleosome stability across diverse human cell types. Genome Res. 2013;23(7):1118–29. doi: 10.1101/gr.150482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schep AN, et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25(11):1757–70. doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295(5558):1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 67.Harismendy O, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470(7333):264–8. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davison LJ, et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum Mol Genet. 2012;21(2):322–33. doi: 10.1093/hmg/ddr468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–76. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–90. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Getz GS, Reardon CA. Animal models of atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32(5):1104–15. doi: 10.1161/ATVBAHA.111.237693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 76.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helgadottir A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40(2):217–24. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 79.Congrains A, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220(2):449–55. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 80.Cunnington MS, Keavney B. Genetic mechanisms mediating atherosclerosis susceptibility at the chromosome 9p21 locus. Curr Atheroscler Rep. 2011;13(3):193–201. doi: 10.1007/s11883-011-0178-z. [DOI] [PubMed] [Google Scholar]
- 81.Folkersen L, et al. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS One. 2009;4(11):e7677. doi: 10.1371/journal.pone.0007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cunnington MS, et al. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6(4):e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Motterle A, et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet. 2012;21(18):4021–9. doi: 10.1093/hmg/dds224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holdt LM, et al. Expression of Chr9p21 genes CDKN2B (p15(INK4b)), CDKN2A (p16(INK4a), p14(ARF)) and MTAP in human atherosclerotic plaque. Atherosclerosis. 2011;214(2):264–70. doi: 10.1016/j.atherosclerosis.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 86.Jarinova O, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29(10):1671–7. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 87.Pilbrow AP, et al. The chromosome 9p21.3 coronary heart disease risk allele is associated with altered gene expression in normal heart and vascular tissues. PLoS One. 2012;7(6):e39574. doi: 10.1371/journal.pone.0039574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Visel A, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464(7287):409–12. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gonzalez-Navarro H, et al. p19(ARF) deficiency reduces macrophage and vascular smooth muscle cell apoptosis and aggravates atherosclerosis. J Am Coll Cardiol. 2010;55(20):2258–68. doi: 10.1016/j.jacc.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 90.Gizard F, et al. PPAR alpha inhibits vascular smooth muscle cell proliferation underlying intimal hyperplasia by inducing the tumor suppressor p16INK4a. J Clin Invest. 2005;115(11):3228–38. doi: 10.1172/JCI22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fuster JJ, et al. Increased gene dosage of the Ink4/Arf locus does not attenuate atherosclerosis development in hypercholesterolaemic mice. Atherosclerosis. 2012;221(1):98–105. doi: 10.1016/j.atherosclerosis.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 92.Wouters K, et al. Bone marrow p16INK4a-deficiency does not modulate obesity, glucose homeostasis or atherosclerosis development. PLoS One. 2012;7(3):e32440. doi: 10.1371/journal.pone.0032440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuo CL, et al. Cdkn2a is an atherosclerosis modifier locus that regulates monocyte/macrophage proliferation. Arterioscler Thromb Vasc Biol. 2011;31(11):2483–92. doi: 10.1161/ATVBAHA.111.234492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leeper NJ, et al. Loss of CDKN2B promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. Arterioscler Thromb Vasc Biol. 2013;33(1):e1–10. doi: 10.1161/ATVBAHA.112.300399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kojima Y, et al. Cyclin-dependent kinase inhibitor 2B regulates efferocytosis and atherosclerosis. J Clin Invest. 2014;124(3):1083–97. doi: 10.1172/JCI70391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim JB, et al. Effect of 9p21.3 coronary artery disease locus neighboring genes on atherosclerosis in mice. Circulation. 2012;126(15):1896–906. doi: 10.1161/CIRCULATIONAHA.111.064881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miller CL, et al. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014;10(3):e1004263. doi: 10.1371/journal.pgen.1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu J, Richardson JA, Olson EN. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Dev. 1998;73(1):23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 100.Acharya A, et al. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis. 2011;49(11):870–7. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Acharya A, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139(12):2139–49. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Braitsch CM, et al. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol. 2012;368(2):345–57. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest. 2014;124(10):4154–61. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller CL, et al. Dissecting the causal genetic mechanisms of coronary heart disease. Curr Atheroscler Rep. 2014;16(5):406. doi: 10.1007/s11883-014-0406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nagiec MM, et al. Modulators of hepatic lipoprotein metabolism identified in a search for small-molecule inducers of tribbles pseudokinase 1 expression. PLoS One. 2015;10(3):e0120295. doi: 10.1371/journal.pone.0120295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beaudoin M, et al. Myocardial infarction-associated SNP at 6p24 interferes with MEF2 binding and associates with PHACTR1 expression levels in human coronary arteries. Arterioscler Thromb Vasc Biol. 2015;35(6):1472–9. doi: 10.1161/ATVBAHA.115.305534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding Q, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–7. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–7. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]