Summary
Heterotrophic Proteo- and Actinobacteria were isolated from Lake Matano, Indonesia, a stratified, ferruginous (iron-rich), ultra-oligotrophic lake with phosphate concentrations below 50 nM. Here, we describe the growth of eight strains of heterotrophic bacteria on a variety of soluble and insoluble sources of phosphorus. When transferred to medium without added phosphorus (P), the isolates grow slowly, their RNA content falls to as low as 1% of cellular dry weight, and 86-100% of the membrane lipids are replaced with amino- or glycolipids. Similar changes in lipid composition have been observed in marine photoautotrophs and soil heterotrophs, and similar flexibility in phosphorus sources has been demonstrated in marine and soil-dwelling heterotrophs. Our results demonstrate that heterotrophs isolated from this unusual environment alter their mac-romolecular composition, which allows the organisms to grow efficiently even in their extremely phosphorus-limited environment.
Introduction
Phosphorus (P) is essential to biological information storage and transfer, energy metabolism, and membrane integrity. In bacteria, P limitation often results in fewer ribosomes and slower growth rates (Zimmerman et al., 2014), up-regulation of transcripts related to P acquisition and storage (Lamarche et al., 2008; Martín et al., 2012; Palenik, 2015), and altered membrane lipid composition (Van Mooy et al., 2009; Geiger et al., 2010; Carini et al., 2015). Bacteria in environments with chronically low nutrient concentrations may also have streamlined genomes (Dufresne et al., 2005; Giovannoni et al., 2005), more frequent lateral gene transfer (Souza et al., 2008), and modified genome and proteome structures (Elser et al., 2011). Here, we test the hypothesis that heterotrophs isolated from the chronically P-limited Lake Matano, Indonesia, have reduced P requirements and utilize diverse P sources.
To reduce their P requirement, bacteria must synthesize fewer P-rich macromolecules. The major P pools in bacteria are RNA, lipids, and DNA, with smaller pools of free nucleotides (Mitchell and Moyle, 1954). In response to P limitation, microbes may reduce the size of DNA pool by maintaining fewer genome copies (Zerulla et al., 2014); longer-term P starvation often results in microbes with small genomes (Dufresne et al., 2005; Giovannoni et al., 2005). RNA is usually the largest pool of P in cells (Mitchell and Moyle, 1954; Mundry and Kuhn, 1991), so reducing the amount of RNA is common when P is limiting (Elser et al., 2003). The number of ribosomes is correlated with growth rate, and bacterial cells living in nutrient-limited conditions tend to have fewer ribosomes and thus smaller RNA pools (Condon et al., 1995; Elser et al., 2008). The relationship between P availability, P demand, carbon metabolism, and growth efficiency is still not completely understood (Lipson et al., 2008; Godwin and Cotner, 2015; Roller and Schmidt, 2015). Carbon and phosphorus cycling are linked through microbial growth, but flexibility in P demand can change the degree to which they are coupled under different conditions (Keiblinger et al., 2010; Scott et al., 2012; Godwin and Cotner, 2015).
Some bacteria reduce their P demand by synthesizing membrane lipids with P-free head-groups (Hölzl and Dörmann, 2007; Van Mooy et al., 2009; Carini et al., 2015), since as much as 21% of cellular P may be in membrane lipids (Mitchell and Moyle, 1954). Although planktonic marine heterotrophs do not typically alter their lipid headgroups in response to P starvation (Van Mooy et al., 2008, 2009; Van Mooy and Fredricks, 2010; Popendorf et al., 2011), a Pelagibacter strain isolated from the Sargasso Sea replaces phospholipids with amino- and glycolipids when grown with methylphosphonate or low phosphate (Carini et al., 2015). Similarly, a variety of soil-dwelling heterotrophs substitute amino lipids for phospholipids when P is limiting (Geiger et al., 2010; Vences-Guzmán et al., 2012; Parsons and Rock, 2013). Altering the sizes and compositions of P-rich macromolecular pools allows the cells to reduce their P demand and recycle intracellular P (Condon et al., 1995; Vences-Guzmán et al., 2012; Parsons and Rock, 2013).
Lake Matano, Indonesia, is a stratified, ferruginous (iron-rich), ultra-oligotrophic tropical lake, located in South Sulawesi, Indonesia (Crowe et al., 2008). It is ~2-4 million years old, and is located in a steep-sided graben 380 m above sea level on the Matano Fault (Brooks, 1950; Hamilton, 1979). It has a total area of 164 km2, and, with a depth of about 590 m, is one of the ten deepest lakes in the world (Crowe et al., 2008; Vaillant et al., 2011); its depth and steep sides prevent efficient mixing by wind, and because the water temperatures are stable, thermal mixing is rare, though possible (Katsev et al., 2010). Water inflows include three tributaries and groundwater input, but the primary input is precipitation (Katsev et al., 2010). Unlike most other freshwater systems, Lake Matano does not undergo seasonal mixing, so the dissolved P below the chemocline stays trapped (Crowe et al., 2008). Nutrient inputs from land and atmospheric deposition are small (Katsev et al., 2010), and the particulate iron minerals in the surface water adsorb any soluble P (Crowe et al., 2008). The permanent stratification, iron content, and extremely low nutrient inputs make Lake Matano’s P deficiency unusually severe (Crowe et al., 2008; Sabo et al., 2008).
Lakes with P concentrations < 4 μg L-1 (< 129 nM) are classified as “ultra-oligotrophic” (Vollenweider, 1982), and the P in Lake Matano is, at most, 50 nM (Crowe et al., 2008). In deep tropical lakes comparable to Lake Matano, dissolved P ranges from 70 to 760 nM, while in temperate stratified lakes, dissolved P is typically 100 to 160 nM (Hecky et al., 1993). In comparison, the surface water of Lake Matano is chronically P-limited, making this lake an effective natural laboratory for analyzing the effects of long-term nutrient limitation on planktonic microbes.
The extremely low phosphate concentrations in Lake Matano suggest that heterotrophic bacteria there might need additional strategies for acquiring P or reducing the cellular P requirements to survive in the near-absence of this essential nutrient. Additionally, the C:P ratio in freshwater bacteria has been shown to be highly variable (Cotner et al., 2010), indicating that freshwater heterotrophs may have extremely flexible P requirements and a range of growth efficiencies. To examine the P requirements and metabolic flexibility of the heterotrophic microbial community, we isolated heterotrophic bacteria from Lake Matano, characterized their growth on different concentrations and types of P sources, and assessed the P-containing macromolecular pools of cells grown under different conditions. To investigate the effect of P limitation alone, cultures were provided with abundant C and N; the effects of C and N limitation are not discussed here.
Results
Strain isolation and identification
Surface water from Lake Matano was spread on to solid minimal medium amended with 0.1 to 1% organic carbon to isolate aerobic heterotrophs. Based on morphological characteristics and taxonomic identity, eight axenic strains were selected for further study (Table 1). Of these, one (LM-2, most closely related to Microbacterium testaceum) is Gram-positive. Three are Alphaproteobacteria in the Agrobacterium/Rhizobium group (LM-1, LM-5, and LM6-1), but differ in their growth rates and ability to use organophosphates. The strain LM-P is most closely related to Methylobacterium podarium. The remaining three strains are Gammaproteobacteria related to Acinetobacter and Pantoea spp.
Table 1.
16S rRNA identification of strains isolated from Lake Matano surface water.
| Strain ID | Closest cultivated organism in NCBI (accession number) | % identity | Query coverage | Accession number |
|---|---|---|---|---|
| Actinobacteria | ||||
| LM-2 | Microbacterium testaceum (HE716908) | 99% | 100% | KM884890 |
| Alphaproteobacteria | ||||
| LM-1 | Agrobacterium tumefaciens (HQ871877) | 99% | 99% | KM884891 |
| LM-5 | Rhizobium sp. (HG518323) | 99% | 99% | KM884892 |
| LM6-1 | Agrobacterium tumefaciens (HQ871877) | 99% | 99% | KM884893 |
| LM-P | Methylobacterium podarium (AY468363) | 99% | 100% | KM884894 |
| Gammaproteobacteria | ||||
| LM6-2 | Acinetobacter sp. (GU566324) | 99% | 99% | KM884895 |
| 2-LM-22 | Acinetobacter sp. (GU566324) | 99% | 99% | KM884896 |
| LM-Y | Pantoea ananatis (DQ195523) | 99% | 99% | KM884897 |
Growth on dissolved P sources
To identify the types of phosphorus-containing compounds that can serve as P sources for the Lake Matano isolates, cultures were grown in the National Botanical Research Institute’s phosphate growth medium (NBRIP, Nautiyal et al., 1999) with no additional P, 0.2 mM K2HPO4, or 0.2 mM organic phosphorus [para-nitrophenylphosphate (p-NPP), an organic phosphate ester, or 2-amino-ethylphosphonic acid (2-AEP), a phosphonate]. Maxi- mum growth rates varied depending on the P source (Fig. 1A, Fig. S1), but all strains had similar growth yields when supplied with inorganic soluble phosphate or p-NPP (Fig. 1B). Although maximum growth rates and growth yields were much lower in the absence of P, only two strains failed to grow under these conditions, suggesting that most of these isolates need very little phosphorus to grow.
Figure 1. Bacterial growth rates and growth yields with different phosphorus sources.
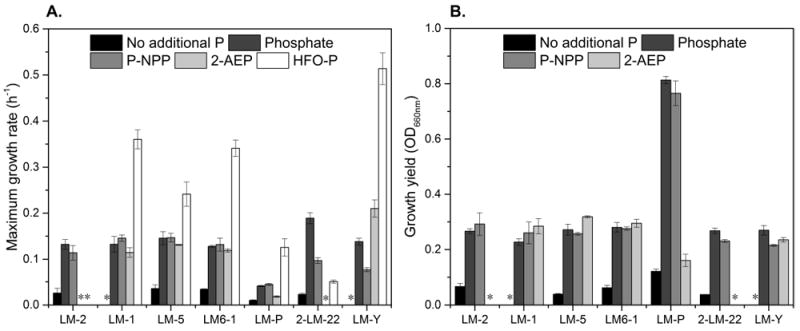
Cells were grown in NBRIP medium without additional P (black bars), with 0.2 mM phosphate (dark gray bars), 0.2 mM p-NPP (gray bars), 0.2 mM 2-AEP (light gray bars), or with hydrous ferric oxide co-precipitated with phosphate (HFO-P; white bars). Measurements are averages of 3 biological replicates for soluble P sources, and 2 biological replicates for HFO-P, and error bars indicate standard deviations. All strains were grown until stationary phase was reached. Growth was monitored by optical density at 660 nm for growth on soluble sources of P, and by plate counting for growth on HFO-P; asterisks indicate conditions in which no growth was observed. The 16S rRNA sequences for isolates LM6-2 and 2-LM-22 are nearly identical, and because they had similar growth patterns, only the results for 2-LM-22 are plotted here. (A) Maximum growth rates calculated from time points taken during exponential growth. See Supplementary Figure 1 for individual growth curves. (B) Growth yields were calculated by subtracting starting optical density from maximum optical density at 660 nm. Because growth on HFO-P was monitored by plate counts, the maximum optical density for those experiments is not included here.
All isolates used p-NPP as a P source, showing that all are capable of cleaving phosphoester bonds (Fig. 1). In fact, the growth rates of all the Rhizobacterial isolates on p-NPP were close to the growth rates on an equivalent amount of soluble inorganic phosphate. Similarly, three strains, LM-1, LM-5, and LM6-1, grow at similar rates when inorganic phosphate, p-NPP, and 2-AEP are available (Fig. 1). LM-Y grows at similar rates on phosphate and 2-AEP, but more slowly on p-NPP. LM-P has similar maximum growth rates on all of these P sources, but its growth yield is much lower on 2-AEP than on the others. Two strains, LM-2 and 2-LM-22, seem unable to utilize 2-AEP as a P source, since the OD660 nm of these cultures did not change over the course of the experiment (Fig. 1B, Fig. S1A-G).
Growth on insoluble P
To determine whether mineral-associated P can be accessed by planktonic microbes in Lake Matano, all strains were grown with ~0.2 mM P provided as phosphate co- precipitated with hydrous ferric oxide (HFO-P). In all cases, growth in medium with HFO-P was much faster than growth with no additional phosphate (Fig. 1A): all strains except LM-2 are capable of obtaining P from HFO-P. As a biological control, E. coli was grown in the same conditions, and solution pH, P, and Fe were quantified after 6 days. The solution pH, P, and Fe in the E. coli culture were similar to those of LM-1 and LM6-1, and higher than those of LM-P and 2- LM-22 (Fig. 2). LM-2 is the only strain apparently unable to acquire P from HFO-P: it neither grows (Fig. 1) nor releases P or Fe into solution, and the pH in this culture stays close to neutral (Fig. 2). In contrast, LM-Y catalyzes rapid dissolution of the mineral and releases large amounts of Fe and P compared to all of the other strains (Fig. 2). To explore whether additional phosphate might inhibit the dissolution of the HFO-P by LM-Y, phosphate (0.2 mM) was added to the medium along with HFO-P. The growth rate of LM-Y was the same with or without additional phosphate (not shown), but ~35% more Fe was released in the presence of excess phosphate (Fig. 2).
Figure 2. Growth of isolates with HFO-P as the only P source.
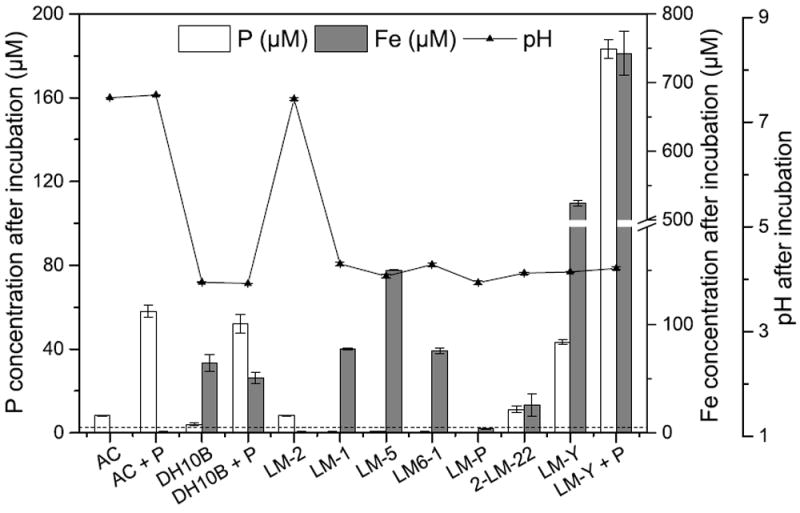
Both P (white bars) and Fe (gray bars) are released from the minerals by all strains, and all strains except LM-2 lower the pH to ~4. Samples for pH, Fe, and P quantification were taken after 6 days’ incubation and were averages of 2 biological replicates; error bars indicate standard deviations between biological replicates. The limits of detection for P and Fe are 2.45 μM and 0.08 μM, respectively. The limit of detection of P is indicated by the dashed line. AC = Abiotic control; DH10B = E. coli strain DH10B. Columns annotated with “+P” had both HFO-P and 0.2 mM K2HPO4 in the solution.
Intracellular polyphosphate storage
Polyphosphate granules may comprise up to 10% of bacterial dry weight (Deinema et al., 2011) or even higher (Nakamura et al., 1995), and storage of P as polyphosphate provides cells with a pool of phosphate that can be easily converted to ATP or GTP (Zhang et al., 2002). After an extended period of P-starvation followed by K2HPO4 addition, polyphosphate accumulation in the Lake Matano isolates was assessed with Neisser staining and light microscopy. Two strains, LM-2 and LM-P, accumulate intracellular polyphosphate granules after this treatment (Fig. 3; see Fig. S3 for images of other strains).
Figure 3. Polyphosphate storage by LM-2 and LM-P.
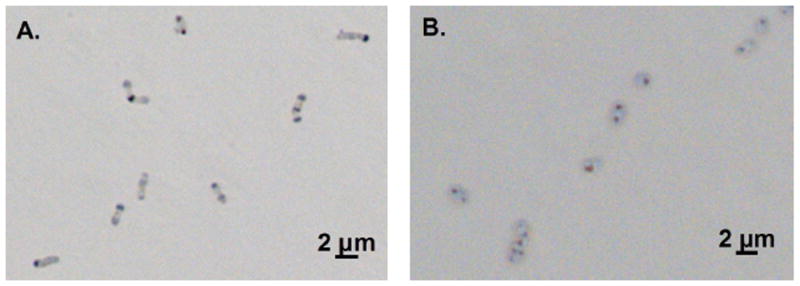
Polyphosphate granules were synthesized during and after P-starvation by LM-2 (A) and LM-P (B). Photos were taken 21.5 and 24.75 hours after transfer into medium with no additional P, respectively. Polyphosphate was stained with methylene blue and appears as dark spots in cells visualized with brightfield microscopy.
Intact polar lipids of P-starved strains
Although the polyphosphate storage pool can be quite large, the major non-nucleotide pool of phosphorus in microbial cells is usually the lipid membrane (Mitchell and Moyle, 1954). When cells were grown with 0.2 mM phosphate in the medium, the membranes of the Lake Matano isolates were 66 to 100% phospholipids (Fig. 4A and Supplementary Table 1). Betaine lipids (DGTS type) comprised 4 to 25% of the lipids in the Alphaproteobacteria and 2-LM-22, a Gammaproteobacterium (Fig. 4A). Under P-replete conditions, LM-2, the only Gram-positive organism isolated, was the only organism in which glycolipids comprised more than 1% of the total lipids (Fig. 4 and Supplementary Table 1).
Figure 4.
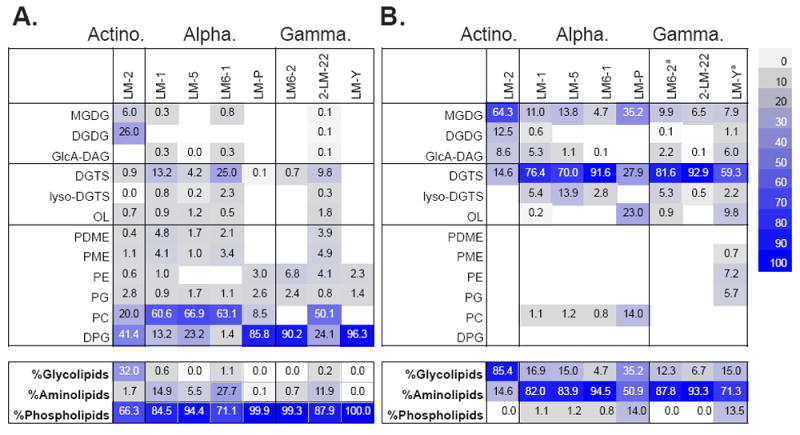
Intact polar lipid (IPL) composition of heterotrophic isolates when grown (A) with 0.2 mM phosphate or (B) without added phosphate (< 2.45 μM). Although all strains are capable of phospholipid production, phospholipids are almost completely absent in cells grown with <2.45 μM P in the medium. Colored boxes indicate percent of total lipids. Data for LM6-2 and LM-Y in panel B was obtained separately from data for other strains, because of low-biomass samples. These samples were analyzed ~6 months apart and some variability within strains was observed between samples. See supplemental tables S1-S3 for details on the differences between the analyses.
Abbreviations: MGDG: monoglycosyl diacylglycerol; DGDG: diglycosyl diacylglycerol; GlcA-DAG: glucuronosyl diacylglycerol; DGTS (betaine): diacylglyceryl-N,N,N-trimethylhomoserine; lyso-DGTS (betaine): monoacylglyceryl-N,N,N-trimethylhomoserine; OL: ornithine lipid; PE: phosphatidylethanolamine; PME: N-methylated PE; PDME: N-dimethylated PE; PG: phosphati-dylglycerol; PC: phosphatidylcholine; DPG: diphosphatidylglycerol.
In contrast, in P-limited medium (P <2.5 μM), no phospholipids were detected in two isolates, and phospholipid content in 6 of 8 strains was less than 2% (Fig. 4B). Instead, lipids in the Gram-negative isolates had primarily betaine headgroups (27 to 93%; Fig. 4B). Glycosylated headgroups such as monoglycosyl diacylglycerol (MGDG) and diglycosyl diacylglycerol (DGDG) were found in every isolate, and constituted the majority of the lipids in LM-2 (Fig. 4B). Only strain LM-P had an appreciable quantity of phospholipids (~14%; Fig. 4B and Figs. S3 and S4). Because the limits of detection for the lipids analyzed here are 1-10 pg (Table S1), anything below those detection limits is likely not synthesized by these cultures. Overall, growing cells without additional P resulted in a change from primarily phospholipids to almost entirely amino-lipids in the Gram-negative isolates, and to primarily glycolipids in the Gram-positive isolate (Fig. 4, bottom panel). All strains are capable of a dramatic reduction of P demand by replacing all or nearly all of the phospholipids with non-phospholipids.
RNA content of P-starved strains
The largest pool of phosphate in bacterial cells is usually the RNA pool, which may have up to 75% of the cellular P (Mitchell and Moyle, 1954). To assess the RNA pools in P-replete and P-starved cultures, isolates were grown with 0.2 mM phosphate or without additional P (< 2.45 μM) until mid-exponential or early stationary phase, and the RNA content of the cells was quantified with GC-MS (Long and Antoniewicz, 2014). For 5 of 7 strains tested, RNA made up 10-18% of the total dry weight of the cells in phosphate-replete conditions (Fig. 5). In the absence of additional P, however, the RNA pool was less than 5% of the dry weight of the cells, a 2- to nearly 6-fold reduction (Fig. 5).
Figure 5. RNA contents of heterotrophic bacteria grown with 0.2 mM (light gray bars) and <2.45 μM (dark gray bars) phosphate.
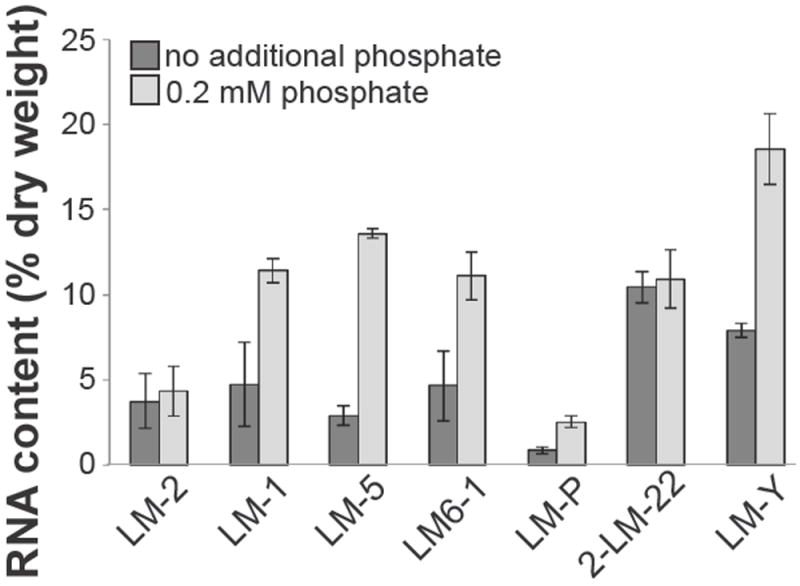
Values reported are the amount of RNA per cell as a percentage of cellular dry weight and are the averages of two independent experiments.
Discussion
Because they can replace all or nearly all of their phospholipids, and reduce the sizes of their RNA pools dramatically, the heterotrophic bacteria isolated from Lake Matano have very low P requirements. Although bacteria with flexible C:P ratios are often oligotrophs (Godwin and Cotner, 2015), we demonstrate that in some Lake Matano isolates, these changes occur whether the strains are copiotrophic or oligotrophic. A better understanding of the relationship between P availability and growth characteristics in individual isolates is crucial to understanding how carbon and P cycling are coupled in natural environments (Godwin and Cotner, 2015).
Changing the type of P available has little effect on growth rates and growth yields of the isolates described here. The potential P-bearing compounds available in Lake Matano include orthophosphate, organophosphates or phosphonates, or mineral-associated phosphate. In fact, in oligotrophic systems, organophosphates and phosphonates may be the most common P sources (Smith and Prairie, 2004; White and Metcalf, 2007). The algae, fish, invertebrates, and microbes present in the lake are potential sources of organic phosphate esters, which are common in lipids and ubiquitous in nucleic acids. Phosphonates are found in all domains of life, in phosphono-lipids, polysaccharides, and proteins, as well as in bioactive small molecules (Metcalf and van der Donk, 2009; McGrath et al., 2013; Yu et al., 2013). Genomic analysis indicates that ~10% of microbes in aquatic environments may synthesize phosphonates, and that representatives of the Proteobacteria, Firmicutes, Actinobacteria, Cyanobacteria, Euryarchaea, and Thaumarchaea encode at least one phosphonate biosynthetic gene (Yu et al., 2013). Similar (meta)genomic analyses have also identified a variety of freshwater bacteria capable of phosphonate uptake (Ilikchyan et al., 2009) and utilization (Livermore et al., 2014). Due to Lake Matano’s high iron content, phosphate-adsorbing iron oxyhydroxides are also present in the Lake Matano surface waters (Crowe et al., 2008; Zegeye et al., 2012). We tested growth on representatives of all of these classes of P sources. The growth rate data indicate that all of the isolates described here can grow using either inorganic or organic P provided in either the +3 or +5 oxidation state (Fig. 1), although two strains, LM-2 and 2-LM-22 (an Actinobacterium and Gammaproteobacterium, respectively) do not grow on 2-AEP, and LM-2 does not grow on HFO-P. Acidification of the medium by normal respiration appears to release enough P from HFO-P to support some growth by most isolates (Fig. 1A, Fig. 2). Uniquely among this group of isolates, LM-Y catalyzes rapid dissolution of HFO-P, releasing much more P than it utilizes. The suite of isolates described here, therefore, have the genes necessary to access a variety of P sources without affecting growth efficiency, and LM-Y may even be able to facilitate P supply to other organisms in its vicinity.
One mechanism for sustained growth when P is limiting is flexibility in P demand. Recent work has demonstrated that freshwater heterotrophs have extremely flexible P requirements (Elser et al., 2003; Cotner et al., 2010; Scott et al., 2012; Godwin and Cotner, 2014, 2015). However, very few of these studies have characterized individual intracellular P pools under different conditions. Here, we demonstrate that both lipid and nucleic acid pools in heterotrophic bacteria isolated from Lake Matano change in response to environmental conditions, providing a physiological explanation for the previously-reported results.
In bacteria isolated from freshwater lakes, Elser et al. demonstrated that 70-97% of cellular P was in the RNA pool (Elser et al., 2003; Makino et al., 2003), implying that phospholipid content was quite low in those strains. More specifically, heterotrophic bacteria isolated from a variety of environments have been shown to replace phospholipids with amino-, sulfo-, or glyco-lipids (Grant, 1979; Batrakov et al., 1996; Geiger et al., 2010; Vences-Guzmán et al., 2012, 2013; Geske et al., 2013; Carini et al., 2015). In the isolates described here, 66% to 100% of their membrane lipids are phospholipids under P-replete conditions (Fig. 4A). All isolates are capable of replacing 86-100% of these phospholipids with amino- or glycolipids when P is limiting (Fig. 4B), though both major lipid classes and the degree of replacement vary between experiments (see tables S2 and S3). This variability between experiments may be due to apparently minor differences in P, which was below our limit of detection in both experiments, differences in inoculum size, or variability in lysis rates, which were not measured. However, even under P-replete conditions, the lipids in bacteria constitute a relatively small fraction (< 21%) of the cellular P (Mitchell and Moyle, 1954). In most environments, the RNA pool is likely the major P pool in the cell.
In fact, the size of the RNA pool drives both P demand and growth rate when P is limiting in freshwater environments (Elser et al., 2003), indicating that changing the size of the RNA pool can have a large effect on the total cellular P demand. Of the isolates characterized here, 5 of the 7 synthesize less RNA when P is limiting. Some of these changes are quite large in magnitude: the RNA content of LM-5 goes from 13.6% of the dry weight of the cell in P-replete conditions to less than 3% in P-limiting conditions, an 80% reduction. Although a relationship between RNA content and growth rate in a variety of organisms has been reported (Elser et al., 2003), there was no apparent correlation of these two factors in our experiments. However, changes in cellular P demand can affect how efficiently cells grow, or how much biomass is produced per unit nutrient (Keiblinger et al., 2010; Sinsabaugh et al., 2013; Roller and Schmidt, 2015). Because it is the largest P pool in the cell, especially in oligotrophic freshwater environments (Elser et al., 2003; Makino and Cotner, 2004; Cotner et al., 2010), and has a large effect on both P requirement and growth rate (Elser et al., 2003), RNA content likely plays a major role in environmental nutrient cycling (Elser et al., 2003).
Growth efficiency is defined as biomass produced per unit resource, and integrates evolutionary history, physiological state, and environmental parameters (Azam and Malfatti, 2007; Keiblinger et al., 2010; Sinsabaugh et al., 2013; Roller and Schmidt, 2015). Both growth rate and growth efficiency have been correlated with P availability (Smith and Prairie, 2004). In general, oligotrophs are more efficient: they grow slowly but consistently over a wide range of conditions and have less RNA (Lauro et al., 2009; Keiblinger et al., 2010). Heterostoichs, organisms with biomass C:P ratios that vary as environmental C:P ratios vary, also typically have low P contents even when P is not limiting (Godwin and Cotner, 2015). Of our isolates, LM-P appears to be an oligotroph and heterostoich, as it has the slowest growth rates but consistently high growth yields (Fig. 1), and low RNA content even in P-replete conditions (Fig. 5). This slow but more efficient strategy is usually more useful in biofilms than in planktonically-growing cells (Lipson et al., 2008; Lauro et al., 2009), which may explain why only one oligotroph was isolated: the water used for isolation was filtered in the field to remove eukaryotes and large particles, where bio-films would be hosted.
In contrast, copiotrophs, although they are less efficient, typically grow rapidly in favorable conditions and have higher RNA contents (Lauro et al., 2009; Keiblinger et al., 2010). They are also often more easily isolated from planktonic populations (Lauro et al., 2009). Copiotrophs are typically homeostoichs, whose biomass C:P ratios stay constant at a range of environmental C:P ratios (Godwin and Cotner, 2015). Almost all of the isolates described here grow quickly for short periods, followed by long, stable stationary phases (Fig. S1). LM-Y and LM-1, which do not grow when no P is added to the medium, may be the most extreme examples of copiotrophs in this suite of isolates. However, all of the isolates described here can alter their lipid profiles, and to some degree their RNA contents. Although they were isolated under conditions that would most likely select for homeostoichs (Godwin and Cotner, 2014, 2015), the P requirements and P uptake characteristics of these strains clearly vary under different conditions.
These variations in efficiency mean that different species may grow better at different times: their niches are separate in time, rather than in space or substrate. Aquatic environments are micro-structured in both space and time (Yawata et al., 2014), and variation of resources in time allows oligotrophs and copiotrophs to coexist (Roller and Schmidt, 2015). Variable growth rates, similarly, would contribute to separation of niches in time (Azam and Malfatti, 2007), while internal storage of P (Fig. 3) could change the time frame over which some strains can utilize P that is only transiently available outside the cell.
We demonstrate here that both oligotrophic and copiotrophic bacteria exist among the planktonic bacteria in Lake Matano. Isolates belonging to the same genus had similar growth phenotypes and lipid compositions (Fig. 1, Fig. S1, Fig. 4). Other work has also found that growth efficiency primarily depends on species identity, not environmental factors (Keiblinger et al., 2010). Despite differences between taxa, isolates from all groups have a low P demand and show remarkable flexibility in the P content of their lipid and RNA pools, characteristics which make growth more efficient in the chronically P-limited Lake Matano. The enhanced ability of one strain to release P from minerals additionally suggests a potential mechanism for sharing of P resources, and the fact that not all strains can use 2-AEP as a P source indicates that some substrate specialization also exists. Taken together, the reduced demand, substrate specialization, and range of growth efficiencies allow a variety of heterotrophic bacteria to coexist in a chronically P-limited environment.
Experimental Procedures
Study site
Lake Matano is one of the ten deepest lakes in the world, located in South Sulawesi, Indonesia. It belongs to the Malili Lakes system, which includes Lakes Matano, Towuti, Mahalona, Masapi, and Lontoa (Vaillant et al., 2011). Water inflows include three tributaries as well as rainwater and groundwater input (Katsev et al., 2010). Water for cultivation, DNA sequencing, and other analyses was collected from 10 m depth at the main sampling station, 2°28’00” S and 121°17’00” E.
Isolation and identification of heterotrophs
Water (100 μL) collected from 10 m depth in Lake Matano was spread on plates containing NBRIP (Nautiyal, 1999), Luria-Bertani medium (Sambrook and Russell, 2001), or Lake Matano synthetic surface water. NBRIP medium contains, per liter, MgCl2•6H2O 5 g, MgSO4•7H2O 0.25 g, KCl 0.2 g, (NH4)2SO4 0.1 g, glucose 10 g, hydroxyapatite 5 g (Nautiyal, 1999). Lake Matano synthetic surface water medium contains, per liter, MgCl2•6H2O 0.133 g, MgSO4•7H2O 0.0037 g, NaHCO3 0.0235 g, CaCl2•2H2O 0.04267 g, Na2SO4 0.0025 g, KCl 0.0003 g and 0.1% (w/v) freshly prepared yeast extract. Where appropriate, medium was solidified with 1.5% agar. Plates were incubated at 30°C until colonies appeared (3-5 days). All colored colonies and several colorless colonies were restreaked on fresh medium until they were pure, as determined by microscopy and 16S rDNA sequencing.
Genomic DNA was extracted from cultures using either the MoBio Power Soil kit (Mo-Bio, Carlsbad, CA, Cat. No. 12888, for LM6-1 only) or the method described in (Frigaard et al., 2004). Following DNA extraction, 16S rRNA genes were amplified, cloned, and sequenced. 16S rDNA genes were amplified using primers 8F and 1492R (Reysenbach et al., 1994). The PCR conditions were as follows: 94°C for 3 min.; 34 cycles of 94°C 30 sec., 48.5°C 30 sec., and 68°C 1:30 min.; final extension at 68°C for 5 min. PCR products were ligated into the TOPO TA vector (Life Technologies, catalog #450071) and the 16S rRNA gene inserts in the plasmids were sequenced by Sanger sequencing at the University of Delaware Sequencing and Genotyping Center. Sequences were compared to the GenBank non-redundant nucleotide database using BLAST (Altschul et al., 1997) and have been deposited in GenBank under accession numbers KM884890 to KM884897. Isolates with unique 16S rRNA sequences were selected for further analysis.
Growth conditions
NBRIP without added P was used as the base medium for all experiments, and all glassware was acid-washed in 0.1N hydrochloric acid to remove trace phosphate. For P-limited medium, no additional P was added, though some may have been introduced via the water used in the medium or as a minor contaminant in other reagents. However, P in this medium was below the detection limit of the ICP-OES system, 0.076 ppm (~2.45 μM). For reference, 1.5-3 μM P has been used as a “limited P” concentration in studies of pure cultures and natural systems (Cotner et al., 2010; Erb et al., 2012; Reaves et al., 2012) and the conditions imposed in our laboratory are P-limited by this standard.
For P-replete liquid media, K2HPO4, para-nitrophenylphosphate (p-NPP), or 2-amino-ethylphosphonic acid (2-AEP) was added to P-free NBRIP to a final concentration of 0.2 mM. The model organophosphate p-NPP is commonly used to demonstrate production of extracellular phosphatases by isolates or in natural environments (e.g. Zhou et al., 2011; Stout et al., 2014). The concentration of free phosphate in a 0.2 mM solution of p-NPP was measured, and is ~2 μM. The model phosphonate 2-aminoethylphosphonate (2-AEP) is often used to test general ability to cleave phosphonate bonds, since it can be degraded by multiple pathways (e.g. Martinez, Tyson, & Delong, 2010; Muscarella, Bird, Larsen, Placella, & Lennon, 2014). The pH of the media was adjusted to 7.0 with a 1 mM solution of NaHCO3 and acetic acid. Cells were grown at 30°C with shaking at ~150 rpm, and growth was monitored by measuring optical density at 660 nm.
Prior to growth measurements, isolates were transferred to the medium to be used for the growth curve and adapted for 2 days. After adaptation, cultures were diluted to an OD660 nm of ~0.04 in the same medium and optical density was monitored until cultures reached stationary phase. Growth rates and yields were calculated from the averages of three replicate samples. Maximum growth rates were calculated based on the difference between OD660 nm at the end of the lag phase and OD660 nm at the end of exponential growth phase (see Fig. S1 for details). Strains were determined to be unable to utilize a substrate if no change in OD660 nm was observed over the course of 1 week.
Phosphorus and iron quantification
Soluble P and Fe concentrations were quantified by inductively coupled plasma optical emission spectrometry (ICP-OES). Solutions or culture supernatants were filtered through 0.2 μm filters and acidified with nitric acid (1% final concentration). Standard curves for P and Fe were made using commercial standards (999 ± 3 mg P L-1; Fluka Analytical, Sigma-Aldrich, or 100.0 ± 0.5 mg Fe L-1; HACH), and absorbance of the standards at 213.618 nm for P, or 238.201 nm for Fe was measured by ICP-OES to determine the relationship between concentration and emission intensity. The limits of detection are 2.45 μM and 0.8 μM for P and Fe, respectively. The ICP-OES system was a Perkin-Elmer Optima 7300 DV.
Co-precipitation of phosphate and hydrous ferric oxide (HFO-P)
The co-precipitation method was modified from Smith et al. (Smith et al., 2008). Briefly, a 2-L solution of 1 mM ferric chloride in 100 μM Na2HPO4 was prepared. The pH was adjusted to 6.0 with NaOH and stirred for 24 hours, then the precipitate was centrifuged and washed 3X with ddH2O. Then ddH2O was then added to the precipitate to a total volume of 30 mL. For cultures that used HFO-P as a P source, 1 mL of the HFO-P solution was added to 30 mL NBRIP medium (final volume), for a final P content of ~0.2 mM. The P and Fe concentrations in the solution supernatants in each step were quantified by ICP-OES, and were all below the detection limit (~2.45 μM and 0.8 μM, respectively).
For growth on HFO-P, colonies were transferred from NBRIP plates to liquid NBRIP without P, washed, resuspended in NBRIP with no additional P, and transferred to flasks with 30 mL NBRIP + HFO-P prepared as described above. All cultures started with ~106 cells mL-1, and growth was monitored by plate counting rather than optical density because the particulate HFO-P increases light-scattering in the culture. The abiotic control was NBRIP medium with HFO-P, and the supernatant of this solution was used to quantify the abiotic dissolution of the minerals under the experimental conditions. To assess the extent of mineral dissolution in the presence of any actively respiring bacteria, E. coli (DH10B) was grown in NBRIP with HFO-P and leucine (20 μg mL-1), with or without additional phosphate (0.2 mM). P or Fe concentrations in the supernatants of all cultures were determined by ICP-OES after a 6-day incubation period.
Growth and staining for polyphosphate analysis
Strains were tested for the ability to store poly-phosphate by incubating them in phosphate-free NBRIP medium for 48 - 90 hours to promote consumption of intracellular phosphate (Harold, 1964), then adding phosphate to a final concentration of 0.2 mM. For Neisser staining (Seki et al., 2013), cell cultures (50-100 μl) were air-dried on slides and fixed with gentle heat. The fixed cells were stained with a solution of two parts solution A (0.1 g methylene blue, 2 mL ethanol, 5 mL acetic acid, 95 mL ddH2O) and one part solution B (0.1 g crystal violet, 1 mL ethanol, 30 mL ddH2O) for 20 seconds, and rinsed with ddH2O. Cells were then stained with chrysoidin (0.3% w/v in ddH2O) for three minutes and rinsed with ddH2O. The slides were air-dried and observed with a light microscope (Zeiss Axio-plan 2). Images were acquired with the AxioCam HR color CCD camera and AxioVision software. All solutions were filtered through a 0.2 μm filter before use.
Quantification of RNA
Cells for RNA quantification were grown in NBRIP with 0.2 mM K2HPO4 at 30°C with shaking (~150 rpm) for 60 h (LM-P only) or 40 h (all other strains), corresponding to mid-exponential to early stationary phase (see Fig. S1). Samples equivalent to 1 mL of OD600=1 were harvested at this time and pelleted for analysis. Simultaneously, 50 mL of culture was concentrated by centrifugation and dried in glass tubes at 80°C to determine dry weight.
The RNA content of cells (as a percentage of dry weight) was measured by GC-MS as described previously (Long and Antoniewicz, 2014) using [U-13C] (fully labeled) E. coli as an internal standard (Leighty and Antoniewicz, 2012). GC-MS analysis was performed on an Agilent 7890B GC system equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 μm-phase thickness; Agilent J&W Scientific), connected to an Agilent 5977A mass spectrometer operating under electron impact ionization (EI) at 70 eV. Helium flow was maintained at 1 mL min-1. The source temperature was maintained at 230°C, the quadrupole temperature at 150°C, the interface temperature at 280°C, and the inlet temperature at 250°C. The RNA in crude cell extracts was processed by acid hydrolysis followed by aldonitrile propionate derivatization of the ribose monomers (Long and Antoniewicz, 2014); 1 μL of the resulting solution was injected into the GC-MS in splitless mode. The column was started at 80°C for 2 min, increased to 280°C at 10°C/min, and held for 12 min. The mass isotopomer distribution of aldonitrile propionate derivative of ribose was obtained by integration of the m/z 173 fragment (Antoniewicz et al., 2011), which contains the last two carbon atoms of ribose. Mass isotopomer distributions were corrected for natural isotope abundances for data analysis (Crown and Antoniewicz, 2013).
Intact polar lipids extraction and analysis
Cells for lipid analysis were grown in NBRIP media with 0.2 mM K2HPO4 until stationary phase, or in NBRIP with no additional P for about 10 days. The cells were pelleted, washed three times with ddH2O and stored at -20°C until analysis. Lipids from each batch were extracted following a modified Bligh & Dyer protocol using PC-C21 (phosphatidylcholine with two C21:0 fatty acid side chains; Avanti Polar Lipids, Alabaster, AL, USA) as an internal standard (Sturt et al., 2004). The total lipid extract was dried under a stream of N2 and stored at -20°C until measurement. Intact polar lipids were quantified by injecting 5-10% of the total lipid extract dissolved in dichloromethane:methanol (9:1, v:v) into a Dionex Ultimate 3000RS ultra-high performance liquid chromatography (UHPLC) system connected to a Bruker maXis Ultra-High Resolution quadrupole time-of-flight tandem mass spectrometer which was equipped with an ESI ion source operating in positive mode (Bruker Daltonik, Bre-men, Germany). Analyte separation was achieved using normal phase UHPLC on an Acquity UPLC BEH Amide column (1.7 μm, 2.1 × 150 mm; Waters Corporation, Eschborn, Germany) maintained at 40°C as described by Wörmer et al. (2013). Lipids were identified by retention time, accurate molecular mass, and MS2 fragmentation as described previously (Sturt et al., 2004; Wörmer et al., 2013). Integration of peaks was performed on extracted ion chromatograms of ±10 mDa width and included the [M+H]+, [M+NH4]+, and [M+Na]+ ions. Lipid abundances were corrected for headgroup-specific response relative to the internal standard using external calibration curves of commercially available standards. The abundances of diacylglycerol (DAG) lipids with phosphatidylglycerol (PG), PE, monomethyl PE (PME) and dimethyl PE (PDME), were corrected by the relative responses of commercial DAG-C16:0/16:0 standards with the respective headgroup (Avanti Polar Lipids Inc., Alabaster, AL, USA). The abundances of diacylglycer-yl-N,N,N-trimethylhomoserine (DGTS), diphosphatidylglycerol DAG (DPG), monoglycosyl DAG (MGDG) and diglycosyl DAG (DGDG) lipids were corrected by the relative responses of DGTS-C16:0/16:0, DPG-C18:1/18:1/18:1/18:1, MGDG-C16:0/16:0-cyclopropyl (Avanti) and DGDG-C18:0/18:0 (Matreya LLC, Pleasant Gap, PA, USA) standards, respectively. Due to a lack of appropriate standards, ornithine and glucuronosyl DAG (GlcA-DAG) lipid abundances were corrected for the response of DGTS-C16:0/16:0 and MGDG-C16:0/16:0-cyclopropyl standards, respectively. The lower limit of detection as determined for authentic standards was in the range of 1 to 10 pg for intact polar lipids, depending on compound class and considering a signal-to-noise ratio of greater than 3 (Table S1).
Supplementary Material
Acknowledgments
We gratefully acknowledge the Indonesian Institute of Sciences (LIPI) for assistance in gaining access to the field site, particularly Dr. Cynthia Hen-ny and Dr. Tri Widiyanto. PT Vale Indonesia Tbk. provided essential logistical support in Soro-wako, Sulawesi. We thank Dr. Deborah Powell at the University of Delaware BioImaging Center for assistance with microscopy, Dr. Brewster Kingham at the University of Delaware Sequencing and Genotyping Center for sequence analysis, and Dr. Jessica Keffer, Dr. Clara Chan, and Dr. Alison Olcott Marshall for helpful discussions. This work was funded by the University of Del-aware Department of Civil and Environmental Engineering and the Danish National Research Foundation; lipid analyses in Bremen were funded through the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (grant HI 616 14-1 to KUH).
Footnotes
The authors declare no conflicts of interest.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–6. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- Batrakov SG, Nikitin DI, Pitryuk IA. Lipid composition of the gram-negative, budding, seawater bacterium Hyphomonas jannaschiana lacking in phospholipids. Biochim Biophys Acta - Lipids Lipid Metab. 1996;1303:39–46. doi: 10.1016/0005-2760(96)00072-0. [DOI] [PubMed] [Google Scholar]
- Brooks JL. Speciation in Ancient Lakes (Concluded) Q Rev Biol. 1950;25:131–176. doi: 10.1086/397539. [DOI] [PubMed] [Google Scholar]
- Carini P, Van Mooy BAS, Thrash JC, White A, Zhao Y, Campbell EO, et al. SAR11 lipid renovation in response to phosphate starvation. Proc Natl Acad Sci U S A. 2015;112:7767–7772. doi: 10.1073/pnas.1505034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–45. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotner JB, Hall EK, Scott T, Heldal M, Scott JT. Freshwater bacteria are stoichiometrically flexible with a nutrient composition similar to seston. Front Microbiol. 2010;1:132. doi: 10.3389/fmicb.2010.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe SA, O’Neill AH, Katsev S, Hehanussa P, Haffner GD, Sundby Bjørn, et al. The biogeochemistry of tropical lakes: A case study from Lake Matano, Indonesia. Limnol Oceanogr. 2008;53:319–331. [Google Scholar]
- Crown SB, Antoniewicz MR. Publishing 13C metabolic flux analysis studies: a review and future perspectives. Metab Eng. 2013;20:42–8. doi: 10.1016/j.ymben.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinema MH, van Loosdrecht M, Scholten A. Some Physiological Characteristics of Acinetobacter spp. Accumulating Large Amounts of Phosphate 2011 [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser JJ, Acharya K, Kyle M, Cotner J, Makino W, Markow T, et al. Growth rate–stoichiometry couplings in diverse biota. Ecol Lett. 2003;6:936–943. [Google Scholar]
- Elser JJ, Acquisti C, Kumar S. Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends Ecol Evol. 2011;26:38–44. doi: 10.1016/j.tree.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser JJ, Sterner RW, Gorokhova E, Fagan WF, Markow TA, Cotner JB, et al. Biological stoichiometry from genes to ecosystems. Ecol Lett. 2008;3:540–550. [Google Scholar]
- Erb TJ, Kiefer P, Hattendorf B, Gunther D, Vorholt JA. GFAJ-1 Is an Arsenate-Resistant, Phosphate-Dependent Organism. Science (80-) 2012;337:467–470. doi: 10.1126/science.1218455. [DOI] [PubMed] [Google Scholar]
- Frigaard NU, Sakuragi Y, Bryant DA. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol Biol. 2004;274:325–340. doi: 10.1385/1-59259-799-8:325. [DOI] [PubMed] [Google Scholar]
- Geiger O, González-Silva N, López-Lara IM, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res. 2010;49:46–60. doi: 10.1016/j.plipres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Geske T, Vom Dorp K, Dörmann P, Hölzl G. Accumulation of glycolipids and other non-phosphorous lipids in Agrobacterium tumefaciens grown under phosphate deprivation. Glycobiology. 2013;23:69–80. doi: 10.1093/glycob/cws124. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–5. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Godwin CM, Cotner JB. Carbon: phosphorus homeostasis of aquatic bacterial assemblages is mediated by shifts in assemblage composition. Aquat Microb Ecol. 2014;73:245–258. [Google Scholar]
- Godwin CM, Cotner JB. Stoichiometric flexibility in diverse aquatic heterotrophic bacteria is coupled to differences in cellular phosphorus quotas. Front Microbiol. 2015;6:159. doi: 10.3389/fmicb.2015.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant WD. Cell wall teichoic acid as a reserve phosphate source in Bacillus subtilis. J Bacteriol. 1979;137:35–43. doi: 10.1128/jb.137.1.35-43.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WB. Tectonics of the Indonesian region. US Govt. Print. Off; 1979. [Google Scholar]
- Harold FM. Enzymic and Genetic Control of Polyphosphate Accumulation in Aerobacter aerogenes. J Gen Microbiol. 1964;35:81–90. doi: 10.1099/00221287-35-1-81. [DOI] [PubMed] [Google Scholar]
- Hecky R, Campbell P, Hendzel L. The stoichiometry of carbon, nitrogen, and phosphorus in particulate matter of lakes and oceans. Limnol Oceanogr. 1993;38:709–724. [Google Scholar]
- Hölzl G, Dörmann P. Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res. 2007;46:225–43. doi: 10.1016/j.plipres.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Ilikchyan IN, McKay RML, Zehr JP, Dyhrman ST, Bullerjahn GS. Detection and expression of the phosphonate transporter gene phnD in marine and freshwater picocyanobacteria. Environ Microbiol. 2009;11:1314–24. doi: 10.1111/j.1462-2920.2009.01869.x. [DOI] [PubMed] [Google Scholar]
- Katsev S, Crowe SA, Mucci A, Sundby Bjørn, Nomosatryo S, Haffner GD, Fowle DA. Mixing and its effects on biogeochemistry in the persistently stratified, deep, tropical Lake Matano, Indonesia. Limnol Oceanogr. 2010;5:763–776. [Google Scholar]
- Keiblinger KM, Hall EK, Wanek W, Szukics U, Hämmerle I, Ellersdorfer G, et al. The effect of resource quantity and resource stoichiometry on microbial carbon-use-efficiency. FEMS Microbiol Ecol. 2010;73:430–40. doi: 10.1111/j.1574-6941.2010.00912.x. [DOI] [PubMed] [Google Scholar]
- Lamarche MG, Wanner BL, Crépin S, Harel J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev. 2008;32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci U S A. 2009;106:15527–33. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. Parallel labeling experiments with [U-13C]glucose validate E. coli metabolic network model for 13C metabolic flux analysis. Metab Eng. 2012;14:533–41. doi: 10.1016/j.ymben.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Lipson DA, Monson RK, Schmidt SK, Weintraub MN. The trade-off between growth rate and yield in microbial communities and the consequences for under-snow soil respiration in a high elevation coniferous forest. Biogeochemistry. 2008;95:23–35. [Google Scholar]
- Livermore JA, Emrich SJ, Tan J, Jones SE. Freshwater bacterial lifestyles inferred from comparative genomics. Environ Microbiol. 2014;16:746–58. doi: 10.1111/1462-2920.12199. [DOI] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Quantifying biomass composition by gas chromatography/mass spectrometry. Anal Chem. 2014;86:9423–7. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino W, Cotner J. Elemental stoichiometry of a heterotrophic bacterial community in a freshwater lake: implications for growth- and resource-dependent variations. Aquat Microb Ecol. 2004;34:33–41. [Google Scholar]
- Makino W, Cotner JB, Sterner RW, Elser JJ. Are bacteria more like plants or animals? Growth rate and resource dependence of bacterial C : N : P stoichiometry. Funct Ecol. 2003;17:121–130. [Google Scholar]
- Martín JF, Santos-Beneit F, Rodríguez-García A, Sola-Landa A, Smith MCM, Ellingsen TE, et al. Transcriptomic studies of phosphate control of primary and secondary metabolism in Streptomyces coelicolor. Appl Microbiol Biotechnol. 2012;95:61–75. doi: 10.1007/s00253-012-4129-6. [DOI] [PubMed] [Google Scholar]
- Martinez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–38. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- McGrath JW, Chin JP, Quinn JP. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat Rev Microbiol. 2013;11:412–9. doi: 10.1038/nrmicro3011. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, van der Donk WA. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Moyle J. The Gram Reaction and Cell Composition: Nucleic Acids and other Phosphate fractions. J Gen Microbiol. 1954;10:533–540. doi: 10.1099/00221287-10-3-533. [DOI] [PubMed] [Google Scholar]
- Van Mooy BaS, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblízek M, et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature. 2009;458:69–72. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- Van Mooy BAS, Fredricks HF. Bacterial and eukaryotic intact polar lipids in the eastern subtropical South Pacific: Water-column distribution, planktonic sources, and fatty acid composition. Geochim Cosmochim Acta. 2010;74:6499–6516. [Google Scholar]
- Van Mooy BAS, Moutin T, Duhamel S, Rimmelin P, Van Wambeke F. Phospholipid synthesis rates in the eastern subtropical South Pacific Ocean. Biogeosciences. 2008;5:133–139. [Google Scholar]
- Mundry C, Kuhn K-P. Modelling and parameter identification for batch fermentations with Streptomyces tendae under phosphate limitation. Appl Microbiol Biotechnol. 1991;35 doi: 10.1007/BF00172717. [DOI] [PubMed] [Google Scholar]
- Muscarella M, Bird K, Larsen M, Placella S, Lennon J. Phosphorus resource heterogeneity in microbial food webs. Aquat Microb Ecol. 2014;73:259–272. [Google Scholar]
- Nakamura K, Hiraishi A, Yoshimi Y, Kawaharasaki M, Masuda K, Kamagata Y. Microlunatus phosphovorus gen. nov., sp nov., a New Gram-Positive Polyphosphate-Accumulating Bacterium Isolated from Activated Sludge. Int J Syst Bacteriol. 1995;45:17–22. doi: 10.1099/00207713-45-1-17. [DOI] [PubMed] [Google Scholar]
- Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- Palenik B. Molecular mechanisms by which marine phytoplankton respond to their dynamic chemical environment. Ann Rev Mar Sci. 2015;7:325–40. doi: 10.1146/annurev-marine-010814-015639. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Rock CO. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res. 2013;52:249–76. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popendorf KJ, Lomas MW, Van Mooy BAS. Microbial sources of intact polar diacylglycerolipids in the Western North Atlantic Ocean. Org Geochem. 2011;42:803–811. [Google Scholar]
- Reaves ML, Sinha S, Rabinowitz JD, Kruglyak L, Redfield RJ. Absence of Detectable Arsenate in DNA from Arsenate-Grown GFAJ-1 Cells. Science. 2012;337:470–473. doi: 10.1126/science.1219861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Wickham GS, Pace NR. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–9. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller BR, Schmidt TM. The physiology and ecological implications of efficient growth. ISME J. 2015;9:1481–1487. doi: 10.1038/ismej.2014.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo E, Roy D, Hamilton PB, Hehanussa PE, McNeely R, Haffner GD. The plankton community of Lake Matano: factors regulating plankton composition and relative abundance in an ancient, tropical lake of Indonesia. Hydrobiologia. 2008;615:225–235. [Google Scholar]
- Sambrook J, Russell DW, editors. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- Scott JT, Cotner JB, Lapara TM. Variable stoichiometry and homeostatic regulation of bacterial biomass elemental composition. Front Microbiol. 2012;3:42. doi: 10.3389/fmicb.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Nitta K, Kaneko Y. Observation of polyphosphate bodies and DNA during the cell division cycle of Synechococcus elongatus PCC 7942. Plant Biol (Stuttg) 2013 doi: 10.1111/plb.12008. [DOI] [PubMed] [Google Scholar]
- Sinsabaugh RL, Manzoni S, Moorhead DL, Richter A. Carbon use efficiency of microbial communities: stoichiometry, methodology and modelling. Ecol Lett. 2013;16:930–9. doi: 10.1111/ele.12113. [DOI] [PubMed] [Google Scholar]
- Smith EM, Prairie YT. Bacterial metabolism and growth efficiency in lakes: The importance of phosphorus availability. Limnol Oceanogr. 2004;49:137–147. [Google Scholar]
- Smith S, Takács I, Murthy S, Daigger GT, Szabó A. Phosphate complexation model and its implications for chemical phosphorus removal. Water Environ Res. 2008;80:428–38. [PubMed] [Google Scholar]
- Souza V, Eguiarte LE, Siefert J, Elser JJ. Microbial endemism: does phosphorus limitation enhance speciation? Nat Rev Microbiol. 2008;6:559–564. doi: 10.1038/nrmicro1917. [DOI] [PubMed] [Google Scholar]
- Stout LM, Joshi SR, Kana TM, Jaisi DP. Microbial activities and phosphorus cycling: An application of oxygen isotope ratios in phosphate. Geochim Cosmochim Acta. 2014;138:101–116. [Google Scholar]
- Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs K-U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry--new biomarkers for biogeochemistry and microbial ecology. Rapid Commun Mass Spectrom. 2004;18:617–28. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- Vaillant JJ, Haffner GD, Cristescu ME. The Ancient Lakes of Indonesia: Towards Integrated Research on Speciation. Integr Comp Biol. 2011;51:634–643. doi: 10.1093/icb/icr101. [DOI] [PubMed] [Google Scholar]
- Vences-Guzmán MÁ, Geiger O, Sohlenkamp C. Ornithine lipids and their structural modifications: from A to E and beyond. FEMS Microbiol Lett. 2012;335:1–10. doi: 10.1111/j.1574-6968.2012.02623.x. [DOI] [PubMed] [Google Scholar]
- Vences-Guzmán MÁ, Guan Z, Bermúdez-Barrientos JR, Geiger O, Sohlenkamp C. Agrobacteria lacking ornithine lipids induce more rapid tumour formation. Environ Microbiol. 2013;15:895–906. doi: 10.1111/j.1462-2920.2012.02867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollenweider RA. Eutrophication of waters: monitoring, assessment and control Organisation for Economic Co-operation and Development 1982 [Google Scholar]
- White AK, Metcalf WW. Microbial Metabolism of Reduced Phosphorus Compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- Wörmer L, Lipp JS, Schröder JM, Hinrichs K-U. Application of two new LC–ESI–MS methods for improved detection of intact polar lipids (IPLs) in environmental samples. Org Geochem. 2013;59:10–21. [Google Scholar]
- Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A. 2014;111:5622–7. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Doroghazi JR, Janga SC, Zhang JK, Circello B, Griffin BM, et al. Diversity and abundance of phosphonate biosynthetic genes in nature. Proc Natl Acad Sci U S A. 2013;110:20759–64. doi: 10.1073/pnas.1315107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegeye A, Bonneville S, Benning LG, Sturm A, Fowle DA, Jones C, et al. Green rust formation controls nutrient availability in a ferruginous water column. Geology. 2012;40:599–602. [Google Scholar]
- Zerulla K, Chimileski S, Näther D, Gophna U, Papke RT, Soppa J. DNA as a phosphate storage polymer and the alternative advantages of polyploidy for growth or survival. PLoS One. 2014;9:e94819. doi: 10.1371/journal.pone.0094819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ishige K, Kornberg A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc Natl Acad Sci U S A. 2002;99:16678–83. doi: 10.1073/pnas.262655199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Song C, Huang D, Liu Y, Cao X, Zhou Y. Isolation and Characterization of Organic Phosphorus-Mineralizing Bacteria in Sediment of a Chinese Large Shallow Eutrophic Lake (Lake Taihu) Geomicrobiol J. 2011;28:660–666. [Google Scholar]
- Zimmerman AE, Martiny AC, Lomas MW, Allison SD. Phosphate supply explains variation in nucleic acid allocation but not C : P stoichiometry in the western North Atlantic. Biogeosciences. 2014;11:1599–1611. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


