Abstract
We have reported that normal human salivary gland-derived epithelial cells exclusively express keratinocyte growth factor receptor (KGFR). In the process of malignant transformation of human salivary gland tumors, KGFR gene expression disappeared concomitantly with the de novo expression of the fibroblast growth factor receptor 1 (FGFR1) and FGFR4 genes. In the present study, we introduced wild-type KGFR cDNA or chimeric KGFR/FGFR1 cDNA, which encoded the extracellular domain of KGFR and the intracellular domain of FGFR1, into the HSY human salivary adenocarcinoma cell line. The KGFR tyrosine kinase suppressed the activity of FGF receptor substrate 2 (FRS2) and inhibited the growth of HSY by inducing differentiation and apoptosis in vitro and in vivo. Our results provided significant insight into the mechanism of KGFR tumor suppression and suggest that KGFR gene therapy might be a viable method of inhibiting human salivary adenocarcinoma growth.
Keratinocyte growth factor (KGF/FGF7) is a member of the fibroblast growth factor (FGF) family (1). Its activity is largely restricted to epithelial cells, which express the KGF receptor (KGFR), a transmembrane tyrosine kinase encoded by the IIIb splice variant of FGFR2 (FGFR2-IIIb) (2, 3). KGFR binds KGF and FGF-1 with high affinity. In contrast, FGFR2-IIIc, another splice variant of FGFR2, binds FGF-1 and FGF-2, but not KGF (3, 4). Analysis of KGF and KGFR expression during embryonic development, including that of mammary glands, has provided evidence that KGF is an important mesenchymal mediator of epithelial growth and differentiation. In normal human skin, KGFR immunostaining localizes to the suprabasal layers (5, 6). The lack of KGFR in basal cell progenitors in skin suggests that KGFR might regulate keratinocyte differentiation (7).
Malignant salivary tumors are highly aggressive neoplasms that readily invade adjacent tissues and metastasize to distant organs at early stages of the disease. Malignant salivary tumors exhibit enhanced expression of both FGF-1 and FGF-2 compared with normal salivary gland (8). FGF-1 and FGF-2 can act in an autocrine manner to stimulate the proliferation of salivary adenocarcinoma cells (8, 9). Normal salivary gland epithelial cells and benign salivary gland tumors exclusively express the KGFR gene. During the malignant transformation of salivary gland epithelial cells, the expression of the KGFR gene is abolished, whereas the FGFR1-IIIc and FGFR4 genes are activated (10). The exclusive expression of the KGFR gene in both normal and premalignant salivary epithelial cells correlates well with the slow-growing and differentiated phenotype of premalignant tumors. In contrast, the loss of the KGFR gene and the expression of both the FGFR1 gene and its specific ligand FGF-2 in malignant tumor cells are associated with the emergence of the malignant phenotype. Thus, the loss of normal KGFR gene function may promote tumorigenicity, whereas KGFR may function to suppress human salivary adenocarcinomas.
The Ras-MAP kinase signaling pathway plays an important role in signaling via FGFRs (11, 12). It is now well established that the lipid-anchored docking protein FGF receptor substrate 2 (FRS2) links FGFR molecules with the Ras-MAP kinase signaling pathway by forming a complex with the N-terminal SH2 domain of the protein tyrosine phosphatase Shp2 and the adaptor protein Grb2. Blocking FRS2 signaling results in weak Ras-MAP kinase phosphorylation (13, 14). In the present study, we found that the wild-type KGFR gene transferred into HSY human salivary adenocarcinoma cells caused a decreased level of tyrosine-phosphorylated activity of FRS2, resulting in apoptosis and differentiation in vitro and in vivo. Our data provide insight into the mechanism of KGFR tumor suppression.
Materials and Methods
Cell Culture and Recombinant DNA Constructs.
Normal salivary gland-derived epithelial (SGE) cells were isolated from human submandibular glands and cultured in serum-free medium as described (15). The human salivary adenocarcinoma cell line HSY (16) was kindly provided by Professor Mitsunobu Sato (Second Department of Oral and Maxillofacial Surgery, Tokushima University School of Dentistry, Tokushima, Japan). HSY was maintained in RD medium (RPMI 1640 medium/DMEM, 1:1) supplemented with 5% calf serum as described (10).
KGFR carrying full-length FGFR2-IIIb cDNA and chimeric KGFR/FGFR1 carrying the extracellular domain of KGFR and the intracellular domain of FGFR1 were cloned into mammalian expression vector pcDNA3.1/zeo (Invitrogen) as described (17).
For transfection, 60% confluent HSY cells were preincubated with serum- and antibiotic-free RD medium for 2 h, then transfected with wild-type KGFR or chimeric KGFR/FGFR1 cDNAs by a liposome-mediated method with the use of Lipofectamine Plus Reagent (GIBCO/BRL and Life Technologies, Rockville, MD). As a control, pcDNA3.1/zeo bearing no cDNA insert was introduced. The cells were then selected with 400 μg/ml of Zeocin (Invitrogen) for 2 weeks, and the colonies of cells emerging from the selection medium were isolated and further selected by RRA with 125I-KGF as described (18). KGFR-overexpressing HSY cells were designated HSYR2-IIIb, KGFR/FGFR1-overexpressing HSY cells were designated HSYR2/R1, and the control transfectants were designated HSYzeo.
Cell Proliferation Assay.
For the study of population growth rates, HSYzeo, HSYR2/R1, and HSYR2-IIIb cells (1 × 104 cells per well) were seeded into type I collagen (Cell Matrix type I-A; Nitta Gelatin Co., Osaka, Japan)-coated 24-well culture plates, respectively, in serum-free RD medium containing 10 μg/ml bovine insulin, 5 μg/ml human transferrin, 10 μM mercaptoethanol, 10 μM 2-aminoethanol, and 10 nM sodium selenite (RD5F) (19). Cell numbers were counted daily for 6 days.
To evaluate the effect of FGF on the growth of the transfectants in serum-free culture, HSYzeo, HSYR2/R1, and HSYR2-IIIb cells (3 × 103 cells per well) were seeded in RD5F and incubated with the indicated concentrations of FGF-1, FGF-2, or KGF for 5 days. FGF-1 was purified from bovine brain, and rat recombinant KGF was prepared as described (20, 21). Human recombinant FGF-2 was purchased from Upstate Biotechnology (Lake Placid, NY). Cell numbers were measured with a Coulter Counter (Coulter Electronics, Hialeah, FL).
Immunocytochemical Analysis.
To determine whether transfectants were undergoing differentiation, the salivary gland differentiation markers α-amylase and lactoferrin were analyzed immunocytochemically. Briefly, the cells were cultured in RD5F without KGF on Lab-Tek II chamber slides (Nalgen Nunc International, Naperville, IL) for 2 days, then the cells were fixed with freshly prepared 2% paraformaldehyde for 30 min. After blocking with 10% normal goat serum for 30 min, the cells were incubated with rabbit anti-amylase antibody (Dako) diluted by 1:100 or rabbit anti-lactoferrin antibody (Dako) diluted by 1:100. Immunodetection of the antigens was performed with the Elite Avidin-Biotin-Immunoperoxidase System (Vector Laboratories) according to the manufacturer's instructions. Staining of the cells was graded as follows: 0–1+, negative to indeterminate cell staining; 2+, weak but clearly positive cell staining; 3+, strong cell staining.
Detection of Apoptotic DNA Fragmentation.
The cells (104 cells per milliliter) were cultured in serum-free RD5F with 20 ng/ml KGF on 10 cm2 dish (10 dishes per experiment point). DNA was extracted from the adherent and floating cells every day for 7 days as described (22). DNA samples were electrophoresed on 2% agarose gel and visualized by ethidium bromide staining to detect DNA fragmentation.
Tumor Cell Implantation and Histochemical Analysis.
One hundred and five female athymic mice (BALB/c, AnNCrj-nu/r, 5 weeks old) were used for this study. The research protocol was approved, and mice were maintained in accordance with institutional guidelines of the Hiroshima University Animal Care and Use Committee. Five mice per clone were used per experiment. In each experiment, 35 mice were used. The cells (2 × 106) suspended in 100 μl of PBS were injected s.c. in the lateral back region. Tumor volume (V) was measured twice a week, according to the formula V = 1/2 × length × width2). Mice were killed by cervical dislocation 4 weeks after inoculation. For histochemical analysis, the tumor tissues were fixed in freshly prepared 4% paraformaldehyde and embedded in paraffin. Tissues were sectioned and stained with hematoxylin/eosin.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP-Biotin Nick-End Labeling (TUNEL) Assay.
Paraffin sections (4–6 μm) of tumor specimen were digested with proteinase K (20 μg/ml) for 5 min at 37°C and then incubated with terminal deoxynucleotidyl transferase enzyme (Wako Pure Chemical, Osaka) for 60 min at 37°C. After being treated with 3% H2O2 for 5 min at room temperature, the sections were incubated with peroxidase-conjugated antibody (Wako Pure Chemical, Osaka) for 60 min at 37°C. The reaction was visualized with diaminobenzidine–H2O2 substrate.
Analysis of FRS2 Phosphorylation.
The cells were nutrition-starved for 24 h and then incubated with 20 ng/ml of FGF-1, FGF-2, or KGF for 5 min at 37°C. The cells were washed three times with cold PBS containing 1 mM Na3VO4 and lysed with lysis buffer (50 mM Hepes/1% Triton X-100/10% glycerol/150 mM NaCl/100 mM NaF/1.5 mM MgCl2/1 mM EGTA/1 mM Na3VO4/1 mM phenylmethylsulfonyl fluoride/10 μg/ml aprotinin). The sonicated and clarified cell lysates (500 μg per sample) were immunoprecipitated with 3 μg of rabbit anti-Grb2 antibody (Transduction Laboratories, Lexington, KY) overnight at 4°C, and then the antigen–antibody complexes were recovered by a 2-h incubation with protein A–Sepharose CL-4B (Amersham Pharmacia) at 4°C. The immunoprecipitated samples were electrophoresed on 7.5% SDS/polyacryamide gel under reducing conditions and transferred to Immobilon-P membrane (Millipore). The membrane was incubated with a 1:1,000 dilution of anti-phosphotyrosine monoclonal antibody 4G10 (Upstate Biotechnology) or a 1:1,000 dilution of goat anti-FRS2 antibody (Santa Cruz Technology) overnight at 4°C. Immunodetection of the antigen was performed with the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium Membrane Phosphatase Substrate System (Kirkegaard & Perry Laboratories) according to the manufacturer's instructions.
Statistical Analysis.
Immunocytochemical results were analyzed with the use of contingency tables and the Kruskal–Wallis rank test. ANOVA and Tukey's test were used for other data. Significance was defined as P < 0.05. The statistical software used was statview 5.0 (SAS Institute, Cary, NC).
Results
Differential Growth of Transfectants.
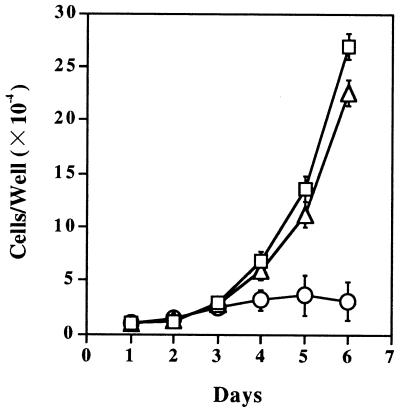
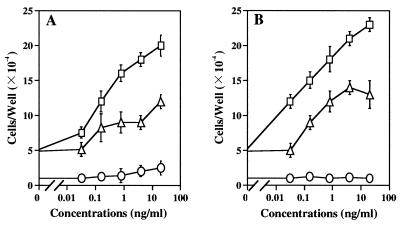
Among 58 Zeocin-resistant HSYR2-IIIb clones and 50 Zeocin-resistant HSYR2/R1 clones, seven HSYR2-IIIb clones and seven HSYR2/R1 clones were selected for analysis because they exhibited higher KGFR numbers (Table 1). The transfected cell line HSYR2-IIIb proliferated with a doubling time of 52 h (SD 1.9), whereas HSYzeo and HSYR2/R1 proliferated with doubling times of 18 h and 19 h (SD 1.2 and 1.7), respectively (Table 1). The growth rate of HSYR2-IIIb cells was significantly lower than those of HSYzeo and HSYR2/R1 (P < 0.001; Fig. 1). FGF-1 at 20 ng/ml showed a 4-fold stimulation of HSYzeo growth and a 2.5-fold stimulation of HSYR2/R1 growth in serum-free culture (Fig. 2A). FGF-2 at 20 ng/ml showed a 4.8-fold stimulation of HSYzeo and a 2.5-fold stimulation of HSYR2/R1 growth in serum-free culture (Fig. 2B). The growth of HSYR2-IIIb was not stimulated by either FGF-1 or FGF-2 (Fig. 2). Although HSYR2/R1 and HSYR2-IIIb expressed KGFR, KGF did not have a mitogenic effect on HSYR2/R1 or HSYR2-IIIb.
Table 1.
The KGFR number, proliferative activity, and tumorigenicity of transfectants
| Transfectant | KGFR number* | Doubling time in vitro, h† | Tumor incidence |
|---|---|---|---|
| HSYR2-IIIb | |||
| c1.1A | 0.8 | 50.6 ± 2.3 | 5/5 |
| c1.2A | 1.2 | 50.5 ± 2.5 | 0/5 |
| c1.3A | 1.6 | 51.8 ± 1.9 | 5/5 |
| c1.4A | 1.8 | 51.8 ± 2.3 | 5/5 |
| c1.5A | 2.8 | 51.9 ± 2.9 | 0/5 |
| c1.6A | 3.3 | 52.5 ± 1.6 | 5/5 |
| c1.7A | 5.7 | 52.9 ± 1.0 | 0/5 |
| HSYR2/R1 | |||
| c1.1B | 0.8 | 19.7 ± 1.4 | 5/5 |
| c1.2B | 0.9 | 18.6 ± 1.9 | 5/5 |
| c1.3B | 0.9 | 19.2 ± 2.7 | 5/5 |
| c1.4B | 1.2 | 18.1 ± 1.5 | 5/5 |
| c1.5B | 1.9 | 19.7 ± 2.8 | 5/5 |
| c1.6B | 2.2 | 19.3 ± 1.5 | 5/5 |
| c1.7B | 4.2 | 18.4 ± 0.4 | 5/5 |
| HSYzeo | |||
| c1.1C | — | 18.0 ± 1.2 | 5/5 |
| c1.2C | — | 17.8 ± 1.5 | 5/5 |
| c1.3C | — | 18.6 ± 1.1 | 5/5 |
| c1.4C | — | 18.6 ± 1.2 | 5/5 |
| c1.5C | — | 19.1 ± 2.0 | 5/5 |
| c1.6C | — | 18.8 ± 0.5 | 5/5 |
| c1.7C | — | 17.5 ± 1.3 | 5/5 |
KGF receptor numbers (×10−5).
Mean ± SD.
Figure 1.
Population growth kinetics of the transfectants in serum-free culture. The cells (1 × 104 cells per well) were seeded in type I collagen-coated 24-well culture plates, and they were counted daily for 6 days. HSYzeo (□) and HSYR2/R1 (▵) had doubling times of 18 h and 19 h, respectively, whereas HSYR2-IIIb (○) proliferated with a doubling time of 52 h. Three independent experiments were performed with duplicate wells. Each point indicates a mean ± SD.
Figure 2.
The effects of FGF-1 and FGF-2 on the growth of the transfectants in serum-free culture. The cells (3 × 103 cells per well) were seeded in type I collagen-coated 24-well culture plates in RD5F and incubated with the indicated concentrations of FGF-1 or FGF-2 for 5 days. (A) FGF-1 at 20 ng/ml showed a 4-fold stimulation of HSYzeo growth (□) and a 2.5-fold stimulation of HSYR2/R1 growth (▵). (B) FGF-2 at 20 ng/ml showed a 4.8-fold stimulation of HSYzeo growth and a 2.5-fold stimulation of HSYR2/R1 growth. Neither FGF-1 nor FGF-2 stimulated the growth of HSYR2-IIIb (○). Mean values of duplicate wells from three independent experiments are shown.
Cellular Differentiation and Apoptosis in HSYR2-IIIb in Vitro.
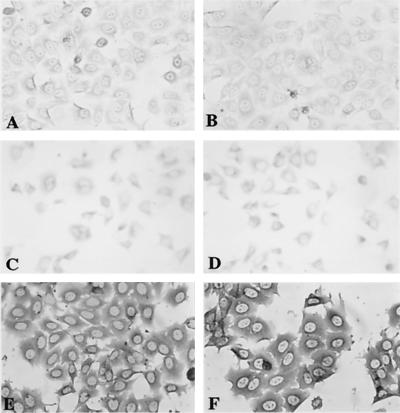
By immunocytochemical analysis HSYzeo and HSYR2/R1 cells exhibited negative to weakly positive staining for α-amylase and lactoferrin, whereas HSYR2-IIIb cells were strongly positive for both α-amylase and lactoferrin in the cytoplasm and perinuclear regions (Fig. 3).
Figure 3.
Immunoperoxidase staining for α-amylase and lactoferrin. HSYzeo and HSYR2/R1 exhibited weakly positive staining for α-amylase (A and C) and lactoferrin (B and D). HSYR2-IIIb showed intensive positive staining for α-amylase (E) and lactoferrin (F) in the cytoplasm and perinuclear regions. The cells shown are representative of seven clones each.
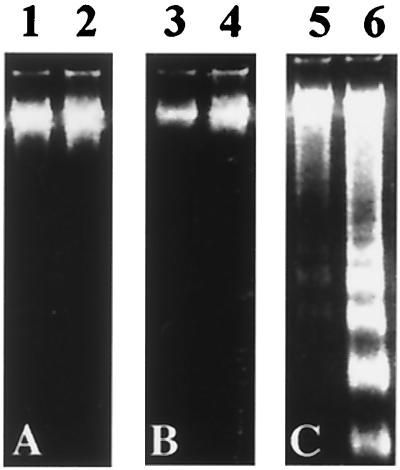
HSYR2-IIIb cells detached from the culture plates over a 4-day serum-free culture period with 20 ng/ml KGF and eventually died. By trypan blue dye exclusion, ≈10% of the cells detached and died on day 2, 15% on day 3, 50% on day 4, and 90% on day 7. TUNEL positive cells were observed in HSYR2-IIIb, although no TUNEL positive cells were detected in HSYzeo or HSYR2/R1. On the third day of culture with KGF, 15% of HSYR2-IIIb was positive for TUNEL staining, and 70% were positive for TUNEL staining on day 7. Compaction and margination of nuclear chromatin, condensation of cytoplasm, and convolution of nuclear and cell outlines were observed in cultured HSYR2-IIIb (data not shown). No detectable fragmentation was observed in DNA derived from HSYzeo and HSYR2/R1 cultured with KGF (20 ng/ml) for 7 days. On the other hand, DNA laddering was observed in DNA derived from HSYR2-IIIb cultured with KGF (20 ng/ml) for 4 days, and it increased markedly over a 7-day period (Fig. 4).
Figure 4.
Detection of DNA fragmentation in HSYzeo (A), HSYR2/R1 (B), and HSYR2-IIIb (C). The cells were incubated with KGF (20 ng/ml) for 4 days (lanes 1, 3, and 5) or 7 days (lanes 2, 4, and 6).
Differentiation and Apoptosis in HSYR2-IIIb in Vivo.
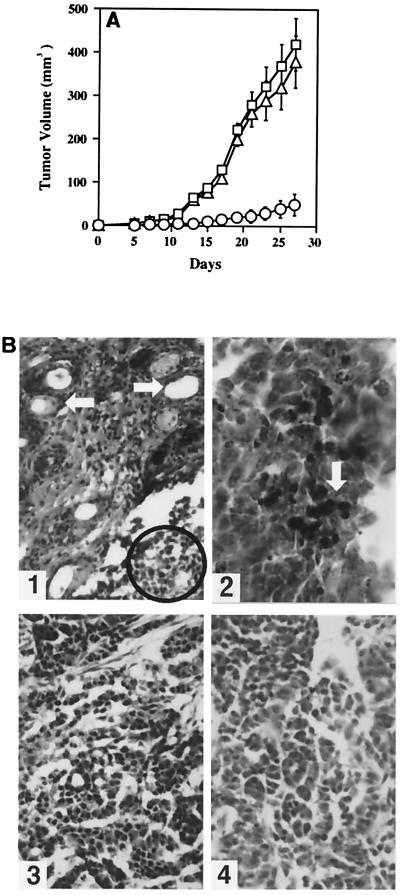
Because KGFR was able to inhibit the growth of HSY in vitro, we attempted to determine whether KGFR expression suppressed the tumorigenicity of HSY cells. There was no significant difference between the tumor growth of HSYzeo and HSYR2/R1 (P > 0.05). In contrast, there was a significant reduction in growth rate of HSYR2-IIIb-derived tumors compared with that of HSYzeo- and HSYR2/R1-derived tumors (P < 0.001, Fig. 5A, Table 1), and three of seven HSYR2-IIIb clones lost their tumorigenicity (Table 1). The HSYR2-IIIb-derived tumors were much smaller, softer, and paler in comparison with HSYzeo-derived tumors; they exhibited rounded structures with smooth surfaces; and they were well encapsulated and demarcated from the surrounding connective tissues. HSYzeo- and HSYR2/R1-derived tumors had an irregular rounded appearance with lobulated or multinodular surfaces. The connective tissue, which is abundant in vascularization, did not completely encapsulate the tumors (data not shown). Histologically, tumors developed from HSYR2-IIIb exhibited prominent duct-like and acinar-like structures surrounded by stromal cells (Fig. 5B1). Furthermore, in some undifferentiated areas, DNA fragmentation and apoptotic bodies were observed in the nuclei of HSYR2-IIIb-derived tumors (Fig. 5B2). HSYzeo- and HSYR2/R1-derived tumors exhibited typical adenocarcinomas with a solid and trabecular pattern (Fig. 5 B3 and B4).
Figure 5.
Analysis of tumors formed in nude mice. (A) The tumorigenicity of HSYzeo, HSYR2/R1, and HSYR2-IIIb. There was no significant difference between the tumor growth of HSYzeo (□) and HSYR2/R1 (▵) (P > 0.05). In contrast, there was a significant reduction in growth rate of HSYR2-IIIb-derived tumors (○) as compared with those of HSYzeo- and HSYR2/R1-derived tumors (P < 0.001). For each group, the mean ± SE was plotted. (B1) Tumors developed from HSYR2-IIIb exhibited predominant gland-like and acinar-like structures surrounded by stromal cells (arrowhead). In some areas, apoptotic bodies were observed in the nuclei of the tumor cells (circled) (original magnification: ×100). (B2) On TUNEL examination, tissue section of HSYR2-IIIb-derived tumor showed positive signals in tumor cells (arrowhead) (original magnification: ×200). The histological appearances of HSYR2/R1-derived (B3) and HSYzeo-derived (B4) tumors exhibited typical adenocarcinomas with a solid and trabecular pattern (original magnification: ×100).
Differential Kinase Phosphorylation.
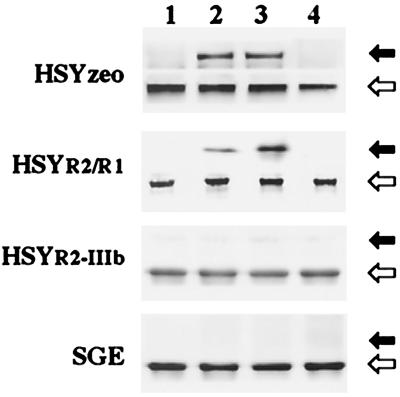
As FRS2 is essential for FGF-induced DNA synthesis (13), we assessed the phosphorylation state of FRS2 after FGF stimulation in different transfectants and SGE cells. FRS2 was identified as a tyrosine-phosphorylated protein in HSYzeo and HSYR2/R1 cells stimulated with FGF1 and FGF2, but not in HSYR2-IIIb and SGE cells (Fig. 6). On stimulation with FGF-1 and FGF-2, p38 MAP kinase was strongly activated in HSYzeo and HSYR2/R1 cells, but not in HSYR2-IIIb or SGE (data not shown). Moreover, the proliferation of HSYzeo and HSYR2/R1 was inhibited significantly in the presence of the specific p38 MAP kinase inhibitor SB203580 (data not shown).
Figure 6.
FGF-stimulated phosphorylation of FRS2. The cell lysates extracted from SGE, HSYzeo, HSYR2/R1, and HSYR2-IIIb were immunoprecipitated with anti-Grb2 antibody and were blotted with anti-phosphotyrosine antibody or anti-FRS2 antibody. Closed arrowheads indicate the position of the phosphorylated form of FRS2, and open arrowheads indicate the position of the unphosphorylated forms of FRS2. Lane 1: Control; lane 2: FGF-1 stimulated; lane 3: FGF-2 stimulated; lane 4: KGF stimulated. Data represent the results of clone 7A and clone 7B, as other clones gave essentially the same results.
Discussion
In the present study, we have demonstrated that the wild-type KGFR gene transferred into HSY human salivary adenocarcinoma cells induced differentiation and apoptosis, while suppressing tumor cell growth in vitro and inhibiting tumor growth in athymic mice. Apoptosis, or programmed cell death, is a fundamental process in organ development and tissue homeostasis in complex organisms (23). The induction of apoptosis has been widely used as an approach to cancer therapy (24). The morphological hallmarks of apoptosis, including membrane blebbing, volume loss, nuclear condensation, and DNA fragmentation, were all observed when a wild-type KGFR gene was transfected into HSY salivary adenocarcinoma cells. The apoptosis of HSYR2-IIIb cells was observed during a 7-day culture in KGF. Furthermore, endogenous DNA fragmentation and apoptotic bodies were observed in HSYR2-IIIb-derived tumors in athymic mice. These results describe a mechanism of blocking tumor growth in which restoration of KGFR tyrosine kinase activity in HSY cells induces apoptosis on stimulation by its ligand KGF.
α-Amylase and lactoferrin are major secretory products of human salivary glands that serve as markers of acinar cell and ductal cell differentiation, respectively (25–27). Immunocytochemical analysis indicated that the expression level of α-amylase and lactoferrin in HSYR2-IIIb was much higher than that in HSYzeo and HSYR2/R1 even without KGF. Histological examination revealed that predominant duct-like, acinar-like structures and apoptotic cells were always observed closely opposed to stromal cells in HSYR2-IIIb-derived tumors. These results suggest that the tyrosine kinase of KGFR is very important for cellular differentiation and that the communication between stromal cells and epithelial cells was involved in the induction of cellular differentiation.
The growth of HSYzeo and HSYR2/R1 was stimulated by either FGF-1 or FGF-2, but not by KGF. In contrast, the growth of HSYR2-IIIb was not stimulated by FGF-1, FGF-2, or KGF. We found that FGFR1 and FGFR4 were still expressed in HSYR2-IIIb by reverse transcription–PCR analysis (data not shown). How does KGFR tyrosine kinase alter the signaling of FGF-1 and FGF-2 to inhibit tumor growth? Like many other growth factor receptors, KGFR, when overexpressed, may be easily autophosphorylated in the absence of ligand (28, 29). Our results showed that the differentiation and apoptosis of HSYR2-IIIb cells occurred even in the absence of KGF and were further accelerated in the presence of KGF. Stimulation of cell proliferation and differentiation by FGF receptors depends on the Ras-MAP kinase signal pathway (30, 31). FRS2 plays an important role in linking FGF with Ras-MAP kinase signaling. In response to FGF stimulation, FGF receptors bind to Grb2 to become tyrosine-phosphorylated by FRS2. It has been shown that FGF-induced DNA synthesis can be inhibited by microinjection of anti-FRS2 antibody (13). Moreover, an FRS2 mutant impaired Grb2 binding and failed to induce neuronal differentiation of PC12 cells (14). In the present study, either FRS2 or p38 MAP kinase was identified as a phosphorylated protein in HSYzeo and HSYR2/R1, but not in HSYR2-IIIb or SGE cells on stimulation by FGFs. These results demonstrated that the tyrosine kinase of KGFR suppresses the phosphorylation of FRS2 and blocks p38 MAP kinase signaling. Moreover, the specific p38 MAP kinase inhibitor SB203580 inhibited the proliferation of HSYzeo and HSYR2/R1, suggesting that the blocking of p38 MAP kinase may be one mechanism by which the restoration of KGFR tyrosine kinase inhibits adenocarcinoma cell growth.
We have reported that FGF-1 and KGF stimulated the growth of SGE cells (10); however, tyrosine phosphorylation of FRS2 was not observed in SGE. The result implies that FGF stimulates proliferation of normal epithelial cells and malignant cells by distinct signaling pathways.
The progressive loss of KGFR expression has been implicated in the malignant progression of many tumors, including human bladder tumors (32) and prostate tumors (33). It has also been demonstrated that restoration of KGFR tyrosine kinase induced the differentiation of malignant cells and inhibited the growth of human bladder carcinomas and malignant rat prostate carcinomas (32, 34). However, the mechanism by which the KGFR gene inhibits malignant cell growth was not elucidated. Our findings demonstrate that the restoration of KGFR tyrosine kinase in human adenocarcinoma cells caused decreased levels of FRS2 phosphorylation and led to induction of apoptosis and differentiation. These results suggest that KGFR gene therapy might be a viable approach to inhibiting the growth of salivary adenocarcinomas.
Acknowledgments
This work was supported in part by a grant from Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists to Y.Z.; by a grant-in-aid from the Japanese Ministry of Education, Science and Culture to T.O.; and by a grant from the Smoking Research Foundation to T.O. J.D.S. is supported by grant DAMD 17-98-1-8348 from the U.S. Army Medical Research and Material Command and by Grant P40 RR15452 from the National Institutes of Health (NIH). M.K. and W.L.M. were supported in part by NIH Grants DK 35310, DK 47039, and CA 59971.
Abbreviations
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- KGF
keratinocyte growth factor
- KGFR
keratinocyte growth factor receptor
- FRS2
FGF receptor substrate 2
- Grb2
growth factor receptor-bound protein 2
- MAP
mitogen-activated protein
- SGE cells
salivary gland-derived epithelial cells
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick-end labeling
References
- 1.Finch P W, Rubin J S, Miki T, Ron D, Aaronson S A. Science. 1989;245:752–755. doi: 10.1126/science.2475908. [DOI] [PubMed] [Google Scholar]
- 2.Rubin J S, Osada H, Finch P W, Taylor W G, Rudikoff S, Aaronson S A. Proc Natl Acad Sci USA. 1989;86:802–806. doi: 10.1073/pnas.86.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battaro D P, Rubin J S, Ron D, Finch P W, Florio C, Aaronson S A. J Biol Chem. 1990;265:12767–12770. [PubMed] [Google Scholar]
- 4.Miki T, Fleming T P, Bottaro D P, Rubin J S, Ron D, Aaronson S A. Science. 1991;251:72–75. doi: 10.1126/science.1846048. [DOI] [PubMed] [Google Scholar]
- 5.Marchese C, Chedid M, Dirsch O R, Csaky K G, Santanelli F F, Latini C, Larochelle W J, Torrisi M R, Aaronson S A. J Exp Med. 1995;182:1369–1376. doi: 10.1084/jem.182.5.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Rochelle W J, Dirsch O R, Finch P W, Cheon H G, May M, Marchese C, Pierce J H, Aaronson S A. J Cell Biol. 1995;129:357–366. doi: 10.1083/jcb.129.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchese C, Sorice M, Stefano C D, Frati L, Torrisi R. Cell Growth Differ. 1997;8:989–997. [PubMed] [Google Scholar]
- 8.Myoken Y, Myoken Y, Okamoto T, Sato J D, Kan M, McKeehan W L, Nakahara M, Takada K. J Pathol. 1996;178:429–436. doi: 10.1002/(SICI)1096-9896(199604)178:4<429::AID-PATH495>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Myoken Y, Myoken Y, Okamoto T, Kan M, McKeehan W L, Sato J D, Takada K. Int J Cancer. 1996;65:650–657. doi: 10.1002/(SICI)1097-0215(19960301)65:5<650::AID-IJC15>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, Sakamoto A, Zhang Y, Toratani S, Kan M, Okamoto T. Tissue Culture Res Commun. 1997;16:207–212. [Google Scholar]
- 11.Bar-Sagi D, Feramisco J R. Cell. 1985;42:841–848. doi: 10.1016/0092-8674(85)90280-6. [DOI] [PubMed] [Google Scholar]
- 12.Noda M, Ko M, Ogura A, Liu D G, Amano T, Takano T, Ikawa Y. Nature (London) 1985;318:73–75. doi: 10.1038/318073a0. [DOI] [PubMed] [Google Scholar]
- 13.Kouhara H, Hadari Y R, Spivak-Krozman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 14.Hadari Y R, Kouhara H, Lax I, Schlessinger J. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myoken Y, Okamoto T, Sato J D, Kan M, McKeehan W L, Fujihara M, Takada K. In Vitro Cell Dev Biol Anim. 1995;31:84–86. doi: 10.1007/BF02633966. [DOI] [PubMed] [Google Scholar]
- 16.Yanagawa T, Hayashi Y, Naganine S, Yoshida S, Yura Y, Sato M. Virchows Arch B Cell Pathol. 1986;51:185–195. doi: 10.1007/BF02899028. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara A, Kan M, Feng S, McKeehan W L. Cancer Res. 1998;58:1509–1514. [PubMed] [Google Scholar]
- 18.Kan M, Shi E G, McKeehan W L. Methods Enzymol. 1991;198:158–171. doi: 10.1016/0076-6879(91)98017-z. [DOI] [PubMed] [Google Scholar]
- 19.Sato J D, Kawamoto T, Okamoto T. J Exp Med. 1987;165:1761–1766. doi: 10.1084/jem.165.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang J H, Wang F, Kan M. In Vitro Cell Dev Biol Anim. 1997;33:819–824. doi: 10.1007/s11626-997-0162-7. [DOI] [PubMed] [Google Scholar]
- 21.McKeehan W L, Crabb J W. Anal Biochem. 1987;164:563–569. doi: 10.1016/0003-2697(87)90534-3. [DOI] [PubMed] [Google Scholar]
- 22.Gavrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 24.Kerr J P R, Winterford C M, Biol A D A, Harmon B V. Cancer (Philadelphia) 1993;73:2013–2026. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 25.Przbyla A E, MacDonald R J, Harding J D, Pictet R L, Rutter W J. J Biol Chem. 1979;254:2154–2159. [PubMed] [Google Scholar]
- 26.MacDonald R J, Crerar M M, Swain W F, Pictet R L, Thomas G, Rutter W J. Nature (London) 1980;287:117–122. doi: 10.1038/287117a0. [DOI] [PubMed] [Google Scholar]
- 27.Shirasuna K, Morika S, Watatani K, Hayashido Y, Furusawa H, Sugiyama M, Okura M, Matsuya T. Cancer Res. 1988;48:2819–2824. [PubMed] [Google Scholar]
- 28.Vikkula M, Boon L M, Carraway K L, III, Calvert J T, Diamonti A J, Goumnerov B, Pasyk K A, Marchuk D A, Warman M L, Cantley L C, et al. Cell. 1996;87:1181–1190. doi: 10.1016/s0092-8674(00)81814-0. [DOI] [PubMed] [Google Scholar]
- 29.Mohammadi M, Dikic I, Sorokin A, Burgess W H, Jaye M, Schlessinger J. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J K, Xu H, Li H C, Goldfarb M. Oncogene. 1996;13:721–729. [PubMed] [Google Scholar]
- 31.Klint P, Kanda S, Kloog Y, Welsh L C. Oncogene. 1999;18:3354–3364. doi: 10.1038/sj.onc.1202680. [DOI] [PubMed] [Google Scholar]
- 32.Medina S G, Dominique C, Ahmed E, Annie D, Wiliam J L, Anras H, Claude A, Stuart A A, Jean P T, Francois R. Oncogene. 1997;14:323–330. [Google Scholar]
- 33.Russ P C, James E, Hannah R K, Phillip J W, Mariano A G-B. Oncogene. 1997;15:3059–3065. [Google Scholar]
- 34.Feng S, Wang F, Matsubara A, Kan M, McKeehan W L. Cancer Res. 1997;57:5369–5378. [PubMed] [Google Scholar]