Abstract
The dorsal–ventral polarity of the somite is controlled by antagonistic signals from the dorsal neural tube/surface ectoderm, mediated by WNTs, and from the ventral notochord, mediated by sonic hedgehog (SHH). Each factor can act over a distance greater than a somite diameter in vitro, suggesting they must limit each other's actions within their own patterning domains in vivo. We show here that the growth-arrest specific gene 1 (Gas1), which is expressed in the dorsal somite, is induced by WNTs and encodes a protein that can bind to SHH. Furthermore, ectopic expression of Gas1 in presomitic cells attenuates the response of these cells to SHH in vitro. Taken together, these data suggest that GAS1 functions to reduce the availability of active SHH within the dorsal somite.
Somites are the fundamental units of the segmental body plan of vertebrate embryos. They bud off from unsegmented presomitic mesoderm (psm) and become organized into epithelial spheres. The ventral cells differentiate into sclerotome, and the dorsal cells differentiate into dermomyotome under the influence of signals produced by adjacent tissues (1). In the mouse, sonic hedgehog (SHH) and Noggin, expressed in the notochord, are thought to be the ventralizing signals for sclerotome induction (2–4). WNT proteins produced by the surface ectoderm and dorsal neural tube are presumed to help establish the dorsal domain (dermomyotome) of the somites (5). The phenotypes of Shh (6), Noggin (4), and Wnt1/Wnt3a (7) homozygous null mutant embryos support the roles of the corresponding proteins initially defined by in vitro assays. Tissue extirpation, transplantation, and ectopic expression experiments in the chicken embryo have led to similar conclusions (8–13).
When placed adjacent to psm in vitro, SHH or notochord cells can act over a distance of 150–250 μm to activate the sclerotomal markers Pax1 and Twist (2). Likewise, dorsal neural tube or WNT1 protein can act over 120 μm to activate dermomyotomal markers such as Pax3 and Pax7 (2, 5). An early somite is only 80–100 μm in diameter, suggesting that every cell in the somite is within the reach of both signals. Nevertheless, in vivo, cells closest to the WNT signals invariably differentiate into dermomyotome, whereas cells closest to the SHH signal give rise to sclerotome. Moreover, when applied at opposite sides of the explanted psm in varying concentrations, the two signals pattern the psm into mutually exclusive sclerotome and dermomyotome domains, the sizes of which vary according to the relative levels of the input signals (5). These results together support the hypothesis that there is competition between the two signals to subdivide the naive somitic mesoderm into dorsal and ventral domains. This model is also supported by many elegant ablation and misexpression studies in chick embryos (8–13). However, the mechanism(s) by which WNT and SHH competitively limit the range of each other's action is not known. Here we provide evidence that GAS1, the protein encoded by the growth arrest-specific gene 1 (Gas1), can mediate the antagonistic effect of WNT proteins on SHH signaling in the developing somite.
Materials and Methods
cDNA Library Screen.
A cDNA library was constructed in pMT21 with the use of poly(A)+ RNA isolated from the caudal quarter of 180 embryonic day (E)9.5 CD1 mouse embryos. COS cells were transfected with 174 pools (1,000 cDNAs per pool) of the library with Lipofectamine (GIBCO/BRL) and screened for SHH-N-alkaline phosphatase (SHH-N-AP) binding activity (14). Positive pools were rescreened to obtain single clones. SHH-N-AP and SHH-N-Fc were fusions of SHH-N (amino acids 1–198) to AP and to Ig constant region (Fc) (15). Vectors containing these fusions were transfected into 293EBNA cells (Invitrogen), and the ligands were produced and collected in conditioned media according to the methods described in ref. 15.
Dissociation Constant (Kd) Measurement.
Kds were determined according to the methods described in ref. 15, with the use of SHH-N-AP with 100 ng/ml heparin. 293EBNA cells stably expressing GAS1 were used for binding. SHH-N-AP and SHH-N had the same activity in the psm explant assay (2). SHH-N-AP displayed 76% of the AP activity measured for unmodified AP at the same molarity. Activity is expressed as OD405 unit/h and measured by the absorbance of p-nitrophenol converted from p-nitrophenyl-phosphate (1 pmol of AP = 30 OD405 unit/h).
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed with an Altermax kit (Promega). The altered amino acids residues are stated in the text. The hemagglutinin (HA) tag was inserted between amino acid 188 and amino acid 189 of GAS1 (GAS1-HA).
Immunoprecipitation/Immunofluorescence/Western Blot.
Immunoprecipitation, immunofluorescence, and Western blot were performed as described in ref. 16. Rabbit Abs to GAS1 were described (17). Anti-AP (Medix), anti-Fc (Jackson ImmunoResearch), and anti-SHH-N (5E1) Abs were used to detect fusion ligands. Immunoprecipitations of GAS1 with SHH-N-Fc and SHH-N with GAS1-HA were performed with cells expressing GAS1 and GAS1-HA, respectively. After incubation with 1 μg/ml of SHH-N-Fc or SHH-N in the media, cells were rinsed with PBS and lysed in PBS/0.5% Nonidet P-40. Protein A or 12CA5/protein A beads were added, incubated for 30 min, washed three times, and eluted in sample buffer at 80°C. Samples were subjected to SDS/PAGE and Western blot with anti-GAS1 or 5E1 and visualized by AP-2° Abs coupled with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color development. Live-cell labeling was performed with 0.02% of Na azide at 4°C. Tetramethylrhodamine B isothiocyanate, and FITC-2° Abs were from Sigma.
In Situ Hybridization.
Whole-mount or sectioned embryos of specified stages were subjected to in situ hybridization with the use of [35S]UTP or digoxigenin-UTP-labeled probes (18). For radioactive in situ hybridizations, silver granules were photographed as dark-field images, phase images were taken, and the two were overlaid. Adjacent sections were used for comparison. Shh and Ihh probes were provided by A. McMahon, Harvard University, Cambridge, MA.
Generation of Adenovirus Vectors.
Adeno-X vector (CLONTECH) was used to construct cytomegalovirus promoter-driven Gas1- internal ribosomal entry site-green fluorescent protein (GFP) or GFP expression vehicles. Viruses were produced and concentrated to 5 × 1010 plaque-forming units/ml according to the manufacturer's instructions. A total of 1 × 108 plaque-forming units was used to infect one psm explant (≈4,000 cells) in 250 μl of culture media. Twenty hours after infection, explants were further treated as indicated.
BrdUrd Assay.
Explants were incubated with 10 μM BrdUrd for 6 h after 20 h of viral infection, or for 1 h after overnight treatment with SHH-N (50 ng/ml) or basic fibroblast growth factor (10 ng/ml). BrdUrd-positive cells were detected by anti-BrdUrd and TRITC-2° Abs (Sigma). Six explants of each treatment were assessed. The statistical differences are based on P values less than 0.005–0.01 by t test.
Results
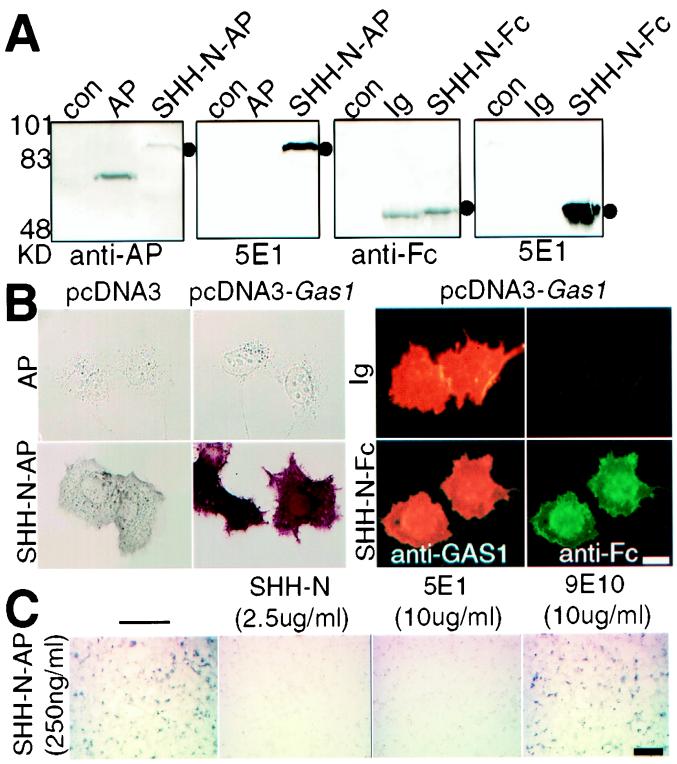
One possible mechanism used by WNT to antagonize SHH action is for WNT to induce in the dorsal somite an antagonist to SHH that limits its availability to surface receptors. To investigate this possibility, we performed a screen for genes whose products interact with SHH-N (the N-terminal fragment of SHH) on the cell surface. An expression cDNA library was made from the caudal 1/4 of E9.5 mouse embryos containing the presomitic mesoderm (psm) and early somites. COS7 cells were transfected with pools of cDNA expression plasmids and screened for cell surface binding of SHH-N. To aid in the visualization of binding-positive cells, SHH-N-AP and SHH-N-Fc fusion proteins were used as ligands (14, 15). These fusion proteins were of the predicted sizes (Fig. 1A) and were functional in vitro (not shown). The first identified SHH-N-AP binding-positive pool was subjected to sib selection to isolate a single cDNA clone. This clone was found to correspond to Gas1 (19). GAS1 is a 45-kDa glycosylphosphatidylinositol-linked membrane glycoprotein (20), originally identified by its ability to arrest the cell cycle in cultured cells (17). The following data demonstrate the specificity of the surface binding activity observed: (i) SHH-N-AP but not AP alone binds to GAS1-expressing cells (Fig. 1B); (ii) SHH-N-Fc but not human IgG (Ig) alone binds to GAS1-expressing cells; (iii) GAS1 and surface-bound SHH-N-Fc are strictly colocalized on the same cells (Fig. 1B); (iv) high concentrations of anti-GAS1 Abs block SHH-N-AP and SHH-N-Fc binding (not shown); (v) binding of SHH-N-AP and SHH-N-Fc (not shown) to GAS1-expressing cells can be blocked by a 50-fold excess of SHH-N added simultaneously (Fig. 1C); (vi) a variety of cell types, including 293, 10T1/2, 3T3 cells, and chick embryonic fibroblasts expressing GAS1 also display SHH-N-AP binding activities (not shown). These results indicate that the SHH-N of the fusion reporters is involved in binding to the surface of the GAS1-expressing cells. The binding withstands up to 1 μg/ml of heparin, indicating that it does not involve a nonspecific ionic interaction. Binding of SHH-N-AP to GAS1-expressing cells can be blocked by the SHH-N functional blocking Ab, 5E1 (21) (Fig. 1C), suggesting that the binding is physiologically relevant.
Figure 1.
SHH-N binds to COS cells expressing GAS1. (A) Western blots using Abs to AP (anti-AP), to Fc (anti-Fc), and to SHH-N (anti-SHH-N, 5E1) confirmed that the SHH-N-AP and SHH-N-Fc fusion proteins were of the expected sizes. con (control) and AP, media from 293EBNA cells transfected with vector without insert and vector with AP. As controls, 50 ng of AP and human IgG (Ig) was used. Markers are in kDa. (B) (Left) COS cells transfected with pcDNA3 did not bind AP and bound low levels of SHH-N-AP. COS cells transfected with pcDNA3-Gas1 did not bind AP but did bind SHH-N-AP. (Right) Labeling of SHH-N-Fc and GAS1 on the same cells. (Upper) Cells expressing GAS1 (red, detected by anti-GAS1 and Cy3-2° Abs) do not bind human IgG (Ig, green, detected by FITC-anti-Fc). (Lower) Cells expressing GAS1 bind SHH-N-Fc; 1 μg/ml of fusion ligands was used for binding. (Scale bar = 10 μm.) (C) GAS1-SHH-N-AP binding was competed by the simultaneous addition of a 50-fold excess of recombinant SHH-N or 5E1, but not 9E10 Ab. −, no other reagent was included. (Scale bar, 100 μm.)
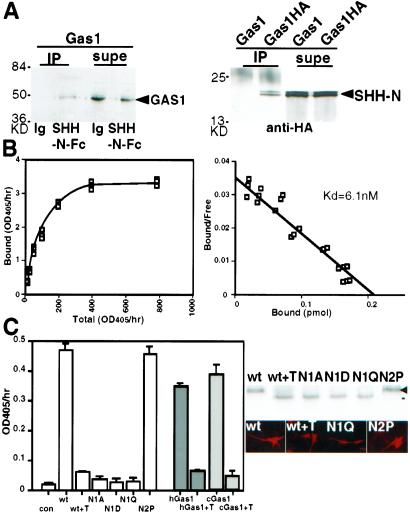
To demonstrate that GAS1 protein forms a complex with SHH-N, we performed coimmunoprecipitation experiments. Because 5E1 interferes with binding and anti-GAS1 Ab is ineffective in immunoprecipitation, we opted to use HA-tagged GAS1 (GAS1-HA) and SHH-N-Fc proteins. As shown in Fig. 2A, SHH-N-Fc, but not IgG, formed a complex with GAS1 that was precipitated by protein A beads. Reciprocally, with the use of anti-HA Ab, SHH-N was precipitated with GAS1-HA but not with untagged GAS1. These results demonstrate that GAS1 forms a physical complex with SHH-N. With the use of SHH-N-AP and 293EBNA cells expressing GAS1, we have determined the dissociation constant (Kd) between GAS1 and SHH-N to be 6.1 nM (Fig. 2B). In a side-by-side comparison, we obtained a Kd of 1.4 nM for PTC1 and SHH-N-AP, similar to the 0.5–1 nM reported by others (22–24). Thus, GAS1-SHH-N has a Kd distinct from those reported for PTC1-SHH-N and HIP1-SHH-N (14). A specific mutation of SHH-N, H134Y, was speculated to be an activating mutation (25). We found that this mutant form of SHH-N binds to PTC1 with the same affinity, but it binds to GAS1 with 4.5-fold lower affinity (Kd = 27.4 nM). This finding further confirms that the binding between GAS1 and SHH is unique and distinct from that between PTC1 and SHH-N.
Figure 2.
SHH-N binds GAS1 and the binding depends on the N-glycosylation of GAS1. (A) GAS1 and SHH-N immunoprecipitate each other. (Left) GAS1 expressed on 293EBNA cells was precipitated by SHH-N-Fc but not by IgG (Ig) with protein A. The supernatant (supe) was lysate containing unbound GAS1. (Right) GAS1-HA precipitated recombinant SHH-N in the presence of anti-HA. The supernatant contained the unbound SHH-N. GAS1-HA and SHH-N (arrowheads) were detected by Western blots. (B) The binding data (Left) and Scatchard plot (Right) show the Kd between SHH-N-AP and GAS1 to be 6.1 nM. (C) (Left) Cells expressing wild-type mouse GAS1 (wt), human GAS1 (hGas1, gray), and chick GAS1 (cGas1, slashed lines) display SHH-N-AP binding. The binding was reduced after 25 ng/ml of tunicamycin treatment for 4 h (+T). Change of the first N-glycosylation site of GAS1 to A, D, or Q (N1A, N1D, and N1Q, respectively) reduced SHH-N-AP binding. (Right) N1A, N1D, N1Q, or tunicamycin-treated proteins were produced at the ≈50% level of the wt GAS1 and were detected on the cell surface by live cell labeling. Tunicamycin-treated GAS1 migrated to the same position as N1A, N1D, and N1Q (black line). GAS1 with the second predicted glycosylation site changed to a proline (N2P) behaved identically to the wild type.
The association between PTC1 and SHH-N depends on the N-glycosylation of PTC1 (23, 24). We attempted to determine whether N-glycosylation was also required for the interaction between GAS1 and SHH-N. When cells expressing mouse GAS1 were treated with tunicamycin, they displayed much reduced SHH-N-AP binding (Fig. 2C). Cells expressing human GAS1 or chick GAS1 also displayed tunicamycin-sensitive binding to SHH-N-AP. There are two predicted N-glycosylation sites in the mouse GAS1. When the first predicted site (N342) was mutated to alanine, glutamic acid, or glutamine (N1A, N1D, and N1Q, respectively), less than 10% of the SHH-N-AP binding activity was observed (Fig. 2C). Mutant and tunicamycin-treated proteins were produced at about half the level of the wild-type protein and were transported to the cell surface, as assessed by live cell surface labeling (Fig. 2C Left). Importantly, mutant proteins had the same electrophoretic mobility as the tunicamycin-treated protein. When the second predicted glycosylation site, which is not conserved, was changed to a proline (N2P), neither the glycosylation of GAS1 nor its binding to SHH-N-AP was affected. SHH-N-AP and SHH-N-Fc did not bind to other N-glycosylated proteins such as L1 and TAG1 (not shown). It is thus likely that both the carbohydrate moiety and the polypeptide chain of GAS1 contribute to the formation of a complex with SHH-N.
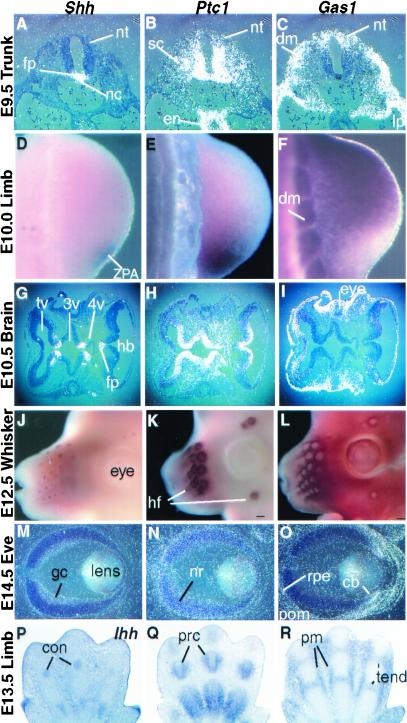
We have documented Gas1 expression during mouse embryogenesis (26). The expression pattern of Gas1 is in striking contrast to those described for Ptc1 and Shh. Here we compare their expression patterns by in situ hybridization on adjacent sections and whole-mount in situ hybridization of embryos of various stages. Gas1 expression in the E9.5 trunk is enriched in the dermomyotome and the dorsal neural tube and skirts the ventrally localized expression of Ptc1 (Fig. 3 A–C). Gas1 is transcribed in the psm at low levels and is up-regulated in the dorsal region as soon as the somite forms, before dermomyotome formation (26). Similar complementary yet overlapping expressions between Ptc1 and Gas1 are observed in the E10.5 limb bud with respect to the expression of Shh in the zone of polarizing activity (Fig. 3 D–F). Opposing patterns between Ptc1 and Gas1 are also seen, relative to the midline expression of Shh, in the brain ventricles and head mesenchyme at E10.5 (Fig. 3 G–I). In the vibrissae, Shh is expressed in the epithelium at the base of the follicle; Ptc1, in the adjacent mesenchyme; and Gas1, in a ring outside of the Ptc1 domain (Fig. 3 J–L). In the E14.5 eye, Shh is expressed in ganglion cells; Ptc1, in the neural retina layer; and Gas1, in the cilliary body, retinal pigmented epithelium, and the perioptic mesenchyme (Fig. 3 M–O). Finally, in the E13.5 limb skeletal elements, Ihh is expressed in condensing chondrocytes; Ptc1, in perichondrocytes; and Gas1, in the surrounding mesenchyme and future tendon regions (Fig. 3 P–R). In light of this limb expression pattern, we found that GAS1 binds IHH-N-AP with an affinity similar to that with which it binds SHH-N-AP (not shown). Thus, Gas1 expression is consistently associated with sources of Shh and Ihh expression. Moreover, its expression pattern suggests that the protein modulates SHH-N activity via a mechanism distinct from that of PTC1.
Figure 3.
Gas1 expression is associated with the Shh and Ihh expression centers and complementary to Ptc1 expression. Comparative studies were carried out with radioactively labeled or digoxigenin-labeled Shh, Ihh, Ptc1, and Gas1 probes for in situ hybridization on adjacent sections (A–C, G–I, M–R) or whole-mount embryos (D–F, J–L). The embryo stages and tissues are labeled to the left. (A–C) Transverse sections through E9.5 trunk. nc, notochord; nt, neural tube; fp, floorplate; sc, sclerotome; dm, dermomyotome; lp, lateral plate. (D–F) Dorsal views of E10.5 hindlimbs. ZPA, zone of polarizing activity. (G–I) horizontal sections of E10.5 head. tv, telencephalic vesicle; 3v, third ventricle; 4v, fourth ventricle; hb, hindbrain. (J–L) E12.5 vibrissae. wf, whisker follicles. (M–O) E14.5 eyes. gc, ganglionic cells; nr, neural retina; rpe, retinal pigmented epithelium; pom, perioptic mesenchyme; cb, cilliary body. (P–R) E13.5 hand plate. c, condensing chondrocyte; prc, perichondrium; pm, perimesenchyme; tend, future tendon region. Note that P is hybridized with the Indian hedgehog (Ihh) probe.
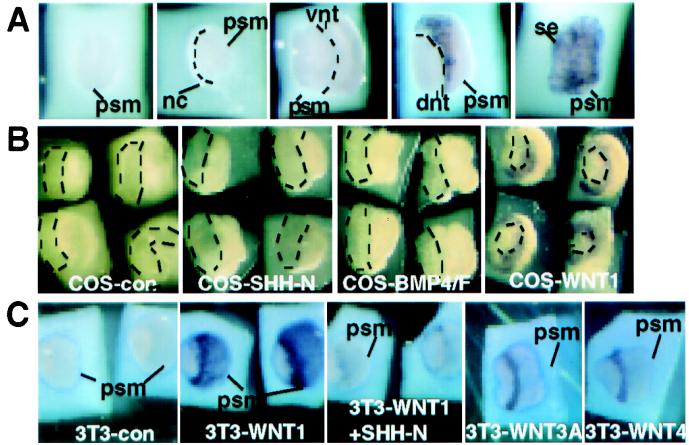
Dorsal neural tube and surface ectoderm provide signals to the psm that compete with the ventralizing activity of SHH-N and establish the somitic dorsal domain (2). Gas1 expression in the dorsal somite is consistent with its regulation by dorsal structures. To test this hypothesis, we used the psm explant system (2). Psm explants cultured alone for 24 h expressed low or no Gas1 transcript when assessed by whole-mount in situ hybridization (Fig. 4A). As predicted, coculture of psm with surface ectoderm and dorsal neural tube induced Gas1 expression in the psm. In contrast, notochord and ventral neural tube had no effect. The molecular identities of the dorsalizing signals have been assigned to members of the WNT family (5). In support of this conclusion, Gas1 expression was robustly activated by WNT1-producing COS cells but not by control COS cells (Fig. 4B). BMP4 is also expressed in the dorsal tissues and can antagonize SHH-N (4). However, neither COS cells expressing BMP4 with Furin (27) nor recombinant BMP2, BMP4, or BMP7 (not shown) activated Gas1 expression. SHH-expressing COS cells also did not induce Gas1 (Fig. 4B). Among the WNT-expressing cells tested, WNT3A and WNT4 cells also induced Gas1 (Fig. 4C), but WNT7A and WNT7B cells did not (not shown). Intriguingly, WNT1-induced Gas1 expression could be repressed by high concentrations of SHH-N (500 ng/ml), as evidenced by both reduced signal intensity and expression domain size. Consistently, Gas1 expression in the somites extends to the ventral domain in Shh mutant embryos (data not shown). Thus, the regulated Gas1 expression documented in vitro is consistent with its somitic expression pattern observed in vivo.
Figure 4.
Gas1 expression in the paraxial mesoderm is regulated by WNT and SHH. (A) E9.5 mouse psm explants were cultured in collagen gels. The inductive tissues were isolated from E2.5 chick. (Left to Right) psm was cultured alone, next to a notochord (nc), a ventral neural tube (vnt), a dorsal neural tube (dnt), or inside a surface ectoderm (se). Dashed lines, the boundary between psm and the chick tissues. Explants were assessed for Gas1 expression. (B) (Left to Right) psms were cocultured with control COS cells (COS-con) or cells expressing SHH-N (COS-SHH-N), BMP4/Furin (COS-BMP4/Furin), and WNT1 (COS-WNT1). Explants were assessed for Gas1 induction. Four samples are represented by each image. (C) Parental 3T3 cells (3T3-con) did not induce Gas1. WNT1-expressing cells (3T3-WNT1) induced Gas1 over 200 μm. The induction was reduced in the presence of 500 ng/ml SHH-N. WNT3A- and WNT4-expressing 3T3 cells also induced Gas1.
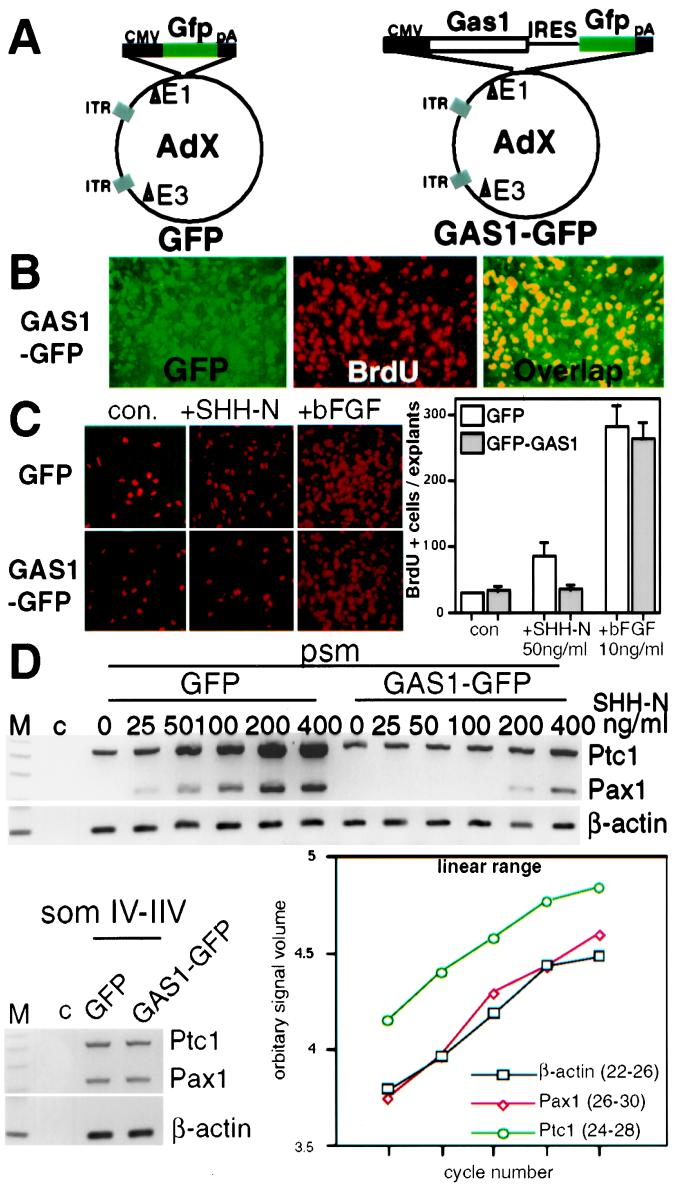
The expression, regulation, and SHH-N binding activity of GAS1 all strongly suggest that the protein normally functions to physically prevent SHH-N from activating its downstream response pathways in the dorsal somitic cells. To test this model, we assessed whether ectopic Gas1 expression in the psm could render cells resistant to SHH-N signaling. To effectively deliver expression constructs to mouse psm in vitro, we used adenoviral vectors containing cytomegalovirus-Gas1-internal ribosomal entry site-GFP (GAS1-GFP) and cytomegalovirus-GFP (GFP) (Fig. 5A). In psm infected with GAS1-GFP virus, GAS1 protein was expressed at high levels, and there was a complete correspondence between GAS1 and GFP protein expression in infected cells (not shown). At least 107 plaque-forming units were needed to infect all cells within a psm explant (≈2.5 × 103 plaque-forming units per cell). Explants were treated with reagents after 20 h of infection when GAS1 was already at high levels. Although Gas1 can block cell cycle progression in some cell types, GAS1-GFP-infected psm cells readily incorporated BrdUrd (Fig. 5B) to the same level as observed in noninfected or control GFP-infected explants (not shown), indicating that Gas1 does not inhibit psm cell proliferation. This observation allowed us to test whether GAS1 interfered with the stimulation of cell proliferation by SHH-N in psm (3). When a moderate concentration of SHH-N (50 ng/ml) was added, 3-fold more BrdUrd-positive cells were observed in GFP-infected or uninfected (not shown) explants than in explants with no added SHH-N. In contrast, GAS1-GFP-infected explants incorporated BrdUrd at the basal level, even in the presence of 50 ng/ml of SHH-N (Fig. 5C). Importantly, GAS1-GFP-infected psm was responsive to a moderate concentration of basic fibroblast growth factor (10 ng/ml) to the same level as the GFP control. Thus, GAS1 specifically blocks the growth-stimulating activity of SHH-N in the psm.
Figure 5.
GAS1 renders psm resistant to SHH-N. (A) Cytomegalovirus-driving GFP (GFP, as control) and Gas1-internal ribosomal entry site-GFP (GAS1-GFP) were constructed in pAdeno-X vector. (B) BrdUrd incorporation. Explants were cultured in 10 μM BrdUrd for 6 h after infection. Anti-BrdUrd and TRITC-2° Abs were used. GAS1-GFP cells (green) incorporated BrdUrd (red) readily (overlap, yellow). (C) psm cells infected with GAS1-GFP did not respond to SHH-N (50 ng/ml)-induced proliferation but responded to basic fibroblast growth factor (10 ng/ml). (Right) The graph of BrdUrd counts of each treatment. The bars represent standard errors. (D) Pax1 and Ptc1 induction was assessed by quantitative reverse transcription–PCR. psms were infected with virus for 20 h, and SHH-N ranging from 12.5 ng/ml to 400 ng/ml was added. β-actin was used as a control. (Lower Left) GAS1-GFP and GFP infection did not repress Pax1 and Ptc1 in mature somites (somIV–VII). (Lower Right) The amplification ranges for the quantitative reverse transcription–PCR of Ptc1, Pax1, and β-actin. The midamplification cycles were used. PCR products were resolved on 2% agarose gels, stained with ethidium bromide, and quantified by imagequant.
We next examined the possibility that ectopic GAS1 expression also diminished the patterning activity of SHH-N in the psm by assessing Pax1 and Ptc1 expression (Fig. 5D). When quantitative reverse transcription–PCR was performed, we found that GAS1-GFP-infected psm was less responsive to exogenously applied SHH-N than the control GFP-infected psm. This GAS1-mediated attenuation was overcome by supplying SHH-N at concentrations above 200 ng/ml. Similarly, the attenuation of proliferation mediated by GAS1 described above was also overcome at these high concentrations of SHH-N (not shown). These results strongly argue that the GAS1 sites on the cell surface are saturable and that these cells retain their potential to respond to SHH-N. Furthermore, mature somites infected with GAS1-GFP or GFP virus expressed Pax1 and Ptc1 to the same level as controls, indicating that GAS1 itself does not nonspecifically interfere with expression of reporter genes in infected cells (Fig. 5D). GAS1-GFP virus also does not induce dorsal somitic marker expression (not shown). This finding is consistent with a model that GAS1 protein only serves as a physical sink for SHH-N.
Discussion
The WNT-induced expression of Gas1 and the SHH-N binding activity of its encoded protein support a model in which GAS1 functions as a mediator of WNTs' antagonism to SHH-N in the dorsal somite. The boundary of SHH-N diffusion was thought to be controlled locally through a self-imposed negative feedback loop involving the binding proteins PTC1 and HIP1 (14, 28). Recently, a long-range diffusible form of SHH-N has been identified in the anterior limb (29), where little or no Ptc1 and Hip1 is detected. Because Ptc1 expression and Hip1 expression serve as markers for SHH signaling activity, the detection of the long-range form of SHH-N implies that it is subjected to negative modulation at a distance from its source. Here we show that Gas1 is expressed at a distance from the SHH source (including the anterior limb) and distal to the Ptc1 expression domains. Moreover, the protein is capable of negatively regulating the availability of SHH-N activity with somitic mesoderm as an assay system. Taken together, these results suggest that GAS1 binding serves as a general mechanism to reduce the actions of both SHH and IHH at a distance.
The negative and positive regulation, respectively, of Gas1 and Ptc1 by SHH implies that multiple mechanisms control the patterning of the uncommitted psm by SHH (1). The combination of local feedback inhibitors of SHH activated by itself (via PTC1 and HIP1) and inhibition by GAS1 at a distance suggests an exquisite modulation of SHH availability. Intriguingly, there is no amino acid sequence homology between PTC1, HIP1, and GAS1. Analogously, Chordin, Noggin, and the DAN family of proteins have no clear homology, but in vitro expression studies suggest that they all act as inhibitors of BMP signaling (30). Although the phenotype of mouse embryos lacking Chordin does not reveal its role in gastrulation, Chordin/Noggin double-mutant embryos do show the importance of their combined action during mouse gastrulation (30). Gas1 null mutant mice have been generated and display defects in the eye (31), cerebellum (32), limb, and vertebra/ribs (unpublished observations). Although these tissues are known to be influenced by SHH signaling, the phenotypes cannot be explained simply by the antagonism between GAS1 and SHH, possibly due to compensatory activities of PTC1, HIP1, and/or SHH in the mutant. Thus, to determine precisely how PTC1, HIP1, and GAS1 work in concert to control SHH-N activity in vivo, it may be necessary to analyze double and triple null mutant combinations. Whereas we present data here that WNTs induce GAS1 in the dorsal somite to limit the activity of SHH, there is also evidence that SHH induces a frizzle-like protein, SFRP2, in the ventral somite to limit the activity of WNTs (33). It appears that the concerted action of inhibitory molecules of different structures, affinities, and regulated expression patterns is the recurrent theme in refining the boundaries and activities of signaling molecules during development.
Interference of the binding of SHH-N to GAS1 and to PTC1 by 5E1 suggests that SHH uses the same or overlapping interface for binding to PTC1 and GAS1 and may not be able to interact with both proteins at the same time, consistent with a binding-competition model. It is noted that high levels of mRNA for Smo [encoding the signaling receptor of SHH-N (34)] in the dorsal somite region correspond to the Gas1-positive domain, where little Ptc1 is detected (22). Given that the stability of Drosophila Smo protein is regulated by Hh (34), we cannot exclude the possibility that GAS1 may negatively regulate the stability of the vertebrate SMO protein in the dorsal somite.
One surprising aspect of our findings is that GAS1 is a SHH-N-binding protein. GAS1 was initially identified by its ability to activate growth arrest in cultured cells through an obligatory p53 pathway (35), which appears to be ligand-independent. The expression pattern of Gas1 does not correlate with its documented growth inhibition function (26, 36). Moreover, ectopic GAS1 expression in primary cells (36) and psm does not cause growth arrest. Thus, GAS1 appears to have two independent functions: one involves SHH binding and the other does not. We show here that the tumorigenic mutant form of SHH(H134Y) has reduced affinity for GAS1. It is plausible that this mutation is selected during tumor progression for its reduced affinity for GAS1 rather than for increased affinity for PTC1. Reduced affinity for Gas1 would increase the availability of SHH and lead to excessive SHH signaling, resulting in dysregulated growth. Although GAS1 can suppress tumor cell growth and is deleted in some myeloid tumors (37, 38), it has not been directly implicated in basal cell carcinoma, a condition frequently associated with excessive SHH signaling (25, 39–41). It is not yet known whether Gas1 plays a role in Shh-related tumorigenesis.
Acknowledgments
We thank A. McMahon for reagents, M. Tessier-Lavigne for encouragement, D. Koshland and J. Wilhelm for reading the manuscript, A. Pinder for sequencing, and C. Jewell for graphic assistance. This work is supported by the Arnold and Mabel Beckman Young Investigator Award, the Damon Runyon Cancer Research Scholarship, and National Institutes of Health Grant RO1HD/GM35596 to C.-M.F.
Abbreviations
- SHH
sonic hedgehog
- GAS1 and Gas1
growth arrest-specific gene 1 (protein and gene, repectively)
- psm
presomitic mesoderm
- E
embryonic day
- AP
alkaline phosphatase
- HA
hemagglutinin
- GFP
green fluorescent protein
References
- 1.Ordahl C, editor. Somitogenesis, Parts I and II. San Diego: Academic; 2000. [Google Scholar]
- 2.Fan C M, Tessier-Lavigne M. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 3.Fan C M, Porter J, Chiang C, Chang D T, Beachy P A, Tessier-Lavigne M. Cell. 1995;81:457–465. doi: 10.1016/0092-8674(95)90398-4. [DOI] [PubMed] [Google Scholar]
- 4.McMahon J A, Takada S, Zimmerman L B, Fan C M, Harland R M, McMahon A P. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan C M, Lee C S, Tessier-Lavigne M. Dev Biol. 1997;191:160–165. doi: 10.1006/dbio.1997.8713. [DOI] [PubMed] [Google Scholar]
- 6.Chiang C, Lee Y, Lee E, Young K E, Corden J L, Westphal H, Beachy P A. Nature (London) 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 7.Ikeya M, Takada S. Development (Cambridge, UK) 1998;125:4969–4976. doi: 10.1242/dev.125.24.4969. [DOI] [PubMed] [Google Scholar]
- 8.Pourquie O, Coltey M, Teillet M A, Ordahl C, Le Douarin N M. Proc Natl Acad Sci USA. 1993;90:5242–5246. doi: 10.1073/pnas.90.11.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand-Saberi B, Epensberger C, Wilting J, Balling R, Christ B. Anat Embryol. 1993;188:239–245. doi: 10.1007/BF00188215. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R D, Laufer E, Riddle R D, Tabin C J. Cell. 1994;79:1161–1174. doi: 10.1016/0092-8674(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 11.Kuratani S, Martin J, Wawarsik S, Lilly B, Eichele G, Olson E. Dev Biol. 1994;161:357–369. doi: 10.1006/dbio.1994.1037. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich S, Schubert F R, Lumsden A. Development (Cambridge, UK) 1997;124:3895–3908. doi: 10.1242/dev.124.19.3895. [DOI] [PubMed] [Google Scholar]
- 13.Capdevila J, Tabin C, Johnson R L. Dev Biol. 1998;193:182–194. doi: 10.1006/dbio.1997.8806. [DOI] [PubMed] [Google Scholar]
- 14.Chuang P T, McMahon A P. Nature (London) 1999;463:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 15.Flanagan J G, Cheng H. Methods Enzymol. 2000;327:198–210. doi: 10.1016/s0076-6879(00)27277-7. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 17.Del Sal G, Ruaro M, Philipson L, Schneider C. Cell. 1992;70:595–607. doi: 10.1016/0092-8674(92)90429-g. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson D G. In Situ Hybridization: A Practical Approach. Oxford: Oxford Univ. Press; 1992. [Google Scholar]
- 19.Schneider C, King R, Philipson L. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 20.Stebel M, Vatta P, Ruaro M E, Del Sal G, Parton R G, Schneider C. FEBS Lett. 2000;481:152–158. doi: 10.1016/s0014-5793(00)02004-4. [DOI] [PubMed] [Google Scholar]
- 21.Briscoe J, Ericson J. Semin Cell Dev Biol. 1999;10:353–362. doi: 10.1006/scdb.1999.0295. [DOI] [PubMed] [Google Scholar]
- 22.Stone D M, Hynes M, Armanini M, Swanson T A, Gu Q, Johnson R L, Scott M P, Pennica D, Goddard A, Phillips H, et al. Nature (London) 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 23.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Nature (London) 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 24.Oro A E, Higgins K, Hu Z, Bonifas J M, Epstein E H, Jr, Scott M P. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 25.Lee C S, Fan C M. Mech Dev. 2001;101:293–297. doi: 10.1016/s0925-4773(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 26.Constam D B, Robertson E. J Cell Biol. 1999;144:139–149. doi: 10.1083/jcb.144.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrich L V, Johnson R, Milenkovic L, McMahon J A, Scott M P. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 28.Zeng X, Goetz J, Suber L M, Scott W J, Jr, Schreiner C M, Robbins D J. Nature (London) 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 29.De Robertis E M, Wessely O, Oelgeschlager M, Brizuela B, Pera E, Larrain J, Abreu J, Bachiller D. Int J Dev Biol. 2001;45:189–197. [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C S, May N R, Fan C-M. Dev Biol. 2001;236:17–29. doi: 10.1006/dbio.2001.0280. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, May N R, Fan C-M. Dev Biol. 2001;236:30–45. doi: 10.1006/dbio.2000.0146. [DOI] [PubMed] [Google Scholar]
- 32.Lee C S, Buttitta L, May N R, Kispert A, Fan C M. Development (Cambridge, UK) 2000;127:109–118. doi: 10.1242/dev.127.1.109. [DOI] [PubMed] [Google Scholar]
- 33.Murone M, Rosenthal A, de Sauvage F J. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 34.Alcedo J, Zou Y, Noll M. Mol Cell. 2000;6:457–465. doi: 10.1016/s1097-2765(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 35.Del Sal G, Ruaro E M, Utrera R, Cole C N, Levine A J, Schneider C. Mol Cell Biol. 1995;15:7152–7160. doi: 10.1128/mcb.15.12.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K K, Leung A, Tang M K, Cai D Q, Schneider C, Brancolini C, Chow P H. Dev Biol. 2001;234:188–203. doi: 10.1006/dbio.2001.0249. [DOI] [PubMed] [Google Scholar]
- 37.Evdokiou A, Webb G, Peters G B, Dobrovic A, O'Keefe D S, Forbes I J, Cowled P A. Genomics. 1993;13:731–733. doi: 10.1016/s0888-7543(05)80388-x. [DOI] [PubMed] [Google Scholar]
- 38.Evdokiou A, Cowled P. Int J Cancer. 1998;75:568–577. doi: 10.1002/(sici)1097-0215(19980209)75:4<568::aid-ijc13>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Xie J, Murone M, Luoh S M, Ryan A, Gu Q, Zhang C, Bonifas J M, Lam C W, Hynes M, Goddard A, et al. Nature (London) 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 40.Hahn H, Wicking C, Zaphiropoulous P G, Gailani M R, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden A B, Gillies S, et al. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 41.Johnson R L, Rothman A, Xie J, Goodrich L V, Bare J W, Bonifas J M, Quinn A G, Myers R M, Cox D R, Epstein EH, Jr, et al. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]