Significance
Ion channels are sophisticated proteins that exert control over a plethora of body functions. Specifically, the members of the Kv7 family are prominent components of the nervous systems, responsible for the ion fluxes that regulate the electrical signaling in neurons and cardiac myocytes. Albeit its relevance, there are still several questions, including the Ca2+/calmodulin (CaM)-mediated gating mechanism. We found that Ca2+ binding to CaM triggers a segmental rotation that allosterically transmits the signal from the cytosol up to the transmembrane region. NMR-derived analysis of the dynamics demonstrates that it occurs through a conformational selection mechanism. Energetically, CaM association with the channel tunes the affinities of the CaM lobes (calmodulation) so that the channel can sense the specific changes in [Ca2+] resulting after an action potential.
Keywords: ion channel, calmodulin, calcium regulation, Kv7 potassium channel, M-current
Abstract
The Kv7.2 (KCNQ2) channel is the principal molecular component of the slow voltage-gated, noninactivating K+ M-current, a key controller of neuronal excitability. To investigate the calmodulin (CaM)-mediated Ca2+ gating of the channel, we used NMR spectroscopy to structurally and dynamically describe the association of helices hA and hB of Kv7.2 with CaM, as a function of Ca2+ concentration. The structures of the CaM/Kv7.2-hAB complex at two different calcification states are reported here. In the presence of a basal cytosolic Ca2+ concentration (10–100 nM), only the N-lobe of CaM is Ca2+-loaded and the complex (representative of the open channel) exhibits collective dynamics on the millisecond time scale toward a low-populated excited state (1.5%) that corresponds to the inactive state of the channel. In response to a chemical or electrical signal, intracellular Ca2+ levels rise up to 1–10 μM, triggering Ca2+ association with the C-lobe. The associated conformational rearrangement is the key biological signal that shifts populations to the closed/inactive channel. This reorientation affects the C-lobe of CaM and both helices in Kv7.2, allosterically transducing the information from the Ca2+-binding site to the transmembrane region of the channel.
Voltage-gated K+ channels are responsible for the ion fluxes that regulate the electrical signaling in excitable cells such as neurons, cardiac myocytes, auditory cells, or vascular smooth muscle cells (1, 2). In turn, such channels are often controlled by the temporal and spatial modulation of the intracellular (i) Ca2+ concentration, which can vary from 100 nM in resting cells up to 1–10 μM (local [Ca2+]i) (3). Calcium regulation of a K+ channel may occur through direct Ca2+ association with a cytosolic domain of the channel (3, 4), or, more frequently, it can be mediated by calmodulin (CaM), the primary Ca2+ sensor in eukaryotic cells (5, 6).
CaM regulation of ion channels (calmodulation) is a process well understood thermodynamically, which often involves tuning Ca2+ affinities from the CaM lobes upon complexation (7). From the structural point of view, an allosteric mechanism is required to propagate Ca2+ signaling from the cytosol up to the transmembrane region (8). The rich complexity of functional states accessible for ionic channels suggests that measurable conformational transitions are likely responsible for calmodulation signal propagation. However, despite the large repository of high-resolution structures of CaM bound to ion channel fragments (6), the mechanism for this allosteric event has not yet been reported for any channel.
Here, we have investigated the Ca2+-dependent conformational landscape of the voltage-gated and Ca2+-regulated K+ channel Kv7.2. In combination with Kv7.3, it conforms the molecular components of the M-current (9), which is opened at subthreshold membrane potentials, providing a brake for membrane excitation (10, 11). To close the channel and regulate the M-current, Kv7.2 is sensitive to the Ca2+ release after bradykinin activation in sympathetic neurons (12, 13). Kv7.2 is sensitive to Ca2+ only after it binds to CaM at the specific binding motif located in helix A (IQ motif) (Fig. S1A), which can occur in the presence or absence of the metal ion (14, 15). Thus, Kv7.2 regulation by Ca2+ is a representative example of ion channel modulation by CaM.
The Kv7 family has been investigated using X-ray crystallography: The structure of the chimeric Kv7.2/7.3 cytosolic region (hA and hB in Fig. S1A) complexed with CaM (16) and the structures of equivalent moieties for the homologous channels Kv7.1 (17) and CaM complexed with helix hB of Kv7.4 (18) are available. Despite the obvious interest of such structural instances, a thorough study of the conformational landscape of Ca2+ gating is missing. Using NMR spectroscopy, we have characterized the structure and dynamics of hAB of Kv7.2 in complex with CaM (263 residues in total) at different Ca2+ saturation states. In the absence of added Ca2+, the protein complex structure is loaded with two Ca2+ ions in the N-lobe of CaM. In this intermediate Ca2+ state, the complex undergoes extensive dynamics on the millisecond time scale, affecting backbone and side chains. Intriguingly, Ca2+ saturation and/or depletion results in the disappearance of the dynamics and is accompanied by a structural rearrangement, particularly noticeable in the Ca2+-loaded state. Collective analysis of the structural instances, the modulated Ca2+ affinities of CaM upon binding Kv7.2-hAB, and the protein dynamics provides an unprecedented view of the energy landscape of calmodulation and a structural model for the allosteric rearrangement associated with the Ca2+ gating of a Kv7 channel.
Results
Finding Soluble and Functional Constructs of Kv7.2-hAB.
The expression of the cytosolic moiety of Kv7.2 covering helices hA and hB (Fig. S1B) results in an insoluble and aggregated protein. We identified a hydrophobic region prone to be unstructured and to suffer from proteolysis and generated a construct of the Kv7.2 C terminus that encloses helices hA and hB (residues Q310–R532) with the linker deletion ΔR374–K493 (Kv7.2-hAB[Δ6L]; Fig. S1B). Bacterial coexpression of Kv7.2-hAB[Δ6L] with CaM results in a monodisperse protein complex. Experimental evidence supports the biological relevance of this construct: The interlink region between helices hA and hB is not required for channel function or assembly (19), and electrophysiological recordings of cells expressing Kv7.2-hAB[WT] and Kv7.2-hAB[Δ6L] demonstrate that the average current density and Boltzmann parameters are comparable in both channels (Fig. S1 C and D). Altogether, it can be concluded that the Kv7.2-hAB[Δ6L] construct (henceforth Kv7.2-hAB) represents a bona fide and meaningful model that retains all of the functional properties of the channel fragment.
Solution Structure of the CaM/Kv7.2-hAB Complex in the Absence of Added Ca2+.
The solution structure of the CaM/Kv7.2-hAB complex in the absence of added Ca2+ was determined by NMR spectroscopy. First, chemical shift assignments were achieved using orthogonal isotopic labeling (Fig. S2) to alleviate signal overlap, and more than 95% of the 1H, 15N, and 13C (backbone) resonances were assigned [BioMagResBank (BMRB) accession no. 34097; Fig. S2].
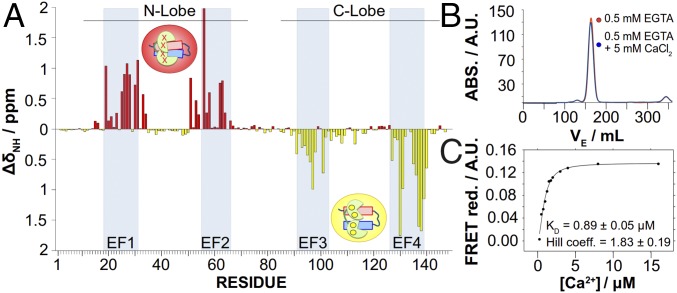
The Ca2+ association state with the CaM/Kv7.2-hAB complex was investigated using the 1H, 15N chemical shift perturbations (CSPs) as reporters of the ion effect. Upon Ca2+ saturation, the vast majority of the CaM-perturbed residues belong to the EF hands located in the C-lobe of CaM, with no significant CSPs in the N-lobe (Fig. 1A, yellow bars). Ca2+ depletion of the complex with EGTA produces exactly the mirror effect (Fig. 1A, red bars). Thus, in the absence of added Ca2+, Kv7.2-hAB associates with CaM at an intermediate Ca2+ state in which only its N-lobe is loaded. This is consistent with the crystallographic structure of the CaM/Kv7.1-hAB complex (17), where CaM is also Ca2+-loaded just in the N-lobe. We will refer to this partially Ca2+-loaded complex as “intermediate” intCaM/Kv7.2-hAB.
Fig. 1.
CSPs induced by Ca2+ over CaM complexed with helices A and B. (A) CSP analysis of residues forming CaM. The apoCaM/Kv7.2-hAB complex shows large CSPs in the EF1 and EF2 hands, whereas holoCaM/Kv7.2-hAB shows large CSPs in the EF3 and EF4 hands, demonstrating that intCaM/Kv7.2-hAB is Ca2+-loaded in the N-lobe, whereas the C-lobe is Ca2+-free. ΔδNH, absolute amide chemical shift differences. (B) Size exclusion chromatography profiles after injecting samples with and without Ca2+ in a Superdex 26/60 column. Elution peaks for the apo- and holoCaM/Kv7.2-hAB complexes (30.27 kDa) are almost identical (162.66 mL and 163.896 mL, respectively), corresponding to a theoretical molecular mass of 31.13 kDa and 30.3 kDa, respectively. ABS., absorbance; VE, eluted volume. (C) Ca2+ titration curve for the CaM/Kv7.2-hAB complex. Ca2+ affinity is measured by FRET ratio reduction between the mTFP1 (donor) and mVenus (acceptor) fluorophores located in the N and C termini of the Kv7.2-hAB construct complexed with CaM. The EC50 value (equivalent to an apparent Kd for the C-lobe) is 0.89 ± 0.05 μM, and the Hill coefficient is h = 1.83 ± 0.19 as obtained from the fitting to a Hill equation: where EC50 corresponds to the concentration of Ca2+ at which FRET change is half-maximal, h is the Hill coefficient, and is the FRET change found at a large excess of free calcium.
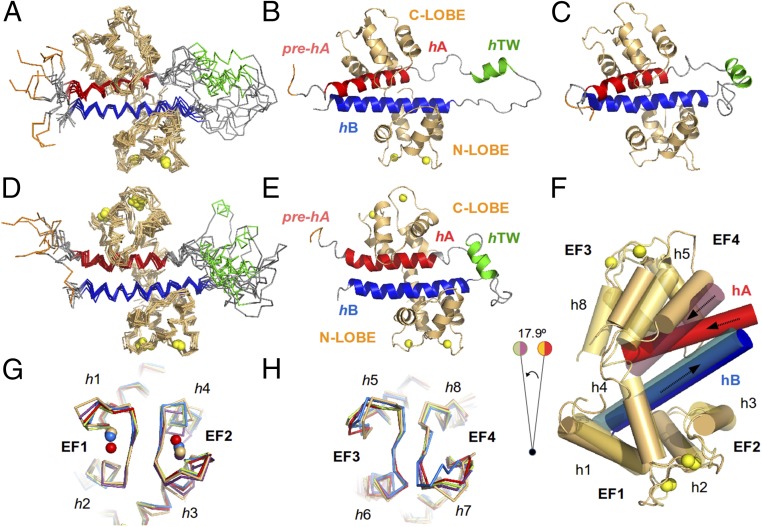
The 3D structure of the intCaM/Kv7.2-hAB complex in solution [Protein Data Bank (PDB) ID code 6FEG; Fig. 2 A and B and Fig. S4] was calculated employing 5,981 NOEs, 306 dihedral angles, and 134 residual dipolar couplings (RDC) restraints (Table S1). The protein complex shows helices hA and hB wrapped by CaM (Fig. 2 A and B). A third helix composed of residues H357–R365 (hTW in Fig. 2B) is located in the flexible linker between helices hA and hB, and it does not make significant contacts with the rest of the protein complex. The pre-hA region (16, 17) is partially unstructured in intCaM/Kv7.2-hAB (Fig. 2B), but this is likely due to the channel truncation. The complex is structurally similar to the available PDB entries of CaM complexed to other Kv7 subunits (Fig. S3), showing rmsd values of 1.07 Å with the CaM/Kv7.1-hAB complex (17) and 1.4 Å with the CaM/Kv7.2-hA/Kv7.3-hB chimera (16), while it shows fewer similarities with the Ca2+-loaded CaM/Kv7.4-hAB complex (18) (rmsd = 8.1 Å, mostly due to a segmental rearrangement).
Fig. 2.
Structure of the CaM/Kv7.2-hAB complex at different calcification states. An overlay of the 10 lowest energy conformers representing the 3D solution structure of the intCaM/Kv7.2-hAB (A) or holoCaM/Kv7.2-hAB (D) complex is shown. A ribbon representation of the intCaM/Kv7.2-hAB (B) or holoCaM/Kv7.2-hAB (E) complex shows the structural elements: CaM protein (orange); the prehelix A(pre-hA), which connects segment 6 of the pore with the rest of the intracellular C terminus of the Kv7.2 channel (purple); helix hA (red); helix hTW (green); and helix hB (blue). Ca2+ ions are represented as spheres. The same color code is used for the ribbon representations of the apoCaM/Kv7.2-hAB complex. (C) Ribbon representation of the structural model of the apoCaM/Kv7.2-hAB complex. (F) Alignment of intCaM/Kv7.2-hAB and holoCaM/Kv7.2-hAB by the center of mass of the N-lobe of CaM displays the rotation (17.9°) of a segment of the complex upon Ca2+ binding to the C-lobe of CaM. The involved structural elements are h5 and h8 in CaM and hA in Kv7.2-hAB. (G) Alignment of the EF-hand motifs of the N-lobe of intCaM/Kv7.2-hAB (orange) with the equivalent motif in other reported (Ca2+-loaded) CaM structures: 4RJD (purple), 4UMO (blue), 4GOW (red), and 5J03 (green), with rmsd values of 1.12 Å, 1.22 Å, 1.26 Å, and 1.39 Å, respectively. (H) Alignment of the EF-hand motifs of the C-lobe of intCaM/Kv7.2-hAB (orange) with the equivalent motif in other reported (apo) CaM structures: 4V0C (blue), 4E50 (red), 4JQ0 (purple), and 5J03 (green), with rmsd values of 0.93 Å, 0.94 Å, 1.18 Å, and 1.25 Å, respectively.
The Structural Basis of CaM/Kv7.2-hAB Ca2+ Gating.
Ca2+ saturation of the CaM/Kv7.2-hAB complex results in CSPs in the Kv7.2-hAB moiety as well. To investigate the putative conformational changes as a function of Ca2+, we have investigated the structural properties of CaM/Kv7.2-hAB at two other concentrations of this ion. The “apo” form (devoid of Ca2+, apoCaM/Kv7.2-hAB) and the fully loaded Ca2+ complex (holoCaM/Kv7.2-hAB) were purified and found to be stable over time. The hydrodynamic radius and the stoichiometry of the complexes do not vary with [Ca2+], as shown by the elution profile from size exclusion chromatography (Fig. 1B), indicating that the ion-dependent structural variations are subtle, in agreement with the moderate CSPs of CaM/Kv7.2-hAB with [Ca2+] (Fig. S2). A structural model from apoCaM/Kv7.2-hAB was obtained based on these chemical shift changes combined with 1J(N-H) residual dipolar couplings in an aligned medium. The resulting apoCaM/Kv7.2-hAB model (Fig. 2C) is similar to the partially Ca2+-loaded intCaM/Kv7.2-hAB complex, showing variations in the flexible regions (including the EF-hand loops as discussed below) and, to a lesser extent, in the hAB interhelical orientation.
The solution structure of the holoCaM/Kv7.2-hAB complex was also determined by NMR spectroscopy (Fig. 2 D and E and Fig. S4). The assigned chemical shifts (94% of the 1H, 15N, and 13C; BMRB accession no. 34226), a set of NOEs (n = 5,287), and RDCs (n = 131) were used to resolve the 3D fold of the protein complex (PDB ID code 6FEH). Even though the main structural features are maintained, the helices hA and hB have changed their relative disposition and holoCaM/Kv7.2-hAB is conformationally different from the existing structures, with rmsd values of 3.3 Å, 3.7 Å, and 9.3 Å for the CaM/Kv7.1-hAB complex (17), CaM/Kv7.2-hA/Kv7.3-hB chimera (16) and CaM/Kv7.4-hAB complex (18), respectively (Fig. S3). More importantly, holoCaM/Kv7.2-hAB also significantly differs from intCaM/Kv7.2-hAB (rmsd = 3.1 Å), but the two structures can be reconciled by a segmental rearrangement (Fig. 2F): Ca2+ association with CaM produces a 17.9° rotation of the EF hands, which also pulls helices h8 and h5 of CaM and helix hA of Kv7.2. Remarkably, as a result of the segmental solid-rigid rotation, the hAB helices have changed the interhelical angle and their relative orientation with respect to CaM. Altogether, such torsion is the necessary (and sufficient) rearrangement to explain the allosteric transmission from the Ca2+-binding site of the CaM’s C-lobe up to the helix hA (and to the pre-hA) of Kv7.2.
All of the experiments were run at 120 mM KCl, and under these close-to-physiological conditions, the N-lobe of CaM in the complex binds Ca2+ with extremely high affinity. Actually, apoCaM/Kv7.2-hAB can only exist in the presence of a large excess of a strong Ca2+ chelating agent (i.e., EGTA), and the Ca2+ affinity for the N-lobe is estimated to be subnanomolar (Kd < 10−3 μM). This is in contrast to free CaM, where the C-lobe shows the highest affinity for the ion (Kd[C-lobe] = 3.4 μM, Kd[N-lobe] = 14 μM) (20). On the other hand, the Ca2+ affinity for the C-lobe in intCaM/Kv7.2-hAB has been determined by fluorescence resonance energy transfer (FRET): When two independent fluorophores are attached to Kv7.2-hAB, the above-mentioned Ca2+-gated conformational rearrangement produces a change in the FRET intensity (Fig. 1C). The FRET intensity as a function of the free Ca2+ concentration (as determined by fura-2) (21) provides an apparent affinity constant of 0.89 ± 0.05 μM, consistent with other determinations in similar constructs (22). The Hill coefficient of 1.83 suggests that the event is monitoring the saturation of CaM’s C-lobe with two ion units.
Both CaM lobes in CaM/Kv7.2-hAB show altered affinity with respect to free CaM (calmodulation), and we focused on the conformation adopted by the EF-hand loops, since they are responsible for Ca2+ association. Compared with the available crystallographic (Fig. 2G) and solution (Fig. S5) structures of CaM, the N-terminal EF-hand loops of apoCaM/Kv7.2-hAB agree better with Ca2+-bound conformations. Moreover, the interhelical angle analysis (6) (Table S2) also suggests that the lobe is already in a “holo” (Ca2+-bound) conformation, providing a structural explanation for the increase in affinity. However, this mechanistic explanation is constrained by the validity of the structural model of apoCaM/Kv7.2-hAB, based on a limited number of experimental restraints (chemical shifts and RDCs). On the other hand, the EF hands of the C-lobe of intCaM/Kv7.2-hAB have interhelical angles characteristic of a partially open conformation (Table S2), and the conformation is similar to apoCaM structures (Fig. 2H and Fig. S5). These results are consistent with the moderate affinity increase and the micromolar Kd range observed for the C-lobe in the complex.
The Conformational Changes Are also Observed upon Tetramerization.
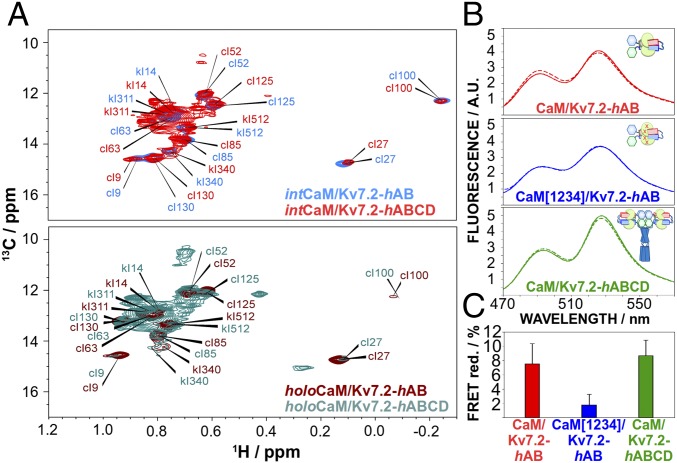
To validate whether the observed changes are also present in a tetrameric architecture, we looked for Ca2+-dependent conformational changes in a channel moiety that includes the regions responsible for tetramerization (helices hC and hD of the cytosolic C-lobe of Kv7.2; Fig. S1A). The 1H,15N–transverse relaxation optimized spectroscopY (TROSY)–heteronuclear single quantum spectroscopy (HSQC) of the CaM/Kv7.2-hABCD (Fig. S2) shows only a few peaks corresponding to residues from flexible regions, in good agreement with the spectrum of a tetrameric structure of more than 120 kDa. Instead, the methyl-TROSY spectrum (23) of the 13C–δ-Ile methyl groups of intCaM/Kv7.2-hABCD displays the vast majority of residues. Despite the signal overlap, the superposition of intCaM/Kv7.2-hABCD and intCaM/Kv7.2-hAB spectra (Fig. 3A) allows the assignment of the hAB δ-Ile residues in intCaM/Kv7.2-hABCD. Noticeably, the spectrum for holoCaM/Kv7.2-hABCD shows the same Ca2+-induced CSP as holoCaM/Kv7.2-hAB (Fig. 3A), indicating that the ion-dependent conformational changes are maintained in the tetrameric complex.
Fig. 3.
Conformational changes are also observed in tetrameric channel constructs. (A) 13C–δ-Ile region of the methyl-TROSY spectrum for intCaM/Kv7.2-hAB (blue, Top), intCaM/Kv7.2-hABCD (red, Top), holoCaM/Kv7.2-hAB (brown, Bottom), and holoCaM/Kv7.2-hABCD (green, Bottom). The monomeric complex (CaM/Kv7.2-hAB) and the tetrameric assembly (CaM/Kv7.2-hABCD) experience the same conformational changes triggered by Ca2+. The assignment of the methyl peaks in intCaM/Kv7.2-hAB was achieved by a combination of multiple 3D experiments, while the same peaks in intCaM/Kv7.2-hABCD are estimated by spectra overlap. (B) mTFP1 (Donor) and mVenus (Acceptor) are localized in the N and C termini of helices AB, respectively. CaM/Kv7.2-hAB (Top, red), CaM[1234]/Kv7.2-hAB (Middle, blue), and CaM/Kv7.2-hABCD (Bottom, green) emission spectra in the absence (full line) and presence (dashed line) of Ca2+-saturating concentrations are shown. (C) FRET reduction ratios relative to the maximum excitation (∼492 nm) and emission (∼526 nm) peaks. Ca2+ addition induces noticeable conformational changes (similar in magnitude) in the monomeric version (CaM/Kv7.2-hAB) or the tetrameric version (CaM/Kv7.2-hABCD), but not for the CaM[1234]/Kv7.2-hABCD tetramer.
FRET experiments provide further evidence that the Ca2+-dependent conformational changes are also present in the tetrameric complex. Fig. 3 B and C shows how FRET analysis is sensitive to the conformational change occurring in the transition between apoCaM/Kv7.2-hAB (solid red line) and holoCaM/Kv7.2-hAB (dashed red line). The magnitude of such change is maintained in the intCaM/Kv7.2-hABCD complex (Fig. 3C and green lines in Fig. 3B), while no change in the FRET signal can be detected when using a CaM mutant unable to bind Ca2+ (CaM[1234]; blue lines in Fig. 3 B and C).
intCaM/Kv7.2-hAB Shows Excursions to an Excited State, Compatible with holoCaM/Kv7.2-hAB.
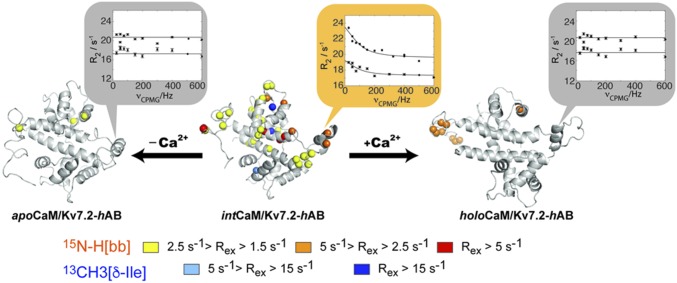
Analysis of the conformational ensemble obtained after the experimental restraint refinement indicates that the intCaM/Kv7.2-hAB protein complex may undergo intermolecular flexibility. NMR is especially suitable for the characterization of protein motions at multiple time scales, and for motions in the microsecond-to-millisecond time scale, the relaxation-dispersion (RD) experiment allows a full description of the exchange process, providing an estimation for the exchange rate (kex) and the population (pB)/chemical shift (Δδ) of the excited state (24). Backbone amide RD experiments collected at two magnetic fields identified pervasive dynamics in CaM’s C-lobe region of intCaM/Kv7.2-hAB, with measurable line broadening ranging between 1.5 and 20 Hz (Fig. 4). The individual fitting of the RD profiles shows a high degree of consistency in the obtained parameters (homogeneous kex and pB values), so a collective fitting was attempted. Dynamic residues could be initially clustered into three different groups (groups I–III; Fig. S6) with robust statistics (Table S3). Group I entails five residues included in the construct but not corresponding to coding regions of the channel, so it is not considered further. Group II encloses 15 residues belonging to helices hA, hB, and hTW of Kv7.2. Most of the residues in group II face the C-lobe of CaM, whose dynamic residues are clustered in group III. Remarkably, group II and III residues can be interpreted using equivalent fitting parameters (Table S3), suggesting that the intCaM/Kv7.2-hAB may undergo intermolecular concerted dynamics. Groups II and III are well described by a single motional event of 1.1 ± 0.2 ms, with an excited state of 1.5 ± 0.6% (Table S3).
Fig. 4.
Millisecond dynamics of intCaM/Kv7.2-hAB populate the holoCaM/Kv7.2-hAB state to a low extent. In the structure, the spheres pinpoint the 15N-H backbone (bb) amide groups and 13C–δ-Ile methyl groups that are experiencing microsecond-to-millisecond dynamics for the three conformations under consideration. The magnitude of the exchange line broadening is color-coded as indicated. The vignettes show a representative example of the RD profiles (Val366). The solid lines correspond to the fitting to the Carver–Richards equation, simultaneously, of the two static magnetic field datasets (600 and 800 MHz).
The region of the complex that experiences conformational dynamics (Fig. 4) agrees well with the segmental motion upon Ca2+ association with the C-lobe of CaM/Kv7.2-hAB (Fig. 2F). Moreover, the RD-fitted Δδ values show an excellent correlation with the CSP upon Ca2+ addition to intCaM/Kv7.2-hAB (Fig. S6D). Equivalent experiments were repeated for the apo- and holoCaM/Kv7.2-hAB states. In both cases, the dynamic residues conforming the conformational exchange (dynamic residues from groups II and III) are abrogated. Thus, we conclude that intCaM/Kv7.2-hAB undergoes excursions toward an excited state compatible with holoCaM/Kv7.2-hAB. This conformational exchange is not originated from the in and out transfer of residual Ca2+ ions to the CaM-binding site since equivalent line broadening is obtained when the RD experiments are repeated in the presence of an excess of EGTA (Fig. S6G). Instead, the collective motion seems to be originated by an incomplete electrostatic cancellation from the acidic CaM and the basic Kv7.2-hAB subunits. In this hypothesis, line broadening is drastically attenuated when the ionic strength is increased (up to 500 mM KCl; Fig. S6F). This electrostatic complementarity has also been previously observed for CaM binding to its binding domain in the SK channel (25), among other systems (26).
We also investigated the dynamic properties of the δ-Ile methyl groups, which are widespread throughout the protein complex (Fig. 3A). There are three methyl groups (out of 12) showing dynamics in the intCaM/Kv7.2-hAB complex, but not in the apo- or holoCaM/Kv7.2-hAB complex (Fig. 4), indicating that side chains are equally sensing the collective dynamics of the complex. Thus, our results demonstrate that changes in Ca2+ concentration result in modulation of the dynamic behavior of the protein complex, with two resting states (apoCaM/Kv7.2-hAB and holoCaM/Kv7.2-hAB) and a highly dynamic and interconverting state (intCaM/Kv7.2-hAB).
Discussion
Ca2+ gating of K+ channels mediated by CaM is responsible for the tight control of a plethora of subcellular functions (27). However, the Ca2+ regulation mechanism for such voltage-dependent channels has remained, to the best of our knowledge, ill-defined since the first CaM-binding epitopes were identified. So far, the most accurate structural description of a Ca2+-gated regulation mechanism comes from the SK2 channels (25, 28, 29), where it has been demonstrated that Ca2+ binding to CaM produces a large conformational change that includes bridging different channel subunits. In Kv7 channels, it has been long hypothesized that Ca2+ regulation is also driven by conformational changes derived from the interaction between Ca2+ and CaM, and Minor and coworkers (18) suggested that CaM’s conformational change could be the mechanical switch required to open/close the pore depending on the Ca2+ levels. In this paper, we provide extensive experimental evidence that such conformational change does indeed occur, providing an estimation of the energy landscape and a structural model for the allosteric rearrangement associated with the Ca2+ gating of the Kv7.2 channel.
We report the conformational instances of the CaM/Kv7.2 complex with atomic resolution at the two calcification states plus a structural model of a third state, covering all of the intracellular physiological concentrations, from 10–100 nM at the resting state up to 1–10 μM after chemical or electrical signaling: apoCaM/Kv7.2-hAB (Ca2+ unloaded), intCaM/Kv7.2-hAB (loaded in the N-lobe of CaM), and holoCaM/Kv7.2-hAB (full Ca2+-loaded). In the three cases, the structures show a very intimate complex in a 1:1 stoichiometry where CaM wraps the Kv7.2 helices that are forming an antiparallel coiled-coil: Helix hA is bound to the apo–C-lobe of CaM, whereas helix hB is making contact with the Ca2+-loaded N-lobe. This result is largely consistent with the crystallographic structures of the CaM/Kv7.1-hAB and CaM/Kv7.3-hA/Kv7.2-hB complexes reported by Hirsch and coworkers (16, 17).
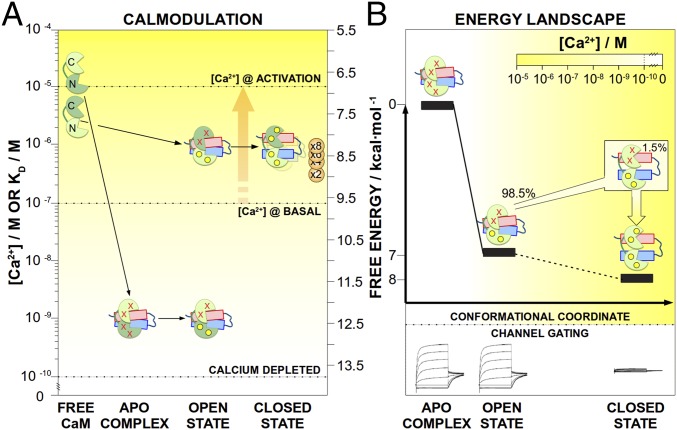
The reported conformational instances conceal the dynamic transitions required to define the conformational/energy landscape associated with Ca2+ gating of the channel (energy landscape of Fig. 5 and Movie S1). The apoCaM/Kv7.2-hAB complex exists only in the presence of strong chelating agents. Moreover, apoCaM/Kv7.2-hAB results in an open complex according to in cellulo M-current recordings with a CaM variant unable to chelate the ion (CaM1234; Fig. 5). Since the basal Ca2+ concentration of the neuronal cytosol is around 100 nM, we conclude that this conformation should be only theoretical and not found intracellularly. On the other hand, intCaM/Kv7.2-hAB is structurally rather similar and, energetically at least, 7 kcal⋅mol−1 more stable than apoCaM/Kv7.2-hAB (as estimated from the change in Kd values), also maintaining the channel in the open state. The existence of different conformations that result in an open channel underscores that the conformational plasticity in the N-lobe may be required for putative interactions with other secondary messengers (i.e., phosphatidylinositol biphosphate) (30) to further modulate the gating of the channel.
Fig. 5.
Energy landscape and calmodulation of Kv7.2. The energy landscape and calmodulation connect the two different states of the channel: the open state (apoCaM/Kv7.2-hAB and intCaM/Kv7.2-hAB complexes) and the closed state (holoCaM/Kv7.2-hAB complex). In both panels, [Ca2+] is visualized by the yellow color intensity, as indicated in the inset scale. (A) Cartoons representing the different molecules are located in the ordinate axis according to the affinities of the dark green lobes. For instance, the N-lobes of CaM change their Kd for Ca2+ from 14 μM (free) to a Kd of less than 10−3 μM after complexation with Kv7.2-hAB (calmodulation). The horizontal dotted lines indicate the two different intracellular concentrations (the basal concentration and the maximum concentration induced by bradykinin), as well as a level that can be considered ion-free (Ca2+ depleted). The bradykinin-mediated Ca2+ release (orange arrow) triggers the transition from the open state toward the closed state of the channel. The apparent affinity of the channel for Ca2+ depends on the number of ions required to trigger the conformational change (from one to eight), as indicated by the orange circles. (B) Ca2+ affinities are used to estimate the relative energy levels of the different conformations, underlining the progressive stabilization of the complex with the Ca2+-binding coordinate. In the intCaM/Kv7.2-hAB complex, the main population is in the open state, while a small population (1.5%) is already in the closed state.
From the functional point of view, the key step is the transition from intCaM/Kv7.2-hAB to holoCaM/Kv7.2-hAB: Ion binding to the C-lobe concertedly displaces a segment of CaM (helices h5 and h8), which is allosterically transmitted to the channel moiety by modifying the orientation of helices hA and hB (Fig. 2F). We hypothesize that this conformational change is mechanically transmitted to the intramembranous region of the channel through a change in orientation of the pre-hA element, gating the closing of the channel and, ultimately, resulting in M-current depletion. Such change is energetically favored at about 1 kcal⋅mol−1, which ensures the reversibility of the system upon disappearance of the signaling Ca2+ concentration.
The above-mentioned states are not isolated conformations only connected by the (increasing) Ca2+ concentration; rather, they are also intrinsically coupled through thermal motions. The RD experiments show pervasive dynamics in CaM’s C-lobe of intCaM/Kv7.2-hAB, which can be adequately described as a collective motion of the complex. In the most simple and widespread interpretation of the RD experiment, the two-state model chemical shift analysis demonstrates that intCaM/Kv7.2-hAB is sensing a low-populated state (1.5% at 303 K), which corresponds to the holoCaM/Kv7.2-hAB state. Such dynamics are likely originated by an insufficient electrostatic complementarity between the (acidic) CaM and the (basic) Kv7.2-hAB in intCaM/Kv7.2-hAB, which becomes fully satisfied (and the millisecond dynamics largely quenched) when an excess of Ca2+ locks the complex in the holoCaM/Kv7.2-hAB state. From the functional point of view, the preexisting equilibrium of populations is a consequence of the (low) energy barrier between the states, and Ca2+ association with intCaM/Kv7.2-hAB implies a conformational selection mechanism instead of a more canonically induced fit. Conformational selection is widely used by nature, and has been reported in many enzymes (31) and binding proteins (32), including CaM (33), and it is most efficient to produce an allosteric response after a stimulus (34).
The results presented here are also able to explain the role of CaM and the calmodulation mechanism associated with the Ca2+ gating of the Kv7.2 channel (Fig. 5). At any ion concentration, the Kv7.2-hAB complex subunit cannot be purified in the absence of CaM due to aggregation followed by precipitation. Moreover, CaM’s affinity for the channel is so high that the complex cannot be dissociated by any of the in vitro-tested conditions. Likely, this is also the case in vivo, due to the high intracellular concentration of CaM (10−6–10−5 M) (5). Together, these results suggest that CaM association with Kv7.2 may become consubstantial to the channel’s integrity once both molecules interact. This situation is similar to the homologous channel Kv7.1, where a constitutive binding role was hypothesized (17), and to other proteins like phosphorylate kinase (5). In any case, CaM association with Kv7.2 results in a significant alteration of the Ca2+ affinities of the two lobes, sensitizing the CaM-effector complex to the specific [Ca2+] in the different neuronal states. At the structural level, such changes in affinity are consistent with the conformational changes observed in the EF hands of CaM/Kv7.2-hAB, compared with free CaM (Fig. 2 F and G and Table S2). The N-lobe of free CaM has a low affinity for calcium (20), which is drastically increased upon association with Kv7.2 (Kd < 10−3 μM in apoCaM/Kv7.2-hAB; Fig. 5). Considering that the basal Ca2+ concentration in the neuron is around 100 nM, the N-lobe should always be Ca2+-loaded. On the other hand, the C-lobe of CaM also alters its affinity for Ca2+ upon binding to Kv7.2 (Fig. 5), lowering its apparent Kd from 3.4 μM (for free CaM and at this ionic strength) (20) down to 0.9 μM in the intCaM/Kv7.2-hAB complex. This affinity tuning is required for the complex to rest inactive until an agonist-induced increase in the intracellular Ca2+ concentration occurs (up to 1–10 μM; Fig. 5). The higher [Ca2+] shifts the equilibrium toward the holoCaM/Kv7.2-hAB conformation, with the subsequent conformational change that is allosterically transmitted up to the pre-hA element and likely to the transmembrane region, ultimately resulting in the closing of the channel.
Finally, it is worth considering the implications that channel tetramerization may have on the mechanisms introduced here. The methyl-TROSY spectra of the CaM/Kv7.2-hABCD complex demonstrate that the same conformational states are maintained after channel tetramerization of the C terminus (Fig. 3A), while FRET experiments prove that Ca2+ triggers a conformational change in the tetrameric protein. According to our model, four CaM units will bind the cytosolic moiety of a full channel, generating a double number of putative loci for Ca2+ association (in the C-lobe). An unsolved question is how many Ca2+ units are needed to trigger the allosteric motion that will close the channel. Depending on the required number of ion units, the effective sensitivity of the channel for the ion concentration varies within the range of 0.9 μM (eight Ca ions per channel unit) up to 200 nM (two Ca ion per channel unit), as shown in Fig. 5.
In summary, in this contribution, we have presented a plethora of experimental data to support an unprecedented model for the Ca2+ gating of the potassium channel Kv7.2, which includes a complete description of the conformational landscape, the calmodulation mechanism, and a hypothesis on how Ca2+ can regulate the M-current in neurons. It is our belief that these results are of broader applicability to other Ca2+-gated K+ channels as well.
Methods
Expression and Purification of the CaM/Kv7.2-hAB Complex.
The Kv7.2-hAB segment (residues 316–532) with the deletion ΔR374-K493 [Δ6L] was purified as previously described (20).
NMR Spectroscopy and Structure Calculation.
NMR data were collected at 303 K on an 800-MHz Bruker Avance III spectrometer equipped with a cryoprobe and on a 600-MHz Bruker Avance III US2 spectrometer. Resonance assignments were obtained by the combined use of 15N-HSQC, 13C-HSQC, HNCA, HNcoCA, HNcaCB, HNcocaCB, HNCO, HNcaCO, HNHA, HNcaHA, HccoNH, and HNCccoNH experiments. NOE connectivities were obtained using 15N–NOESY-HSQC and 13C–NOESY-HSQC (120-ms mixing time, 16-ns rotation correlation time). Residual dipolar coupling was extracted by using TROSY 15N-HSQC and semi-TROSY 15N-HSQC experiments in isotropic and anisotropic (C12E5/1-hexanol) conditions. Protein complex structure was determined using ARIA2 (35). Transversal RD experiments were always acquired at two fields (600 MHz and 800 MHz) using a relaxation compensated-pulse Carr-Purcell-Meiboom-Gill sequence (CPMG) sequence (80 μs of total CPMG time) and variable effective fields: 25, 50(×2), 75, 100, 125, 150, 200, 250, 325, 400(×2), 500, 600, 700, 800, 900, and 1,000 Hz.
Electrophysiological Recordings.
For the WT constructs, all experiments were carried out using HEK293T cells (HEK 293T/17, CRL-11268; American Type Culture Collection). The data were acquired at a sampling rate of 1 kHz and filtered at 100 Hz, and were analyzed using pCLAMP software (version 8.2; Axon Instruments) and plotted using SigmaPlot.
FRET Experiments.
All FRET experiments were carried using an AMINCON Bowman series 2 luminescence fluorimeter. The fluorescence emission spectra of the proteins at 500 nM were collected over 470–570 nM after excitation at 458 nm (4-nm bandwidth). The FRET index was calculated as the ratio between the emission at 485–490 nm for the blue donor and the emission at 520–525 nm for the yellow acceptor. The total protein in each condition was assessed by direct excitation of the yellow protein at 520 nm, collecting the emission at 520–570 nm.
Supplementary Material
Acknowledgments
Support was provided from the Department of Industry, Tourism and Trade of the Government of the Autonomous Community of the Basque Country (Grant Elkartek BG2015) and from the Ministry of Science and Technology Ministerio de Economía, Industria y Competitividad (Grants CTQ2015-68756-R, BFU2015-66910-R and CSD2008-00005).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 34097 and 34226) and in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6FEG and 6FEH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800235115/-/DCSupplemental.
References
- 1.Jespersen T, Grunnet M, Olesen S-P. The KCNQ1 potassium channel: From gene to physiological function. Physiology (Bethesda) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 2.Sanguinetti MC, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 3.Berkefeld H, et al. BKCa-Cav channel complexes mediate rapid and localized Ca2+-activated K+ signaling. Science. 2006;314:615–620. doi: 10.1126/science.1132915. [DOI] [PubMed] [Google Scholar]
- 4.Miranda P, Giraldez T, Holmgren M. Interactions of divalent cations with calcium binding sites of BK channels reveal independent motions within the gating ring. Proc Natl Acad Sci USA. 2016;113:14055–14060. doi: 10.1073/pnas.1611415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin D, Means A-R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 6.Villarroel A, et al. The ever changing moods of calmodulin: How structural plasticity entails transductional adaptability. J Mol Biol. 2014;426:2717–2735. doi: 10.1016/j.jmb.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Tadross M-R, Dick I-E, Yue D-T. Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell. 2008;133:1228–1240. doi: 10.1016/j.cell.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haitin Y, Attali B. The C-terminus of Kv7 channels: A multifunctional module. J Physiol. 2008;586:1803–1810. doi: 10.1113/jphysiol.2007.149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jentsch T-J. Neuronal KCNQ potassium channels: Physiology and role in disease. Nat Rev Neurosci. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez C-C, Zaika O, Tolstykh G-P, Shapiro M-S. Regulation of neural KCNQ channels: Signalling pathways, structural motifs and functional implications. J Physiol. 2008;586:1811–1821. doi: 10.1113/jphysiol.2007.148304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soldovieri M-V, Miceli F, Taglialatela M. Driving with no brakes: Molecular pathophysiology of Kv7 potassium channels. Physiology (Bethesda) 2011;26:365–376. doi: 10.1152/physiol.00009.2011. [DOI] [PubMed] [Google Scholar]
- 12.Cruzblanca H, Koh D-S, Hille B. Bradykinin inhibits M current via phospholipase C and Ca2+ release from IP3-sensitive Ca2+ stores in rat sympathetic neurons. Proc Natl Acad Sci USA. 1998;95:7151–7156. doi: 10.1073/pnas.95.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamper N, Shapiro M-S. Calmodulin mediates Ca2+-dependent modulation of M-type K+ channels. J Gen Physiol. 2003;122:17–31. doi: 10.1085/jgp.200208783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bal M, Zaika O, Martin P, Shapiro M-S. Calmodulin binding to M-type K+ channels assayed by TIRF/FRET in living cells. J Physiol. 2008;586:2307–2320. doi: 10.1113/jphysiol.2008.152777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosenko A, Hoshi N. A change in configuration of the calmodulin-KCNQ channel complex underlies Ca2+-dependent modulation of KCNQ channel activity. PLoS One. 2013;8:e82290. doi: 10.1371/journal.pone.0082290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strulovich R, Tobelaim W-S, Attali B, Hirsch J-A. Structural insights into the M-channel proximal C-terminus/calmodulin complex. Biochemistry. 2016;55:5353–5365. doi: 10.1021/acs.biochem.6b00477. [DOI] [PubMed] [Google Scholar]
- 17.Sachyani D, et al. Structural basis of a Kv7.1 potassium channel gating module: Studies of the intracellular c-terminal domain in complex with calmodulin. Structure. 2014;22:1582–1594. doi: 10.1016/j.str.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q, Chang A, Tolia A, Minor D-L., Jr Structure of a Ca(2+)/CaM:Kv7.4 (KCNQ4) B-helix complex provides insight into M current modulation. J Mol Biol. 2013;425:378–394. doi: 10.1016/j.jmb.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aivar P, et al. Surface expression and subunit specific control of steady protein levels by the Kv7.2 helix A-B linker. PLoS One. 2012;7:e47263. doi: 10.1371/journal.pone.0047263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linse S, Helmersson A, Forsén S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 21.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 22.Alaimo A, et al. Pivoting between calmodulin lobes triggered by calcium in the Kv7.2/calmodulin complex. PLoS One. 2014;9:e86711. doi: 10.1371/journal.pone.0086711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesner S, Sprangers R. Methyl groups as NMR probes for biomolecular interactions. Curr Opin Struct Biol. 2015;35:60–67. doi: 10.1016/j.sbi.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Ortega G, Pons M, Millet O. Protein functional dynamics in multiple timescales as studied by NMR spectroscopy. Adv Protein Chem Struct Biol. 2013;92:219–251. doi: 10.1016/B978-0-12-411636-8.00006-7. [DOI] [PubMed] [Google Scholar]
- 25.Schumacher M-A, Rivard A-F, Bächinger H-P, Adelman J-P. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 26.André I, Kesvatera T, Jönsson B, Linse S. Salt enhances calmodulin-target interaction. Biophys J. 2006;90:2903–2910. doi: 10.1529/biophysj.105.068718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adelman J-P, Maylie J, Sah P. Small-conductance Ca2+-activated K+ channels: Form and function. Annu Rev Physiol. 2012;74:245–269. doi: 10.1146/annurev-physiol-020911-153336. [DOI] [PubMed] [Google Scholar]
- 28.Halling D-B, Kenrick S-A, Riggs A-F, Aldrich R-W. Calcium-dependent stoichiometries of the KCa2.2 (SK) intracellular domain/calmodulin complex in solution. J Gen Physiol. 2014;143:231–252. doi: 10.1085/jgp.201311007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Pascal J-M, Zhang J-F. Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation. Proc Natl Acad Sci USA. 2013;110:4828–4833. doi: 10.1073/pnas.1220253110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tobelaim W-S, et al. Competition of calcified calmodulin N lobe and PIP2 to an LQT mutation site in Kv7.1 channel. Proc Natl Acad Sci USA. 2017;114:E869–E878. doi: 10.1073/pnas.1612622114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenmesser E-Z, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 32.Ortega G, Castaño D, Diercks T, Millet O. Carbohydrate affinity for the glucose-galactose binding protein is regulated by allosteric domain motions. J Am Chem Soc. 2012;134:19869–19876. doi: 10.1021/ja3092938. [DOI] [PubMed] [Google Scholar]
- 33.Hoang J, Prosser R-S. Conformational selection and functional dynamics of calmodulin: A (19)F nuclear magnetic resonance study. Biochemistry. 2014;53:5727–5736. doi: 10.1021/bi500679c. [DOI] [PubMed] [Google Scholar]
- 34.Vega S, Abian O, Velazquez-Campoy A. On the link between conformational changes, ligand binding and heat capacity. Biochim Biophys Acta. 2016;1860:868–878. doi: 10.1016/j.bbagen.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Rieping W, et al. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.