Summary
Plants modify their development to adapt to their environment, protecting themselves from detrimental conditions such as chilling stress by triggering a variety of signaling pathways; however, little is known about how plants coordinate developmental patterns and stress responses at the molecular level.
Here, we demonstrate that interacting transcription factors OsMADS57 and OsTB1 directly target the defense gene OsWRKY94 and the organogenesis gene D14 to trade off the functions controlling/moderating rice tolerance to cold.
Overexpression of OsMADS57 maintains rice tiller growth under chilling stress. OsMADS57 binds directly to the promoter of OsWRKY94, activating its transcription for the cold stress response, while suppressing its activity under normal temperatures. In addition, OsWRKY94 was directly targeted and suppressed by OsTB1 under both normal and chilling temperatures. However, D14 transcription was directly promoted by OsMADS57 for suppressing tillering under the chilling treatment, whereas D14 was repressed for enhancing tillering under normal condition.We demonstrated that OsMADS57 and OsTB1 conversely affect rice chilling tolerance via targeting OsWRKY94.
Our findings highlight a molecular genetic mechanism coordinating organogenesis and chilling tolerance in rice, which supports and extends recent work suggesting that chilling stress environments influence organ differentiation.
Keywords: cold tolerance, D14, gene network, organogenesis, OsMADS57, rice (Oryza sativa), trade‐off, WRKY94
Introduction
Rice (Oryza sativa) is a major staple crop for over half of the world's population. One of the key issues in rice production is its limited adaptability to local environmental stresses such as chilling, as it evolved in tropic and temperate regions (Kovach et al., 2007; Sang, 2011). Improvement of plant chilling tolerance could enable the expansion of rice cultivation to more northerly latitudes.
Chilling tolerance is a complex agronomic trait controlled by genetic networks and signal transduction pathways (Thomashow, 1999; Yamaguchi‐Shinozaki & Shinozaki, 2006; Zhu et al., 2007; Zhang et al., 2014; Zhao et al., 2015). Interactions between COLD1 (CHILLING TOLERANCE DIVERGENCE 1) and RGA1 (rice G‐protein α subunit 1) enable cold signaling to confer chilling tolerance in japonica rice (Ma et al., 2015; Shi & Yang, 2015) through the subsequent calcium signaling triggered in response to downstream stress response networks of C‐repeat‐binding factor (CBF) transcription factors(Sangwan et al., 2002; Knight & Knight, 2012; Zhu, 2016). However, less is known about how plants coordinate the stress response with developmental patterning to adapt to their environment over the longer term.
Developmental plasticity enables plants to respond to abnormal ambient temperatures by reprogramming their gene expression to adapt their architecture. Cold temperatures can disrupt the intrinsic signal network in the shoot apical meristem (SAM), and set a dormancy cycling at the SAM to enhance stress tolerance (van der Schoot & Rinne, 2011). The protection of root stem cell niche forms a sacrifice‐for‐survival mechanism from chilling stress (Hong et al., 2017). This altered development in response to temperature is crucially important for the maintenance of meristematic activity and the proper organization of differentiated cells (Komaki & Sugimoto, 2012). Under chilling treatment, several specific genes involved in the cell cycle are activated by transcription factors such as OsMYB3R‐2 to keep the cell mitotic for their tolerance (Ma et al., 2009). It suggests that the capability of maintaining cell behavior and activity is a resource to enhance survival and growth during and/or after chilling stress.
Shoot branching is controlled by the formation and subsequent outgrowth of axillary buds in the axils of the leaf primordia. LAX PANICLE 1 (LAX1) and MONOCULM1 (MOC1) control axillary bud initiation(Komatsu et al., 2003; Li et al., 2003).The DWARF genes, including D3, D10, D14, D17, D27 and D53, play critical roles in the strigolactone biosynthesis and signaling pathways during axillary bud outgrowth, which is required for tillering in rice(Ishikawa et al., 2005; Arite et al., 2007, 2009; Yan et al., 2007; Jiang et al., 2013).OsTB1/FC1 acts downstream of the DWARF genes to repress the outgrowth of axillary buds in rice (Takeda et al., 2003; Minakuchi et al., 2010). OsTB1 interacts with OsMADS57 to reduce its inhibitory impact on D14, a gene encoding strigolactone receptor that controls the organogenesis of tillers(Zhao et al., 2013; Yao et al., 2016), enabling it to regulate axillary bud initiation(Guo et al., 2013). Axillary buds are indeterminate structures that can be developmentally controlled in response to endogenous or environmental cues (Domagalska & Leyser, 2011; Janssen et al., 2014).However, the regulatory network influencing axillary bud development in response to stress is unknown.
In the present study, a network with a core OsMADS57 was identified to trade off chilling tolerance and axillary bud for adaptation to cold environment. We present evidence that OsMADS57 represents a new class of regulator, functioning in conjunction with OsTB1 to optimize chilling stress tolerance in rice seedlings. Our investigation revealed that OsMADS57 and OsTB1 can both directly target a transcription factor OsWRKY94 and axillary bud regulated gene D14 for adaptation to cold, thus providing evidence that OsMADS57 acts as a molecular link between the developmental response and chilling stress tolerance in rice.
Materials and Methods
Plant material and growth conditions
The osmads57‐1 and d14 mutants, as well as the OsMADS57 overexpression (OsMAD57‐OE) and antisense (OsMADS57‐AS) lines, were described previously (Guo et al., 2013). The T‐DNA insertion mutant lines PFG_3A‐15619.R (osmads57‐2) and PFG_3A‐13305.R (oswrky94‐1) in the Oryza sativa L. ssp. japonica cv Dongjin background were obtained from RiceGE, the Rice Functional Genomics Express Database of Korea. The ostb1 mutants were a gift from Professor Qian Qian, China National Rice Research Institute.
Chilling stress treatment
Rice plants were grown in a temperature‐controlled glasshouse with 30°C : 28°C, day : night cycles in Kimura B nutrient solution (Kato‐Noguchi & Ino, 2005) for 2 wk, then placed in a 4°C circulating water bath for the chilling treatment for various periods of time. The evaluation of chilling tolerance with survival rate was performed as described by Ma et al. (2015).After the chilling treatment, the plants were frozen in liquid nitrogen for further analyses.
Gene expression analysis
Plant total RNA was isolated using a TRIzol RNA Extraction Kit (Invitrogen) and treated with RNase‐free DNase I (MBI Fermentas, Waltham, MA, USA). A 2‐μg aliquot of total RNA was used to synthesize cDNA using AMV Reverse Transcriptase (Promega). The cDNA was diluted 1 : 50 into 15 μl SYBR Green quantitative PCR Master Mix (Toyobo, Osaka, Japan), according to the manufacturer's instructions. Quantitative reverse transcription polymerase chain reaction (RT‐PCR) was performed on a Mx3000P instrument (Stratagene, La Jolla, CA, USA). The gene expression levels were normalized to that of UBIQUITIN (UBQ). Each experiment included three technical replicates and at least three biological replicates. The values represent means ± SD of three technical replicates. Primers used are listed in Supporting Information Table S1.
Yeast one‐hybrid assays
Yeast one‐hybrid assays were used to check the binding of OsMADS57 and OsTB1 to the promoter of OsWRKY94. The coding sequences of OsMADS57 and OsTB1 were independently cloned into the EcoRI–XhoI site of the pJG4‐5 vector as prey. The effectors contained the GAL4‐activation domain. DNA fragments corresponding to the promoters of OsWRKY94 and D14 were independently cloned into the pLacZi plasmid as bait. Primers used for cloning are listed in Table S1. These constructs were transformed into the Saccharomyces cerevisiae strain EGY48. The transformed yeast was selected on a synthetic complete medium lacking Ura and Leu.
Protoplast transformation and transient expression assays
In order to assess the transient expression of OsWRKY94 driven by OsMADS57 and OsTB1, the cDNAs of full‐length OsMADS57, OsMADS57N and OsTB1 were fused into the pBI221 vector driven by the 35S promoter as effectors. The reporter plasmids OsWRKY94p::LUC and D14p::LUC were generated, and the 35S::GUS plasmid was used as a normalization control (LUC, luciferase; GUS, β‐glucuronidase). A transcriptional activity assay was carried out in the transiently transformed Arabidopsis thaliana protoplast system (Lin et al., 2007). The values represent means ± SD of three technical replicates. Co‐transformation of OsMADS57 and OsTB1 was performed to identify the effect of OsTB1 in the transient assay. As a control, OsTB1 (OsTB1V159QWL162L163) was replaced with OsTB1m (OsTB1A159QWA162A163) to disturb its interaction with OsMADS57 (Heery et al., 1997; Guo et al., 2013). After transformation, the Arabidopsis protoplasts were incubated at 22°C for 18 h followed by a treatment of 25°C or 4°C for an additional 30 min. Relative LUC activity (LUC/GUS) was calculated to determine OsWRKY94 promoter activity (Lin et al., 2007).
For the OsWRKY94 transcriptional activity assay, a GAL4 binding domain (BD)‐OsWRKY94 fusion protein was generated, which can bind to the GAL4 DNA‐binding sites of the GUS reporter. The transcription suppressor HOS15 and a transcription activator, ARF5M, were used as the controls (Zhu et al., 2008). A GUS reporter containing four upstream GAL4 DNA‐binding sites (GAL4(4X)‐ D1‐3(4X)‐ GUS) and the 35S::LUC internal control were co‐transformed with GAL4 BD‐OsWRKY94 into Arabidopsis protoplasts. After cell lysis, a 5‐μl extract was mixed with 45 μl LUC Assay Substrate (Promega) to determine LUC activity. For the GUS activity assay, a 5‐μl extract was incubated with 45 μl 4‐methylumbelliferyl β‐d‐glucuronide assay buffer (50 mM sodium phosphate pH 7.0, 1 mM 4‐methylumbelliferyl β‐d‐glucuronide, 10 mM EDTA, 10 mM β‐mercaptoethanol, 0.1% sarkosyl and 0.1% Triton X‐100) at 37°C for 30 min, after which the reaction was terminated by adding 950 μl 0.2 M Na2CO3. The GUS : LUC ratio was calculated as the relative reporter expression level.
Electrophoretic mobility shift assays
GST‐OsMADS57 and GST‐OsTB1 recombinant proteins were expressed in the Escherichia coli BL21 (DE3) strain and purified using Glutathione Sepharose 4B beads (GE Healthcare, Stockholm, Sweden) (GST, Glutathione S‐transferase). An electrophoretic mobility shift assay (EMSA) was performed using a LightShift Chemiluminescent EMSA Kit (Pierce, Waltham, MA, USA), according to the manufacturer's protocol with some previously described modifications (Ma et al., 2009). Oligonucleotides complementary to different motifs of the OsWRKY94 promoter were synthesized, annealed, and labeled using a Biotin 3′ End DNA Labeling Kit (Pierce). The oligonucleotide sequences are shown in Table S1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously (Bowler et al., 2004; Liu et al., 2014). The FLAG‐OsMADS57 line and the wild‐type (WT) ZH10 (Oryza sativa L. ssp. japonica cv Zhonghua 10), were grown for 2 wk and used to carry out the ChIP assay. Root‐dislodged seedlings (2 g) were cross‐linked for 20 min with 1% formaldehyde under a vacuum. Chromatin complexes were isolated and fragmented as described previously (Liu et al., 2014). An anti‐FLAG polyclonal antibody (Abcam, Cambridge, UK) and Protein A agarose/salmon sperm DNA (Millipore, Darmstadt, Germany) were used for immunoprecipitation. After reverse cross‐linking and protein digestion, the DNA products were analyzed using quantitative RT‐PCR. The enrichment was calculated as the ratio of FLAG‐OsMADS57 to ZH10. Values are the means ± SD of three independent experiments. The primers used in the ChIP assays are listed in Table S1.
Surface plasmon resonance analysis
All experiments were performed using a Biacore 3000 instrument and streptavadin‐coated biosensor chips (SA‐Chip; GE Healthcare) (Henriksson‐Peltola et al., 2007; Wang et al., 2015). All buffers were freshly prepared, filtered using 0.22‐μm syringe filters and de‐gassed. The instrument was first primed three times with reaction buffer (10 mM Hepes PH 7.9, 50 mM KCl, 1 mM EDTA, 0.5 mM DTT, 5 mM MgCl2, 10% Glycerol, 0.005% surfactant P20) and flow cell 1 (FC1) was used as the reference flow cell, which was unmodified and lacked the oligonucleotide ligand. Flow cell 2 (FC2) was used for the immobilization of the oligonucleotide. It was conditioned with three consecutive 1‐min injections of 50 mM NaOH in 1 M NaCl, in accordance with the manufacturer's instructions. The biotin‐labeled oligonucleotide (the same as was used in the EMSA) was then injected over a 1‐min period at a flow rate of 5 μl min−1, followed by the extraclean feature. Oligonucleotide immobilization levels of 200–300 RU were routinely observed under these conditions. Protein‐DNA binding assays were performed in the reaction buffer at the relatively high flow rate of 10 μl min−1 to avoid or minimize any mass‐transport limitation effects. Protein solutions were injected for 120 s followed by a dissociation in reaction buffer for 280 s. At the end of the dissociation period, the sensor chip was regenerated to remove any remaining bound material by injecting reaction buffer, containing 50 mM NaOH and 1 M NaCl, at 30 μl min−1 for 30 s.
Results
OsMADS57 is required for chilling tolerance in rice
The gain‐of‐function rice mutant osmads57‐1, in which the T‐DNA was inserted in the 3′terminus of OsMADS57, exhibits an enhanced outgrowth of axillary buds that increases its number of tillers (Guo et al., 2013). The osmads57‐2 mutant line (PFG_3A‐15619.R)(Jeong et al., 2002) contains a T‐DNA insertion in the first exon of OsMADS57, 61 bp from the transcriptional initiation site, which results in a loss of the full‐length transcript(Figs 1a,d, S1a–d). Therefore, it is a knockdown mutant on the genome.
Figure 1.
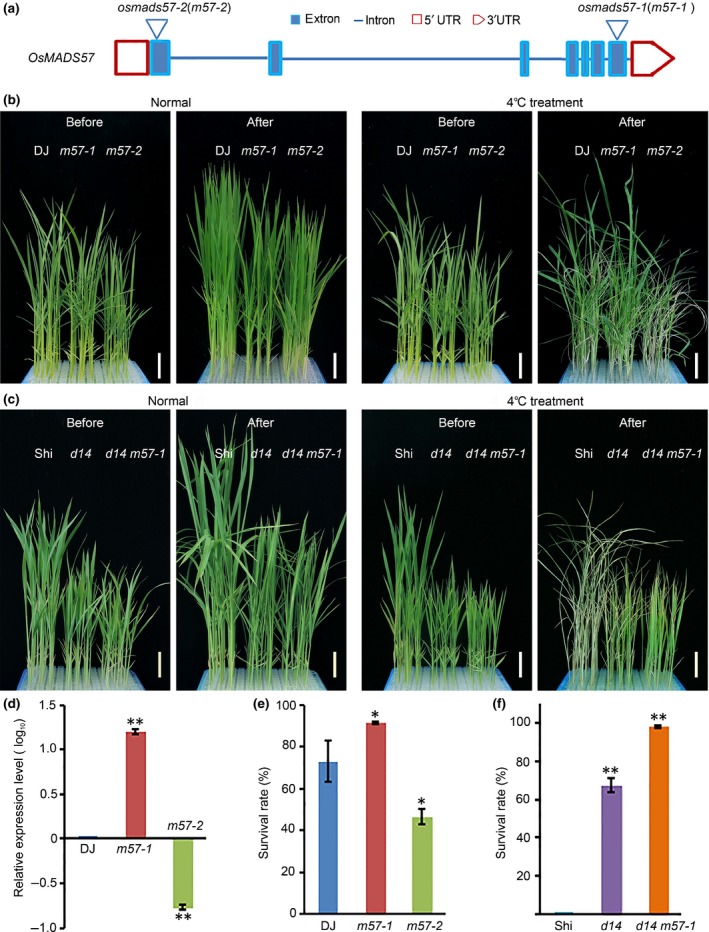
OsMADS57 is required for chilling tolerance. (a) Diagram of OsMADS57 gene structure. The triangles represent the T‐DNA insertion sites in osmads57‐1 (m57‐1) and osmads57‐2 (m57‐2) mutants. (b) Chilling tolerance phenotype of the gain‐of‐function mutant osmads57‐1 (m57‐1), the loss‐of‐function mutant osmads57‐2 (m57‐2) and the wild‐type (WT) Dongjin (DJ, Oryza sativa ssp. japonica cv Dongjin). Seedlings were incubated at 4°C for 84 h, then transferred back to the normal condition for recovery. Bars, 2.5 cm. (c) Chilling tolerance phenotype of the loss‐of‐function mutant d14, the double mutant d14 osmads57‐1 (d14 m57‐1) and the WT Shiokari (Shi, Oryza sativa ssp. japonica cv Shiokari). Seedlings were incubated at 4°C for 96 h, then transferred back to the normal condition for recovery. Bars, 2.5 cm. (d) Expression analysis of OsMADS57 in osmads57‐2 (m57‐2) using quantitative reverse transcription polymerase chain reaction. Values are expressed as the mean ± SD, n = 3 (three technical replicates per biological repeat). **, P < 0.01 (Student's t‐test). (e) Survival rate (percentage of seedlings surviving) of seedlings from (a) following the chilling treatment. Values are expressed as mean ± SD, n = 3, *, P < 0.05 (Student's t‐test). (f) Survival rate of seedlings from (b) following the chilling treatment. Values are expressed as mean ± SD, n = 3, **, P < 0.01 (Student's t‐test).
At the transcription level, OsMADS57 expression was significantly induced during abiotic stresses such as salt, drought, abscisic acid and chilling (Fig. S1e)(Arora et al., 2007). To examine the genetic function of OsMADS57 in response to chilling, seedlings of the osmads57 mutants were exposed to chilling temperatures (4°C) and subsequently returned to normal conditions (30°C : 28°C, day : night) for recovery. Rice seedlings with chilling tolerance were defined as those that could differentiate new leaves or continue leaf growth during the recovery treatment (Ma et al., 2015). After 7 d of recovery from the 84‐h chilling treatment, 91% of the osmads57‐1 seedlings survived, in contrast to 73% of WT Dongjin (DJ, Oryza sativa L. ssp. japonica cv Dongjin). Only 46% of osmads57‐2 plants survived after the chilling treatment (Fig. 1b,e). The data suggest that OsMADS57 is required for chilling tolerance in rice.
OsMADS57 negatively regulates D14 transcription expression, affecting tiller production (Guo et al., 2013). To assess the possibility of D14 being involved in the chilling response genetic network, the d14 mutant was exposed to a chilling treatment.The d14 seedlings exhibited an obviously more tolerant phenotype (67% of the seedlings survived) than the WT Shiokari (Shi, Oryza sativa L. ssp. japonica cv Shiokari)(none of the seedlings survived), whereas the double mutant d14 osmads57‐1 showed a higher survival rate (98% of the seedlings survived) than the d14 single mutant (Fig. 1c,f). This suggests that the regulation of D14 by OsMADS57 might be involved in the chilling stress response.
During the recovery period following the chilling treatment, the leaves of the osmads57‐2 mutant seedlings withered, which was particularly noticeable in the younger leaves (Fig. S2a). This withering was less pronounced in the WT, whereas the leaves of the osmads57‐1 plants were green and grew well, with new leaves being formed. After > 14 d of recovery, the osmads57‐1 mutants had notable new tiller growth, in contrast to the WT and the osmads57‐2 mutants (Fig. S2b,c, Methods S1).The mitotic index in the root apical meristem did not differ significantly between the WT and the osmads57 mutants when grown in normal temperatures. By contrast, when subjected to chilling conditions, the mitotic index of the gain‐of‐function mutant osmads57‐1 was higher than that of the WT, whereas that of the loss‐of‐function mutant osmads57‐2 was lower than that of the WT (Fig. S3a,b).This suggests that OsMADS57 could maintain cell division under chilling stress.
The overexpression line of OsMADS57 (OsMAD57‐OE) (Guo et al., 2013) showed a higher survival rate (70%) than its WT, ZH10 (37%), after the chilling treatment. By contrast, the antisense line (OsMADS57‐AS) was sensitive to chilling stress, and its survival rate (25%) was significantly lower than ZH10 (Fig. S4a,b). After > 14 d of recovery from the chilling treatment, the OsMADS57‐OE plants showed obvious tiller outgrowth, in contrast to the WT, whereas the OsMADS57‐AS plants that survived showed no obvious difference in tiller number compared with the WT (Fig. S4c). This result implies that OsMADS57 may play a role in both tillering and cold tolerance.
OsMADS57 directly activates OsWRKY94 expression in response to chilling
A previous microarray assay of the osmads57‐1 mutant revealed the candidate targets of OsMADS57 (Guo et al., 2013). These include WRKY genes predicted to function in plant developmental processes and in the response to abiotic stresses. By using the putative OsMADS57‐binding motif to search all of the WRKY gene promoters in rice, two potential OsMADS57‐binding CArGboxes, site 1 [CTTTTTATAG] and site 2[CTTTAAAAAG], were identified 1895–1886 bp and 1068–1059 bp upstream of the OsWRKY94 transcription start site, respectively (Fig. 2a). Site 2 comprised the same sequence as the OsMADS57‐binding motif in D14. To test whether OsMADS57 binds to OsWRKY94, an EMSA was used. As shown in Fig. 2(b), a clear OsMADS57‐dependent mobility shift was identified, and incubation with the OsMADS57 protein caused strong mobility shift bands in the probes for site 2 (lanes3–6) as compared with the probe alone (lane 8), or the GST protein alone (lane7). The unlabeled WRKY94S2 probe competed for the binding of labeled WRKY94S2 to MADS57 protein. As a negative control, the mutant probe did not show any shifted band when incubated with labeled WRKY94S2 probe (Fig. 2b).To further determine whether OsMADS57 directly associates with OsWRKY94 in vivo, a ChIP assay was performed using transgenic FLAG‐OsMADS57 lines. An assay using the FLAG antibody showed that the ‘S2’ region of the OsWRKY94 promoter, involving the site 2 CArG motif, was enriched in FLAG‐OsMADS57 compared with the WT, ZH10 (Fig. 2c). By contrast, there were no obvious enrichment for regions ‘S1’ and ‘S4’.The activities of OsMADS57 on OsWRKY94 also were tested in yeast one‐hybrid assays via the expression of the reporter gene LacZ driven by the OsWRKY94 promoter‐Pcyc1 (WRKY94p). The strains treated with OsMADS57 to induce OsWRKY94 expression grew well and were blue (Fig. 2d). By contrast, yeast cells treated with OsMADS57, but lacking the OsWRKY94 promoter, did not turn blue. D14 was used as a positive control for the CArG box, and yeast cells treated with OsMADS57 to drive D14 expression turned blue. WRKY94p ▵, a truncated form of the OsWRKY94 promoter that lacked the CArG box, was used as a negative control. Thus, OsMADS57 targets the CArG cis‐elementat site 2, which was present in the promoter of OsWRKY94. Our results suggested that OsMADS57 could directly target OsWRKY94.
Figure 2.
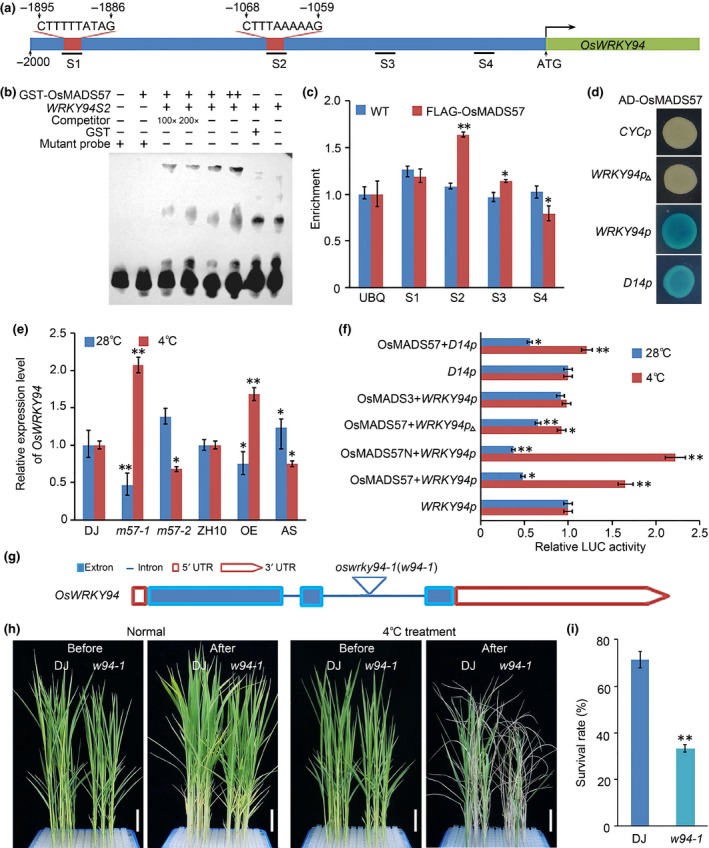
OsMADS57 directly upregulates OsWRKY94 expression during chilling treatment. (a) Schematic of the OsWRKY94 promoter showing the CArGboxes in red (S1 and S2). The S1 sequence is CTTTTTATAG and the S2 sequence is CTTTAAAAAG. Black lines S1–S4 indicate the sequences tested in the chromatin immunoprecipitation (ChIP) assays. (b) In vitro electrophoretic mobility shift assay (EMSA) showing that the GST‐OsMADS57 fusion protein binds to the OsWRKY94 promoter (WRKY94S2). The oligos were synthesized by fusing three copies of the S2 motif and their flanking sequence. The mutant probe (AGGGCCCCCT) served as the negative control. (c) ChIP to measure OsMADS57 occupancy at the OsWRKY94 promoter in vivo. The DNA isolated using ChIP was analyzed using quantitative PCR. Values are expressed as mean ± SD, n = 3. The UBIQUITIN (UBQ) promoter was used as a negative control.*, P < 0.05; **, P < 0.01 (Student's t‐test). (d) Yeast one‐hybrid analysis. D14p represents the positive control. WRKY94p ▵ is the truncated promoter of OsWRKY94 in which the CTTTAAAAAG sequence is absent. CYCp is the negative control of the blank vector. (e) OsWRKY94 expression in response to the chilling treatment in lines expressing various OsMADS57 constructs. Values are expressed as mean ± SD, n = 3. *, P < 0.05;**, P < 0.01 (Student's t‐test). (f) Transient transcriptional assay of OsWRKY94 driven by OsMADS57 in Arabidopsis protoplasts.*, P < 0.05; **, P < 0.01 (Student's t‐test). (g) Diagram of OsWRKY94 gene structure. The triangles represent the T‐DNA insertion sites in oswrky94‐1 (w94‐1) mutant. (h) Phenotype and (i) survival rate of oswrky94‐1 (w94‐1) and the wild‐type, Dongjin (DJ, Oryza sativa ssp.japonica cv Dongjin) following 84 h of the chilling treatment. Values are expressed as mean ± SD, n = 3, **, P < 0.01 (Student's t‐test). Bars, 2.5 cm.
Transcription of OsWRKY94 was upregulated in either osmads57‐1 or OsMADS57‐OE seedlings subjected to the chilling treatment, but was repressed under normal growth conditions (Fig. 2e). Conversely, OsWRKY94 expression was downregulated in either osmads57‐2 or OsMADS57‐AS plants following chilling. This suggested that the expression of OsWRKY94 is positively regulated by OsMADS57 under chilling stress in planta.
In order to confirm the effect of OsMADS57 on OsWRKY94 transcription, we carried out a transient expression assay in Arabidopsis mesophyll protoplasts. The data showed that OsMADS57 repressed the expression of the LUC reporter gene driven by the OsWRKY94 promoter under normal temperatures (25°C) (Fig. 2f). When the transformed protoplasts were treated with low temperatures (4°C) for 30 min, the expression of OsWRKY94 was strongly activated by OsMADS57 rather than being repressed, and to a greater extent than its repression in normal growth conditions. The activation of OsWRKY94 by OsMADS57 was diminished by the use of a truncated promoter missing the site 2 CArG box. The activity of the OsMADS57 N‐terminal peptide (170 aminoacid residues; OsMADS57N) was analyzed in the assay. OsMADS57N repressed OsWRKY94 expression under normal conditions, and activated OsWRKY94 expression under the chilling stimulus. It is consistent with our previous study that truncated OsMADS57 protein lacking the C‐terminus is sufficient for its function (Guo et al., 2013). D14, the positive control, was repressed by OsMADS57 under normal conditions but the activity showed little activation under chilling stress. OsMADS3, the negative control, showed no significant change in LUC expression either under low temperatures or normal growth conditions. The data suggest that OsMADS57 can act on OsWRKY94 either as a repressor or as an activator in a temperature‐dependent manner.
OsWRKY94 positively regulates chilling tolerance
The oswrky94‐1 mutant contains a T‐DNA insertion 946 bp from the transcription initiation site of OsWRKY94, in the second intron, identified by genome sequencing (Figs 2g, S5a,b)(Jeong et al., 2002).This T‐DNA insertion results in the lack of the conserved WRKY domain for OsWRKY94. A homozygous oswrky94‐1 mutant was identified using specific primers from rice GE(Fig. S5c), and transcription expression assays using semiquantitative and quantitative PCRs showed that oswrky94‐1 is a knockdown mutant in the genome (Fig. S5d,e). A phylogenetic analysis revealed that OsWRKY94 shares a higher sequence similarity with OsWRKY121 than with other homologs of WRKY proteins in rice and Arabidopsis (Fig. S5f, Methods S1).
Clear differences in survival rate were shown between oswrky94‐1 and its WT, DJ, after the chilling treatment (Fig. 2h). After a 7‐d recovery from the chilling treatment, 33% of oswrky94‐1 seedlings had survived, in contrast to 71% of the WTplants (Fig. 2i), suggesting that OsWRKY94 is involved in chilling tolerance in rice.
The loss‐of‐function mutant oswrky94‐1 and osmads57‐2 showed a decreased chilling tolerance, and the double mutant oswrky94‐1 osmads57‐2 exhibited a more significant sensitivity to chilling stress than either single mutant (Fig. 3a,b). Meanwhile, the expression level of OsWRKY94 was more dramatically decreased in the oswrky94‐1 osmads57‐2 double mutant than the oswrky94‐1 single mutant under normal conditions (Fig. 3c), suggesting that OsMADS57 and OsWRKY94 may act in the same genetic pathway to regulate the chilling response.
Figure 3.
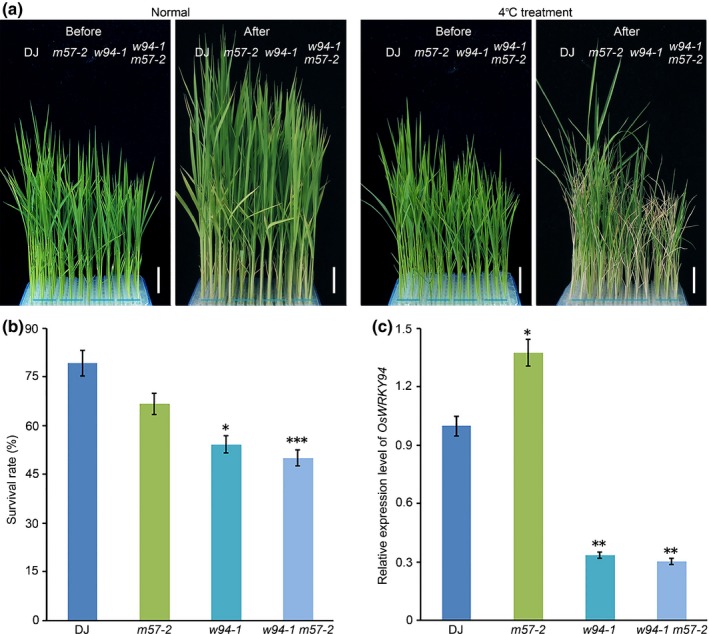
The oswrky94‐1 osmads57‐2 (w94‐1 m57‐2) double mutant is sensitive to chilling stress. (a) Phenotype of oswrky94‐1 osmads57‐2 (w94‐1 m57‐2) double mutant following 72 h of chilling treatment. Bars, 2.5 cm. (b) Survival rate of the oswrky94‐1 osmads57‐2 (w94‐1 m57‐2) double mutant following the chilling treatment. Values are expressed as mean ± SD, n = 3, *, P < 0.05; ***, P < 0.001. Student's t‐test. (c) OsWRKY94 expression level in the oswrky94‐1 osmads57‐2 (w94‐1 m57‐2) double mutant. Values are expressed as mean ± SD, n = 3. *, P < 0.05; **, P < 0.01 (Student's t‐test).
Quantitative RT‐PCR assays showed that the transcript level of OsWRKY94 was low in the leaves, but high in the root, shoot, culm, leaf sheath and spike (Fig. S6a). OsWRKY94 expression was markedly induced by the chilling treatment (Fig. S6b). Transcriptional activity assays using protoplasts revealed that OsWRKY94 repressed 42% of the expression of the GUS gene which reports activity of OsWRKY94 compared with BD alone (set to 100%). The repressor HOS15 was used as a positive control, and reduced GUS expression to 11% of that with BD alone. The negative control, ARF5M, activated GUS expression by 2.6‐fold compared with BD only (Fig. S6c) (Zhu et al., 2008; Guo et al., 2013). Subcellular localization assays showed that OsWRKY94‐GFP fluorescence completely merged with H2B‐mCherry nuclear protein marker in rice protoplast (Fig. S6d, Methods S1). These results demonstrated that chilling‐induced OsWRKY94 expression was required for its function in chilling tolerance.
Moreover, the transcription expression patterns of WRKY paralogs that could be putative targets of OsMADS57 were investigated. OsWRKY8, OsWRKY50, OsWRKY62, OsWRKY71 and OsWRKY108 all showed induced expression patterns in the WT following a chilling treatment. In osmads57‐1, OsWRKY71 expression was upregulated. By contrast, expression of the WRKY paralogs also was induced in osmads57‐2 in response to chilling (Fig. S6e). The WRKY paralogs may therefore display some functional redundancy.
OsTB1 reduces chilling tolerance by directly downregulating OsWRKY94
OsTB1 interacts with OsMADS57 to modulate tillering (Guo et al., 2013). The potential transcription factor binding motif of TCP (TEOSINTE BRANCHED1 in Zea mays, CYCLOIDEA in Antirrhinum majus, and PROLIFERATING CELL FACTOR1 in Oryza sativa)(Aggarwal et al., 2010; Manassero et al., 2013)was searched in the OsWRKY94 promoter, revealing two potential sites, site 1(TGGTCC) and site 2(TGGGCC), located 1386–1392 bp and 269–275 bp upstream of the initiation site of OsWRKY94, respectively(Fig. 4a). EMSA data revealed that site 2 (P2), but not site 1 (P1), was bound by the GST‐OsTB1 recombinant protein, whereas both sites were not bound by the GST protein alone (Fig. 4b). Addition of excess unlabeled probes reduced the binding. Addition of OsMADS57‐GST had no effect on the binding ability of OsTB1 to the promoter of OsWRKY94. A yeast one‐hybrid system assessing the activities of OsTB1 on the OsWRKY94 promoter‐Pcyc1 (WRKY94p) showed that the strains containing the OsWRKY94 promoter grew well and became blue. By contrast, yeast cells treated with OsTB1 but lacking the OsWRKY94 promoter did not turn blue. Under the same assay conditions, OsTB1 was found not to bind to D14 (Fig. 4c).
Figure 4.
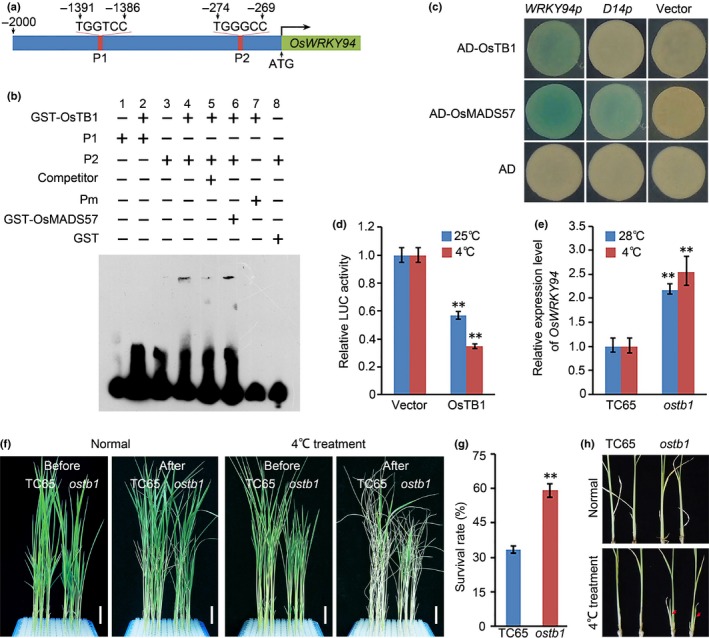
OsTB1 directly targets OsWRKY94. (a) Schematic of theOsWRKY94 promoter showing the OsTB1 binding motif. The P1 sequence is TGGTCC and the P2 sequence is TGGGCC. (b) Electrophoretic mobility shift assay (EMSA). The GST protein and the mutant probe (Pm) served as the negative control. In Pm, TGGGCC was mutated to AAAAAA. Excessive amounts (×50) of unlabeled oligos were added as the competitors. (c) Yeast one‐hybrid analysis. The effectors contained the GAL4 activation domain. (d) Expression activity assay of OsWRKY94 driven by OsTB1 in Arabidopsis protoplasts. **, P < 0.01. Student's t‐test. (e) Quantitative reverse transcription polymerase chain reaction analyses of OsWRKY94 transcript levels in seedlings of ostb1 and the wild‐type, Taichung 65 (TC65, Oryza sativa L. ssp. japonica cv Taichung 65) following 24 h of chilling treatment. The expression levels in TC65 both before and after chilling treatment were defined as 1. Values are expressed as mean ± SD, n = 3. **, P < 0.01 (Student's t‐test). (f) Phenotype and (g) survival rate of ostb1 following 84 h of chilling treatment. Values are expressed as mean ± SD, n = 3, **, P < 0.01 (Student's t‐test). Bars, 2.5 cm. (h) Chilling treatment increases tiller growth in ostb1. The arrows indicate the tillers that developed after the chilling treatment.
A transcriptional regulation activity assay in protoplasts revealed that OsTB1 suppressed the expression of LUC driven by the OsWRKY94 promoter under either normal temperature or the chilling treatment (Fig. 4d). By contrast, the chilling treatment enhanced directly the repression activity of OsTB1 on OsWRKY94. An expression pattern assay showed that OsWRKY94 transcription was increased in the ostb1 mutant compared with the WT. Chilling treatment enhanced the expression of OsWRKY94 in ostb1 compared with that in the WT (Fig. 4e). OsTB1 therefore directly suppresses OsWRKY94 expression in response to change in the ambient temperature.
A phenotypic assay demonstrated that 59% of ostb1 mutants were alive after the chilling treatment (Fig. 4f,g). By contrast, the WT Taichung 65 (TC65), had a 33% survival rate. After 14 d of recovery following the chilling treatment, the ostb1 mutant showed obvious tiller outgrowth, which was not seen in the WT (Fig. 4h).
OsMADS57 interacts with OsTB1 to coordinate rice growth and chilling tolerance
An EMSA revealed that OsTB1 enhanced OsMADS57 binding to the promoter of OsWRKY94 (Fig. 5a). The expression of OsWRKY94::LUC was significantly induced by the co‐transformation of OsMADS57 and OsTB1 (Fig. 5b). The LxxLL motif that facilitates the interaction between various proteins (Heery et al., 1997) was mutated in OsTB1; its sequence, VQWLL, which enables its interaction with OsMADS57, was mutated to AQWAA. When OsMADS57 and the mutated version of OsTB1 (OsTB1A159QWA162A163) were co‐expressed, the expression activity pattern of OsWRKY94::LUC was decreased at low temperatures compared with plants in which the WT versions of both proteins were co‐expressed (Fig. 5b). In contrast, the temperature‐dependent expression pattern of D14 was significantly independent of the interaction between OsMADS57 and OsTB1 in the same LUC activity system. The co‐localization assay showed that the fluorescence signal for OsMADS57‐mCherry overlapped with that for GFP‐OsTB1 in the nucleus (Fig. S7, Methods S1).These results demonstrate that OsTB1 enhances the binding of OsMADS57 to the promoter of OsWRKY94, enhancing its promotional effect on transcription expression.
Figure 5.
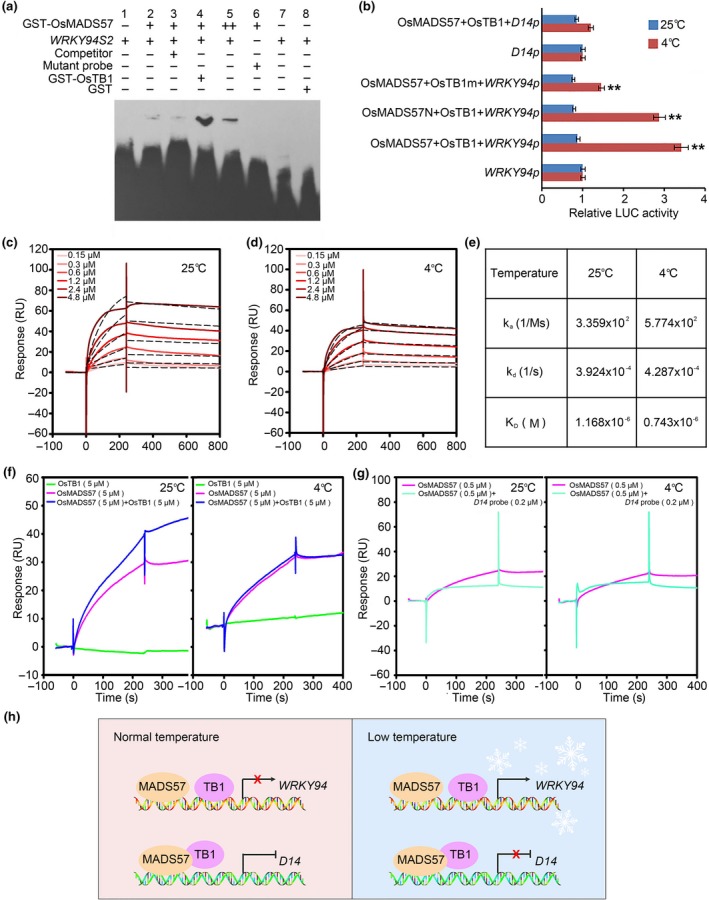
OsMADS57 interacts with OsTB1 to coordinate rice growth and chilling tolerance. (a) Electrophoretic mobility shift assay (EMSA) of the effect of OsTB1 on OsMADS57 binding to the OsWRKY94 promoter. Excess amounts (×50) of unlabeled oligos were added as the competitors. GST protein served as the negative control. (b) Effect of OsTB1 and OsMADS57 on the transcriptional regulation of OsWRKY94 in Arabidopsis protoplasts. OsTB1m was used as the negative control. **, P < 0.01 (Student's t‐test). (c, d) Kinetic binding of OsMADS57 to the OsWRKY94 promoter subfragment WRKY94S2 under (c) 25°C and (d) 4°C. The raw data curves are shown in red and the fitted curves in dotted black lines. (e) Kinetic and association constants of OsMADS57 binding to the promoter of OsWRKY94; ka, association rate constant; kd, dissociation rate constant; KD, dissociation constant; M, mol l−1; s, second. (f) OsTB1 effect on OsMADS57 binding to WRKY94S2 at 25°C and 4°C. A mixture of 1 μM OsMADS57 and 5 μM OsTB1 was placed in an ice bath for 30 min then passed over the biosensor chip. (g) Assay of D14 as a competitor for OsMADS57 binding to WRKY94S2 at 25°C and 4°C. The D14 probe contained the CATTAAAAAG CArG‐box. (h) Temperature‐dependent regulation of OsWRKY94 and D14 by OsMADS57 interacting with OsTB1 to balance rice growth and chilling tolerance. OsMADS57 activates OsWRKY94 under chilling stress, and represses D14 under normal conditions.
In order to confirm the temperature dependence of OsMADS57 binding to the promoter of OsWRKY94, a surface plasmon resonance (SPR) biosensor technique was used to perform a kinetic assay with different protein concentrations. The purified recombinant OsMADS57 and OsTB1 proteins were checked, and the WRKY94S2 oligonucleotide probe was immobilized on the SA‐chip (Fig. S8a,b). The relative responses of the binding of OsMADS57 to the promoter of OsWRKY94 were dependent on temperature changes, and positively correlated to the protein concentration (0.15–4.80 μM) (Fig. 5c,d). The dissociation rate constant K D was 1.2 × 10−6M at 25°Cand 0.7 × 10−6 M at 4°C (Fig. 5e).Temperature change showed an effect of OsMADS57‐OsTB1 heterodimerization on its binding to the promoter of OsWRKY94. When recombinant OsTB1 was added to the reaction of OsMADS57 and OsWRKY94S2, the transcription factor had an increased binding affinity compared with OsMADS57 alone at 25°C; however, no significant change of affinity was observed at 4°C (Fig. 5f). The D14 probe was mixed into the binding reaction to assess whether it affected the OsMADS57 transcription factor binding to the promoter of OsWRKY94. The D14 probe decreased the binding of OsMADS57 to OsWRKY94S2 at 25°C, whereas at 4°C, the binding response showed a slight change when D14 was added (Fig. 5g).These suggested that the affinity of OsMADS57 binding to the promoter of OsWRKY94 was dependent on experiencing chilling temperatures. D14 as one of the targets competed with OsWRKY94 for OsMADS57 binding, which was dependent on temperature.Our data indicate that the chilling‐dependent induction of OsMADS57 expression together with OsTB1 optimizes organ differentiation via D14 and the chilling response via OsWRKY94, thereby determining the tolerance of rice growth to environmental temperature changes (Fig. 5h).
Discussion
OsMADS57 maintains the capacity for chilling tolerance via its network of stragolactone signaling pathways
Hormones such as stragolactone modulate differentiation of axillary buds in the axils of the leaf primordia for plant development (Guo et al., 2013; Ha et al., 2014).OsMADS57 together with OsTB1 modulate regulation of axillary buds via D14 as a receptor‐mediating signaling pathway (Guo et al., 2013; Zuo & Li, 2014). The meristem, including axillary meristem, is a source for organogenesis in plant development, its tolerance capacity is based on developmental regulation networks, such as OsMADS57‐OsTB1‐D14. Our data suggest that OsMADS57 might be a pivotal component to coordinate OsWRKY94‐mediated stress responses with D14‐mediated leaf organogenesis to adapt to cold environments. Decreased OsMADS57 expression in the cold1‐1 mutant during chilling (Fig. S9) suggests that OsMADS57 is involved in the COLD1‐RGA1 sensing pathway for cold tolerance. This result may hint that cold signaling via sensors such as COLD1/RGA1 triggers a series of signals that maintain developmental capability. The gain‐of‐function mutant osmads57‐1 and the OsMADS57‐OE line had improved chilling tolerance, whereas the loss‐of‐function mutant osmads57‐2 and the OsMADS57‐AS line were sensitive to chilling (Figs 1b, S4a). Overexpression of OsMADS57 increased the tiller number by regulating the outgrowth of axillary buds (Guo et al., 2013) and exhibited a strong adaptive response to cope with chilling stress. The outgrowth of axillary buds was visibly increased in osmads57‐1 after the chilling treatment (Fig. S2b, S4c). Meanwhile, the d14 mutant, as well as the double mutant d14 osmads57‐1, had improved chilling tolerance (Fig. 1c). It means that the genes involved in the strigolactone network, such as OsMADS57, OsTB1 and D14, may influence plant abiotic stresses. The network may well link the phenotypic adaptation of plants to certain growth environments through its action on meristem initiation and differentiation in response to different stimuli. The chilling tolerant line osmads57‐1 showed newly differentiated leaves and tillers following the chilling treatment. The increased mitotic index in the gain‐of‐function mutant osmads57‐1 under the chilling treatment indicates that OsMADS57 probably maintains the cell division capability under stress conditions.
OsMADS57 triggers its temperature‐dependent targets controlling trade‐off between stress responses and plant development
MADS‐box proteins with DNA binding motif CArG box (CC(AT)6GG) target a series of genes for their transcription expression(De Bodt et al., 2003; de Folter & Angenent, 2006; Gramzow & Theissen, 2010).The genes for development and stress response are involved in the targets of MADS proteins. The cis‐element at which OsMADS57 binds the promoter of OsWRKY94 was found to be identical to the CArG‐box motif previously reported for D14 (Guo et al., 2013), suggesting that OsMADS57 might act to link the organogenesis and stress response networks. Data from the chromatin immunoprecipitation (ChIP), electrophoretic mobility shift (EMSA) and yeast one‐hybrid assays, as well as the phenotype of oswrky94‐1, support the hypothesis that OsWRKY94 is a direct target of OsMADS57 in the stress response. The affinity of OsMADS57 for OsWRKY94 was dependent on chilling temperatures, as revealed by dissociation rate constant (K D) changes at cold temperatures (Fig. 5e). The data suggested a change of OsMADS57 activity on OsWRKY94 from transcriptional repression at normal temperature to activation at the chilling temperature. This reveals that OsMADS57 can act as a transcriptional activator or repressor in plant development to enable tolerance to the ambient temperature. The affinity of OsMADS57 to the targets and the transcriptional dual regulation were dependent on temperature change, which might in turn be based on temperature‐dependent protein modification, such as phosphorylation (Molkentin et al., 1996; Badodi et al., 2015). The diverse functions of MADS‐box proteins are most likely achieved through interaction with other proteins. However, our data suggested that OsTB1 interacts with OsMADS57 to enhance OsWRKY94 transcription in protoplasts under a chilling treatment. In the gain‐of‐function mutant osmads57‐1, OsWRKY94 expression was increased relative to the wild‐type (WT) at cold temperatures, whereas it was decreased in the loss‐of‐function mutant osmads57‐2 (Fig. 2e).The osmads57‐1 mutant and the loss‐of‐function mutant ostb1 both improved chilling tolerance, whereas the loss‐of‐function mutant oswrky94‐1 exhibited chilling sensitivity. Alternative transcription factors, such as OsTB1, may form complexes with the target genes and/or OsMADS57 in the different environmental conditions. Homo‐ or heterodimerization of the component of OsMADS57 and OsTB1 might regulate the target genes flexibly and diversely in response to the ever‐changing environment. OsTB1 suppressed OsWRKY94 expression in the chilling treatment, which might be explained as a fine regulation for defense and development. Moreover, the temperature‐dependent alternative splicing of MADS‐box transcription factors in response to ambient temperature variation may result in different isoforms that could compete to interact with other factors and affect the DNA binding activity (Pose et al., 2013). The two transcription factors OsMADS57 and OsTB1 could therefore converge on the OsWRKY94 promoter to ensure the fine‐tuned control of OsWRKY94 expression, coordinating chilling tolerance with the outgrowth of axillary buds (Fig. 5h). OsMADS57 may function as a molecular switch in the cross‐talk between endogenous developmental cues and external signals to coordinate plant development with the response to chilling stress.
In nature, all species may be exposed to environmental stresses at some point in their lifecycle. Our findings support a model in which OsMADS57 acts as a key regulator, enabling the plant to adapt to its environment by balancing cell differentiation, cell division and chilling stress tolerance. OsMADS57 and its interaction partner OsTB1 coordinate the regulation of D14 for organogenesis and OsWRKY94 for the stress response. Our elucidation of the molecular roles of OsMADS57 in OsMADS57/OsTB1‐OsWRKY94/D14 signaling provides novel insights into the complicated regulatory network controlling plant developmental responses to chilling environments.
Author contributions
L.C. and K.C. designed the experiments; L.C. performed experiments, data analysis and wrote the manuscript; Y.Z., S.X. and Z.Z. contributed to assist in performing part of the experiments; Y.X. and J.Z. contributed to assist in analyzing and discussing the data; and K.C. supervised the project and revised the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Molecular identification of the osmads57‐2 mutant and the OsMADS57 expression patterns under different abiotic stresses.
Fig. S2 Developmental responses of osmads57‐1, osmads57‐2, and the wild‐type Dongjin to the chilling treatment.
Fig. S3 Flow cytometry assay of cell division in osmads57‐1, osmads57‐2, and the wild‐type Dongjin following chilling treatment.
Fig. S4 Chilling response of OsMADS57 overexpression and antisense lines.
Fig. S5 Molecular identification of the oswrky94‐1 mutant and the phylogenetic analysis of OsWRKY94.
Fig. S6 Expression patterns and regulation activity of OsWRKY94 and five other WRKY genes in the osmads57 mutant.
Fig. S7 Co‐localization of OsMADS57 and OsTB1 in rice protoplasts.
Fig. S8 Characterization of purified proteins and quantification of their binding to DNA using SPR.
Fig. S9 Relative OsMADS57 expression in the cold1‐1 mutant treated at 4°C for 1 or 5 h.
Table S1 List of primers and accession numbers of genes used in this study
Methods S1 Materials and methods.
Acknowledgements
We thank: Fengqin Dong (Institute of Botany, Chinese Academy of Sciences) for kindly providing help on histological analyses; Siyi Guo, Yuda Niu and Xiaoxia Wang (Institute of Botany, Chinese Academy of Sciences) for assistance in gene transformation; and Wei Luo, Yongyan Tang and Bo Wang (Institute of Botany, Chinese Academy of Sciences) for their technical assistance. This work was funded by the National Key Research and Development Program of China Grant (2016YFD0100901), the CAS Strategic Priority Research Programs A (XDA08010205) and CAS‐CSIRO Project (151111KYSB20160052).
References
- Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U. 2010. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis . Plant Cell 22: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant Journal 51: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone‐insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiology 50: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. 2007. MADS‐box gene family in rice: genome‐wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badodi S, Baruffaldi F, Ganassi M, Battini R, Molinari S. 2015. Phosphorylation‐dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle 14: 1517–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. 2004. Chromatin techniques for plant cells. Plant Journal 39: 776–789. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Van de Peer Y, Theissen G. 2003. And then there were many: MADS goes genomic. Trends in Plant Science 8: 475–483. [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nature Review Molecular Cell Biology 12: 211–221. [DOI] [PubMed] [Google Scholar]
- de Folter S, Angenent GC. 2006. Trans meets cis in MADS science. Trends in Plant Science 11: 224–231. [DOI] [PubMed] [Google Scholar]
- Gramzow L, Theissen G. 2010. A hitchhiker's guide to the MADS world of plants. Genome Biology 11: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K. 2013. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14 . Nature Communications 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CV, Leyva‐Gonzalez MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Yamaguchi S, Dong NV et al 2014. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences, USA 111: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. 1997. A signature motif in transcriptional co‐activators mediates binding to nuclear receptors. Nature 387: 733–736. [DOI] [PubMed] [Google Scholar]
- Henriksson‐Peltola P, Sehlen W, Haggard‐Ljungquist E. 2007. Determination of the DNA‐binding kinetics of three related but heteroimmune bacteriophage repressors using EMSA and SPR analysis. Nucleic Acids Research 35: 3181–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Savina M, Du J, Devendran A, Kannivadi Ramakanth K, Tian X, Sim WS, Mironova VV, Xu J. 2017. A sacrifice‐for‐survival mechanism protects root stem cell niche from chilling stress. Cell 170: 102–113. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. 2005. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiology 46: 79–86. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Drummond RS, Snowden KC. 2014. Regulation of axillary shoot development. Current Opinion in Plant Biology 17: 28–35. [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G. 2002. T‐DNA insertional mutagenesis for activation tagging in rice. Plant Physiology 130: 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y et al 2013. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato‐Noguchi H, Ino T. 2005. Possible involvement of momilactone B in rice allelopathy. Journal of Plant Physiology 162: 718–721. [DOI] [PubMed] [Google Scholar]
- Knight MR, Knight H. 2012. Low‐temperature perception leading to gene expression and cold tolerance in higher plants. New Phytologist 195: 737–751. [DOI] [PubMed] [Google Scholar]
- Komaki S, Sugimoto K. 2012. Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiology 53: 953–964. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J. 2003. LAX and SPA: major regulators of shoot branching in rice. Proceedings of the National Academy of Sciences, USA 100: 11 765–11 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR. 2007. New insights into the history of rice domestication. Trends in Genetics 23: 578–587. [DOI] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F et al 2003. Control of tillering in rice. Nature 422: 618–621. [DOI] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. 2007. Transposase‐derived transcription factors regulate light signaling in Arabidopsis . Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K. 2014. OsmiR396d‐regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4 . Plant Physiology 165: 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X et al 2009. Enhanced tolerance to chilling stress in OsMYB3R‐2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology 150: 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D et al 2015. COLD1 confers chilling tolerance in rice. Cell 160: 1209–1221. [DOI] [PubMed] [Google Scholar]
- Manassero NG, Viola IL, Welchen E, Gonzalez DH. 2013. TCP transcription factors: architectures of plant form. Biomolecular Concepts 4: 111–127. [DOI] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S et al 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant and Cell Physiology 51: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Li L, Olson EN. 1996. Phosphorylation of the MADS‐Box transcription factor MEF2C enhances its DNA binding activity. Journal of Biological Chemistry 271: 17 199–17 204. [DOI] [PubMed] [Google Scholar]
- Pose D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M. 2013. Temperature‐dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417. [DOI] [PubMed] [Google Scholar]
- Sang T. 2011. Toward the domestication of lignocellulosic energy crops: learning from food crop domestication. Journal of Integrative Plant Biology 53: 96–104. [DOI] [PubMed] [Google Scholar]
- Sangwan V, Orvar BL, Beyerly J, Hirt H, Dhindsa RS. 2002. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant Journal 31: 629–638. [DOI] [PubMed] [Google Scholar]
- van der Schoot C, Rinne PL. 2011. Dormancy cycling at the shoot apical meristem: transitioning between self‐organization and self‐arrest. Plant Science 180: 120–131. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yang S. 2015. COLD1: a cold sensor in rice. Science China Life Sciences 58: 409–410. [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi‐Tanaka M, Ashikari M, Matsuoka M, Ueguchi C. 2003. The OsTB1 gene negatively regulates lateral branching in rice. Plant Journal 33: 513–520. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Wang H, Ramakrishnan A, Fletcher S, Prochownik EV. 2015. A quantitative, surface plasmon resonance‐based approach to evaluating DNA binding by the c‐Myc oncoprotein and its disruption by small molecule inhibitors. Journal of Biology Methods 2: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Yan H, Saika H, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. 2007. Rice tillering dwarf mutant dwarf3 has increased leaf longevity during darkness‐induced senescence or hydrogen peroxide‐induced cell death. Genes & Genetic Systems 82: 361–366. [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L et al 2016. DWARF14 is a non‐canonical hormone receptor for strigolactone. Nature 536: 469–473. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen Q, Wang S, Hong Y, Wang Z. 2014. Rice and cold stress: methods for its evaluation and summary of cold tolerance‐related quantitative trait loci. Rice 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Lang Z, Zhu JK. 2015. Cold responsive gene transcription becomes more complex. Trends in Plant Science 20: 466–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Wu ZS, Yi W, Xu Y, Li S, Xu TH, Liu Y, Chen RZ, Kovach A et al 2013. Crystal structures of two phytohormone signal‐transducing α/β hydrolases: karrikin‐signaling KAI2 and strigolactone‐signaling DWARF14. Cell Research 23: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Dong CH, Zhu JK. 2007. Interplay between cold‐responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology 10: 290–295. [DOI] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ et al 2008. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proceedings of the National Academy of Sciences, USA 105: 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2016. Abiotic stress signaling and responses in plants. Cell 167: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Li JY. 2014. Molecular dissection of complex agronomic traits of rice: a team effort by Chinese scientists in recent years. National Science Review 1: 253–276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Molecular identification of the osmads57‐2 mutant and the OsMADS57 expression patterns under different abiotic stresses.
Fig. S2 Developmental responses of osmads57‐1, osmads57‐2, and the wild‐type Dongjin to the chilling treatment.
Fig. S3 Flow cytometry assay of cell division in osmads57‐1, osmads57‐2, and the wild‐type Dongjin following chilling treatment.
Fig. S4 Chilling response of OsMADS57 overexpression and antisense lines.
Fig. S5 Molecular identification of the oswrky94‐1 mutant and the phylogenetic analysis of OsWRKY94.
Fig. S6 Expression patterns and regulation activity of OsWRKY94 and five other WRKY genes in the osmads57 mutant.
Fig. S7 Co‐localization of OsMADS57 and OsTB1 in rice protoplasts.
Fig. S8 Characterization of purified proteins and quantification of their binding to DNA using SPR.
Fig. S9 Relative OsMADS57 expression in the cold1‐1 mutant treated at 4°C for 1 or 5 h.
Table S1 List of primers and accession numbers of genes used in this study
Methods S1 Materials and methods.


