Abstract
Purpose
The majority of bladder cancer patients present with localized disease and are managed by transurethral resection. However, the high rate of recurrence necessitates lifetime cystoscopic surveillance. Developing a sensitive and specific urine-based test would significantly improve bladder cancer screening, detection, and surveillance.
Experimental Design
RNA-seq was used for biomarker discovery to directly assess the gene expression profile of exfoliated urothelial cells in urine derived from bladder cancer patients (n=13) and controls (n=10). Eight bladder cancer specific and 3 reference genes identified by RNA-seq were quantitated by qPCR in a training cohort of 102 urine samples. A diagnostic model based on the training cohort was constructed using multiple logistic regression. The model was further validated in an independent cohort of 101 urines.
Results
418 genes were found to be differentially expressed between bladder cancer and controls. Validation of a subset of these genes was used to construct an equation for computing a probability of bladder cancer score (PBC) based on expression of 3-markers (ROBO1, WNT5A, and CDC42BPB). Setting PBC=0.45 as the cutoff for a positive test, urine testing using the 3-marker panel had overall 88% sensitivity and 92% specificity in the training cohort. The accuracy of the 3-marker panel in the independent validation cohort yielded an area under the curve of 0.87 and overall 83% sensitivity and 89% specificity.
Conclusions
Urine-based molecular diagnostics using this 3-marker signature could provide a valuable adjunct to cystoscopy and may lead to a reduction of unnecessary procedures for bladder cancer diagnosis.
Keywords: RNA-sequencing, urinary biomarkers, RNA, bladder cancer, molecular diagnostics
Introduction
Bladder cancer is the fifth most common cancer with about 74,000 new cases and 16,000 disease-specific deaths in 2015 in the United States (1). The majority of cases are non-muscle invasive bladder cancer (NMIBC) at diagnosis and are primarily managed with transurethral resection (TUR). With a recurrence rate of up to ~70% at 5 years, bladder cancer requires lifelong cystoscopic surveillance (2). Due to the invasiveness of cystoscopy, there are strong interests to develop non-invasive urine-based diagnostics. A reliable urine test could improve surveillance strategies by prioritizing high-risk patients to undergo cystoscopy and biopsy, while reducing procedural frequency in low-risk patients. Despite inadequate diagnostic sensitivity for low grade (LG) and high grade (HG) cancer at ~20% and ~80%, respectively, urine cytology is commonly performed due to its high specificity (>95%) and positive predictive value (3). Other FDA-approved urine tests including single-biomarker immunoassays, fluorescent immunohistochemistry, and fluorescence in-situ hybridization (4,5) are available. However, these tests have not been widely adopted due to insufficient diagnostic performance (6).
Emerging bladder cancer molecular diagnostics have focused on development of multi-biomarker panels ranging from 2 to 18 targets (7-11). Most biomarker discovery efforts have depended on microarray-based screening of the bulk mass of tumor tissues. However, challenges of lower specificity than cytology and low sensitivity for LG tumors have hindered adoption of these test (8,12). To identify biomarkers for urine-based molecular diagnostics, exfoliated urothelial cells may be a better starting material given the continuous contact of bladder tumors with urine (13). RNA sequencing (RNA-seq) is a next generation sequencing technology that offers unbiased identification of known and novel transcripts, single base-pair resolution, high sensitivity and high specificity, broad dynamic range of over 8000-fold for gene expression quantification and ability to detect rare and low-abundance genes (14).
In this study, we applied RNA-seq as a discovery tool for bladder cancer-specific urinary RNA markers. Deep sequencing of RNA extracted from urine sediments from bladder cancer patients and controls resulted in an average of 100 million sequencing reads per sample. Genes selected based on the RNA-seq analysis were evaluated using quantitative real-time polymerase chain reaction (qPCR) in a training cohort. This data was used to select a 3-marker panel consisting of two cancer-specific genes (ROBO1, WNT5A) and one reference gene (CDC42BPB). The diagnostic accuracy of the 3-marker panel was evaluated in an independent patient cohort and compared favorably to urine cytology.
Methods
Study design
The study protocol was approved by the Stanford University Institutional Review Board and Veterans Affairs Palo Alto Health Care System (VAPAHCS) Research and Development Committee. All patients were recruited from VAPAHCS. The study was divided into 3 parts: 1) biomarker discovery, 2) construction of the diagnostic model, and 3) validation of the diagnostic model (Figure 1). For each part, urine samples were collected from bladder cancer and control subjects. Patients of both genders ≥ 18 years old were eligible for enrollment. Patients with other urological cancer were excluded. For biomarker discovery, urine samples were collected from 23 subjects (13 bladder cancer and 10 controls) for RNA-seq. To construct the diagnostic model, expression of candidate genes identified by RNA-seq was analyzed in urinary RNA extracts from a training cohort of 102 urines samples (50 bladder cancer and 52 controls) using qPCR. The 3-marker diagnostic panel was then validated in 101 urine samples (47 bladder cancer and 54 controls) to determine assay diagnostic sensitivity and specificity. The diagnostic performance of the 3-marker panel was also compared to urine cytology collected per routine clinical care in a subset of samples.
Figure 1. Study design.
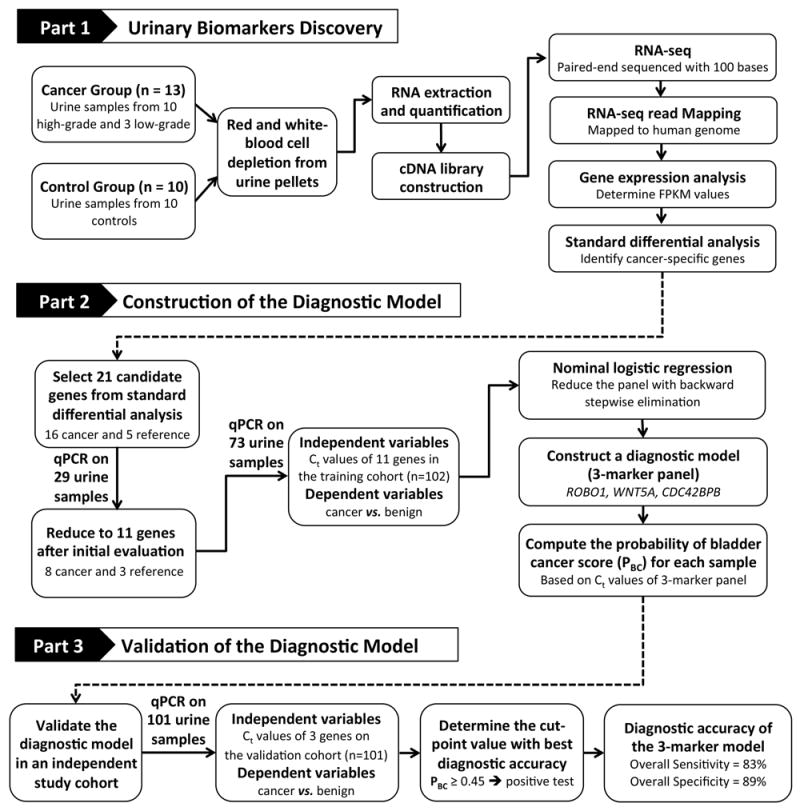
For the biomarker discovery in part 1, urine samples from 13 bladder cancer patients and 10 control subjects were collected for RNA-seq analysis. For model construction in part 2, a subset of genes that were differentially expressed in bladder cancer compared to controls was selected for qPCR validation in 102 urine samples. A model for computing a probability of bladder cancer score (PBC) based on the gene expression of the 3-marker panel in urine was constructed using multivariate logistic regression. For model validation in part 3, the diagnostic performance of the 3-marker panel was evaluated in an independent study cohort of 101 urine samples.
Patient population and samples
The “bladder cancer-evaluation” group included patients with no prior history of bladder cancer who were undergoing urological work-up, primarily for hematuria. The “bladder cancer-surveillance” group included patients with prior history of bladder cancer undergoing routine surveillance. The “control” group included patients with non-neoplastic urological diseases and healthy volunteers ≥ 35 years old. Urine was collected prior to cystoscopy or tumor resection for bladder cancer-evaluation and surveillance groups and mid-day for the control group. Urine samples were categorized as cancer or benign based on corresponding tissue histopathology from TUR or cystoscopic biopsy when available. For urine samples without a matching tissue sample from bladder cancer evaluation or surveillance patients, diagnosis was based on cystoscopic findings. Urine samples from patients with non-neoplastic urological diseases (e.g. kidney stones) and healthy control groups that did not undergo cystoscopy were presumed negative for bladder cancer based on clinical history (Table 1). Cytology results were considered positive when reported as suspicious or malignant, and negative when reported as atypical or negative.
Table 1.
Demographic and clinicopathogic features of the study cohorts.
| Biomarker Discovery | Diagnostic Model | Validation | |||||
|---|---|---|---|---|---|---|---|
| Demographic features | Benign (n = 10) | Cancer (n = 13) | Benign (n = 52) | Cancer (n = 50) | Benign (n = 54) | Cancer (n = 47) | |
| Average age (range)a | > 35 | 72.8 (58-90) | 67.3 (30-89) | 71.8 (53-93) | 70.8 (29-100) | 71.4 (55-91) | |
| Gender: male/female, n | 10/0 | 13/0 | 52/0 | 50/0 | 53/1 | 47/0 | |
| BC-evaluation | - | 8 | 15 | 23 | 22 | 15 | |
| BC-surveillance | - | 5 | 23 | 27 | 31 | 32 | |
| Healthy/other controls | 10 | - | 14 | - | 1 | - | |
| Clinicopathologic featuresb | Cancer (n = 13) | Cancer (n = 50) | Cancer (n = 47) | ||||
| Grade | Lowc | 3 | 19 | 29 | |||
| High | 10 | 31 | 18 | ||||
| Clinical Stage | Papillary | ||||||
| Ta | 8 | 28 | 36 | ||||
| T1 | 1 | 10 | 4 | ||||
| ≥ T2 | 2 | 3 | 5 | ||||
| Papillary + CIS | |||||||
| Ta | - | 1 | 2 | ||||
| T1 | 1 | - | - | ||||
| T2 | 1 | 2 | - | ||||
| CIS | - | 6 | - | ||||
Abbreviation: CIS, carcinoma in situ.
Average age and range does not include healthy controls as specific ages were not collected for this group.
Clinicopathologic features are available only for bladder cancer patients.
All LG cancer samples were with stage pTa.
Urine sample preparation
For RNA-seq, urine samples (10 - 750 ml) were processed within two hours of collection. Urine sediment was collected by centrifugation for 15 minutes at 500xg and pellets were washed 3 times with PBS. Washed urine sediment was depleted of red and white blood cells (RBCs and WBCs). RBCs were selectively lysed by addition of 1 ml of 10-fold diluted RBC lysis solution (Miltenyi Biotec). Remaining cells were collected by centrifugation at 300xg for 5 minutes and cell pellets were washed 3 times with PBS. To deplete WBCs, cells were incubated for 15 minutes at 4°C with 80 μl of magnetic-activated cell sorting (MACS) buffer (PBS, 0.5% bovine serum albumin, and 2 mM EDTA) and 20 μl of anti-CD45 magnetic micro-beads. Then 1 ml of MACS buffer was added and cells collected by centrifuged at 300xg for 15 minutes at 4°C. The cells were re-suspended in 500 μl MACS buffer and applied to a MACS LD column (Miltenyi Biotec). The column was washed twice with 1 ml MACS buffer and the total effluent was collected. For RNA extraction, urothelial cells were collected by centrifugation and resuspended in 1 ml TRIzol® (Invitrogen) and stored at -80°C. Total RNA from the urotheilal cells was extracted with TRIzol® reagent followed by DNA degradation with RQ1 RNase-free DNase (Promega) then purification on RNeasy® MinElute Cleanup columns (Qiagen) according to the manufacturer’s instructions. An Agilent 2100 Bioanalyzer and RNA Pico chips were used for total RNA quantification and qualification analysis. RNA concentration and RNA integrity number (RIN) were determined for each sample.
Library preparation and RNA-seq
cDNAs were synthesized from samples with total RNA ≥ 6 ng in 12 μl of nuclease-free water using the Ovation RNA Seq System V2 kit (NuGEN Technologies) according to manufacturer’s instructions. cDNAs were fragmented with S-Series Focused-ultrasonicator (Covaris). To enrich for cDNAs >300 bases in length, cDNAs were size fractionated by incubating with 0.8 volume of Agencourt AMPure XP beads (Beckman Coulter) for 10 minutes followed by bead separation on 96S super magnet plate (Alpaque) for 10 minutes. Beads were then washed three times with 80% ethanol and air-dried for 15 minutes on the magnetic plate. cDNA products were eluted with 102 μl of RNase-free water and quantity was measured by spectrophotometry (NanoDrop). Barcoded sequencing libraries were prepared using a NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs) and cDNA libraries were enriched with the Agencourt AMPure XP beads (Beckman Coulter) as described above and eluted with 30 μl of buffer EB (Qiagen). Sequencing libraries were paired-end sequenced with reads of 100 bases long on the Illumina HiSeq 2000 at Stanford Stem Cell Institute Genome Center.
RNA-seq gene expression analysis and candidate selection
RNA-seq reads were mapped to the human genome (GRCh38) using TopHat. Mapped reads were assembled and gene expression analysis was performed using Cufflinks software tools. The sequence fragments were normalized to take into account both gene length and mapped reads for each sample, to measure the relative abundance of genes based on fragments per kilobase of exon per million fragments mapped (FPKM). Standard differential analysis based on the FPKM values was performed to compare gene expression profiles of control, bladder cancer, HG, and LG using Cuffdiff software to identify and prioritize cancer-specific genes by the fold-change of genes with a false discovery rate (q-value) ≤ 0.05. To select against candidate markers also highly expressed in blood cells, the gene expression profiles of potential candidate genes in blood cells were examined using gene expression commons, an open platform for absolute gene expression profiling in the human hematopoietic system (15).
qPCR gene expression analysis
For qPCR analysis, total urine sediments were collected and RNA was extracted, purified and quantitated as described above, but without blood cell depletion. cDNAs for all samples were generated using the Ovation RNA Seq System V2 kit (NuGEN Technologies) according to manufacturer’s instructions, and in 4 samples (1 LG, 1 HG, and 2 controls in the training cohort) cDNA synthesis was also carried out with High-Capacity RNA-cDNA kit (Applied Biosystems) for comparison. cDNAs were enriched for > 300 base fragments with the AxyPrepMag™PCR Clean-up bead solution (AxyPrep™) and bead separation on 96S super magnet plate (Alpaque), eluted and quantitated as described above for RNA-seq analysis. cDNA products were amplified in single reactions using TaqMan® Gene Expression Assays (Applied Biosystems). The TaqMan® primers and probes were selected to span an exon-exon junction without detecting genomic DNA. qPCR reactions were performed in triplicate. For each reaction, 10 ng cDNA in 9 μl was mixed with 10 μl 2X TaqMan® Gene Expression Master Mix (Applied Biosystems) and 1 μl 20X TaqMan® Gene Expression Assay solution in a final volume of 20 μl and amplified in an ABI Prism® 7900 HT Sequence Detection System (Applied Biosystems). Reactions were heated to 50°C for 2 minutes and 95°C for 10 minutes before being cycled 40 times at 95°C for 15 seconds and 60°C for 1 minute. qPCR results were processed with SDS 2.4 and RQ manager software packages (Applied Biosystems). An automated threshold and baseline were used to determine the cycle threshold value (Ct). The mean of the triplicate measurements of Ct was used for data analysis. For genes with undetermined Ct values, Ct value of 45 was assigned. Samples with Ct ≥ 37 for 2 of 3 reference genes (QRICH1, CDC42BPB, and DNMBP) in the training cohort and the 1 reference gene (CDC42BPB) in the validation cohort were excluded from analysis due to insufficient RNA quantity or quality.
Statistical analysis
For initial diagnostic model construction, 21 markers were tested with 29 urine samples. The relative expression level of cancer genes was evaluated as the geometric average of the Ct of 5 reference genes – Ct of the cancer gene (ΔCt). The initial panel was narrowed to 11 markers (8 cancer and 3 reference) for testing of an additional 73 urine samples. The Ct values of the 11-marker panel were used for statistical analysis with JMP Pro 12 (SAS Institute Inc.). Univariate logistic regression was used to study the predictive ability of the 11 markers on the cancer status with the odds ratios (ORs) with 95% confidence intervals (CIs), area under the receiver operation characteristic (ROC) curve (AUC), and p-value. Multiple logistic regression with backward stepwise elimination using stopping rule of entering p-value = 0.25 and leaving p-value = 0.05 was performed to reduce the panel of markers. A reference marker was included in the model as a sample adequacy control and to normalize cell numbers. Ct values of 3-marker signature (ROBO1, WNT5A, and CDC42BPB) were used for calculating the probability of bladder cancer score (PBC) of each sample: PBC = exp [A]/(1+exp [A]) with A = 19.82 - 0.43 × ROBO1 Ct - 0.56 × WNT5A Ct + 0.33 × CDC42BPB Ct. Receiver operating characteristic (ROC) curve and AUC for the 3-marker panel were generated and calculated with the JMP Pro 12 software. Empirical ROC curve for the cytology report was estimated from ordinal empirical data with 4 categories (negative, atypical, suspicious, and malignant) (16). Sensitivity and specificity for each category was determined and the ROC curve was generated with 4 sets of data point connected by straight line. AUC of the ROC curve was calculated using R software. The statistical significance of the difference between two AUCs was evaluated as described (17).
Results
Study participants
Between 2013 and 2016, 186 human subjects were recruited and 226 urine samples were collected and processed. Subject demographic and clinicopathologic characteristics are shown in Table 1. Urine samples were collected from 1) patients undergoing bladder cancer evaluation (BC-evaluation) who presented with hematuria (n=78), suspicious urine cytology (n=2) or suspicious mass in computer tomography (n=3); 2) patients with known history of bladder cancer undergoing surveillance cystoscopy (BC-surveillance, n=118); 3) patients with non-neoplastic urological diseases including benign prostatic hyperplasia (n=2), urolithiasis (n=2), urinary tract infections (n=1) and indwelling ureteral stents (n=3) (other non-neoplastic urological diseases); and 4) healthy male volunteers age > 35 years with no prior history of cancer or active urological issues (healthy controls, n=17).
Urinary biomarker discovery
To identify candidate urinary biomarkers, RNA-seq was applied to 10 urine samples from patients with HG bladder cancer, 3 samples from patients with LG bladder cancer, and 10 control samples (Table 2). To reduce non-urothelial cell sequences related to the blood-cell-associated transcriptome, RBCs and WBCs were depleted prior to total RNA isolation for sequencing. Notably, more RNA was extracted from cancer samples (mean concentration 0.98 ng/ml) than from controls (0.08 ng/ml). This difference is likely due to a higher concentration of urothelial cells in urine of cancer patients. The RNA integrity number (RIN) ranged from 2.5 to 9.5 independent of sample type. As shown in Table 2, 41-313 million paired-reads were generated per sample and 13-72.5% of the reads could be mapped to human genome. Two control samples had a low percentage of mapped reads, sample 4 with 13% and sample 9 with 27%, suggestive of sample contamination and were excluded from further analysis. Standard differential analysis based on FPKM values was performed for pairwise comparison of the gene expression profiles of control, HG, LG and combined HG and LG bladder cancer. Comparison of control and combined bladder cancer RNA-seq data identified 418 differentially expressed genes, 281 over-expressed and 137 under-expressed in bladder cancer. Comparison of control and HG samples yielded 105 differentially express genes, 74 over-expressed and 31 under-expressed in HG. Comparison of control and LG samples identified, 17 differentially express genes, 8 over-expressed and 9 under-expressed in LG. When comparing LG to HG samples, 3 genes were over-expressed in HG. The full panel of differentially expressed genes, prioritized by fold change of FPKM value is listed in Supplementary Table S1-4.
Table 2.
Summary of urine samples used for RNA-seq transcriptome profiling.
| Sample Number | Clinicopathologic features | Urine volume (ml) | Total RNA concentration (ng) | RIN (1-10)a | Number of reads | % of mapped reads |
|---|---|---|---|---|---|---|
| 1 | Control | 200 | 7.1 | 9.4 | 88,922,624 | 35.3 |
| 2 | Control | 75 | 13.6 | 5.3 | 84,926,624 | 37.8 |
| 3 | Control | 190 | 6.1 | 9.2 | 82,152,466 | 58.3 |
| 4 | Control | 150 | 11.6 | 2.5 | 202,122,232 | 13.0 |
| 5 | Control | 200 | 11.1 | 6.6 | 211,454,432 | 47.6 |
| 6 | Control | 435 | 51.0 | 5.6 | 261,575,308 | 48.4 |
| 7 | Control | 327 | 27.5 | 2.7 | 313,362,582 | 44.3 |
| 8 | Control | 750 | 17.4 | 3.7 | 59,641,782 | 54.8 |
| 9 | Control | 460 | 13.3 | 4.9 | 54,411,982 | 27.0 |
| 10 | Control | 500 | 81.7 | 6.6 | 73,908,808 | 52.9 |
| 11 | Ta HG | 50 | 9.7 | 9.5 | 77,299,864 | 35.2 |
| 12 | Ta HG | 176 | 57.4 | 7.6 | 100,212,450 | 72.5 |
| 13 | Ta HG | 140 | 8.8 | 3.1 | 76,097,546 | 54.1 |
| 14 | Ta HG | 125 | 28.0 | 2.7 | 90,664,046 | 59.5 |
| 15 | Ta HG | 82 | 72.8 | 3.8 | 57,041,308 | 59.8 |
| 16 | T1 HG | 110 | 128.8 | 7.0 | 41,764,318 | 64.6 |
| 17 | T2 HG | 60 | 63.7 | 6.7 | 95,286,642 | 70.1 |
| 18 | T2 HG | 115 | 110.2 | 8.6 | 67,061,502 | 68.1 |
| 19 | T1 HG +CIS | 125 | 101.4 | 6.9 | 91,298,356 | 39.0 |
| 20 | T2 HG + CIS | 133 | 603.8 | 6.2 | 70,401,502 | 65.9 |
| 21 | Ta LG | 80 | 79.4 | 7.7 | 58,109,042 | 65.3 |
| 22 | Ta LG | 215 | 110.0 | 6.3 | 55,461,696 | 46.8 |
| 23 | Ta LG | 68 | 67.9 | 6.2 | 48,072,074 | 61.8 |
Abbreviations: CIS, carcinoma in situ.
The RNA integrity number (RIN) is an algorithm for evaluating the integrity of RNA with a value of 1 to 10, with 10 being the least degraded.
Biomarker selection based on RNA-seq
To minimize false positive signals due to hematuria and inflammation, genes known to be highly expressed in blood cells were excluded as candidate biomarkers (15). Sixteen candidate cancer-specific genes were chosen from the control vs. HG and control vs. combined HG + LG comparisons. Fifteen of the candidate genes selected (CP, PLEKHS1, MYBPC1, ROBO1, RARRES1, WNT5A, AKR1C2, AR, IGFBP5, ENTPD5, SLC14A1, FBLN1, SYBU, STEAP2, and GPD1L) were overexpressed in HG samples with fold-change above control ranging from 3.10 to 7.39. One bladder cancer specific gene, BPIFB1, identified in control vs. combined HG + LG comparison had a 6.65-fold increase in cancer. All of the candidate cancer specific genes were among the top 30 genes in the control vs. combined HG + LG comparison. The Cuffdiff output for the 16 bladder cancer-specific genes selected for the validation in the training study cohort is shown in Supplementary Table S5. In order to find a suitable reference gene to control urinary RNA quantity, 5 genes (QRICH1, CDC42BPB, USP39, ITSN1, and DNMBP) with uniform expression level, mean FPKM value ~ 4, and standard deviation (SD) ≤ 0.25 among all RNA-seq samples were selected for investigation (Supplementary Table S6).
Biomarker validation in the training cohort
Candidate biomarkers were validated in a training cohort of cancer and control urine samples to confirm expression level and select a panel with best diagnostic performance for bladder cancer. In contrast to the biomarker discovery phase, sample preparation during the biomarker validation phases were performed with RNA isolation from total urine sediment without blood cell depletion, thereby facilitating future clinical translation. Gene expression of an initial panel of 16 cancer-specific and 5 reference genes was determined by qPCR in 29 urine samples (16 cancer and 15 controls). Uniform expression of the candidate reference genes was evaluated and the qPCR Ct values from control and cancer samples were collected and compiled (Supplementary Figure S1). Among the candidates references genes, QRICH1, CDC42BPB and DNMBP had the most similar Ct values (~28) and least variability (SD range 2.0 to 2.6), indicating they are stably expressed and suitable for data normalization in qPCR experiments. Based on the relative expression of the cancer genes normalized to the reference genes (ΔCt), 8 of the cancer genes (WNT5A, RARRES1, ROBO1, CP, IGFBP5, PLEKHS1, BPIFB1, and MYBPC1) were selected for additional testing. These 8 cancer and 3 reference genes were evaluated in an additional 73 urine samples (34 cancer and 39 controls).
To confirm that qPCR validation results were not biased by the reverse transcriptase method used to generate cDNA from urinary RNA, qPCR experiments with the 11 candidate genes were run on 4 samples (2 cancer, and 2 controls) with cDNAs produced using two different kits (NuGEN Technologies and Applied Biosystems). After the qPCR data were normalized using the geometric average of the 3 reference genes, the relative expression levels of the 8 cancer genes were consistent between methods (data not shown) suggesting reverse transcriptase kit did not introduce bias in the gene expression analysis.
Construction of the diagnostic model
Univariate logistic analysis of Ct values of the 11 candidate genes in the training cohort was performed to evaluate predictive accuracy for bladder cancer for each candidate. The 8 cancer markers were all significant predictors (p < 0.0001) with WNT5A, RARRES1, ROBO1 and CP the strongest predictors of bladder cancer with odds ratios ranging from 1.65 to 2.12 and AUCs ≥ 0.9 (Supplementary Table S7). Although the reference markers were chosen as sample adequacy and reference levels for the number of cells in the sample, two of the reference markers, CDC42BPB (p = 0.0476) and DNMBP (p < 0.0001), were significant predictors of bladder cancer, likely due to higher concentration of urothelial cells in bladder cancer samples.
Multiple logistic regression analysis of Ct values of the 11 candidate genes in the training cohort was used to construct a diagnostic model equation. ROBO1, WNT5A and CDC42BPB were identified as having relevant, non-redundant diagnostic values for constructing an equation to calculate a score for probability of bladder cancer (PBC):
Using this equation, the PBC for each sample in the training cohort was calculated (Figure 2A). A PBC ≥ 0.45-cutoff was designated a positive test as it gave the best overall combination of sensitivity and specificity at 88% and 92% respectively (Table 3). In 81 samples, the diagnostic accuracy of the 3-marker panel using PBC ≥ 0.45 cutoff was compared to cytology. While the overall specificity of the 3-marker panel was modestly lower than cytology, the overall sensitivity was much better, 88% for the 3-marker panel compared to 21% for cytology.
Figure 2. Diagnostic performance of the 3-marker panel for bladder cancer prediction.
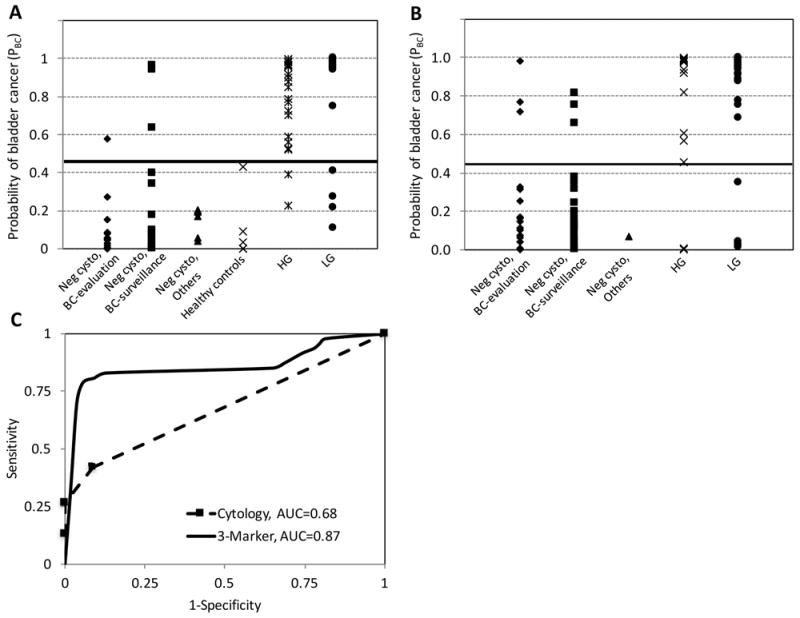
The probability of bladder cancer score (PBC) based on the diagnostic equation using the 3-marker (ROBO1, WNT5A, CDC42BPB) urine assay was measured in A, the training cohort (n=102) and B, the validation cohort (n=101). PBC ≥ 0.45 (the black line in A and B) as the threshold for a positive test gave the best concordance with clinical findings for patients without evidence of bladder cancer (Neg cysto, BC-evaluation; Neg cysto, BC-surveillance; Neg cysto, others (other non-neoplastic urological diseases); and Healthy controls) and patient with bladder cancer (HG and LG). C, comparison of the diagnostic performance of the 3-marker in the validation cohort (n= 101) with cytology on a subset of samples (n=89) using ROC curves resulting in AUCs of 0.87 for the 3-marker panel and 0.68 for cytology. Neg cysto, Negative cystoscopy.
Table 3.
Summary of diagnostic performance for bladder cancer prediction on urine based on the 3-marker panel using ROBO1, WNT5A, and CDC42BPB and cytology in both training and validation cohorts with the cutoff of PBC ≥ 0.45 giving a positive test.
| Training Cohort | Validation Cohort | ||||
|---|---|---|---|---|---|
| 3-Marker Panel | Cytologya | 3-Marker Panel | Cytologya | ||
| Sensitivity | All Cancer | 88% (44/50) | 21% (9/42) | 83% (39/47) | 25 % (10/40) |
| HG | 94% (29/31) | 35% (8/23) | 83% (15/18) | 50 % (7/14) | |
| LG | 79% (15/19) | 5% (1/19) | 83% (24/29) | 12 % (3/26) | |
| Specificity | All Non-Cancer | 92% (48/52) | 97% (38/39) | 89% (48/54) | 100 % (49/49) |
| Negative BC evaluation | 93% (14/15) | 100% (15/15) | 86% (19/22) | 100 % (19/19) | |
| Negative BC surveillance | 87% (20/23) | 96% (22/23) | 90% (28/31) | 100 % (30/30) | |
| Healthy/Other Controls | 100% (14/14) | 100% (1/1) | 100% (1/1) | N/A | |
Cytology was only available for a subset of samples.
Validation of the diagnostic model
The 3-marker panel of ROBO1, WNT5A, and CDC42BPB was evaluated by qPCR in an independent validation set of 101 urine samples (47 cancer and 54 controls) from 86 patients (Table 1, Figure 2B). Similar to sample preparation in the training cohort, total urine sediment was used for RNA isolation and analysis. Using PBC ≥ 0.45 as the threshold for a positive test, the overall sensitivity and specificity for the 3-marker panel was 83% and 89%, respectively (Table 3). Notable the sensitivity for the 3-marker panel was the same for HG and LG disease (83% sensitivity). The diagnostic performance of 3-marker panel was also compared with cytology on a subset of samples (n = 89) with an AUC of 0.87, which was significantly more accurate than the diagnosis by cytology with an AUC of 0.68 (p < 0.01) (Figure 2C). Cytology had high specificity (100%) but lower overall sensitivity (25%). While the 3-maker panel performed equally well for HG and LG, the sensitivity of cytology for HG disease (50%) was considerably better than for LG (12%).
Using the 3-marker panel for bladder cancer surveillance
To explore the potential of using the 3-marker panel urine test for bladder cancer surveillance, we evaluated its test performance in serially collected urine samples from six patients. For each patient, 2 to 4 urine samples were collected over 7 to 18 months. The results from the 3-marker panel were compared with cystoscopic and/or pathologic findings. In all patients, the 3-marker panel was concordant with cystoscopic and/or pathologic results, both in cancer positive and negative scenarios (Figure 3).
Figure 3. Bladder cancer surveillance using the 3-marker urine test.
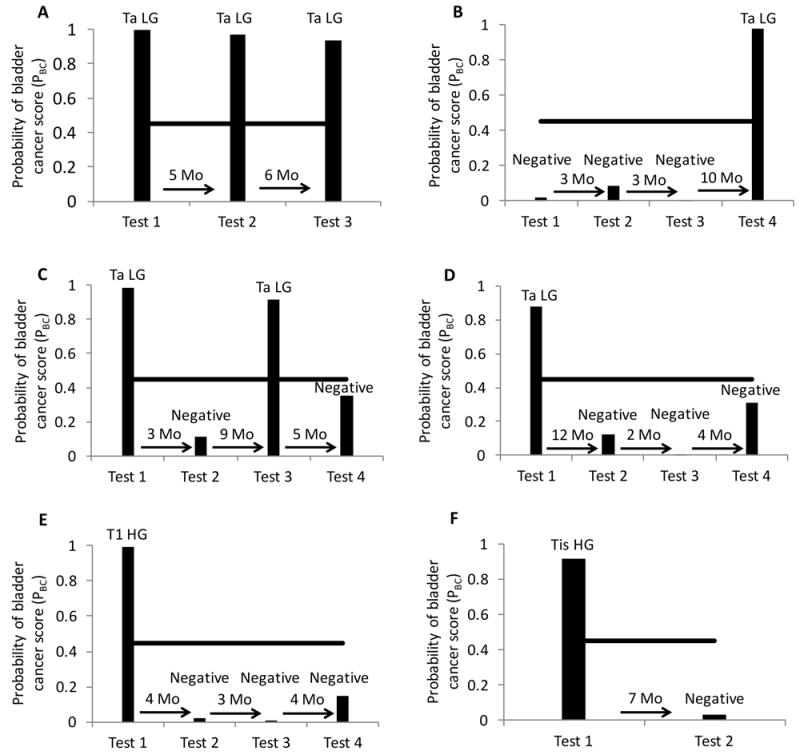
Serial urine samples were collected from 6 patients and the probability of bladder cancer score (PBC) based on the 3-marker (ROBO1, WNT5A, CDC42BPB) diagnostic equation was determined. PBC ≥ 0.45 (black line) was considered positive for bladder cancer. Corresponding bladder cancer pathology (stage, grade) or cystoscopy (if no bladder cancer detected) was indicated above urine test result. A, urine test can accurately detect persistent bladder cancer. Test 1 for bladder cancer evaluation accurately detected bladder cancer as did follow up surveillance tests after 5 months (test 2) and another 6 months (test 3). B, urine test can accurately detect bladder cancer recurrence in patient disease free for >16 months. Test 1 for bladder cancer surveillance was negative consistent with negative cystoscopy, as were tests 2 and 3 at 3 month intervals, test 4 accurately detected bladder cancer recurrence 10 months later. C, urine test was reliable for prediction of alternating pattern of positive and negative tests. Test 1 for bladder cancer evaluation accurately detected bladder cancer, follow up surveillance at 3 months was negative by both urine test and cystoscopy, bladder cancer recurrence was accurately detected after another 9 months, followed by negative results from both urine test and cystoscopy after another 5 months. D, E and F, after initial positive bladder cancer test, the subsequent urine tests accurately predict disease free survival. Test 1 for bladder cancer surveillance (D) or bladder cancer evaluation (E and F) accurately detected bladder cancer, subsequent surveillance tests were negative by both urine test and cystoscopy. LG, low grade; HG, high grade.
In a patient with LG Ta bladder cancer (Figure 3A), the 3-marker panel was positive at the initial diagnosis and two subsequent cancer recurrences, whereas cytology remained negative throughout, indicating that the 3-marker panel is a better adjunct to cystoscopy for this patient. In another patient with prior history of LG with focal HG bladder cancer, the patient had 3 negative cystoscopy and 3 matched negative 3-marker urine tests (Figure 3B). At the time of tissue-confirmed recurrence 16 months later, the 3-marker panel also turned positive. The concordance of the 3-panel marker with cystoscopy suggest that the use of the panel may reduce the frequency of cystoscopic surveillance in selected patients. Similar findings are seen in two other patients with Ta LG cancer (Figure 3C-D), in which the 3-marker panel paralleled negative cystoscopies and biopsy-proven recurrences.
In patients with HG T1 (Figure 3E) and TIS (Figure 3F) at the time of study entry, both cytology and the 3-marker panel were positive at cancer diagnosis and negative during surveillance. Notably, the patient in Figure 3F underwent induction BCG following the diagnosis of TIS. The surveillance cystoscopy following BCG identified an erythematous patch on the anterior bladder wall. The appropriately negative 3-marker panel (Figure 3F, test 2) suggests that in this case the test remained reliable after BCG and did not falsely identify inflammation as bladder cancer.
Discussion
While most bladder cancers are non-muscle invasive at initial diagnosis, the high recurrence rate and potential to progress to invasive disease necessitates frequent surveillance cystoscopy, contributing to bladder cancer as one of the most expensive cancers to treat (18). To date, a non-invasive test with sufficient accuracy to reduce the frequency of cystoscopy in low-risk patients, while providing timely treatment in high-risk patients, has remained elusive. For development of a urine-base bladder cancer test, we reasoned that direct analysis of exfoliated urothelial cells, rather than tissue biopsies, would yield higher translational potential for biomarker discovery. We applied RNA-seq for unbiased gene expression analysis of urinary cells and demonstrated the success of extracting high quality RNA and generating high quality sequencing for identifying a new 3-marker panel (ROBO1, WNT5A and CDC42BPB) for molecular diagnosis of bladder cancer.
Identification of differentially expressed genes between cancer and benign tissues is a common starting point for biomarker discovery. Development of next generation sequencing technologies that allow for high sensitivity, resolution, throughput and speed have advanced research on biomarker discovery for cancer diagnosis, assessing prognosis, and directing treatment monitoring (19-22). RNA-seq has emerged as a powerful tool for unbiased interrogation of gene expression as well as identification of splice variants and non-coding RNAs (14). An innovative aspect of our study lies in the discovery approach of using urine as the starting material for RNA-seq. Direct application of RNA-seq to urine has been limited, given the relatively low cellularity and heterogeneity of urine samples that may impact RNA integrity. To address these issues, we processed the entire volume urine sample within 2 hours of collection to maximize the number of cells and the yield of total RNA. We observed RIN values that spanned almost the entire range of 1 to 10 (Table 2), suggesting variable levels of RNA degradation. By not excluding samples with low RINs, we aimed to improve the translational potential of the assay as the transcripts identified are likely to remain stable even in partially degraded RNA samples. Furthermore, the variable degrees of RNA degradation did not appear to compromise the number of sequencing reads or percentage of mapped reads.
Through the blood cell depletion steps, our sample preparation protocol for RNA-seq analysis was designed to enrich urothelial cells and genes specific to bladder cancer while reducing potentially confounding markers of inflammation commonly found in urine (e.g. urinary tract infection, post-intravesical BCG administration). Additionally, candidate genes identified by RNA-seq that are also known to be highly expressed in blood cells were excluded from marker validation. Targeting transcripts likely to be specific to urothelial cells specific during the discovery phase allowed us to use total urine sediment RNA without blood cell depletion at the validation phases, thereby simplifying the sample preparation and increasing the translational potential.
Our discovery strategy allowed us to concentrate on a small panel of genes with the highest diagnostic yield that are stable in urine from the myriad of differentially expressed genes in bladder tumors. Although this study aims to identify robust urinary diagnostic markers rather than causative markers for cancer biology, several of the bladder cancer specific genes identified in our approach have been linked to bladder and other cancers. CP, which has the highest fold increase in cancer compared to control in our screen (Supplementary table S1), encodes a feroxidase enzyme and was previously identified in a proteomic screen as a urinary biomarker of bladder cancer (23) and as a serum biomarker in other cancers (24). IGFBP5, another top candidate gene, was previously found to be upregulated in bladder cancer by tissue microarray analysis and is part of the Cxbladder 5-marker panel for bladder cancer diagnosis described below (8,9). The two cancer specific genes in our 3-marker panel were also previously implicated in tumor formation and progression. ROBO1 is a promoter of tumor angiogenesis and overexpressed in both human bladder cancer tissue and cultured cell lines (25,26). WNT5A is a secreted glycoprotein that plays an important regulatory role in embryogenesis, including regulation of cell polarity and migration. WNT5A expression decreases after development and upregulation in adult tissue has been implicated in oncogenesis (27). In bladder cancer, WNT5A protein expression correlated positively with the histological grade and pathological stage (28,29).
Several urine tests have been approved for clinical use in bladder cancer. However, due to inadequate sensitivity (particularly for LG cancer) and specificity in inflammatory conditions, current guidelines on NMIBC do not recommend their routine use for surveillance or initial work-up (6,30). Fluorescence in situ hybridization (UroVysion) and immunocytochemistry (ImmunoCyt) incorporate molecular markers with microscopic evaluation of urine cells with overall better sensitivity but lower specificity than conventional cytology (11,31). However, these tests, like cytology, are subject to interobserver differences in interpretation (32). Protein biomarker assays nuclear matrix protein 22 (NMP22) and bladder tumor antigen (BTA) offer the potential for simple, more objective tests (33). Both tests have higher sensitivity but lower specificity than cytology, especially in patients with inflammation and infection in the urinary tract (34,35).
Recent efforts to improve urine-based diagnostics for bladder cancer have focused on multiplex detection of mRNAs that are differentially expressed between cancer and non-cancerous tissues. A general strategy uses microarray analysis of bladder cancer tissue samples for target selection, followed by validation in urine samples. One panel, Cxbladder (Pacific Edge, Dunedin, New Zealand), assays urinary expression of bladder cancer markers CDC2, HOXA13, MDK and IGFBP5, as well as inflammation biomarker CXCR2 to reduce false positive tests (9). In a multicenter prospective study of 485 patients presenting with gross hematuria, the Cxbladder assay had an overall sensitivity of 81% (97% for HG, 69% for LG) and specificity of 85% (8). Another assay under development by BiofinaDX (Madrid, Spain) uses a 2, 5, 10 or 12 gene signature for urinary detection of bladder cancer (12). The 12-gene signature was first identified by microarray analysis of bladder cancer tumor tissue then validated in urine samples (10). In a multicenter prospective study of 525 samples, the 12-marker panel was narrowed to two (IGF2 and MAGEA) with an overall sensitivity of 81% (89% for HG, 68% for LG) and specificity of 91% (7,12).
Improving the diagnostic sensitivity of LG is one of the central goals of urine-based diagnostics, as the majority of bladder cancer patients present with LG disease. While LG tumors are typically not life threatening, the diagnosis and treatment of these lesions is crucial to prevent morbidity and reduce the risk of progression. Our diagnostic model consisting of ROBO1, WNT5A, and CDC42BPB, had an overall sensitivity of 83% and specificity of 89%. Compared to Cxbladder and BiofinaDX, our overall sensitivity was similar and subset analysis showed improved sensitivity 83% for LG cancer compared to 69% for Cxbladder and 68% for BiofinaDX (8,12). The improved sensitivity may be due in part to our urine-based biomarker discovery strategy to target mRNA that are not only differentially expressed in bladder cancer but also maintain stability in urine. Additionally, concentrating the cellular fraction from the entire urine sample may account for superior detection of LG tumors that shed fewer cells.
One strength of our study is the proof of concept demonstration of serial testing for a cohort of patients over their course of bladder cancer surveillance (Figure 3). The consistent results between cystoscopy and the 3-marker panel suggest that the test may be a dependable adjunct for cancer surveillance. This may be especially true in the setting of an initial positive 3-marker urine test indicating that the markers are upregulated in the tumor.
Based on our dataset, we set the threshold for a positive test at PBC ≥ 0.45 in both bladder cancer evaluation and surveillance populations. In the clinical scenario of using the urine test to prescreen patients before cystoscopy, sensitivity may be considered more important than specificity as the clinical outcome of missing cancer is worse than negative cystoscopy. To maximize the sensitivity, the threshold for a positive test may be set lower for surveillance than in evaluation populations as recurrent bladder tumors tend to be smaller than primary tumors (6), which may result in a lower cancer PBC value. For example, using a lower cutoff for bladder cancer surveillance than evaluation was found to improve sensitivity of the NMP22 test (36). Other efforts that may improve the accuracy of bladder cancer diagnostics include integration of the urine tests with the clinical characteristic (37-39). For example, Kavalieris el al. developed an integrated model consisting of both Cxbladder gene expression urine test and patient characteristic variables such as gender, age, smoking history and frequency of gross hematuria for use to triage patients for hematuria workup but with a low probability of bladder cancer (40).
To further evaluate the 3-marker panel in the future, a prospective, multicenter study is required. A broader study will further allow us to assess assay performance under a range of urologic conditions. It will be especially valuable to evaluate the 3-marker panel in patients undergoing BCG where the performance of urine cytology is poor due to an increase of inflammatory cells in urine (6,41). Our approach of selecting against markers of inflammation suggests our 3-marker may be useful for assessing patient response to BCG treatment. Further, as subjects were selected retrospectively for the current study, valid bladder cancer prevalence estimates could not be obtained. A prospective study is necessary to allow us to calculate negative and positive predictive values of the test and set a PBC cutoff to maximize the negative predictive value, which may be useful for reducing the need for cystoscopy. With a larger sample size, we can also assess whether supplementing our gene expression model with a phenotypic model of risk stratification provides an improved resource for clinical decisions, particularly for patients with scores near the PBC threshold (42). Further interrogation of our RNA-seq dataset may yield insights into bladder cancer biology, identify rare splice variants and other RNA targets (e.g. lncRNA) that were enriched through our sample preparation strategy. Lastly, RNA-seq of urinary RNA could be employed to discover urinary biomarkers for differentiation of HG and LG bladder cancer, detection other urinary tract diseases, and evaluating response to treatments.
Conclusions
Using RNA-seq as a discovery tool, we have demonstrated the feasibility of obtaining high quality sequencing data from urine sediments for RNA expression profiling. Through qPCR evaluation and multiple logistic analysis, we generated an equation to predict bladder cancer probability based on the urinary expression of ROBO1, WNT5A and CDC42BPB. The overall sensitivity for both the HG and LG samples was superior to urine cytology. A prospective multicenter clinical study should be conducted to further validate the 3 marker signature for detection, surveillance, and post-BCG populations.
Supplementary Material
Translational Relevance.
While bladder cancer can be managed by transurethral resection in most patients, the high recurrence rate necessitates lifetime cystoscopic surveillance. The need for frequent invasive surveillance contributes to the high cost of treatment for bladder cancer. A urine test with sufficient accuracy to prioritize high-risk patients to undergo timely cystoscopy and reduce procedural frequency for low-risk patients has remained elusive. This study focused on identifying bladder cancer-specific urinary mRNA markers by sequencing RNA extracted from urine sediment using RNA-seq. A model based on gene expression of a 3-marker panel was constructed. Validation of the model demonstrated a similar sensitivity for high grade and an improved sensitivity for low grade cancer compared to other urine tests suggesting the high translational potential of our panel as majority of bladder cancer patients present with low grade disease. Moreover, serial testing with our panel following 6 patients was consistent with cystoscopic and pathologic results indicating our urine test may serve as a complement to cystoscopy.
Acknowledgments
The authors thank Daniel Bui, Aristeo Lopez, Ruchika Mohan, Thomas Metzner for assistance with patient recruitment and sample collection, Norma Neff and Gary Mantalas from Stanford Stem Cell Institute Genome Center (SCIGC) for their technical support on conducting RNA-seq experiments, and Jens-Peter Volkmer and Irving Weissman for valuable suggestions on study design.
Grant Support: J.C.L. acknowledges the support of Stanford University Department of Urology and NIH R01 CA160986. D.S. acknowledges the support of NIH R00 CA151673, Siebel Foundation, Department of Defense W81XWH-10-1-0500 and Bladder Cancer Advocacy Network 2013 Young Investigator Award. R.S. acknowledges the support of Stinehart/Reed Awards and the Ludwig Center at Stanford.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Aldousari S, Kassouf W. Update on the management of non-muscle invasive bladder cancer. Canadian Urological Association journal = Journal de l’Association des urologues du Canada. 2010;4(1):56–64. doi: 10.5489/cuaj.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fantony JJ, Inman BA. It May Be Time to Abandon Urine Tests for Bladder Cancer. J Natl Compr Canc Ne. 2015;13(9):1163–6. doi: 10.6004/jnccn.2015.0141. [DOI] [PubMed] [Google Scholar]
- 4.Cheung G, Sahai A, Billia M, Dasgupta P, Khan MS. Recent advances in the diagnosis and treatment of bladder cancer. BMC medicine. 2013;11:13. doi: 10.1186/1741-7015-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breen V, Kasabov N, Kamat AM, Jacobson E, Suttie JM, O’Sullivan PJ, et al. A holistic comparative analysis of diagnostic tests for urothelial carcinoma: a study of Cxbladder Detect, UroVysion(R) FISH, NMP22(R) and cytology based on imputation of multiple datasets. BMC medical research methodology. 2015;15:45. doi: 10.1186/s12874-015-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang SS, Boorjian SA, Chou R, Clark PE, Daneshmand S, Konety BR, et al. Diagnosis and Treatment of Non-Muscle Invasive Bladder Cancer: AUA/SUO Guideline. The Journal of urology. 2016 doi: 10.1016/j.juro.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 7.Mengual L, Ribal MJ, Lozano JJ, Ingelmo-Torres M, Burset M, Fernandez PL, et al. Validation study of a noninvasive urine test for diagnosis and prognosis assessment of bladder cancer: evidence for improved models. The Journal of urology. 2014;191(1):261–9. doi: 10.1016/j.juro.2013.06.083. [DOI] [PubMed] [Google Scholar]
- 8.O’Sullivan P, Sharples K, Dalphin M, Davidson P, Gilling P, Cambridge L, et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. The Journal of urology. 2012;188(3):741–7. doi: 10.1016/j.juro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Holyoake A, O’Sullivan P, Pollock R, Best T, Watanabe J, Kajita Y, et al. Development of a multiplex RNA urine test for the detection and stratification of transitional cell carcinoma of the bladder. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(3):742–9. doi: 10.1158/1078-0432.CCR-07-1672. [DOI] [PubMed] [Google Scholar]
- 10.Mengual L, Burset M, Ribal MJ, Ars E, Marin-Aguilera M, Fernandez M, et al. Gene expression signature in urine for diagnosing and assessing aggressiveness of bladder urothelial carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(9):2624–33. doi: 10.1158/1078-0432.CCR-09-3373. [DOI] [PubMed] [Google Scholar]
- 11.Urquidi V, Netherton M, Gomes-Giacoia E, Serie D, Eckel-Passow J, Rosser CJ, et al. Urinary mRNA biomarker panel for the detection of urothelial carcinoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribal MJ, Mengual L, Lozano JJ, Ingelmo-Torres M, Palou J, Rodriguez-Faba O, et al. Gene expression test for the non-invasive diagnosis of bladder cancer: A prospective, blinded, international and multicenter validation study. European journal of cancer. 2016;54:131–8. doi: 10.1016/j.ejca.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Street JM, Yuen PS, Star RA. Bioactive exosomes: possibilities for diagnosis and management of bladder cancer. The Journal of urology. 2014;192(2):297–8. doi: 10.1016/j.juro.2014.05.050. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PloS one. 2012;7(7):e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Obuchowski N, McClish D. Statistical Methods in Diagnostic Medicine. Wiley; 2002. p. 592. [Google Scholar]
- 17.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45. [PubMed] [Google Scholar]
- 18.Yeung C, Dinh T, Lee J. The health economics of bladder cancer: an updated review of the published literature. PharmacoEconomics. 2014;32(11):1093–104. doi: 10.1007/s40273-014-0194-2. [DOI] [PubMed] [Google Scholar]
- 19.Shyr D, Liu Q. Next generation sequencing in cancer research and clinical application. Biological procedures online. 2013;15(1):4. doi: 10.1186/1480-9222-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Nykter M, Chen K. Next generation sequencing applications in cancer research. Cancer letters. 2013;340(2):149–50. doi: 10.1016/j.canlet.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Petric RC, Pop LA, Jurj A, Raduly L, Dumitrascu D, Dragos N, et al. Next generation sequencing applications for breast cancer research. Clujul medical. 2015;88(3):278–87. doi: 10.15386/cjmed-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yli-Hietanen J, Ylipaa A, Yli-Harja O. Cancer research in the era of next-generation sequencing and big data calls for intelligent modeling. Chinese journal of cancer. 2015;34(10):423–6. doi: 10.1186/s40880-015-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YT, Chen HW, Domanski D, Smith DS, Liang KH, Wu CC, et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. Journal of proteomics. 2012;75(12):3529–45. doi: 10.1016/j.jprot.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 24.Turecky L, Kalina P, Uhlikova E, Namerova S, Krizko J. Serum Ceruloplasmin and Copper Levels in Patients with Primary Brain-Tumors. Klin Wochenschr. 1984;62(4):187–9. doi: 10.1007/Bf01731643. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Cheng H, Xu W, Tian X, Li X, Zhu C. Expression of Robo protein in bladder cancer tissues and its effect on the growth of cancer cells by blocking Robo protein. International journal of clinical and experimental pathology. 2015;8(9):9932–40. [PMC free article] [PubMed] [Google Scholar]
- 26.Legg JA, Herbert JM, Clissold P, Bicknell R. Slits and Roundabouts in cancer, tumour angiogenesis and endothelial cell migration. Angiogenesis. 2008;11(1):13–21. doi: 10.1007/s10456-008-9100-x. [DOI] [PubMed] [Google Scholar]
- 27.Kumawat K, Gosens R. WNT-5A: signaling and functions in health and disease. Cell Mol Life Sci. 2016;73(3):567–87. doi: 10.1007/s00018-015-2076-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malgor R, Crouser S, Greco D, Brockett C, Coschigano K, Nakazawa M, et al. Correlation of Wnt5a expression with histopathological grade/stage in urothelial carcinoma of the bladder. Diagnostic pathology. 2013;8:139. doi: 10.1186/1746-1596-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endo M, Nishita M, Fujii M, Minami Y. Insight into the role of Wnt5a-induced signaling in normal and cancer cells. International review of cell and molecular biology. 2015;314:117–48. doi: 10.1016/bs.ircmb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Babjuk M, Burger M, Zigeuner R, Shariat SF, van Rhijn BWG, Comperat E, et al. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2013. European urology. 2013;64(4):639–53. doi: 10.1016/j.eururo.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Chou R, Gore JL, Buckley D, Fu R, Gustafson K, Griffin JC, et al. Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Annals of internal medicine. 2015;163(12):922–31. doi: 10.7326/M15-0997. [DOI] [PubMed] [Google Scholar]
- 32.Vriesema JL, Atsma F, Kiemeney LA, Peelen WP, Witjes JA, Schalken JA. Diagnostic efficacy of the ImmunoCyt test to detect superficial bladder cancer recurrence. Urology. 2001;58(3):367–71. doi: 10.1016/s0090-4295(01)01217-1. [DOI] [PubMed] [Google Scholar]
- 33.Vrooman OP, Witjes JA. Urinary markers in bladder cancer. European urology. 2008;53(5):909–16. doi: 10.1016/j.eururo.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Todenhofer T, Hennenlotter J, Kuhs U, Tews V, Gakis G, Aufderklamm S, et al. Influence of urinary tract instrumentation and inflammation on the performance of urine markers for the detection of bladder cancer. Urology. 2012;79(3):620–4. doi: 10.1016/j.urology.2011.10.067. [DOI] [PubMed] [Google Scholar]
- 35.van Rhijn BWG, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: A systematic review. European urology. 2005;47(6):736–48. doi: 10.1016/j.eururo.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Boman H, Hedelin H, Holmang S. Four bladder tumor markers have a disappointingly low sensitivity for small size and low grade recurrence. The Journal of urology. 2002;167(1):80–3. [PubMed] [Google Scholar]
- 37.Lotan Y, Svatek RS, Krabbe LM, Xylinas E, Klatte T, Shariat SF. Prospective external validation of a bladder cancer detection model. The Journal of urology. 2014;192(5):1343–8. doi: 10.1016/j.juro.2014.05.087. [DOI] [PubMed] [Google Scholar]
- 38.Ajili F, Darouiche A, Chebil M, Boubaker S. The impact of age and clinical factors in non-muscle-invasive bladder cancer treated with Bacillus Calmette Guerin therapy. Ultrastructural pathology. 2013;37(3):191–5. doi: 10.3109/01913123.2013.770110. [DOI] [PubMed] [Google Scholar]
- 39.Lotan Y, Capitanio U, Shariat SF, Hutterer GC, Karakiewicz PI. Impact of clinical factors, including a point-of-care nuclear matrix protein-22 assay and cytology, on bladder cancer detection. BJU international. 2009;103(10):1368–74. doi: 10.1111/j.1464-410X.2009.08360.x. [DOI] [PubMed] [Google Scholar]
- 40.Kavalieris L, O’Sullivan PJ, Suttie JM, Pownall BK, Gilling PJ, Chemasle C, et al. A segregation index combining phenotypic (clinical characteristics) and genotypic (gene expression) biomarkers from a urine sample to triage out patients presenting with hematuria who have a low probability of urothelial carcinoma. BMC urology. 2015;15:23. doi: 10.1186/s12894-015-0018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Beltran A, Luque RJ, Mazzucchelli R, Scarpelli M, Montironi R. Changes produced in the urothelium by traditional and newer therapeutic procedures for bladder cancer. Journal of clinical pathology. 2002;55(9):641–7. doi: 10.1136/jcp.55.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotan Y, Shariat SF, Schmitz-Drager BJ, Sanchez-Carbayo M, Jankevicius F, Racioppi M, et al. Considerations on implementing diagnostic markers into clinical decision making in bladder cancer. Urol Oncol-Semin Ori. 2010;28(4):441–8. doi: 10.1016/j.urolonc.2009.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


