Abstract
Many bacteria contain primitive organelles composed entirely of protein. These bacterial microcompartments share a common architecture of an enzymatic core encapsulated in a selectively permeable protein shell; prominent examples include the carboxysome for CO2 fixation and catabolic microcompartments found in many pathogenic microbes. The shell sequesters enzymatic reactions from the cytosol, analogous to the lipid-based membrane of eukaryotic organelles. Despite available structural information for single building blocks, the principles of shell assembly have remained elusive. We present the crystal structure of an intact shell from Haliangium ochraceum, revealing the basic principles of bacterial microcompartment shell construction. Given the conservation among shell proteins of all bacterial microcompartments, these principles apply to functionally diverse organelles and can inform the design and engineering of shells with new functionalities.
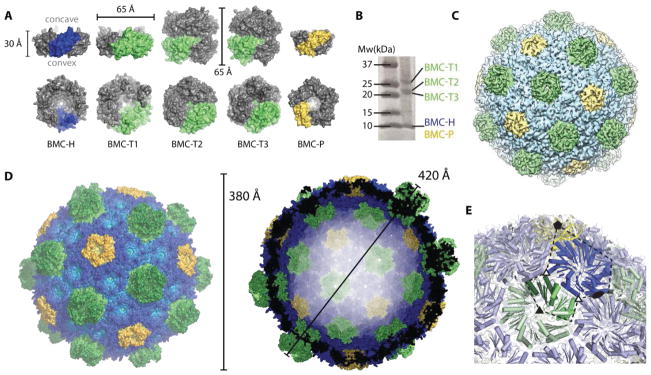
Bacterial microcompartments (BMCs) are large, proteinaceous shells encapsulating enzymes. The first discovered, carboxysomes, enhance carbon fixation (1). The BMC shell is a singular example of a primitive, conserved yet functionally diverse bioarchitecture. Recent bioinformatic surveys of bacterial genomes have revealed the presence of genes encoding shell proteins in 23 different bacterial phyla, encapsulating segments of functionally diverse metabolic pathways (2). The major components of BMC shells are cyclic hexamers with a pronounced concave-versus-convex sidedness (3). These proteins, referred to as BMC-H, contain a single BMC (pfam00936) domain (Fig. 1A, blue). A derivative of BMC-H proteins, BMC-T, is a fusion of two BMC domains forming trimers or pseudohexamers (Fig. 1A, green). Some members of the BMC-T family are known to form tightly appressed, stacked dimers of trimers, containing a central cavity (4, 5) (Fig. 1A, BMC-T2 and BMC-T3). BMC-P proteins belong to pfam03319; they are structurally unrelated to the BMC/pfam00936 domain and form pentamers shaped like a truncated pyramid (6) (Fig. 1A, yellow). Despite detailed structural knowledge of the individual shell components, the architectural principles governing shell self-assembly are unknown.
Fig. 1.
Overview of the components and the overall structure of the BMC shell. (A) Surface representation and dimensions of a side view (top row) and of the concave face (bottom row) of the structures of hexameric BMC-H (blue), trimeric BMC-T (lime) and pentameric BMC-P proteins (yellow) that constitute the shell. The BMC-T2 and BMC-T3 proteins each consist of two closely appressed pseudohexamers. The BMC-P structure was extracted from the whole shell structure, BMC-H and BMC-T1 from previously determined crystal structures (PDB IDs 5DJB and 5DIH, respectively) and BMC-T2 and BMC-T3 are crystal structures determined in this study. (B) SDS-PAGE of purified H. ochraceum BMC shells. (C) Overview of the 8.7 Å cryo electron microscopy structure colored by shell protein. (D) Surface representation of the crystal structure with a color gradient by distance from center (light to dark from inside to outside) (left) and cross-section through the center (right). (E) Close-up of the icosahedral asymmetric unit (dashed line) with symmetry axes indicated with solid symbols, pseudo threefold symmetry with open triangles. Only one stack shown for the BMC-T.
Using a recombinant system containing all of the facet proteins (one BMC-H and three BMC-T paralogs) and one of the three BMC-P proteins of the myxobacterium Haliangium ochraceum BMC (Fig. 1, A and B) (7), we produced homogeneous 40 nm BMC shells with a molecular mass of 6.5 MDa. We have crystallized a complete closed particle and determined its structure to a resolution of 3.5 Å (CC1/2 of 26%, Table S1). A cryo-EM map at a resolution of 8.7 Å (Fig. 1C) was used to place individual structures and phase the crystallographic data. To facilitate the interpretation of our data, we also determined the crystal structures of the pseudohexameric BMC-T2 and BMC-T3 proteins (Table S1).
The co-expressed shell proteins self-assemble into a pseudo T=9 icosahedral shell (designated pseudo because not all subunits are identical), with a diameter of about 400 Å (Fig. 1D). The shell consists of 12 BMC-P pentamers at the vertices; the facets are formed by 60 BMC-H hexamers enclosing 20 BMC-T pseudohexamers of the three different paralogous types (Fig. 1A, green). This stoichiometry is in agreement with what we observe for purified shells on SDS-PAGE (Fig. 1B) and previous analyses (7).
The icosahedral asymmetric unit consists of one BMC-P chain, six BMC-H chains and one BMC-T chain (two chains for the double stacking type) (Fig. 1E). Model building was facilitated by the available high resolution structures of the hexamer (8), the pseudohexamers ((9) and this work) and the 30-fold non-crystallographic symmetry, which collectively resulted in good model fit and geometry (for sample electron density see Fig. S1A). Because three different proteins can occupy the BMC-T positions, this density is representative of a mixture. Due to the structural similarity between all three BMC-T, we can confidently place a protein model (we chose BMC-T2 based on overall fit). The resulting shell facets consist of a single layer with a thickness of 20–30 Å with one of the trimers of BMC-T2 and BMC-T3 protruding to the outside (Fig. 1D). The complete shell structure answers the fundamental questions of the shell being single or double layered, how stacked pseudohexamers are accomodated, as well as the orientations of the individual subunits. For the pentamers, the broader side (the base of the pyramid), faces outward. In the facets, the concave sides of BMC-H and BMC-T1 (pseudo) hexamers (containing the N- and C-termini) face outwards. Likewise, the lower trimers of the double stacking BMC-T2 and BMC-T3 pseudohexamers are in the same (concave out) orientation but due to a circular permutation, their N- and C-termini face the inside. Given that it is the interface with cytosolic metabolism, knowledge of the location of the polypeptide termini and the sidedness of the shell proteins is crucial for understanding and manipulating the function of BMCs in their native context, as well as for engineering synthetic microcompartments.
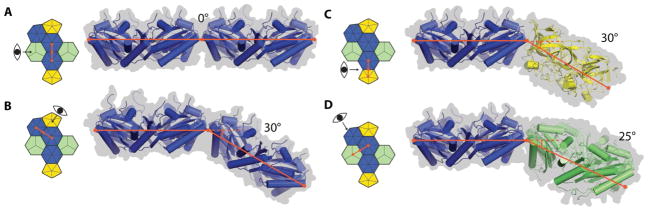
There are four distinct interfaces in the intact shell (Fig. 2): two different hexamer-hexamer interactions (Fig. 2, A and B), the hexamer-pentamer (Fig. 2C) and the hexamer-pseudohexamer interaction (Fig. 2D). The hexamers connecting pentamers between two vertices of the intact shell (Fig. 2A) are in a side-by-side, planar orientation, while the hexamers surrounding the pentamers (Fig. 2B) are tilted by 30°. Considering the high structural conservation among hexamer and pentamer proteins (Fig. S2, A and B) those orientations are likely universal among BMCs. The hexamer in the shell is slightly compressed on the edge adjoining the pentamer, this is apparent by superimposing it on the structure of the hexamer determined in isolation (8) (Fig. S2C and Fig. 1D, where the edge facing the pentamer bulges outwards (darker color)). This distortion illustrates why computationally modelling such a large, multi-protein complex based on individual crystal structures would likely fail to result in an accurate model.
Fig. 2.
Overview of the four distinct interfaces between the pentamer, hexamers and the pseudohexamers. Structures are shown in cartoon view (surface view as grey background) with a pictogram showing their location on the shell. (A) Coplanar hexamer-hexamer interface connecting two pentamer vertices. (B) Hexamer-hexamer interface as observed surrounding the pentamer. (C) Hexamer-pentamer interface. (D) Hexamer-pseudohexamer interface.
Structurally, the pseudohexameric BMC-T proteins are slightly more compact than the BMC-H hexamers, with the BMC domains folded relatively inward on the concave side (Fig. S2D). Placing hexamers in these positions would require significant deformation to enable them to be accommodated. BMC-T pseudohexamers contain two copies of the BMC domain, and in our structure one domain interacts with the coplanar hexamer-hexamer corner and the other with the corner where the two hexamers join at a 30° angle (Fig. 1E). Because the two domains are decoupled on a genetic level, their primary structures have evolved separately so that each domain can fulfil distinct interface roles. Indeed, all characterized BMCs contain at least one BMC-T type protein; in almost all genomes encoding BMCs, including those of unknown function, a gene for a BMC-T protein is present (2), underscoring their structural importance.
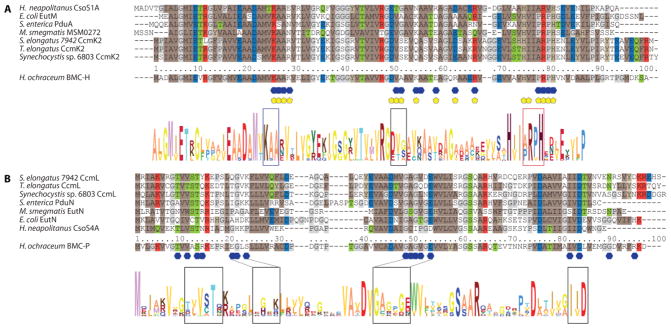
The specific residues involved in the interactions among hexamers and pentamers are located in distinct conserved patches distributed across the primary structure (Fig. 3A). Highly conserved pentamer residues that are involved in intersubunit interactions (Fig. 3A, 4A, Fig. S3) include S13, the GAGxGE motif (48–53) and the I(V/I)D motif (81–83). On the hexamer, the KAA motif at position 25–27 as well as the PRPH motif at position 77–80 play central roles for forming the interface with the pentamer (Fig. 3B, 4A, Fig. S4). Hexamer residues 49–51 (D/E-T/V-A/G/S) are located at the corner between the pentamer and two hexamers, the conservation of a small amino acid at position 51 is crucial; large residues there would likely preclude shell formation. Overall, shape complementarity governs the hexamer-pentamer interactions there are few salt bridges and hydrogen bonds.
Fig. 3.
Sequence alignment of BMC-H and BMC-P of representative species Sequence alignment of representative BMC-H (A) and BMC-P (B) (selected to correspond to characterized, functionally diverse BMCs and with available crystal structures for the isolated subunits) with numbering adjusted to correspond to the H. ochraceum sequences. Interfacing residues are marked by yellow pentagons for pentamer interactions and blue hexagons for hexamer interactions. Coloring of conserved residues according to physical properties (brown: hydrophobic, grey: proline/glycine, red: positively charged, blue: negatively charged, green: polar). Sequence conservation logos of the combined representative types below, with each amino acid colored individually; height of letters corresponds to relative frequency at each position. Additional detail for each type shown in Fig. S3 and Fig. S4.
Fig. 4.
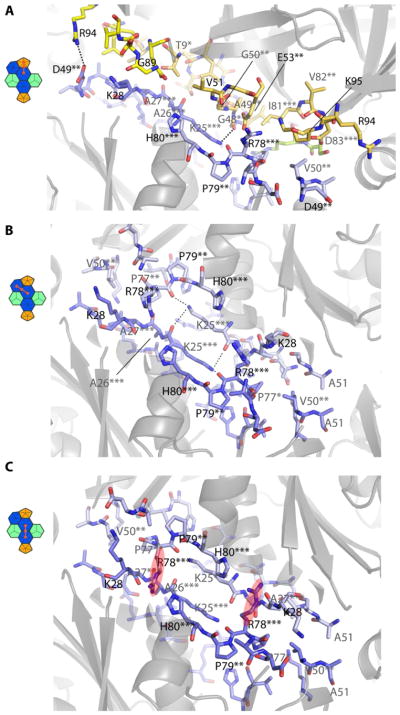
Detailed view of the BMC-H-BMC-P and the two different BMC-H interfaces viewed from the outside. (A) Pentamer-hexamer interface, pentamer in yellow, hexamer residues in blue with conservation indicated with asterisk(s), different chains indicated by color shading, dashed lines indicate hydrogen bonds. (B) Angled hexamer-hexamer interface. (C) Coplanar hexamer-hexamer interface.
For the hexamer-hexamer interface (Fig. 4B), the KAA and PRPH motifs of complementing chains account for most of the interacting surface area. The lysines of the KAA motif are arranged in an antiparallel manner, creating a flat interaction surface with hydrogen bonds between the ε-amino group and the backbone oxygens of the opposite lysine and R78 (Fig. 4B). The coplanar hexamer-hexamer interface maintains the KAA-PRPH motif interactions, but contains an additional structural interdigitation between hexamers: the R78 side chain of the PRPH motif inserts in a pocket between the H80 side chain and backbone oxygens of V24, A27 and V29 of the adjacent hexamer (Fig. 4C, Fig. S1B), creating an interlock. This was previously observed as a crystal packing interaction in the structure of the α-carboxysomal BMC-H protein CsoS1A (10), additional indication of the general structural conservation of the interactions across evolutionarily distant shell proteins (Fig. S4).
The specific sidechains influencing the interaction between the BMC-H hexamer and the BMC-T pseudohexamers are more enigmatic. The ability of three different BMC-T proteins to occupy the same position in the shell indicates a tolerance for a variety of sidechain interactions. The only universally conserved residue is the antiparallel lysine corresponding to the KAA motif in hexamers (Fig. S5 and Fig. S6). Notably, all three BMC-Ts are able to occupy equivalent positions in the shell despite significant sequence divergence, suggesting that in the BMC-H:BMC-T interfaces, the specific interactions mediating assembly are based primarily on shape complementarity.
The surface view of the intact shell (Fig. 1D and Fig. 2) shows it is tightly packed; the only conduits to the interior of the shell are the pores formed at the cyclic symmetry axes of the hexamers and pseudohexamers. The largest channel to the interior is formed by the BMC-T proteins; the pore across the trimer within the facet is at least 5 Å wide with the potential to be larger due to flexibility of the loops surrounding the pore. The crystal structure of isolated BMC-T3 has both trimer pores closed while in the crystal structure of the isolated BMC-T2, one pore is open and the other closed, as has been observed before for carboxysome proteins (4), reminiscent of the alternate access model of some transmembrane transporters of eukaryotic organelles (e.g. BtuCD type ABC transporters (11)).
Using the interactions we see in our structure and the same set of hexamers, pseudohexamers and pentamers, we are able to model larger compartments (T=36, diameter 720 Å) than we have experimentally observed by only slightly changing the angles between hexamers and pseudohexamers while maintaining the coplanar hexamer-hexamer contacts (Fig. S7). The extent of the facets is likely dictated by the interactions between different combinations of distinct BMC domains (i.e. the two different domains in each BMC-T paralog and the BMC-H) while the pentamer could prime the structure for an overall icosahedral shape. The subunits in the BMC-T positions thereby influence the curvature and the final size of the compartment. This differs from previous hypothetical models which proposed specific proteins in forming edges (12, 13). Although the particles appear to have edges in some views and in micrographs (Fig. S7B, S9A), the curvature is distributed over the whole shell; larger BMCs effectively have less curvature per subunit. Accordingly, the structure we have determined describes scalable principles for constructing a range of shell sizes, likely corresponding to the variation in shell sizes observed in BMCs in their native hosts, which range from 55–600 nm (14, 15).
The presence of structurally redundant building blocks suggests that the multiplicity is related to function, not structure, for example, to provide a range of conduits (i.e. differing in size and charge at the cyclic symmetry axes) for different metabolites (substrates and products) to cross the same shell. A second function would be to provide distinct patches on the interior surface to anchor and spatially organize the encapsulated enzymes. When we model the shell with the different BMC-Ts, an electrostatic (inside) surface view shows different regions that could be involved in specific interactions with the cargo proteins (Fig. S8). The distinct convex binding surfaces of the different shell proteins could serve to position the encapsulated enzymes to channel substrates and products between enzymes as well as across the shell.
Our model of the basic architecture of the bacterial micrcompartment shell likely applies to functionally diverse organelles found across the Bacterial Kingdom; it also can inform rational design of engineered microcompartments. For the BMC shell described here, based on an inner diameter of 290 Å and assuming a typical protein density, there is space for approximately 150 copies of a 60 kDa enzyme in the interior, ample volume in which to localize multiple enzymes. Targeting could be achieved by either using specific encapsulation peptides found associated with the native cargo proteins (7, 16) or engineered using the structure of the inner surface as guide. The overall structure of the BMC shell invites comparisons to viral capsids and their engineered functions. BMC shells offer an additional structural and functional feature, selective permeability. Collectively, the atomic resolution model of a BMC shell reveals the construction principles of the membranes of these primitive, protein-based organelles that can be applied to understanding and manipulating their native and engineered functions.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975-01 and the U.S. Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. B.G was supported by an advanced postdoc mobility fellowship from the Swiss National Science Foundation (project P300PA_160983). Use of the Stanford Synchrotron Radiation Light source, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. We thank Bradley Paasch and Jan Zarzycki for their contributions to the BMC-T3 structure determination. We thank Eva Nogales for providing access to the electron microscopy facility at UC Berkeley and the Adams lab at Lawrence Berkeley National Lab for use of the crystallization robot. The X-ray crystallographic coordinates and structure factor files have been deposited in the Protein Data Bank (PDB) under the following accession numbers: 5V74 (complete shell), 5V75 (BMC-T2) and 5V76 (BMC-T3).
Footnotes
References and Notes
- 1.Shively JM, Ball F, Brown DH, Saunders RE. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 2.Axen SD, Erbilgin O, Kerfeld CA. PLoS Comput Biol. 2014;10:e1003898. doi: 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerfeld CA, et al. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 4.Klein MG, et al. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 5.Cai F, et al. J Biol Chem. 2013;288:16055–16063. doi: 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka S, et al. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 7.Lassila JK, et al. J Mol Biol. 2014 [Google Scholar]
- 8.Sutter M, et al. Nano Letters. 2016;16:1590–1595. doi: 10.1021/acs.nanolett.5b04259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aussignargues C, et al. J Am Chem Soc. 2016;138:5262–5270. doi: 10.1021/jacs.5b11734. [DOI] [PubMed] [Google Scholar]
- 10.Tsai Y, et al. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locher KP, Lee AT, Rees DC. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka S, Sawaya MR, Yeates TO. Science. 2010;327:81–84. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- 13.Mallette E, Kimber MS. J Biol Chem. 2017;292:1197–1210. doi: 10.1074/jbc.M116.754093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erbilgin O, McDonald KL, Kerfeld CA. Appl Environ Microbiol. 2014;80:2193–2205. doi: 10.1128/AEM.03887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberton M, Austin JR, 2nd, Berg RH, Pakrasi HB. Plant Physiol. 2011;155:1656–1666. doi: 10.1104/pp.110.165332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aussignargues C, et al. Commun Integr Biol. 2015;8:e1039755. doi: 10.1080/19420889.2015.1039755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess RR. Methods Enzymol. 2009;463:259–282. doi: 10.1016/S0076-6879(09)63017-2. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch W. Acta Cryst D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winn MD, et al. Acta Cryst D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Afonine PV, et al. Acta Cryst D. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suloway C, et al. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Lander GC, et al. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohou A, Grigorieff N. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SHW. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voss NR, et al. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto K, et al. Sci Rep. 2016;6:33170. doi: 10.1038/srep33170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres SHW, Chen S. Nat Meth. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenthal PB, Henderson R. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen EF, et al. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Larkin MA, et al. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Guindon S, et al. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 33.Han MV, Zmasek CM. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.