Abstract
Current genotyping methodologies for transplantation and transfusion management employ multiplex systems that allow for the simultaneous detection of multiple human leukocyte antigens (HLA), human platelet antigens (HPA) and red blood cell (RBC) antigens. The development of high resolution molecular HLA typing has led to improved outcomes of unrelated hematopoietic stem cell transplants by better identifying suitable donors typed at the allele level for HLA-A, B, C, DRB1 and DQB1 antigens. In solid organ transplantation, the combination of high resolution HLA typing along with solid-phase antibody identification and the calculated PRA have shown to be of specific benefit to highly sensitized patients, and have resulted in significant reductions of incompatible crossmatches at the time of organ allocation. This database-driven combined HLA antigen/antibody testing has promoted the routine implementation of the virtual crossmatch, in which an electronic crossmatch is performed, and perhaps even obviates the need for a physical crossmatch. Additionally, DNA-based testing for RBC antigens provides as an alternative typing method that mitigates many of the limitations of hemagglutination-based phenotyping. Although there are many applications of RBC genotyping in various transfusion settings, it has arguably been most useful in the management of transfusion-dependent patients with sickle cell disease (SCD) and thalassemia to minimize alloimmunization. The availability of high-throughput RBC genotyping for both patients and large populations of donors, along with coordinated informatics systems to link patients’ antigen needs with available antigen-negative and/or rare blood-typed donors, offer promise toward improving the efficiency, reliability, and extent of RBC matching for this population.
Genotyping applications for hematopoietic stem cell and solid organ transplantation
Introduction
In the era of modern molecular pathology, patients can have their entire HLA genome sequenced from a single swipe of buccal mucosa or an individual hair root. This technological feat represents a huge advance for a field that emerged in the 1950s, when the first successful live donor human kidney transplant was performed between identical twin brothers.1 Another major milestone occurred in 1957 when a patient with leukemia was successfully treated with radiotherapy and subsequent bone marrow transplant from an identical twin.2 While these identical twin transplants were successful and represented a paradigm shift in the field of transplantation, transplant procedures between non-identical donor-recipient pairs proved more problematic for reasons that were not entirely understood at the time.3,4 The breakthrough in unlocking this mystery came about in 1958 with Jean Dausset’s discovery of the human major histocompatibility complex (henceforth referred to as the HLA complex), which shed much needed light on transplant biology. This key discovery was paramount and ultimately the concept of immunologic donor-recipient compatibility (i.e., histocompatibility) was born.5
Following the work of Dausset, the clinical utility of HLA histocompatibility testing was solidified in 1969 by the seminal observations by Patel and Teraski. They showed that, similar to the use of routine RBC crossmatches to detect recipient antibodies that could cause post-transfusion hemolysis, a positive pre-transplant crossmatch using recipient serum and donor lymphocytes frequently predicted hyperacute renal allograft rejection.6 The white blood cell (WBC) antibodies detected by this test were later confirmed to be HLA-specific. This study concluded that proceeding to kidney transplantation without performing a pre-transplant HLA crossmatch was malpractice.6 To this day, one of the main functions of an HLA laboratory supporting solid organ transplant programs is to identify recipient HLA antibodies that can cause early allograft rejection. To meet this challenge, the laboratory is equipped with an array of new technologies. Beginning with a description of early HLA testing methods, this section will describe the evolution of histocompatibility testing and how these tests are currently utilized to improve patient outcomes.
In the Beginning
Before DNA-based HLA typing and solid-phase antibody detection methodology became the standard of practice, there was serology. Serological testing was critical for discovery of HLA antibodies and the antigens to which they reacted. Reactivity between unknown antisera and cells of known phenotype or between well characterized antisera and cells of unknown phenotype were assessed in early crossmatch assays to determine the specificities of sera and their cognate antigens. A novel antigen would be defined when antisera exhibited a new pattern of reactivity not previously characterized. For example, given serum #1 that reacted with five distinct cells, and serum #2 that reacted with five different cells, a new antigen would be defined when a third serum was found that reacted with a subset of cells from each of the above two groups. In time, antisera were identified that would react with only a subset of cells expressing a previously described specific HLA antigen. This selective reactivity divided the previously known HLA antigen into to new antigen subgroups, or “splits” of the original “parent” antigen.7 Similarly, investigators would identify sera reactive with multiple cells each expressing a previously established, but unique, HLA antigen. For instance, five cells containing distinct HLA antigens (eg, B7, B13, B42, B55, B81) might nonetheless react with a single antiserum because these antigens share a common amino acid sequence. Consequently, any cell containing the same sequence could be used to adsorb out an antibody that recognizes that sequence. So, if a B7 cell were used as the adsorber to remove the antibody, it would eliminate reactivity to the other cells as well (B13, B42, B55, B81). Recognition of such antibody cross reactivity with distinct antigens gave rise to the term cross-reactive groups (CREGs). The cross reactivity reflected shared or “public” epitopes present among different HLA antigens. Thus, to a great extent, early serology relied on the interdependent, and sometimes circular, relationship between antibodies and antigens, each necessary to characterize the other.
Early studies of HLA testing required ingenuity and creativity as neither HLA antigens nor their corresponding antibodies were well defined and the appearance of the aforementioned “splits” and CREGs only added to the confusion. Moreover, the serologic techniques used to assess reactivity, which included relatively crude testing such as agglutination and cytotoxic assays, were of limited value. First, both agglutination and cytotoxicity assays are reliant on cell viability, requiring careful collection, handling and processing. Second, interpretation of data obtained with these methods was subjective and relied heavily on the expertise and experience of the individual reading the reactions. Finally, these early serological methods suffered from both poor sensitivity resulting in false negative reactions, and poor specificity leading to false positive reactions. Clearly more sensitive and specific testing methods were needed.
Two Roads Diverge
Starting from the serological foundation of HLA testing described above, the journey to modern HLA laboratory testing took two distinct roads. One led to the elucidation of HLA antigens using techniques of molecular biology while the other illuminated HLA antibodies by further refinements of serologic methods. Importantly, the directions these two camps took evolved from the differing clinical needs of hematologic and solid organ transplantation. Bone marrow and stem cell transplantation depended on accurate, reliable antigen testing to identify HLA-matched recipient-donor pairs while solid organ transplantation (particularly renal transplantation) required identification of HLA antibodies, particularly donor specific antibodies (DSA), to prevent hyperacute and accelerated antibody-mediated rejection (AMR).8 Each camp travelled their own path to develop tests that addressed their patients’ needs.
HLA Antigen Typing: Hematopoietic Progenitor Cell (HPC) Transplants
Bone marrow transplants from non-HLA identical donors are associated with delayed engraftment, increased incidence of rejection, transplant related mortality, and other complications.9 HLA laboratories historically performed mixed lymphocyte cultures (MLC) and serological HLA typing to identify compatible donor/recipient pairs. This approach was a cumbersome, tedious, and time-consuming (e.g. 7 day turn-around). In order to overcome these challenges, high resolution molecular HLA typing was developed and eventually supplanted MLC testing.10
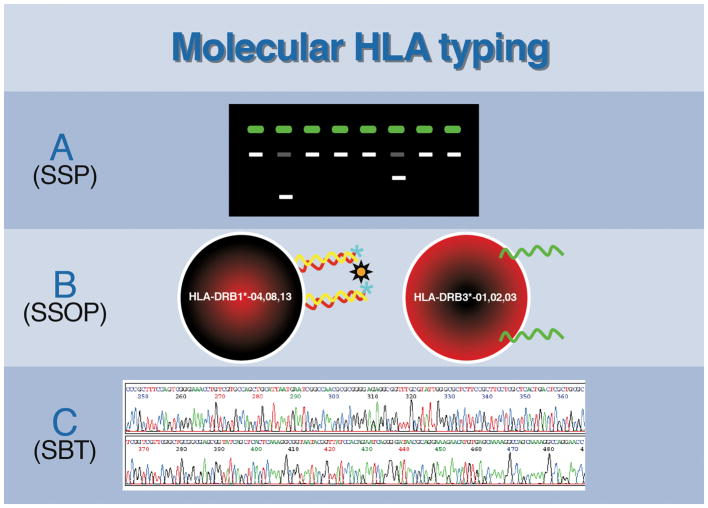
In the mid-1980s, molecular based HLA genotyping was first performed by hybridizing genomic restriction-enzyme fragments with radiolabeled HLA probes to identify restriction-fragment-length polymorphisms (RFLP) associated with specific HLA genotypes.11 Unfortunately, RFLP analysis was cumbersome, required the use of radioactive 32P probes, and took weeks to complete. Consequently, the RFLP method was not widely adapted in clinical HLA laboratories. By 1987, the discovery of the polymerase chain reaction (PCR) revolutionized molecular genotyping. Using PCR, sizeable quantities of target DNA could be amplified and analyzed.12 Moreover, PCR permitted high resolution HLA typing that could distinguish as little as a single nucleotide difference between two alleles, a feat not attainable with serologic testing. Since its inception, many different PCR-based assays have been developed to identify and characterize HLA antigens including: 1) sequence-specific priming (SSP), 2) sequence-specific oligonucleotide probing (SSOP), and 3) DNA sequence based typing (SBT) (Figure 1). Additionally, the development of real-time PCR (RT-PCR) with simultaneous amplification and quantification of amplicons has been extremely useful due to the rapidity in which results can be generated. However, this technique is still not routinely utilized for screening purposes as it is more costly compared to the aforementioned methods.
Figure 1.
The different methods utilized for molecular HLA typing include: A) Sequence Specific Priming (SSP) – PCR products amplified using sequence-specific primers are examined by agarose gel electrophoresis, which displays pattern of positive and negative reactions to determine presence or absence of HLA alleles. Each lane identifies a unique allele or set of alleles and the reaction pattern(s) is analyzed using HLA genotyping software. B) Sequence-specific oligonucleotide probing (SSOP) – PCR amplified products are added to specific oligonucleotide probes affixed to solid-phase matrices (e.g., a microparticle or microwell plate) and binding is assessed colorimetrically or via multiplex flow cytometry. C) Sequence-based testing (SBT) –nucleic acid sequencing of the HLA alleles are performed by Sanger or Next Generation (massively parallel) sequencing to determine the nucleotide sequence of HLA genotypes.
Sequence-Specific Priming
In 1991, SSP started gaining traction when it was employed to subtype HLA DRB1*01; this had not previously been possible using either serology or RFLP approaches.13 SSP makes use of sequence-specific primers to bind and amplify a target DNA sequence.14 If the target sequence is present, an amplicon will be generated, representing a positive reaction. In contrast, if the target sequence is absent, no amplicon is produced. The resulting PCR products are then examined via agarose gel electrophoresis, and the pattern of positive and negative reactions is used to establish the HLA alleles. Multiple PCR reactions can be targeted to specific alleles or groups of alleles. While SSP was found to be more sensitive, specific, and rapid than RFLP, SSP typing is expensive, labor intensive, and requires a large number of primers to achieve high resolution HLA typing.
Sequence-Specific Oligonucleotide Probes
Although a form of sequence-specific oligonucleotide probing (SSOP) had been used in the identification of HLA antigens as early as 1986,15 due to the complexity of the method several years elapsed before SSOP became routine in clinical HLA laboratories. The development of the reverse dot blot, which immobilizes probes as opposed to target DNA, allowed widespread adoption of SSOP. Modern day SSOP employs microarrays of specific oligonucleotide probes affixed to solid-phase matrices (e.g., a microparticle or microwell plate). A generic PCR reaction amplifies all of an individual’s HLA genes, and the resulting amplicons are added to the matrix where they bind to the complementary probes. Hybridization between amplicons and immobilized probes is typically assessed colorimetrically or via multiplex flow cytometry. Microbead array assays have predominated as the assay of choice, as they provide high throughput testing with less sample handling time, and can deliver high resolution typing of HLA Class I and Class II antigens.16 Similar to SSP, a large number of primers are required to obtain high resolution results.
Sequence-Based Typing
Sequence-based testing (SBT) entails nucleic acid sequencing of HLA alleles and is thus considered the “gold standard” for molecular HLA typing. Given the degree of allelic interrogation, SBT not only resolves discrepancies in HLA antigens typed by other methods, but also allows the discovery of novel HLA alleles. Methods as diverse as standard Sanger sequencing and next generation sequencing (NGS) can be used for SBT typing. Even though SBT has advantages over other methods, it is not without its pitfalls. It is an expensive and labor-intensive procedure with longer turn-around times than SSP or SSO. Further, SBT has ambiguities that can make it difficult to determine to which haplotype (maternal or paternal) a given base-pair should be assigned.
Regardless of the actual method employed, molecular HLA typing has been used to accurately and reproducibly assess both prospective HPC donors and recipients, consequently producing steadily better clinical outcomes. The advent of these testing approaches has allowed registries such as the National Marrow Donor Program and Bone Marrow Donors World Wide to accrue the HLA types of over 27 million volunteer donors, which provides a rich pool of potential, nonrelated HLA-matched donors for patients who require lifesaving allogeneic stem cell transplants. Potential donors and recipients are currently allele-level typed molecularly for HLA-A, B, C, DRB1 DQB1 (and more recently, HLA-DP) antigens with resulting improved transplant outcomes.17
Antibody Testing: Solid Organ Transplant
While ABO compatibility is considered the primary immunologic barrier to successful kidney and other solid-organ transplants, HLA compatibility (as opposed to HLA matching) is also critical. This is why recipients and donors are typically HLA-A, B, C, DRB1, and DQB1 typed in addition to being ABO typed. However, solid organ graft allocation is not based on donor: recipient HLA matching like their HPC counterparts. Instead, for solid organ transplantation, the detection of donor-specific antibodies (DSAs) is paramount; an analogy would be the typical RBC transfusion recipient, who receives RBC units compatible with their antibody profile rather than transfusions that are phenotypically-identical to their RBCs. The importance of HLA antibodies has been best studied in renal transplants, where recipient antibodies directed against mismatched donor HLA antigens are associated with antibody mediated rejection and/or graft loss.18 Moreover, numerous studies have also detailed the negative impact of HLA antibodies in other solid organ transplants including heart,19 lungs,20 pancreatic islets cells,21 and recently, liver.22 It was this clinical importance of HLA antibodies in solid organ transplantation that drove the evolution of testing methods from basic serology to modern solid-phase detection.
Serology
The Centers for Medicare & Medicaid Services (CMS) stipulates that an HLA crossmatch be performed before solid organ transplantation.23 Historically, this requirement was met with MLC assays in which recipient serum was tested against lymphocytes from prospective donors to detect either agglutination or cytotoxicity as signs of incompatibilities. As previously stated, these methods suffer from low sensitivity (i.e., the possibility of missing clinically relevant antibodies of low titer or low avidity), and low specificity, in that a positive crossmatch may be the result of non-HLA antibodies and/or nonspecific reactivities that are clinically irrelevant.8
MLC testing characteristics were subsequently improved first through the addition of anti-human globulin (AHG), which enhanced the detection of low-level HLA antibodies, and then later with the introduction of flow cytometric crossmatches (FCXM).24 Nonetheless, even with these advances these tests have limited specificity. Thus, a positive crossmatch may not necessarily be a contraindication to transplantation.8
Another commonly used serologic assay (still in place today) is the panel-reactive antibody (PRA) test. This assay is designed to assess the degree of HLA alloimmunization. Historically, cytotoxicity was assessed against a panel of HLA typed cells from healthy volunteer donors after incubation with recipient serum. Cell death signified a positive result, and the percent of positive reactions corresponded to the breadth of HLA sensitization. The cytotoxic PRA suffers from the same shortcomings as the MLC crossmatch. However, similar to the FCXM, sensitivity of the PRA was greatly improved with the introduction of the flow cytometric PRA.
Solid Phase
A significant step forward in HLA antibody testing occurred with the advent of solid-phase approaches that rely on a plastic matrix coated with purified clusters (phenotypes) of native Class I and class II HLA antigens or recombinant single class I or class II HLA antigens.25 As with HLA typing, the microbead platform has dominated antibody testing methodologies. For example, patient serum is added to microbeads coated with HLA antigens, and corresponding antibodies can be detected by either conventional flow cytometry or the newer LuminexTM technology. Current methodologies employ multiplex systems that allow for the simultaneous detection of multiple antibodies; some configurations contain 100 distinct microbead populations each expressing a unique HLA antigen.24,26,27 Because of this comprehensive HLA coverage, multiplex solid-phase methods have become the most common approach for antibody detection in HLA laboratories. The ability to identify individual antibodies among complex sera is particularly helpful in determining suitable donors for highly sensitized recipients.
Solid-phase technology has also led to the concept of a calculated PRA (cPRA). With the conventional PRA test, the assignment of PRA values varies depending on the antigen composition and distribution of the cell types used to compile a given panel. In contrast, the cPRA is based on antigen frequencies from a population database of more than 12,000 donors and evaluates the likelihood of compatibility between a sensitized recipient candidate against an entire population of previously typed donors.28 When an antibody is detected by solid-phase methodology, the corresponding antigen is listed as unacceptable within the United Network for Organ Sharing website. The cumulative frequency of all listed unacceptable antigens is then used to derive the cPRA. Clinically, the cPRA has shown to be of specific benefit to highly sensitized patients and predicts >90% of positive crossmatches that occur at the time of organ allocation.29 Similarly, this solid-phase methodology is utilized in the evaluation of patients demonstrating platelet transfusion refractoriness, by determining the presence and specificity of HLA class I antibodies which may affect the survival of transfused platelets. Two recent reviews have detailed practical approaches for the selection of platelet products for patients with alloimmune platelet transfusion refractoriness.30,31
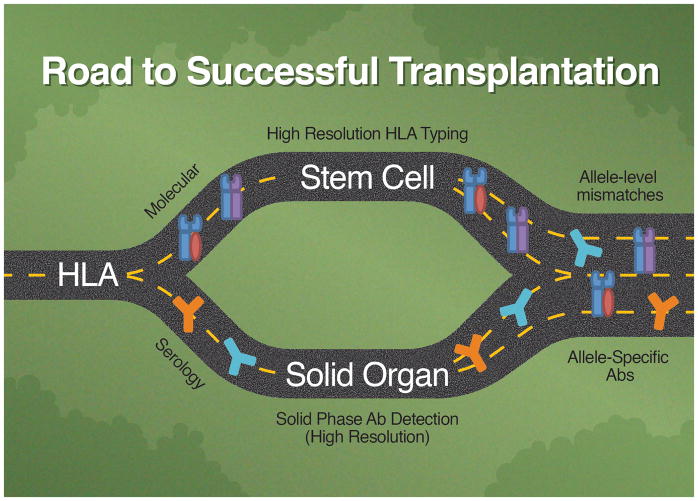
Two Roads Converge
The innovators in HLA testing shaped the field by traveling two distinct pathways: one driven by molecular characterization of HLA antigens, the other driven by advances in antibody identification. Ultimately, antigen testing led to high resolution HLA typing, which identifies individual HLA alleles. In contrast, antibody identification has progressed through development of solid-phase methods that can identify epitopes restricted to distinct HLA alleles.
Importantly, these simultaneous, yet separate, advancements have converged, and once again (as in the early days of HLA typing) modern molecular antigen typing methods are used in conjunction with solid phase antibody testing to optimize patient outcomes. The convergence of the two paths is seen in HLA mismatched stem cell transplants, wherein donors expressing an HLA allele to which the recipient has a corresponding antibody must be considered before moving forward with a transplant since such antibodies can result in failed engraftment. This requires that “high resolution” antibody testing be performed. Similarly, since allele-specific HLA antibodies can be identified in solid organ transplant recipient, high resolution HLA typing of donors will be a requirement to determine compatibility (Figure 2).
Figure 2.
In the beginning, the foundation of HLA testing was serologic in nature with an interdependence of antibodies to identify antigens, and antigens to identify antibodies. However, as time progressed, HLA laboratory testing took two distinct roads, one driven by the need for high resolution molecular characterization of HLA alleles, the other driven by the need to identify and classify HLA antibodies via serology. The directions these two paths took evolved from the clinical needs in stem cell and solid organ transplantation: Bone marrow/stem cell transplantation depends on accurate, reliable allele resolution to identify HLA-matched recipient-donor pairs while solid organ transplantation requires identification of HLA antibodies, specifically donor specific antibodies (DSA). Antigen testing led to high resolution HLA typing, which identifies individual HLA alleles; while antibody identification has progressed using solid-phase methods that now have the ability to identify allele-specific antibodies and characterize unique epitopes restricted to distinct HLA alleles. Eventually, the two roads of histocompatibility testing converged, with antigen and antibody testing coming full circle and once again each relying on the other. This convergence is exemplified in stem cell transplants, wherein donors expressing an HLA allele to which the recipient has a corresponding antibody must be considered before moving forward with a transplant. This requires high resolution antigen typing of the donor as well as high resolution antibody testing of the recipient to be performed. Similarly, since allele-specific HLA antibodies can be identified in solid organ transplant recipient, high resolution HLA typing of donors will be needed to determine compatibility.
The best example of the convergence of state of the art high resolution antigen typing and solid-phase antibody identification is the virtual crossmatch (vXM). In brief, by having a detailed list of unacceptable and acceptable donor antigens (based on the recipient’s HLA antibody profile) and the high resolution HLA type of potential donors, an electronic crossmatch (paper crossmatch or crossmatch in silico) can be performed, perhaps one day obviating the need for a physical crossmatch. Not only does the vXM have a high positive predictive value of compatible physical crossmatches,32 but it has also assisted solid organ transplantation by shortening time-to-transplant and increased access to organs by increasing geographic radius of potential donors.33–35 However, the vXM is critically reliant on the correct HLA typing of donors and antibody assessment of candidate recipients. To this end, there are concerns in both directions. For example, numerous studies have reported that not all antibodies detected by the solid-phase methods are deleterious (or even “real”), but rather may be directed against denatured HLA proteins or contaminated proteins on the microparticles.36–38 Conversely, the error rate for HLA typing exceeds 1% which, when considering a pool of 6000–7000 annual deceased donors, is not a trivial occurrence. Clearly while new technologies has moved the field of HLA significantly forward, there are still issues to be resolved.
The Emory Philosophy and Experience
Undoubtedly, the science of HLA testing has progressed substantially since its inception with basic serologic testing. However, from a cost and workflow perspective, it may not be practical for a laboratory to perform all these cutting-edge tests. Laboratories and their directors have to take into consideration the resources and technical staff they have at their disposal, the patient populations served, and volume of transplants being done. Furthermore, the advantages of different methodologies should be weighed against their drawbacks. For example, historically the Emory HLA laboratory performed SBT in-house for the stem cell transplant program and to resolve discrepancies that arose with SSP and/or SSOP. However, the resources needed perform SBT were extensive, including time, technical expertise, maintenance of competency, and participation in proficiency exercises. Thus, the decision was made to cease in-house SBT. By sending these samples instead to a reference laboratory whose sole business was HLA SBT, we obtained more reliable test results at a lower cost and freed up staff to concentrate on assays that had to be performed in-house.
Currently at Emory, we perform molecular HLA antigen typing (SSP, SSOP, RT PCR) and high-sensitivity antibody testing (flow cytometric PRA, solid-phase antibody testing, flow cytometric crossmatch). Specimens follow an algorithm in which sera from candidate recipients are first screened for HLA antibodies. Sera that test positive are then analyzed by solid-phase testing to determine antibody specificity(ies). The resulting HLA specificities are used to calculate the cPRA and the corresponding antigens are listed on the UNOS website as unacceptable. Once a donor organ becomes available, the unacceptable antigen profile helps determine which candidate recipients have a high likelihood of a negative crossmatch. A flow-cytometric crossmatch is then performed for sensitized patients and, when negative, the patient is eligible to proceed to transplant.39
The Road Ahead
The two roads of innovation have led to the development of highly sensitive and specific testing, and have allowed for the identification of antigens and antibodies down to the allele and even epitope level. Given these advancements, it has been proposed that high resolution typing may eventually be applied to non-sensitized candidates by adopting allele-based compatibility in order to avoid post-transplant sensitization.40 In fact, recent studies suggest HLA allele-matching, and more specifically epitope-matching, results in better long term graft survival through decreased production of DSA compared to established HLA-matching.41 Moreover, with antigen typing able to identify specificity at the allele level, alleles sharing epitopes to which a recipient has antibodies may be avoided. Otherwise a positive crossmatch would not have been predicted because the candidate had never been exposed to the allele in question.40
Nevertheless, it is important to underscore that with new technologies comes new problems and shortcomings. Solid phase antibody testing, for example, has inherent technical characteristics that can lead to discrepant results. Among these potential problematic attributes are variable antigen source,42 antigen configuration,36–38 antigen density on the microbeads,43 varying median fluorescence intensity (MFI) cutoffs for antibody detection,44–46 and the testing platform27 among others. These drawbacks mainly arise from the lack of standardization among manufactures of assays and reagents. Though studies have shown that standardization of protocols is attainable,47 it remains to be seen whether standardization will be implemented. Furthermore, despite advances in molecular technology which have allowed for high resolution typing, discrepancies in donor typing continue to be reported to UNOS due to sample mix-ups and transcriptional errors.35 These errors demonstrate that no matter how advanced the technology, erroneous results can occur. Wherever the road leads, we must remain vigilant of drawbacks and potential pitfalls.
Use of RBC Genotyping for aiding in transfusion management of patients with Hemoglobinopathies
Introduction
As compared to the genotyping diagnostics for hematopoietic stem cell and solid organ transplantation described earlier in this review, the application of genotyping to transfusion medicine has been more recent. In this area, genotyping has been used to improve the safety and efficacy of both platelet and RBC transfusions. Genotyping can, for example, help identify compatible products for patients who are platelet refractory due to antibodies against allogeneic HLA and/or platelet-specific glycoprotein antigens. In addition, human platelet antigen (HPA) genotyping is critical for determining fetal risk of neonatal alloimmune thrombocytopenia (NAIT) in mothers with HPA alloantibodies when the father is found to be heterozygote for the putative HPA.48 When applied to RBC transfusions, genotyping can be used to infer the phenotype of recipient and/or donor RBCs. Predicted RBC phenotypes are useful for fully-characterizing donors with unique/useful blood types, identifying safest components for transfusion of patients with RBC autoantibodies, and solving unusual blood typing problems such as the presence of an anti-Rh(D) antibody in a D+ patient. However, the application of RBC genotyping has arguably been most useful in the management of transfusion-dependent patients with hemoglobinopathies, especially sickle cell disease (SCD).
Current guidelines support transfusion of adults and children with SCD to: raise hemoglobin to 10 g/dL prior to surgical procedures involving general anesthesia (strong recommendation); treat symptomatic severe acute chest syndrome as defined by an oxygen saturation less than 90% despite supplemental oxygen (strong recommendation); address acute splenic sequestration with accompanying severe anemia (strong recommendation); treat acute stroke (moderate recommendation); and manage patients with symptomatic acute chest syndrome who have concomitant reductions in hemoglobin of at least 1 g/dL below baseline (weak recommendation). Further, chronic transfusion therapy is endorsed for primary stroke prevention in children with SCD based on transcranial Doppler (TCD) stroke screening (strong recommendation); and secondary stroke prophylaxis in adults and children with previous clinically overt stroke. In addition, transfusion may also be appropriate in other clinical settings in consultation with an expert on the transfusion support of patients with SCD or other hemoglobinopathies.49
Guidelines also warn against the risks of adverse transfusion events in these patients, including iron overload, hemolysis, hyperviscosity, and most importantly for this discussion the risk of alloimmunization to foreign RBC antigens. Alloimmunization occurs when the transfusion recipient’s immune system produces antibodies against variant (non-self) forms of RBC antigens encountered on transfused RBCs. Once RBC alloantibodies are produced, future transfusions must be limited to the use of donor RBCs that do not express the corresponding antigens. While such blood units are relatively easily identified for patients with a small number of alloantibodies, a proportion of patients with hemoglobinopathies prove to be difficult to provide compatible RBC units for transfusion even with nationwide searches due the development of either a large number of RBC alloantibodies, or complex antibodies, such as anti-Rh alloantibodies due to the high prevalence of RH variants in SCD patients.
As there are few good therapeutic options for management of highly alloimmunized SCD and thalassemia patients presenting with severe anemia, most experts advise using approaches that reduce the risks of alloimmunization. These include eliminating unnecessary transfusions, and prophylactic matching of RBC antigens between donors and recipients.
Prophylactic RBC phenotype matching
In an effort to reduce RBC alloimmunization, many transfusion services have implemented (at a minimum) prophylactic phenotype matching for Rh (C/c, E/e) and K antigens in patients with SCD and thalassemia. Specifically in SCD patients, this transfusion strategy has been associated with a reduction in the prevalence of alloimmunization from 18–75% (rate: 1.7 to 3.9 antibodies/100 units transfused) for ABO/Rh(D)-compatible only transfusions,50–64 to 5–24% (0.26 to 0.50 antibodies/100 units transfused).56,59,61,65,66 Alloimmunization can be further reduced to 0 to 7% (≤ 0.10 antibodies/100 units transfused) when preemptive extended RBC antigen-matching (beyond C/c, E/e and K antigens) is employed (see table 1).67,68 Further efforts to minimize other non-Rh, non-K donor-recipient RBC antigen discrepancies have aimed at providing RBC units from ethnically matched donors due to reports of lower alloimmunization rates in countries with ethnically similar donor and recipient populations.60,69 While identification of donor units with an extended match to the recipient was historically based on RBC phenotyping that used commercial antibody reagents, the availability of genotyping approaches to predict RBC phenotypes in patients and donors has significantly improves the speed, reliability and extent of matching.
Table 1.
RBC antigen matching and alloimmunization worldwide
| Study | N | Age of Patients | Country | Total Units Transfused | Matching | Patient % w/AlloAbs |
|---|---|---|---|---|---|---|
| Orlina et al. 1978 | 50 | Adult | US* | N/A | ABO, D | 36% |
| Coles et al. 1981 | 72 | Pediatric†, Adult | US | N/A | ABO, D | 23.6% |
| Sarnaik et al. 1986 | 245 | Pediatric, Adult | US | N/A | ABO, D | 7.8% |
| Ambruso et al. 1987 | 12 | Pediatric, Adult | US | 492 | ABO, D | 75% |
| Cox et al. 1988 | 73 | Adult | US | N/A | ABO, D | 30% |
| Rosse et al. 1990 | 1044 | Pediatric, Adult | US | N/A | ABO, D | 27% |
| Vichinsky et al. 1990 | 107 | Pediatric, Adult | US | 1,711 | ABO, D | 30% |
| Moreira Jr et al. 1996 | 85 | Pediatric, Adult | Brazil | 1,300 | ABO, D | 12.9% |
| Aygun et al. 2002 | 140 | Pediatric, Adult | US | 3,239 | ABO, D | 37% |
| Castro et al. 2002 | 351 | Adult | US | 8,939 | ABO, D | 29% |
| Sakhalkar et al. 2005 | 387 | Pediatric, Adult | US | 14,263 | ABO, D | 31% |
| Bashwari. 2007 | 350 | Pediatric, Adult | Saudi Arabia | N/A | ABO, D | 13.7% |
| Ameen et al. 2009 | 110 | Pediatric, Adult | Kuwait | N/A | ABO, D | 65.5% |
| Natukunda et al. 2010 | 428 | Pediatric, Adult | Uganda | 3366 | ABO, D | 6.1% |
| Aly et al. 2012 | 42 | Pediatric, Adult | Egypt | N/A | ABO, D | 21.4% |
|
| ||||||
| Vinchinsky et al. 2001 | 61 | Pediatric | US | 1,830 | Limited (C, E, K) | 11% |
| Sakhalkar et al. 2005 | 113 | Pediatric, Adult | US | 2,354 | Limited (C, E, K) | 5% |
| Ameen et al. 2009 | 123 | Pediatric, Adult | Kuwait | N/A | Limited (C, E, K) | 23.6% |
| O’Suoji et al. 2013 | 180 | Pediatric | US | N/A | Limited (C, E, K) | 14% |
| Chou et al. 2013 | 182 | Pediatric, Adult | US | 44,482 | Limited (C, E, K)‡ | 44% |
| DeBaun et al. 2014 | 90 | Pediatric | US | 3,236 | Limited (C, E, K) | 4.5% |
|
| ||||||
| Tahhan et al. 1994 | 40 | Pediatric, Adult | US | 608 | Extended matching¥ | 0% |
| Lasalle-Williams et al. 2011 | 99 | Pediatric, Adult | US | 6,946 | Extended matching§ | 7% |
US = United States; N/A, not available;
Pediatric is defined as any patient ≤ 18 years of age;
Limited matched units from African American donors;
C/c, E/e, K, Fya, Fyb, S;
C/c, E/e, K, Fya, Jka, Jkb.
N/A = not available.
Despite receiving Rh phenotype matched RBCs, many SCD patients still produce Rh antibodies, which often are considered autoantibodies because the patient’s own RBCs type serologically positive for the corresponding Rh antigen. RH genotyping has revealed that many patients with SCD carry alleles encoding partial D, C, and/or e antigens, and that most of these “autoantibodies” are in fact alloantibodies against D, C and e antigens.70,71 A report of SCD patients transfused at the Children’s Hospital of Philadelphia who had received prophylactic C/c, E/e, and K phenotypically matched RBCs from African American donors demonstrated a high prevalence of antibodies to Rh antigens (91 of 146; >60% of all antibodies reported), with no cases of anti-K, and a low rate of anti-Jk, Fy and S antibodies. RH genotyping revealed variant alleles in 87% of individuals, and one-third of the Rh antibodies were associated with laboratory evidence of delayed hemolytic transfusion reactions (DHTRs). Altered RH alleles in both the patients and the African American donors were believed to contribute to Rh alloimmunization because Rh antibodies occurred in both patients whose RBCs were phenotypically positive for the corresponding Rh antigen and in patients who were transfused RBC from donors who were serologically negative for the Rh antigen they lacked.70 Similarly, Sippert et al. identified variant RH alleles in 31 of 48 (65%) Brazilian SCD patients with Rh antibodies, and 42% of the anti-Rh antibodies produced by patients with RH variants were either involved in DHTRs or demonstrated decreased survival of transfused RBCs.71
Complexity of the Rh system: implications on alloimmunization and genotyping
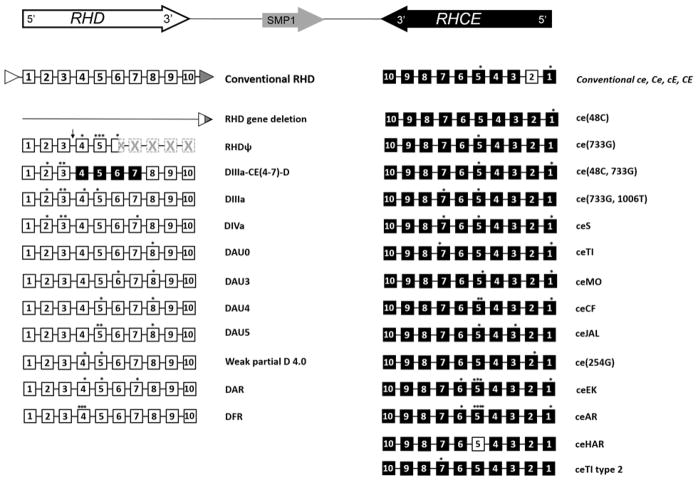
The Rh blood group system consists of numerous antigens in addition to the most common clinically significant D, and C, c, E, e antigens. There have been over 50 Rh antigens identified, all encoded by RHD and RHCE, two genes each with 10 exons in opposite orientation (5′-RhD-3′-3′-RhCE-5′). RHD and RHCE share 92% nucleotide sequence homology and 96% translated amino acid sequence similarity due to the fact that the two genes were evolutionarily created by a duplication event from one gene. The conventional RHD and RHCE genes encode the D antigen and the CE antigens in various combinations (Dce, DcE, DCe, DCE, ce, cE, Ce, or CE), which are found in all ethnic groups, although with different frequencies.72–74 In addition, over 200 RHD and approximately 100 RHCE alleles encoding partial and weak D, and other altered or partial Rh antigens have been described. RH variant alleles may encode Rh proteins with amino acid changes that cannot be distinguished with common serologic reagents (e.g., V, VS, Goa), but can result in allo-sensitization upon exposure in an individual lacking the variant allele. Conversely, individuals with RH variants in homozygous or compound heterozygous form can have RBCs that lack high prevalence Rh antigens (e.g., hrB or hrS), and may make alloantibodies (which often appear to be autoantibodies) to these antigens upon exposure to conventional Rh antigens through transfusion. RHD and RHCE variants are found in < 1–2% of Europeans; however the frequency in individuals of African descent is much higher with certain RH variants being more common in patients with SCD of African ancestry. Figure 3 illustrates the most common RH variants identified in patients with SCD.
Figure 3.
Inverted orientation of the RHD and RHCE genes (top), RHD and RHCE locus structures of conventional RHD and RHCE genes, and the most frequently occurring variant haplotypes in individuals of African descent which complicate transfusion in SCD patients. The 10 coding exons of RHD and RHCE are shown as white and black boxes respectively. The location of nucleotide changes are designated by an asterisk (*). Rhesus boxes are shown as white and gray triangles with a resulting hybrid Rhesus box in individuals with the RHD deletion. The arrow (↓) indicates a 37-bp duplication in intron3/exon 4 junction and the hatched boxes represent exons encoding the untranslated region of the inactive RHD pseudogene (RHDψ) due to the nonsense mutation in exon 6 leading to a premature translation stop codon. Figure references: Ref #70, 73, 74.
RBC genotyping: current methodologies
Since the discovery of the ABO blood group in the early 20th century, more than 300 authenticated blood group antigens have been placed into 35 blood group systems. Moreover, the molecular basis for almost all of the genes responsible for the differences in blood group antigens has been determined, and often this difference is cause by a single nucleotide polymorphism (SNP), a single change in the DNA sequence of the gene.75 Identification of these SNPs has led to the development of several blood group molecular platforms. Most of them currently available high throughput platforms are DNA microarray-based assays, of which only one platform is approved for use by the Food and Drug Administration (FDA).
These assays start with PCR amplification of a number of genetic regions encoding various blood group antigens. Target regions amplified by PCR are then hybridized to RBC antigen allele specific oligonucleotide probes, which have been linked to either glass slides (BLOODchip® ID CORE XT; Progenika Biopharma, Biscay, Spain), colored silica beads assembled on silicon wafers (PreciseType™ HEA; Immucor, Norcross, GA), or fluidic bead suspensions (xMAP®, Luminex Corporation, Austin, TX). Hybridization signals are then analyzed by measuring fluorescence intensities (usually), and a RBC phenotype is predicted based on the genotype result.76–78 Predicting the phenotype from a genotype is relatively straightforward for many RBC antigens encoded by SNPs. However, there still remain some blood group systems where genotyping remains challenging because the genes encode enzymes involved in the modification of carbohydrate chains (e.g. ABO, H, I, GLOB), or blood group antigens are determined by large insertions, deletions, or hybrid genes from recombination of homologous genes (e.g. RHD, RHCE, GYPA, GYPB).78
The platform used at Emory University Center of Transfusion and Cellular Therapies (CTCT) is Immucor’s PreciseType™ HEA, which uses a proprietary elongation-mediated multiplexed analysis of polymorphisms (eMAP®) technology to identify the presence or absence of the selected alleles associated with a given phenotype. PreciseType™ HEA includes 24 polymorphisms associated with 35 human erythrocyte antigens of the Rh (C/c, E/e, V, VS), Kell (K/k, Jsa/Jsb, Kpa/Kpb), Duffy (Fya/Fyb, Fynull due to GATA mutation, Fybweak), Kidd (Jka/Jkb), MNS (M/N, S/s, U), Lutheran (Lua/Lub), Dombrock (Doa/Dob, Hy, Joa), Landsteiner-Wiener (LWa/LWb), Diego (Dia/Dib), Coltan (Coa/Cob) and Scianna (Sca/Scb) blood group systems, and detection of the hemoglobin S mutation in the β-globin gene (which is not intended to make a diagnosis of sickle cell disease). PreciseType™ HEA demonstrates an overall >99.4% agreement to serology, 99.8% concordance with DNA sequencing, and gained FDA approval in May 2014.79 RhD and ABO are not determined by this platform, however we utilize RHD and RHCE variant research use only (RUO) BeadChips, which identify 75 RHD and 35 RHCE variants for elucidation of RH variants in select clinical circumstances (see “Indications for RBC genotyping”).
Other commercially available high-throughput genotyping platforms have also been described such as the BLOODchip® ID CORE XT types ABO (33 haplotypes), RhD (91 haplotypes including various alleles that cause D-negative, partial D, weak D and Del phenotypes), RhCE (9 alleles), Kell (8 alleles), Kidd (4 alleles, including 2 JKnull), Duffy (4 alleles), MNS (9 haplotypes), Diego, Dombrock and Coltan. Validation of this platform demonstrated a global accuracy of 99.8% with the exception of ABO. BLOODchip has been CE-marked in the European Union, however is not FDA approved for diagnostic purposes in the US. 80 Other technologies which have been utilized for high throughput genotyping of a limited number of red cell antigens include: Luminex xMAP®, GenomeLab™ SNPstream® (Beckman Coulter, Fullerton, California), and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS).81 In addition, other customizable high throughput genotyping platforms such as TaqMan® Open Array® (Applied Biosystems, Grand Island, New York) have also be used.76
There are limitations to all current molecular techniques as these platforms have been designed to target only known polymorphisms. As a result, novel alleles are unlikely to be detected. However, novel polymorphisms or known mutations not covered by the assay may influence expression of an antigen, as is the case with the myriad of RH variants not covered in many of the current platforms. Additional SNPs within primer sites may lead to a false-negative result due to failure of an allele to amplify. In difficult cases, DNA sequencing of the blood group gene(s) can mitigate many of these limitations, however sequencing is an expensive and laborious process.77 Target enrichment next-generation sequencing (T-NGS) is an emerging technology that can detect both known and novel SNPs, insertions and deletions, and structural variations, focusing on specified genomic regions. T-NGS for blood group typing may become more widely available in the future as the cost and time required for T-NGS assays continue to decrease.76,77
Indications for RBC genotyping
Based on published literature,82–84 commonly accepted transfusion practices, and our own institutional experience, we routinely perform molecular typing for the following types of patients:
All patients with congenital hemolytic anemias or blood dyscrasias, which affect the erythroid lineage who are likely to receive multiple RBC transfusions, have RBC genotyping performed within the first year of life or on their initial visit if transferring from another institution (unless already performed) (e.g. SCD, thalassemia, Diamond–Blackfan anemia, congenital dyserythropoietic anemias, etc.). Since PreciseType™ HEA is FDA-approved as “test of record,” the RBC phenotype determined by molecular blood group testing does not require confirmation by serologic methods.
Patients with autoantibodies or other serologic reactivities (eg, multiple antibodies, antibodies to high prevalence antigens, and antibodies of undetermined specificity) that obscure detection of clinically significant RBC allo-antibodies.
Patients with suspected antibody against an antigen for which typing antisera are not readily available. Examples include antibodies to: Doa/Dob, Jsa/Jsb, Kpa/Kpb, V or VS.
Patients with a serologic typing discrepancy or weak/inconclusive antigen typing (e.g. RhD serologic typing discrepancy)
Patients with an apparent autoantibody with antigenic specificity (e.g., an anti-C in a C+ patient).
Patients with an unexplained Rh antibodies detected despite antigen matching for RhD, E/e, and C/c, have RH genotyping performed to determine the presence of and characterize the RH variant(s) in the individual.
Our institution utilizes the PreciseType™ HEA, which detects the RHCE*ce(733G,1006T) allele. This RHCE variant allele is commonly linked to the hybrid RHD*DIIIa-CE(4–7)-D gene, and encodes a partial C antigen. Although individuals with this RH haplotype will commonly be serologically C+, they are at risk for alloimmunization to the C antigen (if they are lacking a RHCE gene encoding a conventional C antigen in trans), and therefore receive C–negative blood at our institution.72,85 We also utilize RHD and RHCE variant research use only (RUO) beadchips for elucidation of RH variants in patients with SCD who have complex anti-Rh antibodies since the PreciseType™ HEA does not type for RhD and detects only two SNPs (733G and 1006T) which account for a limited number of RHCE variant haplotypes. The GATA silencing mutation in the Duffy gene, which prevents the transcription of the Fyb antigen on erythrocytes but not tissue cells, is also detected in this array. When it is present in Fyb-negative patients, we permit transfusion of Fyb-positive blood since there is no risk for alloimmunization to the Fyb antigen.86,87
RBC genotyping as an aid to provide extended matched blood for patients with hemoglobinopathies
Although many transfusion services provide RBCs phenotypically matched for Rh (C/c, E/e) and K (limited match), some also endorse prophylactic extended antigen matching to include the Duffy (Fya/Fyb), Kidd (Jka/Jkb), and/or S antigens, citing further reduced alloimmunization rates with this strategy.68 Some further advocate for the recruitment of racially matched, molecularly typed donors for providing extended antigen-matched RBC units for the chronically transfused SCD population. These strategies require the ability to perform high-throughput DNA-based extended antigen phenotyping in both patients and in large (ethnically similar) donor populations. Due to the availability of multiple commercially available high-throughput blood group genotyping systems, the potential to provide extended antigen matched RBC units for large populations of chronically transfused SCD patients may now be considered through expanded use of RBC genotyping in hospitals and blood donor testing centers.
In a feasibility study at BloodWorks (formerly the Puget Sound BloodCenter), Wilkinson et al. enumerated the number of extended matched RBC components available from their inventory of molecularly or serologically typed donors to meet the transfusion needs of molecularly typed SCD patients. From an inventory that included an average of 335 RBC components typed for 11 or more antigens (most molecularly typed), 37.4 extended matched (matched at: E/e, C/c, K, Fya/Fyb, Jka/Jkb, S/s) RBC components per patient were available for 70 SCD patients (28.6% with alloantibodies, 15.7% with warm autoantibodies) when allowing Fyb+ components for patients with FY GATA mutation. Although 6 (8.6%) of the patients had no extended matched components available, the blood donor base was predominantly Caucasian. The authors concluded that recruitment of racially matched, molecularly typed donors may have allowed for a greater ability to provide extended matched components in their patient population.88 Using DNA array analysis (HEA Beadchip, now PreciseType™ HEA), Ribeiro et al. were able to predict compatible donors for a group of multiply transfused SCD patients (29% with alloantibodies) from a pool of 948 donors. They were able to find units matched for ABO, Rh (C/c, E/e), Kell, Fya/Fyb, Jka/Jkb, Ss, in addition to Dombrock and Diego for 134 of 144 SCD patients. The 10 patients for whom they were unable to find compatible units for using this donor pool had unique phenotype combinations or rare phenotypes, such as R2RZ (DcE/DCE), R2R2 (DcE/DcE) and U-, and so likely would have been able to be matched with a larger donor pool.89
A multi-institutional, prospective observational study was conducted at four hospitals to determine the feasibility for hospital transfusion services to maintain an inventory of molecularly typed units to facilitate identification of blood for transfusion at increasingly levels of antigen matching for three groups of potential transfusion recipients: SCD patients (alloimmunized and non-alloimmunized); cardiac surgery patients without previous alloimmunization, and alloimmunized (non-SCD) hematology and cardiac surgery patients requiring antigen-negative units. Approximately 730 donor and 128 patient samples from each of the institutions were molecularly typed using HEA Beadchip. This study demonstrated that by selecting existing inventory of donor units for genotyping, a substantial fraction of RBC requests could be fulfilled (fill fraction) at lower antigen matching levels in most patient groups. However, the hospital with the most SCD patients included the lowest fill fractions for C/c, E/e, K matched units (62%) and extended matched units (31%) for the SCD patient population (which had a mean of 4.4 alloantibodies per patient).90 These results show that supplementing the blood inventory with specially ordered RBC units is necessary for supplying limited or extended matched units in institutions with large numbers of heavily alloimmunized SCD patients.
It is well established that antigen frequencies differ between African American and Caucasian populations, and that alloimmunization to specific antigens can be reduced by using RBCs from racially similar donors in SCD patients. Karafin, et al. evaluated the RBC antigen frequencies in both a cohort of 54 patients from the adult sickle cell transfusion program and 6066 genotyped African American donors.91 They found that the genotype-derived predicted antigen frequencies of the donors were similar to the SCD patients supported by these units. Despite demonstrating that African American sickle-negative donors could support the antigen requirements of their SCD population, they found that a majority of patients received a mix of Caucasian and African American donor units. They reported an overall alloantibody prevalence of 22% in this cohort of chronically transfused SCD patients, and about half of those formed new alloantibodies (9.3%) during their 3-year study interval despite the availability of the genotyped African American donor units. Similar to other reports,70,71 a proportion of the patients developed Rh antibodies due to undetected Rh variants, and alloantibodies to antigens prevalent among African American donors.
RH genotyping has thus been used to identify altered RH alleles and to predict whether Rh antibodies are autoantibodies or allo-antibodies, and is beginning to play an important role in improving transfusion therapy in SCD patients by expanding the ability of providing true Rh antigen matched RBCs for those patients with RH variant haplotypes who lack a conventional Rh antigen. This is only possible with expansion of large-scale donor molecular screening to identify donors with RH variants for genotype matching. Systematic RHD and RHCE molecular analysis performed on African blood donors in France (established by Fy(a-b-) phenotype) provided indirect evidence that the transfusion needs of patients with SCD and anti-Rh antibodies may potentially be met by screening very large populations of donors for RH variant phenotypes.92 Therefore, incorporating RH genotyping into select patient and donor testing may improve transfusion therapy of SCD patients by allowing better donor and recipient matching at the Rh level.
Gaps in knowledge and further research
The prerequisite for providing extended antigen-negative RBCs to patients with SCD, who commonly are negative for the C, E, K, Fya, and Jkb antigens, is creating and maintaining large inventories of African-American donors typed for conventional blood group antigens. Automated DNA extraction and the ability to test both patients and large groups of donors on high-throughput RBC genotyping platforms are now readily accessible. Combined with database-driven RBC matching, blood group molecular phenotyping may facilitate the identification of antigen-matched RBCs and improve transfusion support of patients with SCD in the near future. Although mass screening for antigen negative and rare phenotype blood donors is now possible, large-scale donor genotyping implementation and future feasibility studies are necessary to confirm the positive impact that RBC genotyping may have on patients with hemoglobinopathies in this regard. Although high-resolution RH genotyping is currently largely limited to reference molecular immunohematology laboratories, it is increasingly being employed for distinguishing alloantibodies from autoantibodies, and for identifying patients at risk of producing Rh antibodies despite phenotypically matching for Rh antigens. However, future studies are needed to address whether providing RH genotype matched RBCs for patients with SCD: a) is feasible, b) can prevent the Rh alloimmunization, and c) will improve transfusion safety in a cost-effective manner. Lastly, the advantages over the current blood group molecular typing platforms make T-NGS an attractive prospect in the future toward advancing our knowledge of blood group genetics and increasing our ability to provide extended antigen matched RBCs to patients with hemoglobinopathy if costs continue to decrease to a more competitive level.
References
- 1.Guild WR, Harrison JH, Merrill JP, Murray J. Successful homotransplantation of the kidney in an identical twin. Transactions of the American Clinical and Climatological Association. 1955;67:167–73. [PMC free article] [PubMed] [Google Scholar]
- 2.Henig I, Zuckerman T. Hematopoietic stem cell transplantation-50 years of evolution and future perspectives. Rambam Maimonides medical journal. 2014;5:e0028. doi: 10.5041/RMMJ.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calne RY, Loughridge L, MacGillivray JB, Swales JD. Further observations on renal transplants in man from cadaveric donors. British medical journal. 1966;2:1345–51. doi: 10.1136/bmj.2.5526.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speck B. Bone marrow transplantation-clinical results and problems. Blut. 1973;27:297–301. doi: 10.1007/BF01631040. [DOI] [PubMed] [Google Scholar]
- 5.Dausset J. Iso-leuko-antibodies. Acta haematologica. 1958;20:156–66. doi: 10.1159/000205478. [DOI] [PubMed] [Google Scholar]
- 6.Patel R, Terasaki PI. Significance of the positive crossmatch test in kidney transplantation. The New England journal of medicine. 1969;280:735–9. doi: 10.1056/NEJM196904032801401. [DOI] [PubMed] [Google Scholar]
- 7.Dausset J. The genetics of transplantation antigens. Transplantation proceedings. 1971;3:8–14. [PubMed] [Google Scholar]
- 8.Gebel HM, Bray RA, Nickerson P. Pre-transplant assessment of donor-reactive, HLA-specific antibodies in renal transplantation: contraindication vs. risk. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2003;3:1488–500. doi: 10.1046/j.1600-6135.2003.00273.x. [DOI] [PubMed] [Google Scholar]
- 9.Beatty PG, Clift RA, Mickelson EM, et al. Marrow transplantation from related donors other than HLA-identical siblings. The New England journal of medicine. 1985;313:765–71. doi: 10.1056/NEJM198509263131301. [DOI] [PubMed] [Google Scholar]
- 10.Speiser DE, Tiercy JM, Rufer N, et al. High resolution HLA matching associated with decreased mortality after unrelated bone marrow transplantation. Blood. 1996;87:4455–62. [PubMed] [Google Scholar]
- 11.Marcadet A, Cohen D, Dausset J, Fischer A, Durandy A, Griscelli C. Genotyping with DNA probes in combined immunodeficiency syndrome with defective expression of HLA. The New England journal of medicine. 1985;312:1287–92. doi: 10.1056/NEJM198505163122004. [DOI] [PubMed] [Google Scholar]
- 12.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods in enzymology. 1987;155:335–50. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 13.Olerup O, Zetterquist H. HLA-DRB1*01 subtyping by allele-specific PCR amplification: a sensitive, specific and rapid technique. Tissue antigens. 1991;37:197–204. doi: 10.1111/j.1399-0039.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 14.Welsh K, Bunce M. Molecular typing for the MHC with PCR-SSP. Reviews in immunogenetics. 1999;1:157–76. [PubMed] [Google Scholar]
- 15.Angelini G, de Preval C, Gorski J, Mach B. High-resolution analysis of the human HLA-DR polymorphism by hybridization with sequence-specific oligonucleotide probes. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:4489–93. doi: 10.1073/pnas.83.12.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrik J. Microarray technology: the future of blood testing? Vox sanguinis. 2001;80:1–11. doi: 10.1046/j.1423-0410.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 17.Mickelson EM, Petersdorf E, Anasetti C, Martin P, Woolfrey A, Hansen JA. HLA matching in hematopoietic cell transplantation. Human immunology. 2000;61:92–100. doi: 10.1016/s0198-8859(99)00151-2. [DOI] [PubMed] [Google Scholar]
- 18.Dunn TB, Noreen H, Gillingham K, et al. Revisiting traditional risk factors for rejection and graft loss after kidney transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:2132–43. doi: 10.1111/j.1600-6143.2011.03640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kittleson MM, Kobashigawa JA. Antibody-mediated rejection. Current opinion in organ transplantation. 2012;17:551–7. doi: 10.1097/MOT.0b013e3283577fef. [DOI] [PubMed] [Google Scholar]
- 20.Lobo LJ, Aris RM, Schmitz J, Neuringer IP. Donor-specific antibodies are associated with antibody-mediated rejection, acute cellular rejection, bronchiolitis obliterans syndrome, and cystic fibrosis after lung transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2013;32:70–7. doi: 10.1016/j.healun.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Campbell PM, Salam A, Ryan EA, et al. Pretransplant HLA antibodies are associated with reduced graft survival after clinical islet transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2007;7:1242–8. doi: 10.1111/j.1600-6143.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 22.O’Leary JG, Kaneku H, Susskind BM, et al. High mean fluorescence intensity donor-specific anti-HLA antibodies associated with chronic rejection Postliver transplant. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:1868–76. doi: 10.1111/j.1600-6143.2011.03593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.42 C.F.R. § 493:1278 Standard: Histocompatibility. Centers for Medicare & Medicaid Services.
- 24.Gebel HM, Bray RA. The evolution and clinical impact of human leukocyte antigen technology. Current opinion in nephrology and hypertension. 2010;19:598–602. doi: 10.1097/MNH.0b013e32833dfc3f. [DOI] [PubMed] [Google Scholar]
- 25.Bray RA, Gebel HM. Strategies for human leukocyte antigen antibody detection. Current opinion in organ transplantation. 2009;14:392–7. doi: 10.1097/mot.0b013e32832d31c7. [DOI] [PubMed] [Google Scholar]
- 26.Tait BD, Susal C, Gebel HM, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. 2013;95:19–47. doi: 10.1097/TP.0b013e31827a19cc. [DOI] [PubMed] [Google Scholar]
- 27.Gebel HM, Liwski RS, Bray RA. Technical aspects of HLA antibody testing. Current opinion in organ transplantation. 2013;18:455–62. doi: 10.1097/MOT.0b013e32836361f1. [DOI] [PubMed] [Google Scholar]
- 28.Cecka JM. Calculated PRA (CPRA): the new measure of sensitization for transplant candidates. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2010;10:26–9. doi: 10.1111/j.1600-6143.2009.02927.x. [DOI] [PubMed] [Google Scholar]
- 29.Cecka JM, Kucheryavaya AY, Reinsmoen NL, Leffell MS. Calculated PRA: initial results show benefits for sensitized patients and a reduction in positive crossmatches. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:719–24. doi: 10.1111/j.1600-6143.2010.03340.x. [DOI] [PubMed] [Google Scholar]
- 30.Kopko PM, Warner P, Kresie L, Pancoska C. Methods for the selection of platelet products for alloimmune-refractory patients. Transfusion. 2015;55:235–44. doi: 10.1111/trf.12921. [DOI] [PubMed] [Google Scholar]
- 31.Stanworth SJ, Navarrete C, Estcourt L, Marsh J. Platelet refractoriness--practical approaches and ongoing dilemmas in patient management. British journal of haematology. 2015;171:297–305. doi: 10.1111/bjh.13597. [DOI] [PubMed] [Google Scholar]
- 32.Nikaein A, Cherikh W, Nelson K, et al. Organ procurement and transplantation network/united network for organ sharing histocompatibility committee collaborative study to evaluate prediction of crossmatch results in highly sensitized patients. Transplantation. 2009;87:557–62. doi: 10.1097/TP.0b013e3181943c76. [DOI] [PubMed] [Google Scholar]
- 33.Zangwill S, Ellis T, Stendahl G, Zahn A, Berger S, Tweddell J. Practical application of the virtual crossmatch. Pediatric transplantation. 2007;11:650–4. doi: 10.1111/j.1399-3046.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- 34.Ferrari P, Fidler S, Wright J, et al. Virtual crossmatch approach to maximize matching in paired kidney donation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:272–8. doi: 10.1111/j.1600-6143.2010.03313.x. [DOI] [PubMed] [Google Scholar]
- 35.Gebel HM, Bray RA. HLA antibody detection with solid phase assays: great expectations or expectations too great? American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14:1964–75. doi: 10.1111/ajt.12807. [DOI] [PubMed] [Google Scholar]
- 36.Poli F, Benazzi E, Innocente A, et al. Heart transplantation with donor-specific antibodies directed toward denatured HLA-A*02:01: a case report. Human immunology. 2011;72:1045–8. doi: 10.1016/j.humimm.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Pereira S, Perkins S, Lee JH, et al. Donor-specific antibody against denatured HLA-A1: clinically nonsignificant? Human immunology. 2011;72:492–8. doi: 10.1016/j.humimm.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Jacob EK, De Goey SR, Gandhi MJ. Positive virtual crossmatch with negative flow crossmatch results in two cases. Transplant immunology. 2011;25:77–81. doi: 10.1016/j.trim.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Bray RA, Nolen JD, Larsen C, et al. Transplanting the highly sensitized patient: The emory algorithm. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2006;6:2307–15. doi: 10.1111/j.1600-6143.2006.01521.x. [DOI] [PubMed] [Google Scholar]
- 40.Duquesnoy RJ, Kamoun M, Baxter-Lowe LA, et al. Should HLA mismatch acceptability for sensitized transplant candidates be determined at the high-resolution rather than the antigen level? American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15:923–30. doi: 10.1111/ajt.13167. [DOI] [PubMed] [Google Scholar]
- 41.Wiebe C, Pochinco D, Blydt-Hansen TD, et al. Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:3114–22. doi: 10.1111/ajt.12478. [DOI] [PubMed] [Google Scholar]
- 42.Bray RA, Tarsitani C, Gebel HM, Lee JH. Clinical cytometry and progress in HLA antibody detection. Methods in cell biology. 2011;103:285–310. doi: 10.1016/B978-0-12-385493-3.00012-7. [DOI] [PubMed] [Google Scholar]
- 43.Congy-Jolivet N, Drocourt D, Portet S, Tiraby G, Blancher A. Production and characterization of chimeric anti-HLA monoclonal antibodies targeting public epitopes as tools for standardizations of the anti-HLA antibody detection. Journal of immunological methods. 2013;390:41–51. doi: 10.1016/j.jim.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Lefaucheur C, Loupy A, Hill GS, et al. Preexisting donor-specific HLA antibodies predict outcome in kidney transplantation. Journal of the American Society of Nephrology : JASN. 2010;21:1398–406. doi: 10.1681/ASN.2009101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gloor JM, Winters JL, Cornell LD, et al. Baseline donor-specific antibody levels and outcomes in positive crossmatch kidney transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2010;10:582–9. doi: 10.1111/j.1600-6143.2009.02985.x. [DOI] [PubMed] [Google Scholar]
- 46.Zachary A, Reinsmoen NL. Quantifying HLA-specific antibodies in patients undergoing desensitization. Current opinion in organ transplantation. 2011;16:410–5. doi: 10.1097/MOT.0b013e32834899b8. [DOI] [PubMed] [Google Scholar]
- 47.Reed EF, Rao P, Zhang Z, et al. Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:1859–70. doi: 10.1111/ajt.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arinsburg SA, Shaz BH, Westhoff C, Cushing MM. Determination of human platelet antigen typing by molecular methods: Importance in diagnosis and early treatment of neonatal alloimmune thrombocytopenia. American journal of hematology. 2012;87:525–8. doi: 10.1002/ajh.23111. [DOI] [PubMed] [Google Scholar]
- 49.Yawn BP, Buchanan GR, Afenyi-Annan AN, et al. Management of sickle cell disease: summary of the 2014 evidence-based report by expert panel members. Jama. 2014;312:1033–48. doi: 10.1001/jama.2014.10517. [DOI] [PubMed] [Google Scholar]
- 50.Coles SM, Klein HG, Holland PV. Alloimmunization in two multitransfused patient populations. Transfusion. 1981;21:462–6. doi: 10.1046/j.1537-2995.1981.21481276005.x. [DOI] [PubMed] [Google Scholar]
- 51.Orlina AR, Unger PJ, Koshy M. Post-transfusion alloimmunization in patients with sickle cell disease. American journal of hematology. 1978;5:101–6. doi: 10.1002/ajh.2830050204. [DOI] [PubMed] [Google Scholar]
- 52.Sarnaik S, Schornack J, Lusher JM. The incidence of development of irregular red cell antibodies in patients with sickle cell anemia. Transfusion. 1986;26:249–52. doi: 10.1046/j.1537-2995.1986.26386209381.x. [DOI] [PubMed] [Google Scholar]
- 53.Ambruso DR, Githens JH, Alcorn R, et al. Experience with donors matched for minor blood group antigens in patients with sickle cell anemia who are receiving chronic transfusion therapy. Transfusion. 1987;27:94–8. doi: 10.1046/j.1537-2995.1987.27187121485.x. [DOI] [PubMed] [Google Scholar]
- 54.Cox JV, Steane E, Cunningham G, Frenkel EP. Risk of alloimmunization and delayed hemolytic transfusion reactions in patients with sickle cell disease. Archives of internal medicine. 1988;148:2485–9. [PubMed] [Google Scholar]
- 55.Rosse WF, Gallagher D, Kinney TR, et al. Transfusion and alloimmunization in sickle cell disease. The Cooperative Study of Sickle Cell Disease. Blood. 1990;76:1431–7. [PubMed] [Google Scholar]
- 56.Vichinsky EP, Earles A, Johnson RA, Hoag MS, Williams A, Lubin B. Alloimmunization in sickle cell anemia and transfusion of racially unmatched blood. The New England journal of medicine. 1990;322:1617–21. doi: 10.1056/NEJM199006073222301. [DOI] [PubMed] [Google Scholar]
- 57.Aygun B, Padmanabhan S, Paley C, Chandrasekaran V. Clinical significance of RBC alloantibodies and autoantibodies in sickle cell patients who received transfusions. Transfusion. 2002;42:37–43. doi: 10.1046/j.1537-2995.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 58.Castro O, Sandler SG, Houston-Yu P, Rana S. Predicting the effect of transfusing only phenotype-matched RBCs to patients with sickle cell disease: theoretical and practical implications. Transfusion. 2002;42:684–90. doi: 10.1046/j.1537-2995.2002.00126.x. [DOI] [PubMed] [Google Scholar]
- 59.Sakhalkar VS, Roberts K, Hawthorne LM, et al. Allosensitization in patients receiving multiple blood transfusions. Annals of the New York Academy of Sciences. 2005;1054:495–9. doi: 10.1196/annals.1345.072. [DOI] [PubMed] [Google Scholar]
- 60.Bashawri LA. Red cell alloimmunization in sickle-cell anaemia patients. East Mediterr Health J. 2007;13:1181–9. doi: 10.26719/2007.13.5.1181. [DOI] [PubMed] [Google Scholar]
- 61.Ameen R, Al Shemmari S, Al-Bashir A. Red blood cell alloimmunization among sickle cell Kuwaiti Arab patients who received red blood cell transfusion. Transfusion. 2009;49:1649–54. doi: 10.1111/j.1537-2995.2009.02185.x. [DOI] [PubMed] [Google Scholar]
- 62.Natukunda B, Schonewille H, Ndugwa C, Brand A. Red blood cell alloimmunization in sickle cell disease patients in Uganda. Transfusion. 2010;50:20–5. doi: 10.1111/j.1537-2995.2009.02435.x. [DOI] [PubMed] [Google Scholar]
- 63.Aly R, El-sharnoby MR, Hagag AA. Frequency of red cell alloimmunization in patients with sickle cell anemia in an Egyptian referral hospital. Transfus Apher Sci. 2012;47:253–7. doi: 10.1016/j.transci.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 64.Moreira Junior G, Bordin JO, Kuroda A, Kerbauy J. Red blood cell alloimmunization in sickle cell disease: the influence of racial and antigenic pattern differences between donors and recipients in Brazil. American journal of hematology. 1996;52:197–200. doi: 10.1002/(SICI)1096-8652(199607)52:3<197::AID-AJH11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 65.O’Suoji C, Liem RI, Mack AK, Kingsberry P, Ramsey G, Thompson AA. Alloimmunization in sickle cell anemia in the era of extended red cell typing. Pediatric blood & cancer. 2013;60:1487–91. doi: 10.1002/pbc.24530. [DOI] [PubMed] [Google Scholar]
- 66.DeBaun MR, Casella JF. Transfusions for silent cerebral infarcts in sickle cell anemia. The New England journal of medicine. 2014;371:1841–2. doi: 10.1056/NEJMc1411133. [DOI] [PubMed] [Google Scholar]
- 67.Tahhan HR, Holbrook CT, Braddy LR, Brewer LD, Christie JD. Antigen-matched donor blood in the transfusion management of patients with sickle cell disease. Transfusion. 1994;34:562–9. doi: 10.1046/j.1537-2995.1994.34794330008.x. [DOI] [PubMed] [Google Scholar]
- 68.Lasalle-Williams M, Nuss R, Le T, et al. Extended red blood cell antigen matching for transfusions in sickle cell disease: a review of a 14-year experience from a single center (CME) Transfusion. 2011;51:1732–9. doi: 10.1111/j.1537-2995.2010.03045.x. [DOI] [PubMed] [Google Scholar]
- 69.Natukunda B, Schonewille H, van de Watering L, Brand A. Prevalence and specificities of red blood cell alloantibodies in transfused Ugandans with different diseases. Vox sanguinis. 2009 doi: 10.1111/j.1423-0410.2009.01241.x. [DOI] [PubMed] [Google Scholar]
- 70.Chou ST, Jackson T, Vege S, Smith-Whitley K, Friedman DF, Westhoff CM. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood. 2013;122:1062–71. doi: 10.1182/blood-2013-03-490623. [DOI] [PubMed] [Google Scholar]
- 71.Sippert E, Fujita CR, Machado D, et al. Variant RH alleles and Rh immunisation in patients with sickle cell disease. Blood transfusion = Trasfusione del sangue. 2015;13:72–7. doi: 10.2450/2014.0324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chou ST. Transfusion therapy for sickle cell disease: a balancing act. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2013;2013:439–46. doi: 10.1182/asheducation-2013.1.439. [DOI] [PubMed] [Google Scholar]
- 73.Chou ST, Westhoff CM. Molecular biology of the Rh system: clinical considerations for transfusion in sickle cell disease. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2009:178–84. doi: 10.1182/asheducation-2009.1.178. [DOI] [PubMed] [Google Scholar]
- 74.Noizat-Pirenne F, Tournamille C. Relevance of RH variants in transfusion of sickle cell patients. Transfus Clin Biol. 2011;18:527–35. doi: 10.1016/j.tracli.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 75.Denomme GA. Molecular basis of blood group expression. Transfusion and apheresis science : official journal of the World Apheresis Association : official journal of the European Society for Haemapheresis. 2011;44:53–63. doi: 10.1016/j.transci.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 76.Liu Z, Liu M, Mercado T, Illoh O, Davey R. Extended blood group molecular typing and next-generation sequencing. Transfusion medicine reviews. 2014;28:177–86. doi: 10.1016/j.tmrv.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Tilley L, Grimsley S. Is Next Generation Sequencing the future of blood group testing? Transfusion and apheresis science : official journal of the World Apheresis Association : official journal of the European Society for Haemapheresis. 2014;50:183–8. doi: 10.1016/j.transci.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 78.Veldhuisen B, van der Schoot CE, de Haas M. Blood group genotyping: from patient to high-throughput donor screening. Vox sanguinis. 2009;97:198–206. doi: 10.1111/j.1423-0410.2009.01209.x. [DOI] [PubMed] [Google Scholar]
- 79.U.S. Food and Drug Administration; 2014. [Accessed February 20, 2016]. Evaluation of the safety and effectiveness of the Immucor PreciseType™ HEA Molecular BeadChip Assay, manufactured by BioArray Solutions Limited [Transcript from March 18–19, 2014] at http://www.fda.gov/AdvisoryCommittees/CommitteesMeetingMaterials/BloodVaccinesandOtherBiologics/BloodProductsAdvisoryCommittee/ucm386681.htm. [Google Scholar]
- 80.Avent ND. Large-scale blood group genotyping: clinical implications. British journal of haematology. 2009;144:3–13. doi: 10.1111/j.1365-2141.2008.07285.x. [DOI] [PubMed] [Google Scholar]
- 81.Meyer S, Vollmert C, Trost N, et al. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion. 2014;54:3198–207. doi: 10.1111/trf.12715. [DOI] [PubMed] [Google Scholar]
- 82.Sapatnekar S, Figueroa PI. How do we use molecular red blood cell antigen typing to supplement pretransfusion testing? Transfusion. 2014;54:1452–8. doi: 10.1111/trf.12623. [DOI] [PubMed] [Google Scholar]
- 83.Westhoff CM, Sloan SR. Molecular genotyping in transfusion medicine. Clin Chem. 2008;54:1948–50. doi: 10.1373/clinchem.2008.116038. [DOI] [PubMed] [Google Scholar]
- 84.Hillyer CD, Shaz BH, Winkler AM, Reid M. Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfusion medicine reviews. 2008;22:117–32. doi: 10.1016/j.tmrv.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 85.Westhoff CM, Vege S, Halter-Hipsky C, et al. DIIIa and DIII Type 5 are encoded by the same allele and are associated with altered RHCE*ce alleles: clinical implications. Transfusion. 2010;50:1303–11. doi: 10.1111/j.1537-2995.2009.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 87.Meny GM. The Duffy blood group system: a review. Immunohematology / American Red Cross. 2010;26:51–6. [PubMed] [Google Scholar]
- 88.Wilkinson K, Harris S, Gaur P, et al. Molecular blood typing augments serologic testing and allows for enhanced matching of red blood cells for transfusion in patients with sickle cell disease. Transfusion. 2012;52:381–8. doi: 10.1111/j.1537-2995.2011.03288.x. [DOI] [PubMed] [Google Scholar]
- 89.Ribeiro KR, Guarnieri MH, da Costa DC, Costa FF, Pellegrino J, Jr, Castilho L. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox sanguinis. 2009;97:147–52. doi: 10.1111/j.1423-0410.2009.01185.x. [DOI] [PubMed] [Google Scholar]
- 90.Klapper E, Zhang Y, Figueroa P, et al. Toward extended phenotype matching: a new operational paradigm for the transfusion service. Transfusion. 2010;50:536–46. doi: 10.1111/j.1537-2995.2009.02462.x. [DOI] [PubMed] [Google Scholar]
- 91.Karafin MS, Field JJ, Gottschall JL, Denomme GA. Barriers to using molecularly typed minority red blood cell donors in support of chronically transfused adult patients with sickle cell disease. Transfusion. 2015;55:1399–406. doi: 10.1111/trf.13037. [DOI] [PubMed] [Google Scholar]
- 92.Kappler-Gratias S, Auxerre C, Dubeaux I, et al. Systematic RH genotyping and variant identification in French donors of African origin. Blood transfusion = Trasfusione del sangue. 2014;12(Suppl 1):s264–72. doi: 10.2450/2013.0270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]