Abstract
Background
Cyclin Y (CCNY) is a member of the cyclin family of proteins that regulate the cell cycle. The aims of this study were to compare the expression of CCNY in normal liver and human hepatocellular carcinoma (HCC), in normal and HCC cell lines, and in mouse HCC tumor xenografts.
Material/Methods
Tumor tissues from 55 patients diagnosed with HCC were studied for CCNY expression. Human HCC cell lines, SK-Hep1, HepG2, HEP3B, HuH7 and L02 were studied using the MTT cell proliferation assay, cell apoptosis, transwell and wound healing assays. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot were used to measure CCNY expression. Indirect immunofluorescence was used to assess cell apoptosis. In vivo xenograft mouse model was constructed and examined histologically.
Results
Expression of CCNY in human HCC tumor tissues was significantly increased when compared with adjacent normal liver (all P<0.05). HCC cells grown in vitro showed significantly increased expression of CCNY, cell proliferation, and migration, and a reduced rate of apoptosis, compared with cells with CCNY knockdown (siRNA) (all P<0.05). In the xenograft mouse model, tumor volume and weight in the CCNY overexpression group were significantly increased, compared with CCNY knockdown (siRNA) group (all P<0.05).
Conclusions
In tissue samples of human HCC, and human HCC cell lines, increased expression of CCNY was significantly associated with cell proliferation and migration. Further studies are recommended to evaluate the role of CCNY as a potential diagnostic biomarker or target for treatment in human HCC.
MeSH Keywords: Carcinoma, Hepatocellular; Neoplasm Invasiveness; Transcellular Cell Migration
Background
Liver cancer is the seventh most common cancer in the world, with a mortality rate of 19.7 cases per 100,000 populations in China [1]. Hepatocellular carcinoma (HCC) is the main type of liver cancer and has several known causes, including hepatitis virus B virus (HBV) infection, hepatitis C virus (HCV) infection, and other factors such as toxins and alcohol [2]. The causes of HCC may vary geographically. For example, HBV infection is associated with 80% of cases of HCC in East Asia, whereas HCV infection is a major risk factor for HCC in Europe [2]. About 40% of men and 15% of women who have had HBV infection in the perinatal period eventually developed liver cirrhosis or HCC [3].
Because HCC is of major concern for human health, particularly in East Asia, understanding the molecular mechanisms for the pathogenesis, progression, and prognosis of HCC may improve future diagnosis and treatment. For example, in 2015, Chiba et al. reported that abnormal regulation of certain signaling pathways, including p53/RB, Wnt/β-catenin, and the PI3K/PTEN/Akt/mTOR pathways were involved in the development of HCC [4]. Also, several members of the cyclin superfamily have been shown to have effects on the development of HCC [5]. The upregulation of cyclin D3 in HCC tissues has been shown to be an indicator of disease progression and is associated with tumor invasion [6]. Although HCC has been extensively studied, the mechanism of hepatocarcinogenesis remains unclear, and molecular studies on HCC-related genes have yet to lead to the identification of biomarkers in HCC that can be used diagnostically, prognostically, or as targets for therapy in HCC.
The family of cyclin-dependent kinases (CDKs) consists of 20 protein kinases that regulate cell cycle progression, transcription, and cell differentiation [7]. Cyclin Y (CCNY) is a highly conserved cell cycle protein in the superfamily of cyclins and is a regulatory subunit of CDK14, which regulates the cell cycle and transcription [8]. CCNY can phosphorylate and activate the low-density lipoprotein receptor-related protein 6 (LRP6) co-receptor, an important promoting regulator in the Wnt/β-catenin pathway [8]. Furthermore, CCNY facilitates cancer cell proliferation, and down-regulation of expression of CCNY has been shown to decreases cell proliferation and tumor growth of laryngeal cancer cells [9]. CCNY is also reported to have roles in adipogenesis, since it regulates the hepatic insulin signaling pathway, and also has a role in spermatogenesis [10,11]. Oncogenic Wnt/β-catenin signaling is reported to be activated in many types of cancer, including HCC [12]. It has been hypothesized that CCNY is associated with HCC through the non canonical Wnt pathways, and this has been supported by a study in CDK14-CCNY co-transfected cells [13].
However, there are still limited studies on the molecular mechanism associated with CCNY expression and HCC, although the relationship between CCNY and other human cancers have been studied [14–17]. Recent studies have shown that CCNY expression is associated with increased cell proliferation in human non small-cell lung cancer (NSCLC), and serum levels of anti-CCNY antibodies are increased in patients with NSCLC when compared with normal controls [14,16,17]. Expression of CCNY has also been shown to be increased in human colorectal carcinoma cell lines derived from metastatic carcinoma when compared with cell lines derived from non-metastatic colorectal carcinoma, suggesting a possible role of CCNY in tumor invasion and metastasis [15].
The aims of this study were to compare the expression of CCNY in normal liver and human hepatocellular carcinoma (HCC), in normal and HCC cell lines, and in mouse HCC tumor xenografts.
Material and Methods
Patients studied
This study was approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang Chinese Medical University. All patients who participated in the study signed an informed consent document.
Tissue samples from 55 patients who were diagnosed histologically with primary hepatocellular carcinoma (HCC) (Supplementary Figure 1A) were obtained, together with adjacent normal liver tissue as controls. The adjacent normal liver tissues were sampled from macroscopically normal liver and were located >2 cm from the liver tumors and were confirmed to include normal hepatic tissue without malignant cells following histological examination using routine hematoxylin and eosin (H&E) histochemical staining (Supplementary Figure 1B).
Clinical and pathological patient data were obtained during the study period, between January 2014 and June 2014, including gender, age, tumor size, tumor invasion, histological tumor grade, histological tumor subtype, the presence of lymph node metastasis, clinic stage, and histological grade. Patient data were retrieved from the Second Affiliated Hospital of Zhejiang Chinese Medical University.
All patients with HCC in the study underwent surgical tumor excision with HCC diagnosed histologically. According to the TNM staging criteria of HCC, published by the Union for International Cancer Control (UICC) [18], from the 55 HCC tissues in this study, there were 31 cases of grade I–II HCC and 24 cases of grade III–IV HCC. The average age of the patients was 65.0±8.5 years (28 cases ≤50 years; 27 cases >50 years), including 39 men and 16 women. As recommended by the World Health Organization (WHO) histological classification [19], 44 patients were diagnosed as classical HCC, five patients were diagnosed as combined cholangiocarcinoma-HCC, and six patients were diagnosed as mixed cell type HCC. There were 37 cases with lymph node metastasis. All 55 patients received some form of antineoplastic therapy. All sampled tissues were stored in liquid nitrogen until required for the study.
Cell lines
Human HCC cell lines, SK-Hep1, HepG2, HEP3B, HuH7 and the normal liver cell line L02 were obtained from the Institute of Biochemistry and Cell Biology (Shanghai, China). Cells were cultured at 37°C in Dulbecco’s Modified Eagle’s medium (DMEM) (Sigma, USA) with 10% fetal bovine serum (FBS) (Gibco, USA), 100 U/mL penicillin (Yansheng Industrial Co., Ltd, Shanghai, China), and 100 μg/mL streptomycin (Yansheng Industrial Co., Ltd, Shanghai, China) in humidified atmosphere with 5% CO2.
RNA extraction and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using Trizol reagent (Invitrogen, USA). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using the ABI7500 quantitative PCR Amplifier (Applied Biosystems, USA) to detect the cyclin Y (CCNY) mRNA expression level, normalized to the level of the internal control gene (β-actin). The primer sequences of CCNY and β-actin were purchased from Shanghai Sangon Biological Engineering Co., Ltd., China, and β-actin acted as the internal reference. The primer sequences of CCNY and β-actin are listed in Table 1. The reaction condition was set at 95°C for 10 min followed by 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 45 s. The experiments were conducted in triplicate, and the results were evaluated using the relative quantification approach (2−ΔΔCt).
Table 1.
Primers sequences used for quantitative RT-PCR.
| Genes | Primer pair sequences | |
|---|---|---|
| CCNY | F | 5′-GTCAGTCAACCAAACCTCAAG-3′ |
| R | 5′-GTCAGTCAACCAAACCTCAAG-3′ | |
| β-actin | F | 5′-GGCGGCACCACCATGTACCCT-3′ |
| R | 5′-AGGGGCCGGACTCGTCATACT-3′ |
F – forward primers; R – reverse primers.
Western blot
Total protein was extracted from hepatic tissues and cell lines using radioimmunoprecipitation assay (RIPA) buffer. The protein concentration was measured by the bicinchoninic acid (BCA) assay (Pierce, Rockford, IL, USA). The lysates of hepatic tissues and the hepatoma carcinoma cells were loaded onto 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto the polyvinylidene fluoride (PVDF) membrane. The primary goat anti-mouse antibodies at dilution 1: 800 (Beijing Zhongshan Biotechnology Company) included antibodies to CCNY and β-actin and were incubated on the membranes at 4°C overnight and washed for three times in Tris-buffered saline, 0.1% Tween 20 (TBST). The membranes were incubated with horseradish peroxidase-labeled secondary rabbit anti-goat antibodies at a dilution of 1: 2000, (Beijing Zhongshan Biotechnology Company) at room temperature for one hour. The chemiluminescence system (Snibe Co. Ltd., Shenzhen, China) was used to detect the band intensity of each group, with β-actin as the control. All antibodies were purchased from Enzyme-linked Biotechnology Co., Ltd., Shanghai, China.
CCNY expression, plasmid transfection, and RNA interference
CCNY cDNA (synthesized from Molbase, Shanghai, China) was cloned into the pGCsi-1H/Neo/GFP vector (Chemical Technology Co., Ltd., Shanghai, China). HepG2 cells were transfected with CCNY cDNA, empty vector, siRNA against CCNY and siRNA without specific target using Lipofectamine 2000 (Invitrogen) for 48 h. HepG2 cells were then divided into five groups: the control group (without transfection), the CCNY overexpression group (transfection of CCNY expression vector), the vector control group (transfection of empty vector), the CCNY siRNA group (transfection of siRNA against CCNY), and the siRNA control group (transfection of siRNA without specific target). Fluorescence microscopy was performed to assess the transfection efficiency of HepG2 cells. CCNY-siRNA (AAATGTGTCGCTCTTGCAATA) of reduced CCNY expression was selected due to its efficient reduction in CCNY mRNA expression in HepG2 cells, while siRNA (TTCTCCGAACGTGTCACGT) served as the negative control.
Cell proliferation assay
HepG2 cells (1×104 cells in 100 μl medium) from the different groups were added to 96-well plates and incubated for 24 hours under serum-starved conditions. The MTT cell proliferation assay tetrazolium dye (20 μl, 5 mg/ml, Sigma) was placed into each well and cultured for 4 h at 37°C in an incubator with 5% CO2. Then, 150 μl dimethyl sulfoxide (DMSO) (Sigma, St Quentin Fallavier, France) was added to each well, and cells were gently shaken for 10 min to dissolve the crystals. Absorbance was detected at 490 nm by a fluorescence plate reader (Bio-Rad, USA) on day 1, 2, 3, 4, and 5, respectively. Wells with medium only were used as the blank control and cell viability was confirmed by measuring the optical density (OD) ratio between the treated culture group and the untreated control group. The assays were performed in triplicate.
Indirect immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 min and then washed twice with phosphate-buffered saline (PBS) and Dulbecco’s Formula Modified (ICN Biochemicals, UK). TritonX-100 solution (0.2%) was used to permeabilize the cells for 15 min, and then the cells were treated with 3% bovine serum albumin (BSA) (Boehringer GmbH, Mannheim, Germany) at room temperature for 30 min. The cells were incubated with rabbit anti-CCNY antibody (1: 50), and 50–60 μl primary antibody diluent (PBS with 0.2% TritonX-100 and 3% BSA) was added onto each coverslip. Next day, goat-anti-rabbit IgG labeled with fluorescein isothiocyanate (FITC) (1: 50) was added and the cells were incubated for 30 min, and then the nuclear dye was added followed by incubation of the cells in the dark for 20 min. Then, 30 μl of the fluorescence decay resistant agent was added onto the glass slide, followed by the addition of a glass coverslip, slide mounting, and the cells were dried in the dark for 30 min, and stored in the dark −20ºC.
Transwell assay
A transwell chamber (8-mm pore size) (Costar, UK) was placed into the wells of 12-well culture plates coated with Matrigel (BD Bioscience, USA) to measure cell migration (invasion). A total of 2.5×104 cells were added to the upper chamber (Corning, USA) and filled with serum-free medium, while the lower chamber was cultured with the complete medium. After 12-hour incubation, cells from the upper chamber were carefully removed using a cotton swab; cells that migrated or invaded into the lower chamber were fixed and stained with eosin. Invading cells were counted in five random visual fields using microscopy (Olympus, Japan). The assays were performed in triplicate.
Cell apoptosis assay
Apoptosis of transfected cells was detected using the Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (BD Pharmingen, USA) and analyzed using a FACS Calibur flow cytometry (Beckman FC400 MPL, USA), Annexin V-FITC and propidium iodide (PI) staining. Cells with negative Annexin V and negative PI staining represented normal cells; cells with positive Annexin V and negative PI represented cell apoptosis; cells with positive Annexin V and positive PI staining represented cell necrosis. Fluorescence results of stained cells were also obtained via the Olympus FluoView™ 1000 microscope (CME-UFRGS). The assays were performed in triplicate.
Wound healing assay
A wound healing assay was used to analyze cell motility and migration. Cells in each group were seeded into six-well plates at an adjusted density of 2.5×105/mL for 24 h before wounding by scraping. The wound scraping was done in the middle of the cell monolayer using a sterile micropipette tip. After the detached cells were flushed with PBS and removed, the cells were cultured with medium without serum. Images were captured microscopically at 0 and 12 h and analyzed using Image-Pro Plus (version 6.0) to evaluate the length and size of the scratch and healing.
HCC xenograft tumor model
A total of 150 5-week-old female nude mice were purchased from the Shanghai Animal Experiment Center. The 150 HCC xenograft tumor model mice were randomly allocated into 5 groups, each with 30 mice: the control group (HepG2 cells without transfection were injected into the mice); the CCNY overexpression group (HepG2 cells transfected with CCNY expression vector were injected into the mice); the vector control group (HepG2 cells transfected with empty vector were injected into the mice); the CCNY siRNA group (HepG2 cells transfected with CCNY siRNA was injected into the mice); and the siRNA control group (HepG2 cells transfected without specific target was injected into the mice). Then, 5×106 treated HepG2 cells were subcutaneous injected on the back of each mouse. The injection process was repeatedly conducted over a period of 40 days.
Tumor volume measurements were carried out with calipers, every four days, over the 40-day period using the following formula: volume=(A×B2)/2, where A and B were the largest and the smallest diameters, respectively. Mice were sacrificed and tumors were extracted for imaging and weighing. Representative tumors in each group were fixed in formalin and embedded in paraffin, 5-mm sections were cut and stained with hematoxylin and eosin (H&E) and evaluated by light microscopy. Data were collected from fifteen mice, three mice in each group. The experiments complied with the Animal Management Rule of the Chinese Ministry of Health (Documentation 55, 2001), and the protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang Chinese Medical University.
Statistical analysis
All data analysis from experiments performed in triplicate was performed using SPSS 21.0 software (Chicago, Ill, USA). Measurement data were expressed in the form of mean ± standard deviation (SD) and were analyzed using the Student’s t-test or one-way ANOVA analysis of variance. The Chi-squared test was used to analyze between-group comparisons of categorical variables (counted data). P<0.05 was considered to be statistically significance.
Results
Cyclin Y (CCNY) expression in hepatocellular carcinoma (HCC) and patient clinical characteristics
The correlation between the relative mRNA expression level of CCNY and the clinicopathological features of patients with HCC are shown in Table 2. The findings showed that the expression level of CCNY was closely correlated with tumor size, tumor invasion, lymph node metastasis and histological grade (all P<0.05); there was no significant correlation between CCNY expression and age, gender, clinic stage, or pathological types (all P>0.05).
Table 2.
The correlation between CCNY mRNA expression and clinicopathologic features of HCC patients.
| Clinicopathologic features | N | CCNY mRNA level | P-value |
|---|---|---|---|
| Age | |||
| ≤50 | 28 | 3.68±0.73 | 0.307 |
| >50 | 27 | 3.46±0.85 | |
| Gender | |||
| Male | 39 | 3.60±0.79 | 0.675 |
| Female | 16 | 3.50±0.82 | |
| Pathological types | |||
| Hepatocellular type | 44 | 3.58±0.79 | 0.728 |
| Cholangiocarcinoma type | 5 | 3.34±0.67 | |
| Mixed cell type | 6 | 3.71±0.75 | |
| Tumor size | |||
| ≤5 cm | 13 | 3.21±0.65 | 0.038* |
| >5 cm | 42 | 3.68±0.71 | |
| Tumor invasion | |||
| T1+T2 | 17 | 3.30±0.56 | 0.030* |
| T3+T4 | 38 | 3.75±0.74 | |
| Lymph node metastasis | |||
| N0 | 37 | 3.41±0.70 | 0.041* |
| N+ | 18 | 3.87±0.88 | |
| Clinic stages | |||
| I–II | 31 | 3.65±0.84 | 0.406 |
| III–IV | 24 | 3.47±0.72 | |
| Histological grade | |||
| I+II | 33 | 3.40±0.63 | 0.035* |
| III+IV | 22 | 3.83±0.84 | |
HCC – hepatocellular carcinoma; CCNY – Cyclin Y;
P-value significant differences.
CCNY expression was increased in HCC liver tissues
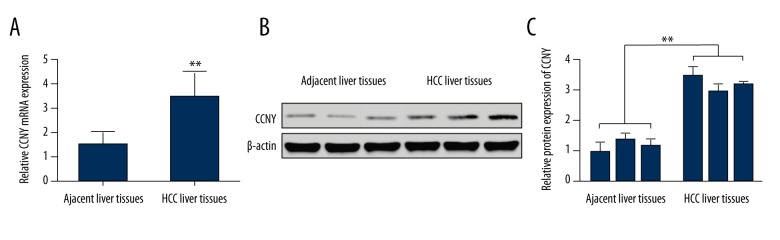
The quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot detected CCNY expression in 55 HCC liver tissues and 55 targeted adjacent normal liver tissues. As shown in Figure 1A, the mRNA level of CCNY in HCC tissues was significantly greater compared with adjacent normal liver tissues (P<0.01). Results from Western blot showed that the CCNY protein level in HCC tissues was increased when compared with adjacent normal liver tissues (Figure 1B). The relative protein expressions of CCNY in HCC tissues was increased when compared with adjacent normal liver tissues (Figure 1C).
Figure 1.
Expression of cyclin Y (CCNY) in human tissue samples of hepatocellular carcinoma (HCC) and adjacent normal liver tissue. (A) The relative mRNA levels of CCNY in HCC liver tissues were measured by qRT-PCR, and adjacent liver tissues were used as controls. ** Significant difference, P<0.01. (B) Western blot showed the expression of CCNY in HCC liver tissues and adjacent liver tissues. (C) The histogram of the relative protein expressions of CCNY in HCC liver tissues, and adjacent liver tissues were used as controls. ** Significant difference, P<0.01
CCNY was upregulated in HCC cells
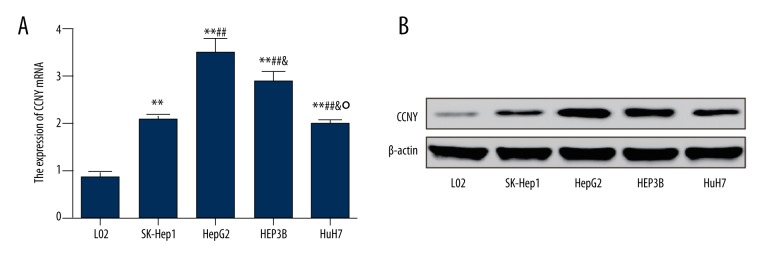
The results of qRT-PCR and Western blot for expression of CCNY differed in in the six cell lines. As shown in Figure 2A, the expression of CCNY mRNA in the cell lines, HepG2, HEP3B, SK-Hep1, and HuH7 were significantly increased compared with the normal liver cell line L02, and the expression of CCNY mRNA in the HepG2 was the highest (all P<0.01). Expressions of CCNY protein in the HepG2, HEP3B, SK-Hep1, and HuH7 cell lines were increased compared with the normal cell line L02 (Figure 2B).
Figure 2.
Expression of cyclin Y (CCNY) in liver cell lines. (A) The relative mRNA levels of CCNY in different cell lines were detected using qRT-PCR. (B) The expression of CCNY in different cell lines was measured by Western blot. Normal liver cell line L02 was used as controls. ** P<0.01 compared with L02 group; ## P<0.01 compared with SK-Hep1 group; & P<0.05 compared with HepG2 group; ° P<0.05 compared with HEPSB group.
These results were consistent with the findings of the expression of CCNY observed in HCC tissue from patients, which further supported the finding that the expression of CCNY was increased in patients with HCC. Since the expression of CCNY in the HepG2 cell line was the highest in comparison to other cell lines, HepG2 cells were selected in the following experiments to investigate the role of CCNY further.
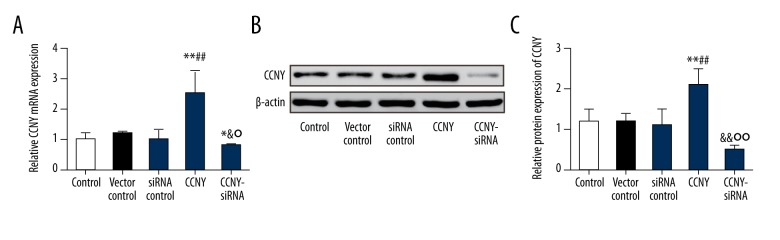
CCNY expression in HepG2 cells following transfection
Fluorescence microscopy was performed to assess the transfection efficiency of HepG2 cells. The transfection efficiency was increased to more than 95% in this study. Also, we conducted qRT-PCR to detect the expression of CCNY mRNA 48 hours following transfection. The mRNA expression level of CCNY in the CCNY siRNA group was reduced compared with that in the siRNA control group, while the mRNA level of CCNY in the CCNY overexpression group was greater than that found in the vector control group (all P<0.01) (Figure 3A). There was no significant difference in the expression of CCNY mRNA between the control group, the siRNA control group, and the vector control group. CCNY protein expression between the different groups showed the same trend as shown by qRT-PCR (P<0.01) (Figure 3B, 3C).
Figure 3.
Expression of cyclin Y (CCNY) in HepG2 cells using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blotting following transfection. (A) The relative CCNY mRNA expression in different groups of HepG2 cells was measured using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). * P<0.05; ** P<0.01 compared with control group; # P<0.05; ## P<0.01 compared with vector control group; & P<0.05 compared with siRNA control group; ° P<0.05 compared with CCNY overexpression group. (B) Expression of CCNY in different groups of HepG2 cells was detected using Western blot, and β-actin was used as a control. (C) The histogram of the relative protein expressions of CCNY in different groups of HepG2 cells, ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; && P<0.01 compared with siRNA control group; °° P<0.01 compared with CCNY overexpression group.
CCNY expression promoted HepG2 cell proliferation
Results from cell proliferation assay are shown in Figure 4. The OD 490 value in the CCNY siRNA group was less than that in the control group and the siRNA control group; the value in the CCNY overexpression group was significantly greater than that in the control group and vector control group (all P<0.01). No significant differences in the cell proliferation of HepG2 were found between the control group, the siRNA control group, and the vector control group.
Figure 4.
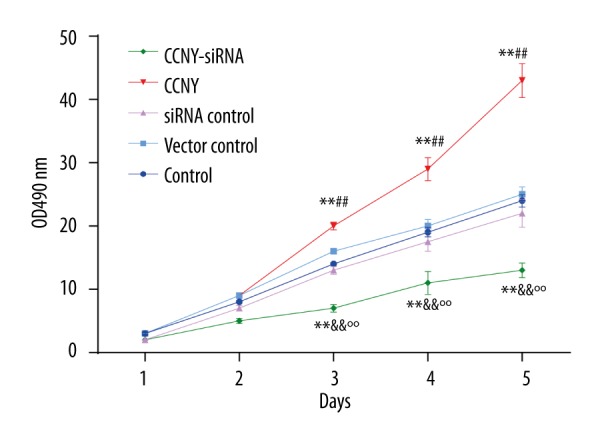
HepG2 cell proliferation in the control group, the cyclin Y (CCNY) overexpression group, the vector control group, CCNY siRNA group, and the siRNA control group. Findings from the cell proliferation assay. The value of the OD490 reflected the number of surviving cells indicating that CCNY overexpression promoted HCC cell proliferation. ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; && P<0.01 compared with siRNA control group; °° P<0.01 compared with CCNY overexpression group.
From the second day, the CCNY overexpression group showed a significant upward trend, while the OD value of the CCNY siRNA group showed a steady uptrend, but was still far lower than that of vector control group and siRNA control group. Cells in the CCNY overexpression group continued to grow substantially, whereas the proliferation was reduced and the growth was stabilized in the CCNY siRNA group on the fifth day. These results indicated that CCNY might effect the proliferation of HCC cells.
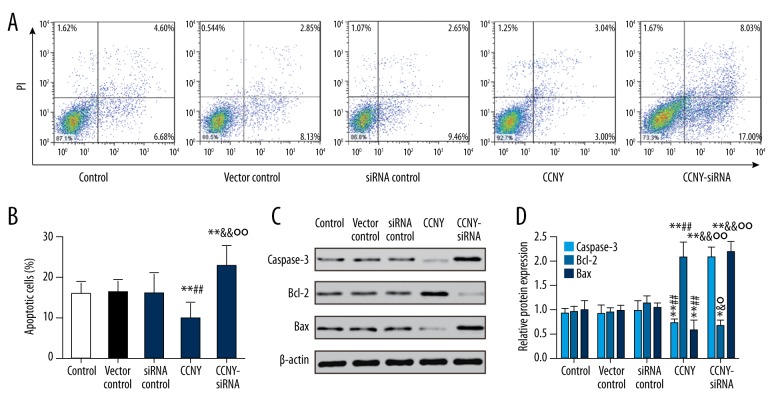
CCNY expression inhibited HepG2 cell apoptosis
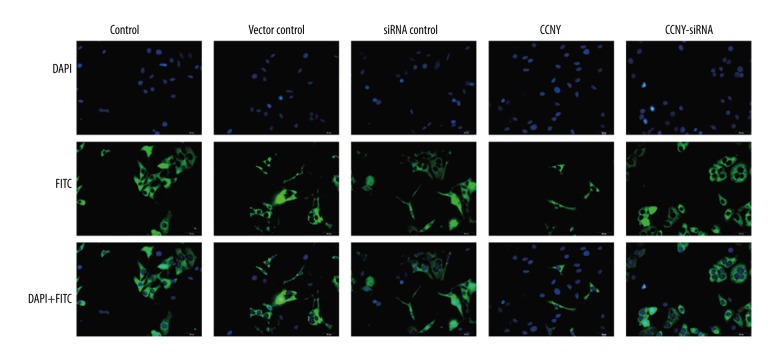
As shown in Figure 5A and 5B, compared with the control group and the vector control group, the CCNY overexpression group had a significantly reduced apoptosis rate (both P<0.01). Furthermore, the apoptosis rate of HepG2 cells was increased in the CCNY siRNA group compared with the control group and the siRNA control group (both P<0.01). Figure 5C and 5D show the expressions of the apoptotic proteins caspase-3 and Bax were much lower in the CCNY overexpression group compared with the control group and the vector control group, whereas CCNY expression was increased in the CCNY siRNA group compared with the control group and the siRNA control group. Compared with the control group and the vector control group, the apoptotic protein Bcl-2 had increased expression in the CCNY overexpression group, whereas Bcl-2 had lower expression in the CCNY siRNA group compared with the control group and the siRNA control group. There was no significant difference in apoptosis rate between the control group, the siRNA control group, and the vector control group. Therefore, CCNY may suppress HCC cell apoptosis. Figure 6 shows the FITC-labeled annexin in the CCNY overexpression group appeared to be less than that in control group, and silencing of CCNY promoted cell apoptosis when compared with the control group.
Figure 5.
Effects of cyclin Y (CCNY) expression on HepG2 cell apoptosis in control group, CCNY overexpression group, vector control group, CCNY siRNA group, and siRNA control group. The flow cytometry findings with Annexin V and propidium iodide (PI) double staining. (A) The effects of CCNY on HepG2 cell apoptosis in different groups. (B) Histogram of the cell apoptosis of HepG2 cells. ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; && P<0.01 compared with siRNA control group; °° P<0.01 compared with CCNY overexpression group. (C) Expression of apoptosis protein in different groups of HepG2 cells was detected using Western blot, and β-actin was used as a control. (D) The histogram of the relative protein expressions of apoptosis protein in different groups of HepG2 cells, * P < 0.05; ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; & P<0.05; && P<0.01 compared with siRNA control group; ° P<0.05; °° P<0.01 compared with the CCNY overexpression group.
Figure 6.
Photomicrographs of the immunofluorescence images of hepatocellular carcinoma (HCC) cells stained with 4′, 6-diamidino-2-phenylindole (DAPI), fluorescein isothiocyanate (FITC), and DAPI + FITC. DAPI stained the cell nuclei (blue), and FITC-labelled Annexin V indicate apoptotic cells (green). Scale bar, 10 μm. HCC – hepatocellular carcinoma; DAPI – 4′, 6-diamidino-2-phenylindole; FITC – fluorescein isothiocyanate. (×50 original magnification).
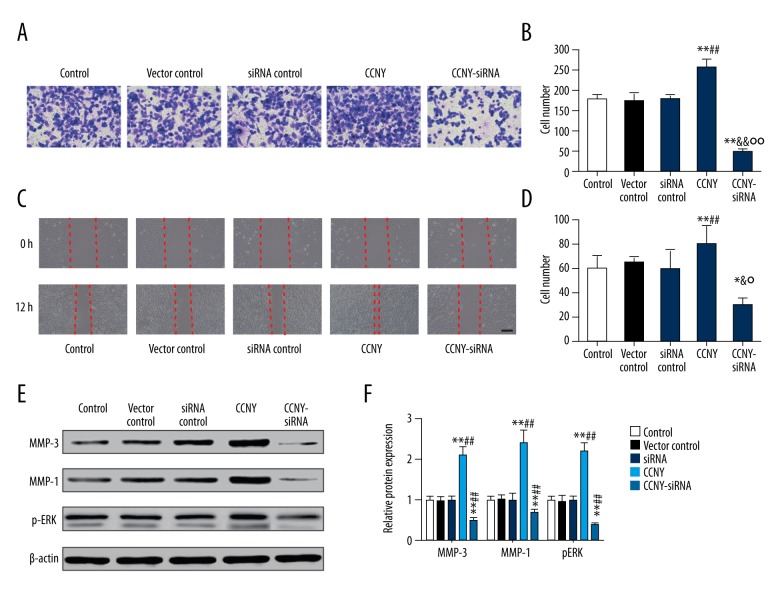
CCNY expression facilitated HepG2 cell invasion and migration
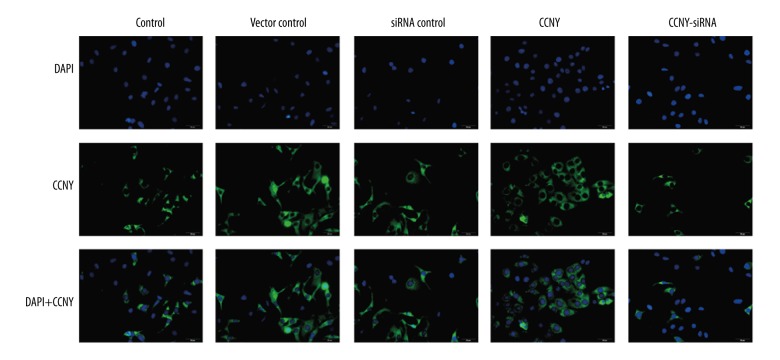
As shown Figure 7A and 7B, the number of migrating (invading) cells in the CCNY overexpression group (265.19±27.94) were significantly increased compared with the control group (180.13±15.14) and the vector control group (170.99±26.13) whereas this finding in the CCNY siRNA group (50.71±13.68) was significantly lower compared with the control group (180.13±15.14) and the siRNA control group (175.00±19.19) (all P<0.01). The closed wound area in the CCNY overexpression group was greater than that in the control group and vector control group, while the closed wound area in the CCNY siRNA group was smaller than that in the control group and vector control group (all P<0.01) (Figure 7C, 7D). Figure 7E and 7F show that the expression of the cell migration and invasion-related proteins were much greater in the CCNY overexpression group compared with the control group and the vector control group, whereas the expressions were reduced in the CCNY siRNA group compared with the control group and the siRNA control group. There was no significant difference between the control group, the vector control group, and the siRNA control group. Also, the number of migrated cells for each group showed similar trends (all P<0.01). The expression of CCNY expressions differed in their distribution within the HCC cell membranes (Figure 8), suggesting that the shapes of the tumor cells in HCC were heterogeneous.
Figure 7.
Effects of cyclin Y (CCNY) expression on invasion and migration of HepG2 cells in the control group, the CCNY overexpression group, the vector control group, the CCNY siRNA group, and the siRNA control group. The findings of the transwell assay at 48 h following transfection. (A) The cell migration was visualized by microscopy. (B) Histogram of the invasion rate of HepG2 cells. ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; && P<0.01 compared with siRNA control group; °° P<0.01 compared with CCNY overexpression group. (C) The cell migration was visualized by microscopy. (D) Histogram of the migration rate of HepG2 cells. ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; & P<0.01 compared with siRNA control group; ° P<0.01 compared with CCNY overexpression group. (E) Expression of migration factor in different groups of HepG2 cells was detected using Western blot, and β-actin was used as a control. (F) The histogram of the relative protein expressions of migration factor in different groups of HepG2 cells, ** P<0.01 compared with control group; ## P<0.01 compared with vector control group.
Figure 8.
Photomicrographs of the immunofluorescence staining showing the heterogeneity of hepatocellular carcinoma (HCC) cells. DAPI stained nuclei (blue) and CCNY was stained in green. Scale bar, 10 μm. HCC – hepatocellular carcinoma; DAPI – 4′, 6-diamidino-2-phenylindole; CCNY – Cyclin Y; FITC – fluorescein isothiocyanate. (×50 original magnification).
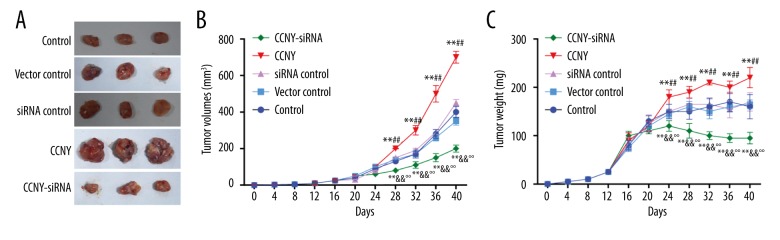
CCNY expression promoted HCC tumor growth in tumor xenografts in vivo
As shown in Figure 9, both tumor volumes and tumor weights in the CCNY overexpression group (698.32±32.53 mm3, 220.21±21.03 mg) were greater than those in the vector control group (350.42±22.37 mm3, 165.17±9.02 mg) after the corresponding treatments were injected over the 40-day period. Tumor volumes and weights in the CCNY siRNA group (200.76±19.79 mm3, 95.19±12.31 mg) and were less than in the siRNA control group (all P < 0.01). From the 28th day, the CCNY overexpression group had an upward trend of the tumor volumes and a decline in the tumor volumes in the CCNY siRNA group from the 24th day compared with the vector control group and siRNA control group. The weight of the tumors increased at the 12th day, and the tumor weights of the CCNY overexpression group were considerably greater, and the tumor weights of the CCNY group showed a decline from the 24th day. There was no significant difference between tumor volumes and weights among the control group, the vector control group, and the siRNA control group.
Figure 9.
Effects of cyclin Y (CCNY) expression on hepatocellular carcinoma (HCC) tumor growth in each xenograft model group. (A) Tumor volumes in control group, CCNY overexpression group, vector control group, CCNY siRNA group and siRNA control group. (B) Tumor weights in the control group, the CCNY overexpression group, the vector control group, the CCNY siRNA group, and the siRNA control group. Compared with the control group, the tumor volumes and weights of CCNY overexpression group were significantly increased; in the CCNY siRNA group tumor volumes and weights were significantly reduced. * P<0.05; ** P<0.01 compared with control group; ## P<0.01 compared with vector control group; & P<0.05; && P<0.01 compared with siRNA control group; °° P<0.01 compared with CCNY overexpression group.
Discussion
The findings of this study to investigate the role of cyclin Y (CCNY) in the proliferation and migration of human hepatocellular carcinoma (HCC) cells showed that the expression of CCNY in human HCC tumor tissues was significantly increased when compared with adjacent normal liver and that HCC cells grown in vitro showed significantly increased expression of CCNY, cell proliferation, and migration, and a reduced rate of apoptosis, compared with cells with CCNY knockdown (siRNA). In a xenograft mouse model, tumor volume and weight in the CCNY overexpression group were significantly increased, compared with CCNY knockdown (siRNA) group.
Carcinoma used to be believed to be due to the abnormal expression of oncogenes and tumor suppressor genes, but there is now increasing evidence that carcinogenesis involves changes in the cell cycle [14]. Cyclin, cyclin-dependent kinase (CDK), and cyclin-dependent kinase inhibitor (CDKI) modulate the cell cycle at the molecular level [20]. The cyclin superfamily of proteins is part of a network that regulates cell growth, proliferation, and apoptosis [7]. All the members of cyclins contain a conserved sequence that includes 100 to 150 amino acids or cyclin box that binds CDK to regulate the cell cycle and cell proliferation [21]. Previously published studies have shown that cyclin participates in a number of processes for tumor formation; cyclin A is reported to be related to the development of glioma and ovarian carcinoma [22,23], while cyclin D overexpression is associated with breast carcinoma and esophageal squamous cell carcinoma, and cyclin E is highly expressed in both bladder carcinoma and colorectal carcinoma [24–27]. Recently, a study conducted by Sun et al. reported that the interaction between CCNY and serine/threonine-protein kinase PFTAIRE-1 (PFTK1) might affect HCC through a non-canonical Wnt signaling pathway [28]. A further study by Yue et al. showed that the expression of CCNY was significantly increased in NSCLC [16]. A previous study has reported that CCNY can enhance cell proliferation, colony formation, cell progression and tumorigenesis in glioma cells [29]. The results from the in vivo xenograft study showed that the volumes and weights of tumors were increased when CCNY was overexpressed, which is a finding supported by the study published by Yue et al., that showed a positive correlation between tumor size and CCNY expression [17]. The findings of the present study showed a significant increase in the expression of CCNY in HCC tissues compared with normal adjacent liver tissues, which add to the recent studies on cyclins, including CCNY, and malignancy.
This study has several limitations. Since only one HCC cell line, HepG2, was filtered in our study, the influence of CCNY on HCC proliferation and apoptosis may not be representative for HCC in the findings using this cell line. Also, our study demonstrated a potential relationship between CCNY and cell proliferation, and cell migration (invasion) of HCC cells. ‘However, the specific mechanisms for the role of CCNY in cell proliferation, invasion, and metastasis for HCC should be studied further.
Conclusions
In tissue samples of human HCC, and human HCC cell lines, increased expression of CCNY was significantly associated with cell proliferation and migration. Further studies are recommended to evaluate the role of CCNY as a potential diagnostic biomarker or target for treatment in human HCC.
Supplementary Figure
Photomicrographs of the histology of the liver tissues. (A) Histology of the light microscopy of the hepatocellular carcinoma (HCC) tissue. Routine staining with hematoxylin and eosin (H&E). (Magnification ×50). (B) Histology of the light microscopy of the normal liver tissue adjacent to the tumor. Routine staining with hematoxylin and eosin (H&E). (Magnification ×50).
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116:544–73. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–63. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 4.Chiba T, Suzuki E, Saito T, et al. Biological features and biomarkers in hepatocellular carcinoma. World J Hepatol. 2015;7:2020–28. doi: 10.4254/wjh.v7.i16.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu C, Lin LI, Cheng YC, et al. Cyclin E1 inhibition can overcome sorafenib resistance in hepatocellular carcinoma cells through Mcl-1 suppression. Clin Cancer Res. 2016;22(10):2555–64. doi: 10.1158/1078-0432.CCR-15-0499. [DOI] [PubMed] [Google Scholar]
- 6.Huang B, Li H, Huang L, Luo C, Zhang Y. Clinical significance of microRNA 138 and cyclin D3 in hepatocellular carcinoma. J Surg Res. 2015;193:718–23. doi: 10.1016/j.jss.2014.03.076. [DOI] [PubMed] [Google Scholar]
- 7.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–39. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Guest S, Finley RL., Jr Why cyclin Y? A highly conserved cyclin with essential functions. Fly (Austin) 2010;4:278–82. doi: 10.4161/fly.4.4.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson G, Niehrs C. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends Cell Biol. 2010;20:453–60. doi: 10.1016/j.tcb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Sode J, Vogel U, Bank S, et al. Genetic variations in pattern recognition receptor loci are associated with anti-TNF response in patients with rheumatoid arthritis. PLoS One. 2015;10:e0139781. doi: 10.1371/journal.pone.0139781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikolcevic P, Sigl R, Rauch V, et al. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol Cell Biol. 2012;32:868–79. doi: 10.1128/MCB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla R, Upton KR, Munoz-Lopez M, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–11. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T, Co NN, Wong N. PFTK1 interacts with cyclin Y to activate non-canonical Wnt signaling in hepatocellular carcinoma. Biochem Biophys Res Commun. 2014;449:163–68. doi: 10.1016/j.bbrc.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda Y, Wakai T, Kubota M, et al. Clinical significance of cell cycle inhibitors in hepatocellular carcinoma. Med Mol Morphol. 2013;46:185–92. doi: 10.1007/s00795-013-0047-7. [DOI] [PubMed] [Google Scholar]
- 15.Ying-Tao Z, Yi-Ping G, Lu-Sheng S, Yi-Li W. Proteomic analysis of differentially expressed proteins between metastatic and non-metastatic human colorectal carcinoma cell lines. Eur J Gastroenterol Hepatol. 2005;17:725–32. doi: 10.1097/00042737-200507000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Yue W, Zhao X, Zhang L, et al. Overexpression of cyclin Y in non-small cell lung cancer is associated with cancer cell proliferation. Sci China Life Sci. 2010;53:511–16. doi: 10.1007/s11427-010-0090-8. [DOI] [PubMed] [Google Scholar]
- 17.Yue W, Zhao X, Zhang L, et al. Cell cycle protein cyclin Y is associated with human non-small-cell lung cancer proliferation and tumorigenesis. Clin Lung Cancer. 2011;12:43–50. doi: 10.3816/CLC.2011.n.006. [DOI] [PubMed] [Google Scholar]
- 18.Ochiai T, Ishii H, Yamamoto Y, et al. Significance of hepatectomy for AJCC/UICC T3 hepatocellular carcinoma. Anticancer Res. 2015;35:2921–28. [PubMed] [Google Scholar]
- 19.Shafizadeh N, Genrich G, Ferrell L, Kakar S. Hepatocellular adenomas in a large community population, 2000 to 2010: reclassification per current World Health Organization classification and results of long-term follow-up. Hum Pathol. 2014;45:976–83. doi: 10.1016/j.humpath.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Gerard C, Tyson JJ, Coudreuse D, Novak B. Cell cycle control by a minimal Cdk network. PLoS Comput Biol. 2015;11:e1004056. doi: 10.1371/journal.pcbi.1004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez I, Dynlacht BD. New insights into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol. 2005;16:311–21. doi: 10.1016/j.semcdb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Miao C, Wang Z, Yang J, et al. Expression and mutation analysis of Cyclin A and Ki-67 in glioma and their correlation with tumor progression. Oncol Lett. 2015;10:1716–20. doi: 10.3892/ol.2015.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arsenic R, Braicu EI, Letsch A, et al. Cancer-testis antigen cyclin A1 is broadly expressed in ovarian cancer and is associated with prolonged time to tumor progression after platinum-based therapy. BMC Cancer. 2015;15:784. doi: 10.1186/s12885-015-1824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czapiewski P, Welnicka-Jaskiewicz M, Seroczynska B, et al. CD99 correlates with low cyclin D1, high topoisomerase 2 status and triple negative molecular phenotype but is prognostically irrelevant in breast carcinoma. Pol J Pathol. 2015;66:269–75. doi: 10.5114/pjp.2015.54961. [DOI] [PubMed] [Google Scholar]
- 25.Guimaraes EP, Carli ML, Sperandio FF, et al. Cyclin D1 and Ki-67 expression correlates to tumor staging in tongue squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20:e657–63. doi: 10.4317/medoral.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rath-Wolfson L, Bergman M, Ori Y, et al. Expression of cyclin E in stage III colorectal carcinoma. Oncol Lett. 2013;5:145–48. doi: 10.3892/ol.2012.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan G, Tang T. Expression of cyclin D1 and cyclin E in urothelial bladder carcinoma detected in tissue chips using a quantum dot immunofluorescence technique. Oncol Lett. 2015;10:1271–76. doi: 10.3892/ol.2015.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, Yuan F, Shen X, et al. Role of COMT in ADHD: A systematic meta-analysis. Mol Neurobiol. 2014;49:251–61. doi: 10.1007/s12035-013-8516-5. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Wang Z, Wang J, et al. Lentivirus-mediated knockdown of cyclin Y (CCNY) inhibits glioma cell proliferation. Oncol Res. 2010;18:359–64. doi: 10.3727/096504010x12644422320582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Photomicrographs of the histology of the liver tissues. (A) Histology of the light microscopy of the hepatocellular carcinoma (HCC) tissue. Routine staining with hematoxylin and eosin (H&E). (Magnification ×50). (B) Histology of the light microscopy of the normal liver tissue adjacent to the tumor. Routine staining with hematoxylin and eosin (H&E). (Magnification ×50).