Abstract
Despite the need for inducible promoters in strain development efforts, the majority of engineering in Saccharomyces cerevisiae continues to rely on a few constitutively active or inducible promoters. Building on advances that use the modular nature of both transcription factors and promoter regions, we have built a library of hybrid promoters that are regulated by a synthetic transcription factor. The hybrid promoters consist of native S. cerevisiae promoters, in which the operator regions have been replaced with sequences that are recognized by the bacterial LexA DNA binding protein. Correspondingly, the synthetic transcription factor (TF) consists of the DNA binding domain of the LexA protein, fused with the human estrogen binding domain and the viral activator domain, VP16. The resulting system with a bacterial DNA binding domain avoids the transcription of native S. cerevisiae genes, and the hybrid promoters can be induced using estradiol, a compound with no detectable impact on S. cerevisiae physiology. Using combinations of one, two or three operator sequence repeats and a set of native S. cerevisiae promoters, we obtained a series of hybrid promoters that can be induced to different levels, using the same synthetic TF and a given estradiol. This set of promoters, in combination with our synthetic TF, has the potential to regulate numerous genes or pathways simultaneously, to multiple desired levels, in a single strain.
Keywords: hybrid promoter, Saccharomyces, strain engineering, synthetic biology
1. INTRODUCTION
The yeast Saccharomyces cerevisiae is one of the most well‐understood and widely used organisms in biological research as well as in bio‐production processes. In addition to natively produced ethanol, S. cerevisiae has been engineered to produce a wide variety of products including pharmaceuticals, fuels and industrial chemicals (Peralta‐Yahya, Zhang, del Cardayre, & Keasling, 2012). For optimizing the production of bulk chemicals, multiple traits, in addition to an optimal biosynthetic pathway, must be developed (Lechner, Brunk, & Keasling, 2016). These necessitate the use of multigenic pathways and genes that modulate other phenotypes, such as for tolerance to the final product (Mukhopadhyay, 2015), pretreatment reagents (Frederix et al., 2014) and optimal carbon uptake (Reider Apel, Ouellet, Szmidt‐Middleton, Keasling, & Mukhopadhyay, 2016). Yet the number of promoters used for genetic engineering in S. cerevisiae has remained limited to a few dozen native promoters, either constitutive promoters or galactose‐inducible promoters that are the staple of the yeast genetic engineer's toolbox (Alper, Fischer, Nevoigt, & Stephanopoulos, 2005; Lee, DeLoache, Cervantes, & Dueber, 2015; Reider Apel et al., 2017). In cases where promoter inducibility is desired, galactose induction is particularly problematic because of the limitations it places on the types of carbon sources that can be used for cultivation.
Both promoter sequences and transcription factors (TF) are modular in nature. The first effort to use this aspect of regulatory regions and DNA binding proteins was a synthetic TF for S. cerevisiae reported in 1993 (Louvion, Havaux‐Copf, & Picard, 1993) that used the Gal4 protein as the scaffold. By simply replacing the ligand binding domain of Gal4 with a human estrogen binding domain and using the VP16 viral activators protein that recruits the RNA Pol II complex, the authors could obtain gratuitous regulation of galactose responsive genes in response to estradiol. Although promising, this system was not used in any reported metabolic engineering efforts, possibly owing to the fact that the TF binds to native Gal responsive promoters, leading to crosstalk and unwanted changes in metabolism. More recently, McIssac and coauthors developed a similar system, where they replaced the Gal4 DNA binding domain (DBD) with zinc‐finger DBDs (McIsaac et al., 2013). Further, using a variable number of operator sequences in the corresponding promoters, they obtained superior dynamic range for the synthetic TF and promoter combinations. Yet the system suffers from low controllability at low‐ to mid‐range expression levels, owing to high basal activity of the zinc fingers (McIsaac, Gibney, Chandran, Benjamin, & Botstein, 2014). Similarly, Ottoz et al. have developed a variation that uses the DBD from the bacterial TF, LexA. The authors tested several variations of the LexA‐based TF where they varied the activating domain to obtain a range of highly regulated TF–promoter combinations, adjusting the output by increasing the number of LexA‐binding sites, without changing basal promoter activity (Ottoz, Rudolf, & Stelling, 2014). Another recent study achieved a similar range of expression levels without the need for externally added inducer compounds (Rantasalo et al., 2016).
Alternative strategies to enable similar regulatory control have focused primarily on modifying the promoter regions for a gene of interest, changing the inducing molecule, or using tunable CRISPR‐based transcription factors (crisprTFs). Examples include Blazeck, Garg, Reed, and Alper (2012), where a large number of native yeast promoters were used as the basis for a hybrid promoter that split the promoter into intact core regions and altered upstream activation sequences, and a similar, but broader, approach that altered both the upstream regions of native promoters as well as the corresponding synthetic DBDs using zinc finger motifs (Khalil et al., 2012). In addition to galactose and estradiol regulated promoters, tetracycline‐inducible and camphor‐repressible versions have been created to regulate gene expression (Cuperus, Lo, Shumaker, Proctor, & Fields, 2015; Garí, Piedrafita, Aldea, & Herrero, 1997; Ikushima, Zhao, & Boeke, 2015). dCas9 has also been used as a RNA‐guided scaffold to recruit different protein effectors, thereby resulting in gene modulation (Gilbert et al., 2013). Targeting crisprTFs to sequences upstream of TATA boxes resulted in gene activation that was further enhanced through addition of multiple operator sites (Farzadfard, Perli, & Lu, 2013).
In this study, we used the LexA DBD, fused with the human estrogen binding domain and the viral activator domain, VP16, as the synthetic‐chimeric TF. Then, by using a combination of operator sequences in promoter scaffolds of native S. cerevisiae promoters, we aimed to generate a series of promoter combinations that, when coupled to the same TF, can be used to modulate gene expression to different extents using the same concentration of the inducer molecule (Figure 1). The potential of such a promoter library would be to induce multiple genes to discrete desired levels, with the same synthetic TF and a small inducer molecule added to the culture medium. Using a combination of the j5 DNA assembly design software (Hillson, Rosengarten, & Keasling, 2012) and the PR‐PR laboratory automation platform (Linshiz et al., 2013; Linshiz et al., 2014), both developed at the Joint BioEnergy Institute (JBEI), and the gene synthesis capability at the Joint Genome Institute, a library of 240 promoter sequences were designed, 154 constructed and tested. The profiles of these regulatory systems are described in this report.
Figure 1.
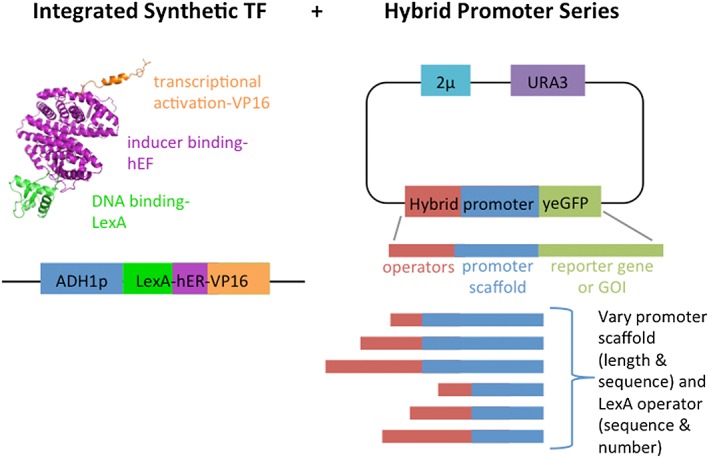
Design of modular transcription factor (TF) and hybrid promoter strains. Schematic of our two‐part control system where we (left) constructed a strain (ZyD1) in which our hybrid transcription factor (ADH1p‐LexA‐hER‐VP16) was integrated into locus YPRC∆15 for stable expression. (right) a series of hybrid promoters were constructed with promoter scaffolds of varying lengths (100 or 250 bp) and identities (GAL1p, LEU2p, SPO13p, TEF1p, HHF2p, GCN4p, CUP1p, HEM13p, ZRT1p, and SSL1p) that were paired with one to three copies of each of four different operator sequences (consensus, uvrA, umuDC and colE1). These 154 promoter combinations were expressed from a pRS426 plasmid and transformed into ZyD1 for analysis [Colour figure can be viewed at wileyonlinelibrary.com]
2. MATERIALS AND METHODS
2.1. Strains and media
Escherichia coli and S. cerevisiae strains, along with their associated information (including annotated sequence files) and sequences of all plasmids constructed are provided through the JBEI public registry (Ham et al., 2012; https://public‐registry.jbei.org/folders/277) and in Table S1 in the Supporting Information.
To produce ZDy1, the construct PADH1‐LexA‐hER‐VP16 was integrated into S. cerevisiae BY4741 {MATa; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0} S. cerevisiae strain at locus YPRC∆15 (Flagfeldt, Siewers, Huang, & Nielsen, 2009; Reider Apel et al., 2017). A complete description of this strain is available at https://public‐registry.jbei.org/entry/9623. ZDy1 was transformed with promoter–reporter library plasmids using the conventional lithium acetate method (Gietz & Woods, 2002), modified for large‐scale transformation.
Experiments were conducted in synthetic defined (0.67%, w/v) yeast nitrogen base without amino acids (VWR International), 0.2% (w/v) complete supplement mixture w/o yeast nitrogen base (Sunrise Science Products) or standard rich media (YP, 1% w/v Bacto yeast extract, 2% w/v Bacto peptone) with 1 or 2% w/v)sugar. E. coli DH10b, used for cloning and plasmid amplification, were grown in LB supplemented with 100 μg/mL carbenicillin.
2.2. Design and construction of synthetic promoters
Designs for the hybrid promoters were constructed using the j5 program (Hillson et al., 2012), by entering one, two or three operator elements followed by the basal promoters. The number of possible combinatorial variants was reduced by specifying Eugene rules (Bilitchenko et al., 2011) so that only one type of operator sequence was present in any given design. The j5 software generated sequences for 240 hybrid promoters resulting from a combination of 3 operator configurations × 4 operator sequences × 10 basal promoters × 2 promoter lengths. Sequences of all hybrid promoter parts are listed in Table S2.
For each hybrid promoter, oligomers for DNA synthesis were designed using the GeneDesign suite (Richardson, Nunley, Yarrington, Boeke, & Bader, 2010); however, for 60 promoters the resulting sequences were deemed too difficult to synthesize owing to the presence of multiple DNA synthesis constraint violations (Oberortner, Cheng, Hillson, & Deutsch, 2017) and were abandoned. For the remaining 180 promoters, ultramer oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA, USA), and DNA synthesis was performed using a 2‐step PCR (Reisinger, Patel, & Santi, 2006) method as described in Heins et al. (2014). Assembled fragments were cloned into the pRS426‐yeGFP vector by chew‐back cloning and transformed into E. coli TOP10 cells. Plating and picking were performed using a QPix 400 system (Molecular Devices). Eight colonies per construct were sequence verified using the PACBIO RSII system (Pacific Biosciences), and variant calling was performed using the GATK software package (McKenna et al., 2010). One‐hundred and fifty‐four successful constructs were recovered from E. coli and used to transform ZDy1 strains.
2.3. Promoter characterization
Growth and fluorescence measurements in the presence of estradiol were conducted in 96‐well plates on a Synergy H4 plate reader equipped with a Bio‐Stack 3 Microplate Stacker (BioTek, Winooski, VT, USA).
Selected yeast clones harbouring the promoter reporter constructs were grown overnight in 24‐well plates in YNB, 1% dextrose and CSM–Ura medium. Cells were diluted into 8 mL of the same medium in six‐well plates and allowed to grow until reaching an OD600 of ~0.2–0.3. Cells were then inoculated into the final 96‐well plates for promoter characterization: 100 μL of each clone was inoculated into each of 24 wells, so that six estradiol concentrations (0, 1, 5, 10, 50 and 100 nm) could be tested in technical quadruplicate. Additionally, this procedure was repeated for three biological clones of each promoter–reporter construct. Estradiol was added individually to each well at the specified concentrations just prior to the start of the experiment. Plates were covered with breathable adhesive plate seals (Thermo, NY, USA).
Prior to growth, plates were incubated at 23°C, without shaking. To measure growth each plate was cycled by shaking for 30 s to resuspend the cells and aerate the culture, and the fluorescence (excitation 485 ± 20 nm; emission 528 ± 20 nm) and OD600 were immediately acquired. Each plate was read every 30 min, and the cycling was repeated 72 times for a total run time of ~36 h.
2.4. Data analysis and methods to generate promoter profile plots
The plate reader data was normalized as follows: (a) the baseline fluorescence of each well was normalized by setting the average of the first five time points after the first two to zero (the first two time points often gave artificially high fluorescence readings and so were ignored); (b) the background fluorescence was measured in a nonfluorescent BY4741 wildtype strain at each time point was subtracted from the readings of each strain in the same condition to eliminate the contributions of autofluorescence to overall fluorescence readings. Analysis of variance and regression analyses were conducted using R [R Core Team (2015), R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org/]. Data used for these plots are provided in the Supporting Information.
3. RESULTS AND DISCUSSION
3.1. Design of hybrid promoters and synthetic‐chimeric TF
Since we wanted to generate a set of novel regulatory elements to enable the precise control of different genes and pathways using the same TF, the key variable in our study was the chimeric promoter sequences. We selected upstream sequences from 10 different native S. cerevisiae genes to act as basal promoter scaffolds: GAL1p, LEU2p, SPO13p, TEF1p, HHF2p, GCN4p, CUP1p, HEM13p, ZRT1p and SSL1p. Promoter scaffolds were chosen with hopes of representing both constitutive and inducible profiles at both low and high expression levels. Generally, 100 bp upstream of a gene is considered the core promoter sequence, which is the minimal stretch of DNA needed to initiate transcription, while 250 bp is thought to contain other cis‐acting elements that are involved in transcriptional regulation (Butler & Kadonaga, 2002; Lubliner et al., 2015; Lubliner, Keren, & Segal, 2013). We selected a shorter (100 bp) and a longer (250 bp) sequence length for each of these promoters to serve as the promoter scaffold.
We replaced the upstream operator region of each promoter with one of four different known LexA operator sequences: consensus, uvrA, umuDC and colE1 (Brent & Ptashne, 1985) using 1×, 2× or 3× repeats of these different LexA binding sequences. In total, we designed 240 plasmid‐based promoter constructs driving a yeGFP that served as the reporter. All 240 promoters can be controlled by the same chromosomally integrated synthetic‐chimeric TF. Our hypothesis was that these promoters would induce the gene of interest, in this case yeGFP, to different levels and have different inducibility, two important parameters for the design of biological circuits.
3.2. Building the promoter library
Out of an initial set of 240 hybrid promoters that were designed, 60 constructs contained DNA synthesis constraint violations that precluded their synthesis and were abandoned at the design stage. DNA synthesis constraints violations are commonly encountered in synthetic biology designs and can be particularly severe when designing regulatory sequences, as they cannot be easily removed by codon shuffling. A strategy to overcome this limitation is to design experiments with sufficient redundancy such that biological insights can be obtained with a subset of the data. Out of the 180 promoters that could be synthesized, 154 were successfully constructed and cloned into the E. coli–S. cerevisiae shuttle vector (pRS426–yeGFP) in order to characterize their functional parameters. This set of 154 promoters covered most of the biological space that we wanted to capture for this experiment (Figure S1 in the Supporting Information).
3.3. Induction and gene regulation using the promoter library
In order to easily test a large number of promoter constructs, we chose to chromosomally integrate a native ADH1 promoter driven copy of the LexA‐hER‐vp16 TF for stable expression. A similar, plasmid expressed, GAL4 estrogen responsive hybrid TF was previously shown to activate reporter genes exclusively in the presence of estradiol (Louvion et al., 1993). Replacement of the GAL4 DBD with the heterologous LexA prevents binding to native GAL4 promoters, thereby minimizing cross‐talk.
For the promoter series, with the exception of TEF1, HHF2 and GCN4, most combinations of basal promoters in two lengths could be built with 1×, 2× or 3× repeats of the four operator sequences. The plasmids with these hybrid promoter:yeGFP cassettes were transformed into the yeast strain with the chromosomally encoded synthetic TF. Estradiol concentrations from 0 to 100 nm were tested for each variant in the promoter library and a profile was generated for each strain. The complete set of plots for all hybrid promoters is provided in the Supporting Information (Figure S2). While good inducibility was observed from 5 to 50 nm of the inducer, 10 nm estradiol was chosen as the inducer concentration that shows the maximal dynamic range for the promoter series (Figure 2).
Figure 2.
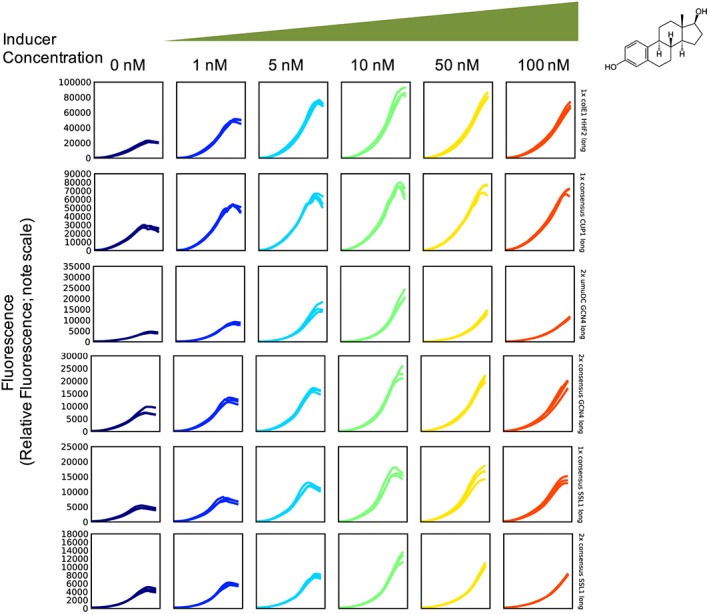
Subset of plots showing interesting hybrid promoter candidates. Florescence output from a subset of yeGFP expressing hybrid promoter strains (promoter names are labelled along right‐hand side) induced at various estradiol concentrations (0, 1, 5, 10, 50 and 100 nm) over a 36 h time course. The plots show the range in maximum levels to which the promoters can be used for protein expression using the same level of estradiol and the range in inducibility. For the complete set of plots see Figure S2 in the Supporting Information [Colour figure can be viewed at wileyonlinelibrary.com]
Some studies have reported toxicity of VP16 and LexA–hER–VP16 in the presence of high levels of inducer (Garí et al., 1997; McIsaac et al., 2011). Both studies that reported toxicity did not observe growth inhibition in constructs with centromeric plasmids or chromosomally integrated versions with low levels of inducer. Consistent with this, with our experimental setup, we did not observe major toxicity at inducer concentrations of 10 nm or less, possibly owing to the use of a chromosomally integrated ADH1‐driven synthetic TF in combination with a hybrid promoter containing plasmid.
The parameters of interest that we quantified for each promoter were the inducibility and response, measured in terms of the fold change in induction from 0 to 100 nm and the maximal level of induction at 10 nm, respectively. Analysis of variance was performed to statistically confirm the impact of each parameter and assess significance in the dataset (Figure 3). Overall, the promoter set with short upstream regions (100 bp) was significantly (p < 0.001) less responsive and did not display a wide range in either maximum fluorescence or inducibility regardless of promoter and operator sequences (Figures S1 and 3: left panels). While only 20% of yeast genes contain TATA elements, a genome‐wide study of their locations revealed that some genes contain TATA boxes further upstream than 100 bp before the beginning of translation (Basehoar, Zanton, & Pugh, 2004). Therefore, it is possible that some of the short promoter scaffolds lack activity owing to the absence of TATA boxes or similar TATA protein‐binding elements. For future design improvements, the lengths of the shorter promoter scaffolds could be individually customized to include functional TATA elements, which could result in better inducibility and range of induction.
Figure 3.
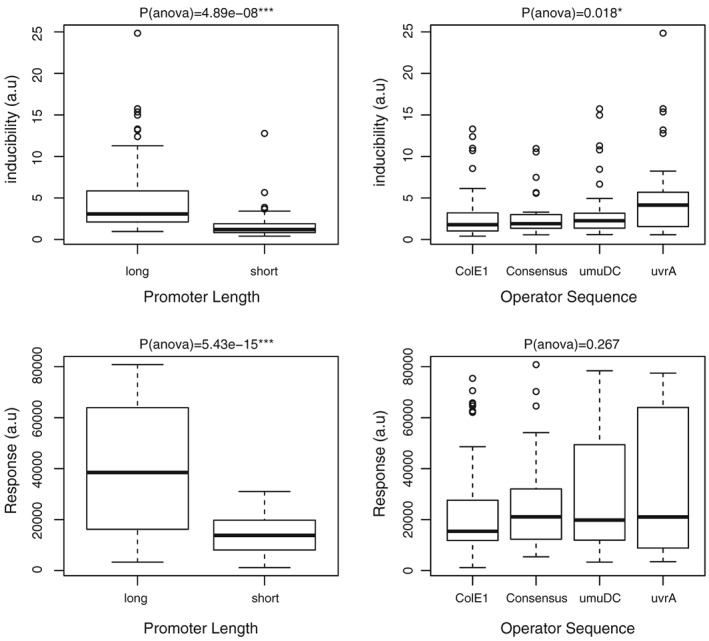
Parameters influencing promoter performance. Analysis of variance was conducted using R to establish inducibility fold (top) and maximum expression (bottom) significance for the dataset. p‐value significance: * <0.05, *** <0.001
Therefore, we focused our analysis on the set of promoters constructed using the longer promoter regions (250 bp). For this set, 84 total designs were constructed and tested. The main parameters, inducibility (fold change in fluorescence with the addition of estradiol) and response (maximal induction of GFP) displayed by the hybrid promoter library are summarized in Figure 4. Of the promoters tested, GAL1 and SPO13 showed the greatest inducibility. The GAL1 promoter also produced the highest fluorescence (response), with the 3× uvrA GAL1 construct being the highest in our entire dataset. A subset of promoters, such as HEM13 and ZRT1 with the uvrA and umuDC operators, gave very high fluorescence but at a constitutive level, that is, high response but low inducibility. Conversely, the SSL1 promoter with the uvrA and umuDC operators, and the LEU2 promoter with the umuDC operator, showed low fluorescence at constitutive levels, that is, both low response and low inducibility. Among the operator sequences, uvrA displayed significantly higher (p < 0.05) inducibility than the consensus and other operator sequences (Figure 3: right panels), without changing maximum expression level for each promoter scaffold. Design improvements can also be envisioned for the longer promoters in this study. Domains in the promoter regions such as the upstream activating sequence could be altered and optimized, especially in the cases where the promoters were not highly inducible. The hybrid promoters based on GAL1 and SPO3 showed the greatest inducibility, but still contained some native upstream activating sequence elements that may allow factors other than the inducer to influence the induction under a subset of growth conditions (Buckingham et al., 1990; Flick & Johnston, 1990).
Figure 4.
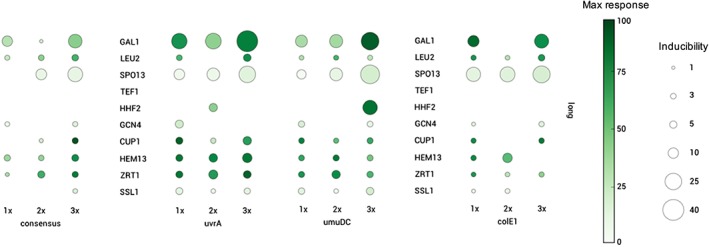
Long promoter scaffolds display a range of inducibility and responsiveness. Bubble plots showing inducibility fold (size of bubble) and maximum responsiveness level (colour of bubble) of yeGFP expression for each of the 250 bp (long) constructed hybrid promoters [Colour figure can be viewed at wileyonlinelibrary.com]
4. CONCLUSIONS
Drawing inspiration from several reported synthetic transcription factors used in yeast, we have constructed a synthetic transcription factor that consists of a chimera of a bacterial DNA binding domain, a mammalian ligand binding domain, and a viral transcriptional activation domain. The gene encoding this transcription factor was integrated into the chromosome and expressed from a constitutive promoter. The synthetic promoters were constructed from the operators that bind the selected DNA binding domain and a native S. cerevisiae basal promoter that will recruit the basal transcription machinery. Transcription is activated by addition of the small molecule ligand, estradiol, which binds the mammalian ligand‐binding domain. We tested the hypothesis that a series of hybrid promoters with varying promoter scaffolds and operator regions will lead to different profiles in gene expression in response to a given level of the estradiol.
The synthetic promoters tested in our study in conjunction with the synthetic TF provide a suite of control systems we set out to compile, wherein in response to the same inducer and inducer concentration, the synthetic promoters can be used to achieve different gene expression profiles such as highly inducible and highly responsive, highly inducible but lowly responsive, constitutively on and highly active as well as constitutively on and lowly activity (Figures 2 and 4). With additional characterization, these and other similar libraries allow a strain engineer to drive many genes and pathways in a cell to different levels using the same TF and same inducer added at a given level. This system has the potential to be useful for engineering heterologous enzymatic pathways in S. cerevisiae and for expressing multiple genes, or finding optimum levels for each gene product in a given pathway.
CONFLICT OF INTEREST
J.D.K. has financial interests in Amyris, Lygos, Constructive Biology, Demetrix and Napigen.
Supporting information
Figure S1: Complete library results of induction and response. Bubble plots showing inducibility fold (size of bubble) and maximum responsiveness level (color of bubble) of yeGFP expression for each of the 250 bp (long; top panel) and 100 bp (short; bottom panel) constructed hybrid promoters.
Figure S2: Complete set of fluorescence output plots. The complete set of plots for promoters shown in Figure S1 showing a wide range of maximum fluorescence and inducibility across a range of inducer concentrations. In all panels, the promoter name is labeled along the right‐hand side. Induction was performed at estradiol concentrations 0, 1, 5, 10, 50 and 100 nM from left to right and over a 36‐hour time course. Data show at least three biological replicate for each strain and condition tested.
Table S1: Descriptions of plasmids used in this study. All plasmids were created for use in this study as described in the materials and methods, and deposited in the public and private instances of the JBEI registry.
Table S2: Sequences of hybrid promoter parts used in plasmid construction. Four different operator sequences and ten different promoter sequences (both long and short) were used in combination to construct the 154 hybrid promoter plasmids used in this study.
ACKNOWLEDGEMENTS
This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) and part of the DOE Joint Genome Institute (http://jgi.doe.gov) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, and was part of the Agile BioFoundry (http://agilebiofoundry.org) supported by the US Department of Energy, Energy Efficiency and Renewable Energy, Bioenergy Technologies Office, through contract DE‐AC02‐05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. The US Government retains and the publisher, by accepting the article for publication, acknowledges that the US Government retains a nonexclusive, paid‐up, irrevocable, worldwide licence to publish or reproduce the published form of this manuscript, or allow others to do so, for US Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe‐public‐access‐plan).
Dossani ZY, Reider Apel A, Szmidt‐Middleton H, et al. A combinatorial approach to synthetic transcription factor‐promoter combinations for yeast strain engineering. Yeast. 2018;35:273–280. https://doi.org/10.1002/yea.3292
REFERENCES
- Alper, H. , Fischer, C. , Nevoigt, E. , & Stephanopoulos, G. (2005). Tuning genetic control through promoter engineering. Proceedings of the National Academy of Sciences of the United States of America, 102, 12678–12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar, A. D. , Zanton, S. J. , & Pugh, B. F. (2004). Identification and distinct regulation of yeast TATA box‐containing genes. Cell, 116, 699–709. [DOI] [PubMed] [Google Scholar]
- Bilitchenko, L. , Liu, A. , Cheung, S. , Weeding, E. , Xia, B. , Leguia, M. , … Densmore, D. (2011). Eugene – A domain specific language for specifying and constraining synthetic biological parts, devices, and systems. PLoS One, 6, e18882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeck, J. , Garg, R. , Reed, B. , & Alper, H. S. (2012). Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnology and Bioengineering, 109, 2884–2895. [DOI] [PubMed] [Google Scholar]
- Brent, R. , & Ptashne, M. (1985). A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell, 43, 729–736. [DOI] [PubMed] [Google Scholar]
- Buckingham, L. E. , Wang, H. T. , Elder, R. T. , McCarroll, R. M. , Slater, M. R. , & Esposito, R. E. (1990). Nucleotide sequence and promoter analysis of SPO13, a meiosis‐specific gene of Saccharomyces Cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 87, 9406–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J. E. F. , & Kadonaga, J. T. (2002). The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Development, 16, 2583–2592. [DOI] [PubMed] [Google Scholar]
- Cuperus, J. T. , Lo, R. S. , Shumaker, L. , Proctor, J. , & Fields, S. (2015). A tetO toolkit to alter expression of genes in Saccharomyces cerevisiae . ACS Synthetic Biology, 4, 842–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard, F. , Perli, S. D. , & Lu, T. K. (2013). Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synthetic Biology, 2, 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagfeldt, D. B. , Siewers, V. , Huang, L. , & Nielsen, J. (2009). Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae . Yeast, 26, 545–551. [DOI] [PubMed] [Google Scholar]
- Flick, J. S. , & Johnston, M. (1990). Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae . Molecular and Cellular Biology, 10, 4757–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederix, M. , Hütter, K. , Leu, J. , Batth, T. S. , Turner, W. J. , Rüegg, T. L. , … Mukhopadhyay, A. (2014). Development of a native Escherichia coli induction system for ionic liquid tolerance. PLoS One, 9, e101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí, E. , Piedrafita, L. , Aldea, M. , & Herrero, E. (1997). A set of vectors with a tetracycline‐regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae . Yeast, 13, 837–848. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D. , & Woods, R. A. (2002). Transformation of yeast by lithium acetate/single‐stranded carrier DNA/polyethylene glycol method. Methods in Enzymology, 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Gilbert, L. A. , Larson, M. H. , Morsut, L. , Liu, Z. , Brar, G. A. , Torres, S. E. , … Qi, L. S. (2013). CRISPR‐mediated modular RNA‐guided regulation of transcription in eukaryotes. Cell, 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, T. S. , Dmytriv, Z. , Plahar, H. , Chen, J. , Hillson, N. J. , & Keasling, J. D. (2012). Design, implementation and practice of JBEI‐ICE: An open source biological part registry platform and tools. Nucleic Acids Research, 40, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins, R. A. , Cheng, X. , Nath, S. , Deng, K. , Bowen, B. P. , Chivian, D. C. , … Deutsch, S. (2014). Phylogenomically guided identification of industrially relevant GH1 β‐glucosidases through DNA synthesis and nanostructure‐initiator mass spectrometry. ACS Chemical Biology, 9, 2082–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillson, N. J. , Rosengarten, R. D. , & Keasling, J. D. (2012). j5 DNA assembly design automation software. ACS Synthetic Biology, 1, 14–21. [DOI] [PubMed] [Google Scholar]
- Ikushima, S. , Zhao, Y. , & Boeke, J. D. (2015). Development of a tightly controlled off switch for Saccharomyces cerevisiae regulated by camphor, a low‐cost natural product. G3 (Bethesda), 5, 1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, A. S. , Lu, T. K. , Bashor, C. J. , Ramirez, C. L. , Pyenson, N. C. , Joung, J. K. , & Collins, J. J. (2012). A synthetic biology framework for programming eukaryotic transcription functions. Cell, 150, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, A. , Brunk, E. , & Keasling, J. D. (2016). The need for integrated approaches in metabolic engineering. Cold Spring Harbor Perspectives in Biology, 8 https://doi.org/10.1101/cshperspect.a023903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. E. , DeLoache, W. C. , Cervantes, B. , & Dueber, J. E. (2015). A highly characterized yeast toolkit for modular, multipart assembly. ACS Synthetic Biology, 4, 975–986. [DOI] [PubMed] [Google Scholar]
- Linshiz, G. , Stawski, N. , Goyal, G. , Bi, C. , Poust, S. , Sharma, M. , … Hillson, N. J. (2014). PR‐PR: Cross‐platform laboratory automation system. ACS Synthetic Biology, 3, 515–524. [DOI] [PubMed] [Google Scholar]
- Linshiz, G. , Stawski, N. , Poust, S. , Bi, C. , Keasling, J. D. , & Hillson, N. J. (2013). PaR–PaR laboratory automation platform. ACS Synthetic Biology, 2, 216–222. [DOI] [PubMed] [Google Scholar]
- Louvion, J. F. , Havaux‐Copf, B. , & Picard, D. (1993). Fusion of GAL4‐VP16 to a steroid‐binding domain provides a tool for gratuitous induction of galactose‐responsive genes in yeast. Gene, 131, 129–134. [DOI] [PubMed] [Google Scholar]
- Lubliner, S. , Keren, L. , & Segal, E. (2013). Sequence features of yeast and human core promoters that are predictive of maximal promoter activity. Nucleic Acids Research, 41, 5569–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubliner, S. , Regev, I. , Lotan‐Pompan, M. , Edelheit, S. , Weinberger, A. , & Segal, E. (2015). Core promoter sequence in yeast is a major determinant of expression level. Genome Research, 25, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac, R. S. , Gibney, P. A. , Chandran, S. S. , Benjamin, K. R. , & Botstein, D. (2014). Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae . Nucleic Acids Research, 42, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac, R. S. , Oakes, B. L. , Wang, X. , Dummit, K. A. , Botstein, D. , & Noyes, M. B. (2013). Synthetic gene expression perturbation systems with rapid, tunable, single‐gene specificity in yeast. Nucleic Acids Research, 41, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac, R. S. , Silverman, S. J. , McClean, M. N. , Gibney, P. A. , Macinskas, J. , Hickman, M. J. , … Botstein, D. (2011). Fast‐acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces Cerevisiae. Molecular Biology of the Cell, 22, 4447–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , … DePristo, M. A. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Research, 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, A. (2015). Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends in Microbiology, 23, 498–508. [DOI] [PubMed] [Google Scholar]
- Oberortner, E. , Cheng, J.‐F. , Hillson, N. J. , & Deutsch, S. (2017). Streamlining the design‐to‐build transition with build‐optimization software tools. ACS Synthetic Biology, 6, 485–496. [DOI] [PubMed] [Google Scholar]
- Ottoz, D. S. M. , Rudolf, F. , & Stelling, J. (2014). Inducible, tightly regulated and growth condition‐independent transcription factor in Saccharomyces cerevisiae . Nucleic Acids Research, 42, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta‐Yahya, P. P. , Zhang, F. , del Cardayre, S. B. , & Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature, 488, 320–328. [DOI] [PubMed] [Google Scholar]
- Rantasalo, A. , Czeizler, E. , Virtanen, R. , Rousu, J. , Lähdesmäki, H. , Penttilä, M. , … Mojzita, D. (2016). Synthetic transcription amplifier system for orthogonal control of gene expression in Saccharomyces cerevisiae . PLoS One, 11, e0148320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider Apel, A. , Ouellet, M. , Szmidt‐Middleton, H. , Keasling, J. D. , & Mukhopadhyay, A. (2016). Evolved hexose transporter enhances xylose uptake and glucose/xylose co‐utilization in Saccharomyces cerevisiae . Scientific Reports, 6, 19512 EP. https://doi.org/10.1038/srep19512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider Apel, A. , d'Espaux, L. , Wehrs, M. , Sachs, D. , Li, R. A. , Tong, G. J. , … Mukhopadhyay, A. (2017). A Cas9‐based toolkit to program gene expression in Saccharomyces cerevisiae . Nucleic Acids Research, 45, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisinger, S. J. , Patel, K. G. , & Santi, D. V. (2006). Total synthesis of multi‐kilobase DNA sequences from oligonucleotides. Nature Protocols, 1, 2596–2603. [DOI] [PubMed] [Google Scholar]
- Richardson, S. M. , Nunley, P. W. , Yarrington, R. M. , Boeke, J. D. , & Bader, J. S. (2010). GeneDesign 3.0 is an updated synthetic biology toolkit. Nucleic Acids Research, 38, 2603–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Complete library results of induction and response. Bubble plots showing inducibility fold (size of bubble) and maximum responsiveness level (color of bubble) of yeGFP expression for each of the 250 bp (long; top panel) and 100 bp (short; bottom panel) constructed hybrid promoters.
Figure S2: Complete set of fluorescence output plots. The complete set of plots for promoters shown in Figure S1 showing a wide range of maximum fluorescence and inducibility across a range of inducer concentrations. In all panels, the promoter name is labeled along the right‐hand side. Induction was performed at estradiol concentrations 0, 1, 5, 10, 50 and 100 nM from left to right and over a 36‐hour time course. Data show at least three biological replicate for each strain and condition tested.
Table S1: Descriptions of plasmids used in this study. All plasmids were created for use in this study as described in the materials and methods, and deposited in the public and private instances of the JBEI registry.
Table S2: Sequences of hybrid promoter parts used in plasmid construction. Four different operator sequences and ten different promoter sequences (both long and short) were used in combination to construct the 154 hybrid promoter plasmids used in this study.


