Abstract
Mammalian cells exhibit complex cellular responses to DNA damage, including cell cycle arrest, DNA repair and apoptosis. Defects in any one of these responses can result in carcinogenesis. Absence of the chromatin remodeling complex Swi/Snf is found in many instances of cancer, and we have investigated its role in the UV damage response. The human carcinoma cell line SW13 is deficient in Swi/Snf and is very sensitive to UV radiation. In contrast, SW13 cells with ectopic Brg1 expression regain active Swi/Snf and become significantly more resistant to UV radiation. Sensitivity to UV light correlates well with dramatic UV induced apoptosis in SW13 cells, but not in SW13 cells expressing Brg1. We show that SW13 cells synchronized at the G1/S border progress into S phase after UV irradiation, and this checkpoint deficiency is corrected after Brg1 expression is restored. Interestingly, Brg1 expression in SW13 cells restores expression of two DNA damage responsive genes, Gadd45a and p21. Furthermore, Gadd45a induction and p21 degradation were observed in the Brg1-expressing SW13 cells after UV irradiation. Our findings demonstrate that Swi/Snf protects cells against deleterious consequences of UV induced DNA damage. These results also indicate that Swi/Snf may modulate checkpoint activation after UV damage via regulation of the two PCNA-binding proteins Gadd45a and p21.
Keywords: Swi/Snf, chromatin remodeling, UV damage response, DNA repair, Gadd45a, p21, BAF complex, checkpoint
Introduction
The Swi/Snf chromatin remodeling complex was first defined in yeast exhibiting mating-type switching and sucrose-nonfermenting defective phenotypes.1–3 Swi/Snf regulates gene expression by modifying nucleosomes through an ATP-dependent mechanism.4,5 The mammalian Swi/Snf complexes contain either of two ATPase subunits, BRM (brahma) or BRG1 (Brahma Related Gene), plus additional proteins called BRG1/BRM Associated Factors (BAFs).6,7 Both BRG1 and BRM proteins are closely related orthologs of the yeast SNF2 protein, which is the catalytic ATPase in the yeast Swi/Snf complex.8,9
Brg1 is frequently deleted or mutated in a variety of tumor cell lines,10,11 implicating Brg1 as a potential tumor suppressor gene. The gene encoding the SNF5/Ini1 core subunit of Swi/Snf is a tumor suppressor in humans and mice.12–15 Snf5 deletion in mouse primary fibroblasts impairs cell proliferation and leads to increased sensitivity to genotoxic stress, suggesting a role for SNF5 in the DNA damage response.16 Chromatin remodeling activities have been shown to play an important role in DNA repair in yeast.17–20 Furthermore, a recent report suggests that the human Swi/Snf (hSwi/Snf) complex facilitates DNA double strand break repair by promoting H2AX phosphorylation.21 The most frequent DNA lesions induced by UV radiation are cis-syn cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts [(6–4)PPs]. These UV lesions, which cause distortions in the DNA duplex, are removed by nucleotide excision repair (NER).22,23 We and others have shown that the yeast Swi/Snf chromatin remodeling activity stimulates NER both in vitro24 and in living cells.25 It is not clear, however, whether the human Swi/Snf plays a similar role in the UV damage response.
In addition to directly repairing DNA lesions, cells may respond to DNA damage by halting cell cycle progression or by undergoing apoptosis. The current study is aimed at exploring the role of hSwi/Snf in the cellular response to UV induced DNA damage. We show that human cells deficient in Swi/Snf are sensitive to UV irradiation. In these cells, high levels of apoptosis are induced by UV and removal of CPDs from their genomic DNA is attenuated. Significantly, restoration of active Swi/Snf suppresses UV induced apoptosis and NER of CPDs proceeds to near completion. In addition, Swi/Snf deficient cells fail to maintain a pause at the G1/S boundary after UV irradiation. Interestingly, we show that expression of two proliferating cell nuclear antigen (PCNA)-binding protein, p21 and Gadd45a, is dependent on active Swi/Snf in these cells. Since GADD45a and p21 have been shown to be actively involved in NER and cell cycle control,26–28 our results indicate that Swi/Snf protects cells against the consequences of UV radiation at least partially by regulating expression of p21 and GADD45a proteins.
Results
Inactivation of Swi/Snf by depletion of the SNF5 subunit decreases NER of CPDs in HeLa cells
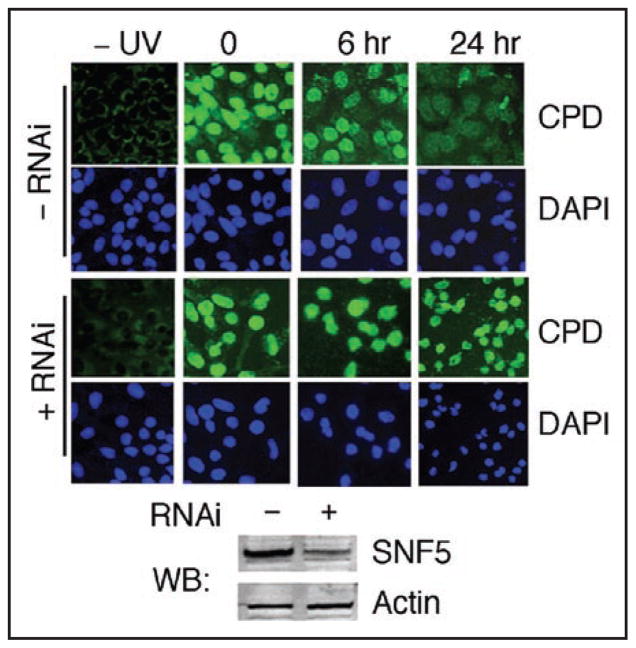
The Swi/Snf chromatin remodeling complex has been shown to stimulate NER both in vitro24 and in living yeast cells.25 To test whether the human Swi/Snf also plays a role in UV damage repair, we examined the effect of Swi/Snf inactivation on repair of CPDs in HeLa cells. To inactivate Swi/Snf, we used siRNAs targeting the Snf5 transcript to knock down the level of SNF5, one of the core Swi/Snf subunits.7,10 We observed that three days after introduction of the siRNAs, SNF5 protein levels in HeLa cells decreased to ~20% of the normal level (Fig. 1). At this point, we exposed cells to UV and examined repair of CPDs by indirect immunofluorescence using a specific anti-CPD monoclonal antibody.29 We observed that SNF5 depleted HeLa cells failed to complete CPD repair (Fig. 1), suggesting that Swi/Snf may play a role in UV damage repair in human cells. However, we also noted that many of the UV irradiated SNF5 depleted cells were rounded with shrunken nuclei, characteristic of apoptosis (Fig. 1). Apoptosis impairs NER of UV damage,30 and the Swi/Snf complex may play a role in the UV damage response by suppressing UV induced apoptosis, which (indirectly) modulates the repair of CPDs. Therefore, we next focused on monitoring UV-induced apoptosis in cells ‘naturally’ depleted of hSwi/Snf remodeling activity.
Figure 1.
Repair of CPDs in HeLa cells. Cells were depleted of SNF5 by RNAi, UV irradiated (10 J/m2), fixed at 6 or 24 hours after irradiation and repair of CPDs was monitored by immunofluorescence microscopy using a specific antibody to CPDs (green signal). Lower panel shows Western blot (WB) of HeLa cell extracts following treatment with (+) or without (−) RNAi depletion of SNF5.
Restoration of Brg1 expression suppresses UV sensitivity in SW13 cells
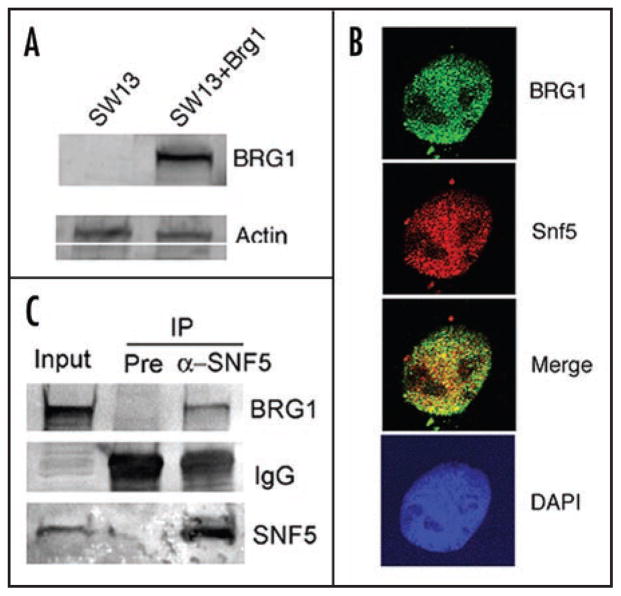
To further explore the role of Swi/Snf chromatin remodeling in the UV damage response in human cells, we utilized the human carcinoma cell line SW13 which lacks a functional Swi/Snf remodeling complex due to the absence of the Swi/Snf ATPases, BRG1 and BRM.31 We established a stable SW13 cell line expressing Brg1, termed SW13 + Brg1 (Fig. 2A). The level of Brg1 protein in SW13 + Brg1 cells is comparable to that of the human fibroblast strain IMR90 (data not shown). Importantly, immunostaining shows that BRG1 localizes in the nucleus and colocalizes with SNF5, a common subunit of the human Swi/Snf complex (Fig. 2B), and this association is confirmed by co-immunoprecipitation (Fig. 2C). Taken together, these findings suggest that Brg1 expressed ectopically is incorporated into the Swi/Snf complex in SW13 cells. Indeed, transient expression of Brg1 has been shown to reconstitute the completely active Swi/Snf complex in SW13 cells and induce formation of flat cells, consistent with a possible role of Brg1 in tumor suppression.31 Moreover, our stable Brg1-expressing SW13 cells show flat cell morphology versus the rounded cell morphology observed for SW13 cells or cells bearing vector alone (data not shown).
Figure 2.
Stable SW13 cell line with constitutive Brg1 expression. (A.) Western blots of BRG1 in SW13 cells. Establishment of the stable SW13 cell line expressing BRG1 (SW13 + Brg1) is described in Materials and Methods. Extracts from SW13 and SW13 + Brg1 cells were analyzed by Western blot using anti-BRG1 antibody and anti-Actin antibody as loading control. (B.) Nuclear localization of the Swi/Snf complex in SW13 + Brg1 cells was examined using antibodies specific to BRG1 and SNF5 and confocal microscopy. (C.) Association of ectopically expressed BRG1 with SNF5. SW13 + Brg1 cells were analyzed for the Swi/Snf complex by immunoprecipitating (IP) with pre-immune serum (Pre) and anti-SNF5 antibody (α-SNF5) and probing the Western blot with anti-BRG1 antibody. IgG in the IP materials was picked up by the secondary antibody in the Western Blot. Membrane was also probed with anti-SNF5 antibody to show the levels of SNF5 in the IP materials and input.
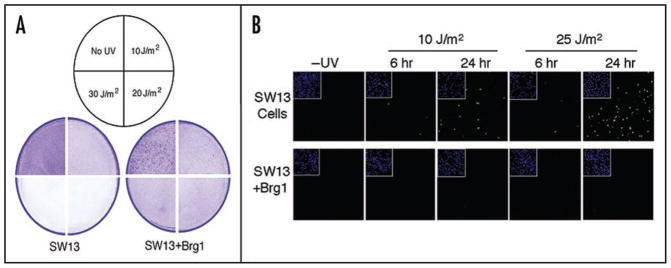
The UV sensitivity of SW13 cells was compared with SW13 + Brg1 cells. We found that SW13 cells are very sensitive to 254 nm UV radiation resulting in extensive cell death and detachment (Fig. 3A). Interestingly, Brg1-expressing SW13 cells are considerably more resistant to UV radiation (Fig. 3A). The observed differential sensitivity to UV radiation was confirmed by a trypan blue exclusion assay (data not shown). These results indicate that restoration of the functional Swi/Snf complex by ectopic expression of Brg1 suppresses the sensitivity of SW13 cells to UV radiation.
Figure 3.
Sensitivity of SW13 and SW13 + Brg1 cells to UV radiation. (A) Cell detachment assay. Monolayers of cells (~80% confluent) were exposed to various doses of UV radiation (shown in sectored circle), incubated for 24 hr and fixed/stained with ethanol/crystal violet. Figures depict quadrants from plates exposed to the different UV doses. (B) Brg1 expression in SW13 cells suppresses UV induced apoptosis. SW13 and SW13 + Brg1 cells were UV irradiated (10 or 25 J/m2), incubated for 6 hr or 24 hr as indicated, and analyzed using the TUNEL assay and confocal microscopy. Insets show DAPI staining of nuclei.
SW13 cells expressing Brg1 are more resistant to UV induced apoptosis
Mammalian cells exhibit complex cellular responses to DNA damage. One well-characterized response, especially following extensive DNA damage, is programmed cell death (or apoptosis). We observed that following UV irradiation SW13 cells, but not SW13 + Brg1 cells, exhibited cell morphology changes characteristic of apoptosis. To compare levels of apoptosis, the two cell lines were analyzed using a TUNEL [Terminal (deoxynucleotidyl transferase biotin)-dUTP Nick End Labeling] assay to detect endonucleolytic cleavage of chromatin DNA, a hallmark of apoptosis. Chromatin cleavage is observed in a small proportion of SW13 cells 6 hr following 10 or 25 J/m2 UV irradiation (Fig. 3B). Significant fractions of SW13 cells, however, undergo apoptosis by 24 hr after UV, and the fraction of cells is UV dose dependent. In contrast, only very few BRG1-expressing SW13 cells showed such an apoptotic response, even 24 hr after 25 J/m2 UV (Fig. 3B). These results indicate that BRG1 plays a role in suppressing UV induced apoptosis in SW13 cells.
Onset of apoptosis attenuates repair of UV photoproducts in cells lacking BRG1
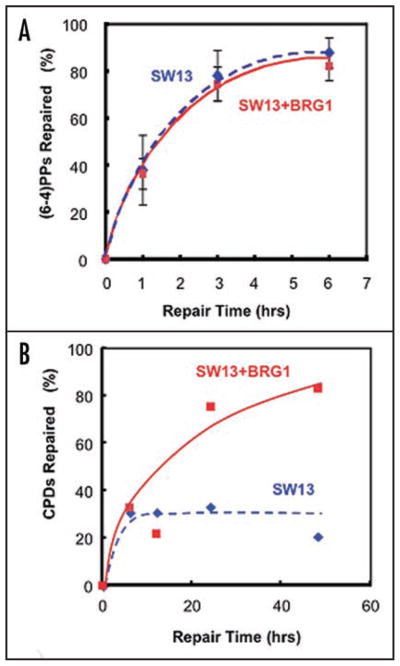
Persistence of unrepaired DNA lesions, especially in replicating cells, can signal the initiation of apoptosis.32 On the other hand, onset of DNA condensation and fragmentation associated with apoptosis would preclude efficient repair from that point on.30 To exclude this latter situation, we measured removal of (6–4)PPs, which is rapid in cells proficient at NER. As seen in Figure 4A, SW13 and SW13 + Brg1 cells repair 6–4 lesions at very similar rates, with approximately 70% of the (6–4)PPs being removed within 3 hr after UV, presumably before the onset of apoptosis. This suggests that Brg1 expression is not directly required for NER of (6–4)PPs. As these UV lesions form more efficiently in nucleosome linker DNA,33–35 we also analyzed repair of CPDs since, compared to (6–4)PPs, these lesions have a different overall distribution in chromatin and are repaired more slowly.36 CPD removal was comparable in the first 6 hr after UV indicating that Brg1 expression was dispensable for repair of readily accessible CPD sites (Fig. 4B). However, whereas the SW13 + Brg1 cells continued to remove CPDs, repair in the SW13 cells was incomplete. Since UV induced apoptosis was detectable during the later repair period, we believe that attenuation of repair in SW13 cells is, for the most part, due to the onset of apoptosis.
Figure 4.
NER of the UV induced lesions in SW13 cells correlates with UV induced apoptosis. (A) Repair of 6–4PPs in SW13 and SW13 + Brg1 cells. Cells were UV irradiated (12 J/m2) and samples collected at the times shown for DNA isolation. Removal of 6-4PPs was analyzed over a 6 hr time period by slot-blot using an antibody specific for these lesions and the signals normalized with the total DNA in each lane (see Materials and Methods). (B) Repair of CPDs in SW13 and SW13 + Brg1 cells. Cells were treated as described in (A). Removal of CPDs from total genomic DNA was analyzed over a 48 hr time period using a T4 endonuclease V/alkaline agarose gel assay (see Materials and Methods).
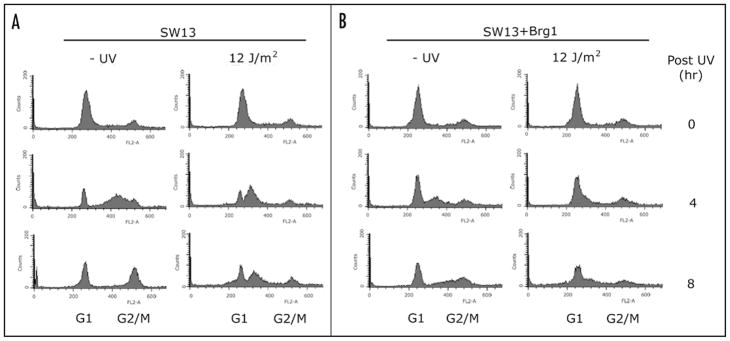
Progression of SW13 cells synchronized at the G1/S border into S phase is insensitive to UV and this checkpoint deficiency is restored by ectopic Brg1 expression
Cellular responses to genotoxic stress include activation of cell cycle checkpoints and apoptosis. Inactivation of checkpoints, whereby cells try to replicate DNA in the presence of damage, is highly deleterious.37 To further examine the high sensitivity of SW13 cells to UV and its suppression by BRG1, SW13 and SW13 + Brg1 cells were synchronized at G1/S border and exposed to UV light. Cell cycle progression was then analyzed by flow cytometry. In the absence of UV damage, both SW13 and SW13 + Brg1 are arrested at the G1/S boundary following treatment with aphidicolin and progress into S and G2/M phases after aphidicolin removal (Fig. 5, −UV). In addition, SW13 cells appear to cycle slightly faster than SW13 + Brg1 cells in the absence of DNA damage (Fig. 5, −UV, 4 hr). Significantly, SW13 cells irradiated with a low UV dose fail to maintain a pause at the G1/S boundary (Fig. 5, SW13). These cells presumably replicate the damaged DNA, since the population decreased rapidly after UV irradiation (Fig. 5, G1 SW13). Interestingly, SW13 cells accumulated near the middle of S phase (Fig. 5). In contrast, SW13 + Brg1 cells maintained a pause at the G1/S border after UV radiation (Fig. 5, SW13 + Brg1). Thus, there is a checkpoint deficiency in SW13 cells in response to UV, and this deficiency is corrected by ectopic expression of Brg1. We note that failure to activate this checkpoint in SW13 cells after UV irradiation correlates well with the high UV sensitivity observed in these cells (Fig. 3). We also noticed that, similar to SW13 cells, a significant amount of SW13 + Brg1 cells appeared at ‘sub-G1’ 24 hrs after UV radiation (Fig. 5). This result seems to conflict with our TUNEL data (Fig. 3B) showing that few SW13 + Brg1 cells undergo apoptosis after UV. We suspect that aphidicolin treatment to synchronize cells at the G1/S border may account for this difference. Indeed, aphidicolin induces replication stress and activates Chk1.38,39
Figure 5.
Effect of Brg1 expression on cell cycle checkpoints. SW13 and SW13 + Brg1 cells were synchronized by treatment with aphidicolin, UV irradiated (12 J/m2) and incubated for various times (indicated on the right), as described in Materials and Methods. Fixed cells were stained with propidium iodide and DNA content determined by flow cytometry.
BRG1 regulates expression of p21 and Gadd45a
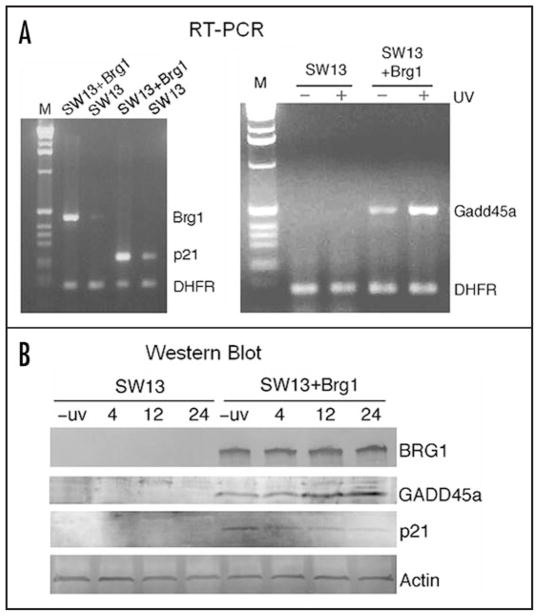
The dominant checkpoint response to DNA damage in mammalian cells is the ATM(ATR)/CHK1(CHK2)-p53/MDM2-p21 pathway.40 The cyclin-dependent kinase (CDK) inhibitor p21CDKN1A plays a fundamental role in the DNA-damage response by inducing cell cycle arrest, and by inhibiting DNA replication through association with PCNA.26–28 It has recently been shown that p21 protein is rapidly recruited to UV-induced DNA-damage sites and co-localizes with PCNA.28 GADD45a has also been shown to bind PCNA, playing a role in UV damage repair and inhibiting entry of cells into S phase.41 We examined the mRNA levels of p21 and Gadd45a in the two SW13 cell lines by multiplex RT-PCR (Fig. 6A). A very low level of both Brg1 and p21 mRNAs was detectable in SW13 cells (Fig. 6A, left). Significantly, the basal level of p21 expression is elevated in SW13 + Brg1 cells. Interestingly, no Gadd45a mRNA is detectable in SW13 cells either before or after UV radiation (Fig. 6A, right). However, low levels of Gaddd45a transcript were detected in SW13 + Brg1 cells and, consistent with earlier reports,42,43 expression of Gadd45a is induced by UV radiation in these cells (Fig. 6A, right). These data indicate that BRG1 upregulates p21 expression and is essential for both basal and UV induced expression of Gadd45a.
Figure 6.
Introduction of Brg1 in SW13 cells stimulates expression of the p21 and Gadd45a genes. (A) RT-PCR analysis of gene expression in SW13 and SW13 + Brg1 cells. RNA was purified and RT-PCR products were analyzed on agarose gels (left, using Brg1 primers, lanes 2 and 3, or p21 primers, lanes 4 and 5; right, using Gadd45a primers). As a control, primers for the DHFR gene were included in each RT-PCR reaction. To measure damage inducible expression of Gadd45a, SW13 and SW13+Brg1 cells were collected for RNA purification 6 hr after 12 J/m2 UV irradiation (+ UV, right panel) or mock irradiation (−UV, right). (B) Protein levels of BRG1, GADD45a and p21. SW13 and SW13 + Brg1 cells were UV irradiated (12 J/m2) and incubated for the indicated times. Cells were collected and cell extracts analyzed by SDS-PAGE and Western blots using specific antibodies.
In agreement with the RT-PCR data, GADD45a protein and its induction by UV are detected only in Brg1 expressing SW13 cells (Fig. 6B). Although we detected low levels of p21 transcript in SW13 cells (Fig. 6A), no p21 protein could be detected by Western blot in these cells (Fig. 6B). In contrast, p21 protein is readily detected in SW13 + Brg1 cells. Interestingly, although the mRNA level of p21 is elevated after UV radiation (data not shown), we found that p21 protein is degraded gradually after SW13 + Brg1 cells were treated with a low dose of UV (Fig. 6B). In agreement with this result, UV-induced p21 degradation has been shown to promote UV damage repair 44 and PCNA ubiquitination.45
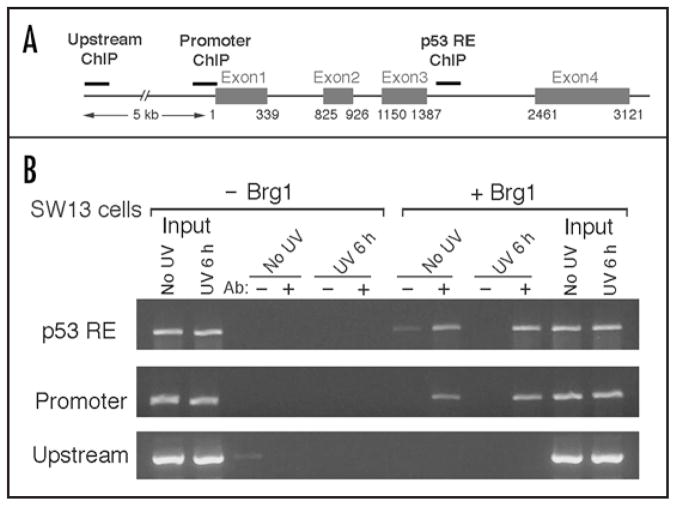
Recruitment of BRG1 to the Gadd45a locus
The BRG1-containing complexes have been demonstrated to directly regulate expression of the CDK inhibitor p21,11,46 where recruitment of BRG1 to the p21 promoter was observed. To test if BRG1-containing Swi/Snf is recruited to the Gadd45a locus to regulate Gadd45a expression, we monitored the accumulation of BRG1 on the Gadd45a gene following UV radiation of SW13 + Brg1 cells. The ChIP analysis in Figure 7 shows that BRG1 is detected on the Gadd45a p53-responsive element (RE) located at the third intron (Fig. 7A) in the absence of UV radiation. Following UV treatment, a somewhat increased amount of BRG1 is bound to the p53 RE (Fig. 7B). Interestingly, a significant amount of BRG1 is also found to be associated with the Gaddd45a promoter region with and without UV radiation (Fig. 7B). Since both p53 and BRCA1 regulate Gadd45a expression by targeting the p53-RE and promoter regions, respectively,43 it will be of interest to examine if the BRG1-containg Swi/Snf complex is involved in p53 and BRCA1 mediated regulation of Gadd45a expression.
Figure 7.
Binding of BRG1 to the Gadd45a gene. SW13 and SW13 + Brg1 cells were UV irradiated (12 J/m2), incubated for 6 hr followed by chromatin immunoprecipitation (ChIP) with anti-BRG1. (A) Schematic of the Gadd45a gene. Thick gray bars denote the p53 response element (p53RE), promoter and upstream regions selected to generate PCR primers (thin solid bars). (B) ChIP analysis. DNA was isolated following ChIP, and PCR products to the regions cited in A were analyzed on an agarose gel. (Ab: −, + indicates absence or presence of anti-BRG1 antibody, respectively, during immunoprecipitation).
Discussion
In this report we demonstrated that reconstitution of the human Swi/Snf chromatin remodeling complex in SW13 cells by ectopic Brg1 expression suppresses UV induced apoptosis. Due to the onset of apoptosis, repair of CPDs, but not (6–4)PPs, is disrupted in SW13 cells without active Swi/Snf. We find that expression of Gadd45a and p21, two PCNA-binding proteins involved in the control of cell cycle progression, is dependent on BRG1. We also showed that the apoptotic response to UV in Swi/Snf deficient cells correlates with these cells’ inability to maintain a pause at the G1/S boundary after UV radiation when cells were first synchronized at the G1/S border. This deficiency is corrected by ectopic Brg1 expression. Our findings establish that Swi/Snf plays an important role in the UV damage response, possibly by regulating Gadd45a and p21 gene expression.
Cell cycle arrest, NER and chromatin remodeling by the Swi/Snf complex
Consistent with our observation that BRG1-containing Swi/Snf plays a role in the UV damage response, Snf5-knockout mouse fibroblasts are sensitive to UV radiation.16 To study why restoration of Brg1 expression in SW13 cells suppresses UV induced apoptosis, several possibilities were considered. We first examined whether Swi/Snf could facilitate NER by remodeling chromatin to increase DNA damage accessibility, given that Swi/Snf copurifies with DNA damage recognition factors in yeast and stimulates NER activity in vivo and in vitro.24,25 However, using a local UV irradiation technique,47 we were unable to demonstrate recruitment of Swi/Snf to sites of CPDs in either HeLa or normal human diploid fibroblast cells (IMR90; data not shown). Since increased sensitivity to UV in SW13 cells could be caused not only by inefficient DNA repair but also by disruption of cell cycle responses to DNA damage, we examined cell cycle progression of SW13 cells following UV radiation. Indeed, we found that SW13 cells fail to maintain at the aphidicolin-synchronized G1/S boundary after UV irradiation. In contrast, SW13 cells expressing Brg1 halt cell cycle progression at the synchronized G1/S border after UV. Based on these results, we believe that the incompletion of CPD repair in both SW13 cells (Fig. 4A) and HeLa cells (Fig. 1) with deficient Swi/Snf is primarily due to the onset of apoptosis. Indeed, the efficient repair of (6–4)PPs in SW13 suggests that NER is functional in both cell lines, at least in the more accessible regions of chromatin. Additionally, there is no significant difference in expression levels of any of the known NER factors, based on microarray analysis (unpublished data). However, as the bulk of the CPDs repaired during early times are in accessible chromatin domains (e.g., transcriptionally active genes),48,49 it remains to be determined if Swi/Snf is required for efficient repair of nucleosome-loaded, highly condensed chromatin domains.
Both basal and UV induced expression of Gadd45a is dependent on Swi/Snf
Consistent with the checkpoint deficiency in SW13 cells to delay cell cycle progression from the G1/S border into S phase, we demonstrated that these cells do not have detectable p21 and GADD45a proteins (Fig. 6B). Both proteins have been suggested to play a role in cell cycle control.40,41,43 Gadd45a is rapidly induced by a variety of genotoxic stresses in almost all mammalian cells.42,43 Several lines of evidence have also implicated Gadd45 gene products in cell cycle arrest, NER, maintenance of genomic stability and apoptosis.43 Cells lacking Gadd45a exhibit NER defects comparable to p53-deficient cells.50 Interestingly, Gadd45a was shown to bind to PCNA and inhibit entry of cells into S phase,41 Thus, the absence of GADD45a protein in SW13 cells may explain the checkpoint deficiency observed in Figure 5 and contribute to the incompletion of CPD repair (Fig. 4B). It remains to be determined if introduction of Gadd45 and/or p21 proteins corrects the cell cycle checkpoint deficiency observed in SW13 cells.
Expression of Gadd45a is regulated by both p53 and BRCA1.43 We have demonstrated in this study that expression of Gaddd45 is dependent on the Swi/Snf complex in the human cancer cell line SW13. Therefore, it is possible that Swi/Snf, BRCA1 and p53 have functional overlap in vivo. Related to this, BRCA1 can directly interact with the BRG1 subunit of the Swi/Snf complex.51 In addition, several subunits of Swi/Snf have been shown to bind p53 in vivo.52 In the present report, accumulation of BRG1 on both the p53 RE and the Gadd45a promoter were demonstrated. It seems likely that the transcription activator BRCA1 and/or p53 may recruit Swi/Snf to upregulate Gadd45a gene expression. Clearly, testing this possibility may further elucidate the complex regulation of Gadd45a expression.
Upregulation of p21 by Swi/Snf and its degradation after UV
When transiently expressed in SW13 cells, BRG1 has been shown to be recruited to the p21 promoter and upregulate its expression in SW13 cells.46 Consistent with this finding, stable expression of Brg1 in SW13 also stimulates p21 expression at both the mRNA and protein levels (Fig. 6). Surprisingly, while UV radiation induces expression of p21 transcripts in SW13 + Brg1 cells (data not shown), we found that the p21 protein level declines following UV treatment (Fig. 6B). This observation may indicate that there is a finely tuned coordination of cell cycle arrest and DNA repair. Indeed, p21 degradation after UV, observed in many human cell lines,53 has been shown to be required for optimal NER.44
Tight cell cycle control in mammalian cells is executed through the concerted efforts of CDKs and their cellular inhibitors. Since p21 functions in a variety of different cellular processes, the consequences of changes in p21 protein level after DNA damage are complex. The p21 protein inhibits cell cycle progression and replication through its interactions with cyclin/CDK complexes and PCNA.54 To explain the possible role of p21 in the control of cell cycle progression from aphidicolin synchronized G1/S border into S phase shown in Figure 5, we propose that UV radiation may lead to disruption of the PCNA-p21 complexes in SW13 + Brg1 cells, possibly by upregulating another PCNA-binding protein GADD45a.41 As a result, more p21 is available to inhibit the activity of CDKs. This will result in hypophosphorylation of the major CDK substrate pRB. Hypophosphorated pRB remains bound to the E2F transcription factor, thereby inhibiting transcription of S-phase genes and blocking cell cycle progression into S phase.55 Indeed, it has been shown that p21 does not bind to PCNA during replication stress; instead, p21 preferentially interacts with CDKs under this stress.56 In addition, transient expression of the CDK inhibitor p21 in SW13 cells is sufficient to cause hypophosphorylation of pRB.46 In any event, further work is needed to examine whether UV radiation modulates protein-protein interactions between p21 and its partners.
In conclusion, our study establishes a role for the BRG1-containing Swi/Snf chromatin remodeling complex in the cellular response to DNA lesions induced by UV irradiation. Our findings highlight the important role of Swi/Snf in maintaining genome stability. Absence of the chromatin remodeling complex Swi/Snf is found in many instances of cancer and we suggest that the tumor suppressor activity of Swi/Snf is, at least in part, through the regulation of Gadd45a and p21.
Materials and Methods
Plasmid constructs, antibodies and RT-PCR
pREP7-BRG1 and its control vector pREP7 were described previously.46 The following antibodies were used in this study: BRG1 antibody,57 p21 (Upstate), GADD45a (Upstate) and CPD and 6–4PP antibodies (generous gifts from Dr. T. Mori, Nara Medical University, Japan). The following primers were used in RT-PCR for detecting expression of GADD45a, BRG1, p21 and DHFR. Gadd45 F1: atgactttggaggaattctcggctg; Gadd45 R1: tcaccgttcagggagattaatc; Brg1 F1: cagcagtggacgtcagctcag
Brg1 R1: atcgtcactcacgaccggcttg; DHFR F1: gagaactcaaggaacctccacaag; DHFR R1: gaactgccaccaactatccagac; P21 F1: gag actctcagggtcgaaaacgg; P21 R1: cagggtatgtacatgaggaggtg.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out as described by Liu et al.,58 using SW13 + Brg1 cells. The following primers were used for detecting p53-RE sequences: GadF-53RE, 5′ggatctgtggtaggtgggggtcagg3′; GadR-53RE, 5′ggaattagtcacgggaggcagtgcag3′. The following primers were used for detecting GADD45a promoter sequences: GAD-PrF1, 5′acacttggcagcctgcttcagcc3′; GAD-PrR1, 5′ctcccagccactgcctaccagc3′. The following were primers for the GADD45a upstream control sequence: GadF-distal, 5′ggagttggagttgtcaggaaaaaggg3′; GadR-distal, 5′ggttgtggtctttcaggcctccacacc3′. Twenty percent of the ChIP DNA was subjected to PCR amplification using Taq polymerase with the following conditions: 94°C, 30 s; 55°C, 30 s; 72°C, 30 s; 30 cycles.
Transfection and establishment of stable cell lines
HeLa and SW13 cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). To establish SW13 + vector and SW13 + BRG1 cell lines, a pREP7 plasmid bearing Brg1 cDNA46 was introduced into SW13 cells using Lipofectamine 2000 (Invitrogen), as per manufacturers’ instructions. Hygromycin-resistant clones were then selected and BRG1 expression was examined by immunoblot. BRG1 was detected in several clones showing flat cell morphology, but not in SW13 cells transfected with the pREP7 plasmid vector. A stable cell line, designated SW13 + Brg1 was established and used in this study. Cells expanded from this clone show moderate flat cell morphology.
DNA repair assays
Monolayers of cells were UV irradiated, harvested and DNA was isolated, as previously described.59,60 Repair of CPDs was measured following T4 endonuclease V digestion, alkaline gel electrophoresis and southern blotting as described in Bespalov et al.61 For CPD removal, gels were visualized on a Storm 840 PhosphorImager (GE Healthcare) and images were analyzed using IMAGE QUANT software (GE Healthcare). Repair of (6–4)PPs was monitored with a monoclonal antibody specific to this lesion29 in a slotblot assay using ECL Plus (GE Healthcare). Briefly, diluted DNA was denatured in 0.4 N NaOH, applied to nitrocellulose in triplicate using a slotblot apparatus and immobilized by treatment with microwaves. After blocking, the membranes were incubated with anti-(6–4)PP antibody followed by anti-mouse HRP conjugate (Bio-Rad). Incubation with ECL Plus generated a signal which was detected on the Storm 840 in fluorescence mode (450 nm). Total DNA on the same membrane was subsequently visualized on the Storm 840 (phosphorImager mode) after Southern blotting with nonspecific probes,61 and data were analyzed as described above.
Proliferation, colony formation and cell cycle assays
For cell cycle analysis, cells were synchronized by treatment with 1 μg ml−1 aphidicolin for 20 hr, washed and UV irradiated (12 J/m2), as described above. Cells were harvested by trypsinization at various repair times, fixed in 70% ethanol, treated with 1mg ml−1 RNase and stained with 10 μg ml−1 propidium iodide. DNA content was analysed on a FACSCalibur flow cytometer (BD Bioscience) using CellQuest software (BD Bioscience).
Apoptosis assay
Cells were grown on glass coverslips, UV irradiated and incubated for various repair periods. Cells were fixed, permeabilized and DNA strand breaks were labeled with fluorescein using the In situ Cell Death Detection Kit (Roche) according to the manufacturer’s instructions.
Acknowledgments
We would like to thank Dr. Keji Zhao for plasmid constructs and Drs. Raymond Reeves and Chengtao Her for helpful discussions. We are grateful to Drs. Anamaria Zavala and Shubho Chaudhuri for critical reading of the manuscript. This study was supported by NIH grants ES02614 and ES04106 from the National Institute of Environmental Health Sciences (to M.J.S.), grant IRG-77-003-26 from the American Cancer Society (to F.G.) and by the Intramural Research Program of the National Institute on Aging, National Institutes of Health (z01 AG000650-08 to W.W.).
References
- 1.Carlson M, Laurent BC. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 2.Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 3.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–91. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 4.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–92. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 5.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–42. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 6.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–18. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 8.Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–4. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–8. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186:136–45. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–76. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97:13796–800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–25. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 14.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 15.Sansam CG, Roberts CW. Epigenetics and cancer: altered chromatin remodeling via Snf5 loss leads to aberrant cell cycle regulation. Cell Cycle. 2006;5:621–4. doi: 10.4161/cc.5.6.2579. [DOI] [PubMed] [Google Scholar]
- 16.Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–74. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Liang B, Qiu J, Laurent BC. ATP-dependent chromatin-remodeling complexes in DNA double-strand break repair: remodeling, pairing and (re)pairing. Cell Cycle. 2005;4:1713–5. doi: 10.4161/cc.4.12.2222. [DOI] [PubMed] [Google Scholar]
- 18.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison AJ, Shen X. Chromatin modifications in DNA repair. Results Probl Cell Differ. 2006;41:109–25. doi: 10.1007/400_008. [DOI] [PubMed] [Google Scholar]
- 20.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 2005;4:884–96. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. Embo J. 2006;25:3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 23.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 24.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol. 2002;22:6779–87. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol. 2006;13:902–7. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 26.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 27.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Perucca P, Cazzalini O, Mortusewicz O, Necchi D, Savio M, Nardo T, Stivala LA, Leonhardt H, Cardoso MC, Prosperi E. Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen. J Cell Sci. 2006;119:1517–27. doi: 10.1242/jcs.02868. [DOI] [PubMed] [Google Scholar]
- 29.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–32. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 30.Vreeswijk MP, Westland BE, Hess MT, Naegeli H, Vrieling H, van Zeeland AA, Mullenders LH. Impairment of nucleotide excision repair by apoptosis in UV-irradiated mouse cells. Cancer Res. 1998;58:1978–85. [PubMed] [Google Scholar]
- 31.Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–30. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 32.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 33.Gale JM, Smerdon MJ. UV induced (6–4) photoproducts are distributed differently than cyclobutane dimers in nucleosomes. Photochem Photobiol. 1990;51:411–7. doi: 10.1111/j.1751-1097.1990.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell DL, Nguyen TD, Cleaver JE. Nonrandom induction of pyrimidine-pyrimidone (6–4) photoproducts in ultraviolet-irradiated human chromatin. J Biol Chem. 1990;265:5353–6. [PubMed] [Google Scholar]
- 35.Suquet C, Mitchell DL, Smerdon MJ. Repair of UV-induced (6–4) photoproducts in nucleosome core DNA. J Biol Chem. 1995;270:16507–9. doi: 10.1074/jbc.270.28.16507. [DOI] [PubMed] [Google Scholar]
- 36.Smerdon MJ, Conconi A. Modulation of DNA damage and DNA repair in chromatin. Prog Nucleic Acid Res Mol Biol. 1999;62:227–55. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 37.Callegari AJ, Kelly TJ. Shedding light on the DNA damage checkpoint. Cell Cycle. 2007;6:660–6. doi: 10.4161/cc.6.6.3984. [DOI] [PubMed] [Google Scholar]
- 38.Marusyk A, DeGregori J. Replicational stress selects for p53 mutation. Cell Cycle. 2007;6:2148–51. doi: 10.4161/cc.6.17.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feijoo C, Hall Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–23. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kastan MB, Bartek J. Cell cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 41.Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O’Connor PM, Fornace AJ., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–80. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 42.Hollander MC, Fornace AJ., Jr Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene. 2002;21:6228–33. doi: 10.1038/sj.onc.1205774. [DOI] [PubMed] [Google Scholar]
- 43.Zhan Q. Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat Res. 2005;569:133–43. doi: 10.1016/j.mrfmmm.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 44.Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25:2829–38. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 46.Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24:1188–99. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–24. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 48.Hanawalt PC, Gee P, Ho L, Hsu RK, Kane CJ. Genomic heterogeneity of DNA repair. Role in aging? Ann N Y Acad Sci. 1992;663:17–25. doi: 10.1111/j.1749-6632.1992.tb38644.x. [DOI] [PubMed] [Google Scholar]
- 49.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci USA. 1986;83:8878–82. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ., Jr p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705–14. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–65. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 52.Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277:22330–7. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 53.Lee H, Zeng SX, Lu H. UV Induces p21 rapid turnover independently of ubiquitin and Skp2. J Biol Chem. 2006;281:26876–83. doi: 10.1074/jbc.M605366200. [DOI] [PubMed] [Google Scholar]
- 54.Dotto GP. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta. 2000;1471:M43–56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 55.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 56.Gottifredi V, McKinney K, Poyurovsky MV, Prives C. Decreased p21 levels are required for efficient restart of DNA synthesis after S phase block. J Biol Chem. 2004;279:5802–10. doi: 10.1074/jbc.M310373200. [DOI] [PubMed] [Google Scholar]
- 57.Liu H, Kang H, Liu R, Chen X, Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol Cell Biol. 2002;22:6471–9. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–18. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 59.Adair JE, Kwon Y, Dement GA, Smerdon MJ, Reeves R. Inhibition of nucleotide excision repair by high mobility group protein HMGA1. J Biol Chem. 2005;280:32184–92. doi: 10.1074/jbc.M505600200. [DOI] [PubMed] [Google Scholar]
- 60.Smerdon MJ, Kastan MB, Lieberman MW. Distribution of repair-incorporated nucleotides and nucleosome rearrangement in the chromatin of normal and xeroderma pigmentosum human fibroblasts. Biochemistry. 1979;18:3732–9. doi: 10.1021/bi00584a014. [DOI] [PubMed] [Google Scholar]
- 61.Bespalov VA, Conconi A, Zhang X, Fahy D, Smerdon MJ. Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environ Mol Mutagen. 2001;38:166–74. doi: 10.1002/em.1068. [DOI] [PubMed] [Google Scholar]