Abstract
Purpose
The incidence of hepatocellular carcinoma (HCC) is increasing in the United States and liver cancer is the second leading cause of cancer-related mortality worldwide. Non-alcoholic steatohepatitis (NASH) is becoming an important risk for HCC and most patients with HCC have underlying liver cirrhosis and compromised liver function, which limit treatment options. Thus, novel therapeutic strategies to prevent or treat HCC in the context of NASH and cirrhosis are urgently needed.
Experimental Design
Constitutive activation of signal transducer and activator of transcription 3 (STAT3) is frequently detected in HCC tumors. STAT3 signaling plays a pivotal role in HCC survival, growth, angiogenesis and metastasis. We identified C188-9, a novel small-molecule STAT3 inhibitor using computer-aided rational drug design. In this study, we evaluated the therapeutic potential of C188-9 for HCC treatment and prevention.
Results
C188-9 showed antitumor activity in vitro in three HCC cell lines. In mice with hepatocyte-specific deletion of Pten (HepPten- mice),C188-9 treatment blocked HCC tumor growth, reduced tumor development, and reduced liver steatosis, inflammation and bile ductular reactions, resulting in improvement of the pathological lesions of NASH. Remarkably, C188-9 also greatly reduced liver injury in these mice as measured by serum AST and ALT levels. Analysis of gene expression showed that C188-9 treatment of HepPten- mice resulted in inhibition of signaling pathways downstream of STAT3, STAT1, TREM-1, and Toll-like receptors. In contrast, C188-9 treatment increased liver specification and differentiation gene pathways.
Conclusions
Our results suggest that C188-9 should be evaluated further for the treatment and/or prevention of HCC.
Keywords: STAT3, NASH, therapeutic and/or prevention effect of HCC
Introduction
Hepatocellular carcinoma (HCC) is the second most common cause of cancer-related deaths worldwide (1). Although the highest rates of liver cancer are found in certain areas of Asia and Africa, liver cancer incidence and mortality rates are increasing strikingly in western countries, including the United States (2, 3). Liver cirrhosis due to hepatitis B virus, hepatitis C virus, high alcohol consumption or non-alcoholic steatohepatitis (NASH) are the main risk factors associated with HCC;most patients with HCC have compromised liver function, which limits treatment options. HCC is a complex and highly heterogeneous disease with a large spectrum of genomic alterations and aberrant activation of cell signaling pathways (4-6). Currently, effective treatment options for HCC are limited. Surgical resection or transplantation represent the only potentially curative therapies for HCC, but most patients are diagnosed at an advanced stage and are not candidates for these approaches. Thus, there is a pressing need for the development of novel approaches to treat and prevent HCC (7, 8).
The pivotal role of signal transducer and activator of transcription 3 (STAT3) in cancer development and progression in many human cancers has led to intense efforts to identify small molecules and other strategies for targeting STAT3 (9-11). STAT3 is a transcription factor that regulates cell proliferation and survival as well as immune responses associated with cancer development and progression including HCC. Nearly 60% of human HCC exhibit activated nuclear STAT3 and STAT3 activation is associated with a poor prognosis (12, 13). The critical role of STAT3 as a regulator of liver cancer development and progression was demonstrated in mice (13, 14). STAT3 has also been linked to liver inflammation, injury and regeneration and to the activation of hepatic stellate cells (15-17).
In a drug development program involving virtual ligand screening, 2-D similarity screening, 3-D pharmacophore analysis, and SAR-based medicinal chemistry, we identified C188-9 as a potent small-molecule that targets the Src-homology (SH) 2 domain of STAT3 (18-21). C188-9 inhibited growth and survival of several cancer cell lines in vitro, including breast cancer (18), acute myeloid leukemia (19), head and neck squamous cell carcinoma (20) and non-small cell lung cancer (21). To test the effects of C188-9 on HCC and underlying liver disease, we herein used mice with hepatocyte-specific deletion of Pten (HepPten- mice). These mice present with hepatomegaly and develop liver disease marked by steatosis, inflammation, and fibrosis characteristic of NASH, which progresses to development of HCC (22-25). This is a highly relevant model for pre-clinical studies of HCC in the context of steatosis, fibrosis or NASH (26, 27).
Materials and Methods
Cell Culture and MTT Viability Assays
Human hepatoma cell lines Huh7 (ATCC), and PLC/PRF/5 and hepatoblastoma cell line HepG2 (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/mL penicillin and 100 μg/mL streptomycin. The hepatic epithelial cells THLE3 (ATCC) were grown and maintained in BEGM+ (Lonza/Clonetics Corporation, Walkersville) but cultured in complete DMEM when assays were performed. For cell viability assays, cells were treated in 96 well plates in medium ± C188-9 (StemMed, Ltd.) for 48 hours and subjected to MTT assays. Briefly, cells were incubated with 0.5mg/ml of the MTT solution (Sigma) for 2hrs and cell viability was measured by optical density (OD) at 590 nm (SpectraMax Plus 384 Microplate Reader, Molecular Devices). Experiments were performed in triplicate and used for IC50 calculation using GraphPad software.
Mice Treatment and MR Imaging
The study was approved by our Institutional Animal Care and Use Committee (IACUC). C57BL/6 mice carrying Pten conditional knockout alleles were crossed with an Albumin (Alb)-Cre-transgenic mouse. For this model, control animals are PtenloxP/loxP; Alb-Cre- while the experimental mice are PtenloxP/loxP; Alb-Cre+. For in vivo treatment, C188-9 was dissolved in DMSO and hepatic Pten null mice (HepPten- mice) (11 month-old) received C188-9 (100mg/kg) or vehicle (DMSO) by intraperitoneal (IP) injection daily for 4 weeks. Each treatment group included 9-12 mice. In a separate prevention study, HepPten- mice (8 month-old) received C188-9 (50mg/kg) or DMSO by IP injection daily for 4 weeks. MR Imaging was performed at the beginning of treatment and repeated after 2 and 4 weeks, on a four-channel, 7T dedicated small animal scanner (Bruker Biospin MRI). For tumor detection, a rapid acquisition with relaxation enhancement (RARE) sequence was used in the coronal and axial planes with a 0.25 mm slice thickness and with the number of slices sufficient to cover the entire liver. For respiration monitoring, a pressure-sensitive pad was placed on the animal bed directly underneath the animal. The compression and decompression of the pad were measured and the generated signal was finally fed to the MRI scanner.
Liver Histopathology, Immunohistochemistry and Liver Function Assays
IACUC-endorsed euthanasia by CO2 was performed followed by necropsy at which time, liver and tumor tissues were collected and snap-frozen in liquid nitrogen or fixed in 10% neutral buffered formalin and paraffin embedded (FFPE). The snap-frozen tissues were pulvarized for RNA or protein extraction. FFPE tissues were sectioned in 4 μm thick histologic sections that were stained with hematoxylin and eosin (H&E) or Masson's trichrome staining. Histopathological analysis was performed blindly by a liver pathologist. A NAFLD Activity Score (NAS) was described for scoring liver biopsies in patients with steatosis/steatohepatitis (28). In most cases of human NASH, the majority of the steatosis is of the macrovesicular type. While focal areas of microvesicular steatosis can be seen, this is not a common component of the steatosis in human disease. In contrast, some mouse models of NASH, including the HepPten- model, have mixed macrovesicular and microvesicular steatosis. We therefore used a modified NAFLD activity score (mNAS) to include both macrovesicular and microvesicular steatosis grades, calculated as an unweighted sum of the grading scores of macrosteatosis (0-3), microsteatosis (0-3), grade of lobular inflammation (0-3), and presence of ballooning (0-2). Bile ductular reaction was assessed on a 1 to 4 scale. Blood samples were also taken at necropsy and processed for serum collection. Serum AST and ALT activities were determined using ACE AXCEL™ clinical chemistry system (Diagnostic Technologies) according to the manufacturer's instructions. Standard controls were run before each determination, and the values obtained for the biochemical parameter were within the expected ranges. Immunohistochemical staining for ki67 was performed using vehicle- and C188-9-treated tumor slides. Slides were scanned and analyzed using Aperio ImageScope software. The proliferation index was determined by calculating the average percentage of ki67 tumor positive cells.
RNA Sequencing and Differentially Expressed Gene (DEG) Analysis
Total RNA was isolated from liver and tumors of the study mice. Following second-strand cDNA synthesis, end repair, adaptor ligation, and PCR amplification, the enriched cDNA libraries were sequenced using Illumina HiSeq 3000. Libraries were run using 50-base-pair single-end reads on Illumina HiSeq 3000 System. Sequence files were generated in FASTQ format, and reads were mapped to mouse genome mm10 and then aligned by TopHat2 (29). Gene expression levels were quantified using Cufflinks (version 1.0.3) in the FPKM unit (fragments per kilobase of exon per million fragments mapped) together with confidence intervals. Cufflinks ran in the default parameters except that the annotated gene set was supplied using the -G option. Subsequently, the edgeR (empirical analysis of digital gene expression data in R) tool was utilized to detect the differentially expressed genes (DEGs) after filtering out genes with counts per million (CPM) under 10 in all samples. CPM values were scaled by the total number of fragments. The multiple comparison P-values were adjusted by using Benjamini-Hochberg method, providing a P-value cutoff for significance controlled by the false discovery rate (FDR). DEGs were selected using the following criteria: FDR < 0.05 and fold change > 1.5. The heat map of DEGs was generated using Pearson correlation and Ward distance in R and CPM values. DEGs were further analyzed using the Ingenuity Pathway Analysis software (IPA, Ingenuity Systems; http://www.ingenuity.com). IPA identifies putative networks, biological functions and canonical pathways overrepresented among the DEGs.
Luminex Assays
Cell and tissue lysates were plated in a 96-well filter plate pre-loaded with beads (Millipore) coupled to antibody against the indicated analytes and incubated overnight at 4°C. Bead-bound analytes were measured using biotinylated detection antibody specific for a different epitope and streptavidin-phycoerythrin (streptavidin-PE) (30, 31). Data were collected and analyzed using the Bio-Plex suspension array system (Luminex 100 system, Bio-Rad Laboratories). GAPDH-normalized pY-STAT1 and pY-STAT3 values from the treated samples were compared to the untreated samples. Serum cytokine levels were measured using Luminex MAP assays (MILLIPLEX MAP Mouse Cytokine/Chemokine, MCYTOMAG-70K, Millipore) and 25 μL of serum samples. The assay was performed on a Luminex 200 instrument (Luminex Corporation, Austin, TX) using Luminex xPONENT 3.1 software. Using the standard curves for the given cytokine or chemokine, we obtained absolute concentrations for the various experimental conditions for all analytes measured.
Quantitative PCR
To quantify selected mRNA levels, equal amounts of total RNA samples were submitted to reverse transcription and real-time PCR using specific primers listed in Supplementary Table S1. PCR amplifications of the respective genes were performed using iTaq SYBR Green Supermix (Bio-Rad) in CFX Connect Real-Time System (Bio-Rad). The Bio-Rad CFX Manager software (version 2.1) was used for the calculation of threshold cycles (Ct)-values and melting curve analysis of amplified DNA. Relative expression of the tested miRNAs and genes was calculated by 2−ΔΔCt method.
Statistical Analysis
Statistical difference between each group was assessed by the non-parametric Mann-Whitney test using Graphpad™ 6.0 software. The high-throughput sequencing data were analyzed using two sample t-test. A value of p<0.05 was considered significant.
Results
C188-9 Reduced Hepatoma Cell Viability
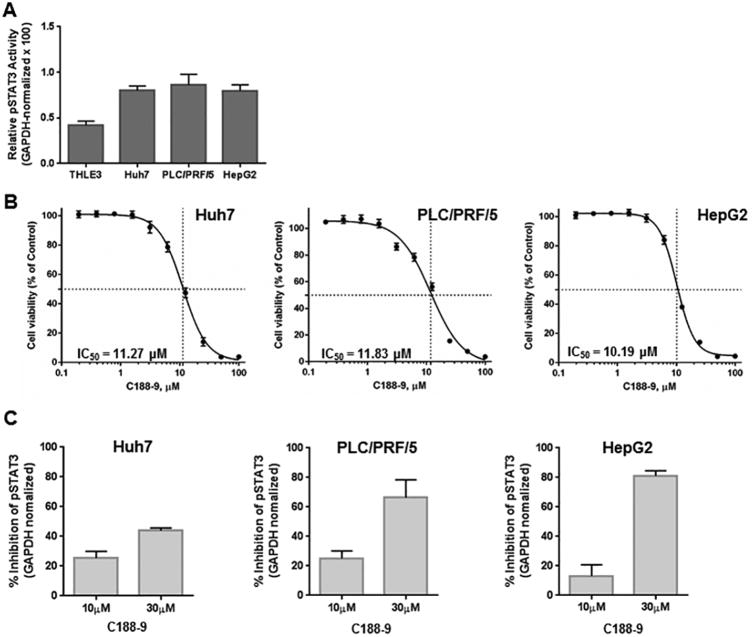
To evaluate the potential of C188-9 as a therapeutic strategy for HCC, we first examined the effect of C188-9, a potent, non-toxic, inhibitor of STAT3, on the viability of the hepatoma cell lines Huh7 and PLC/PRF/5 and the hepatoblast-derived cells HepG2. All three cell lines had increased activated STAT3 (>2-fold), when compared to the normal hepatic epithelial cells THLE3 (Figure 1A). We then treated them with C188-9 at serial concentrations ranging from 0.19 to 100 μM, for 48 hours under anchorage-dependent condition. C188-9 reduced cell viability of all three cell lines with IC50 values of 11.27, 10.19 and 11.83 μM for Huh7, PLC/PRF/5 and HepG2, respectively (Figure 1B). Finally, we confirmed the inhibition of pSTAT3 levels in these cells upon C188-9 treatment. At 10 μM, pSTAT3 levels were reduced by 25.4%, 25.1% and 13.2% and at 30μM, pSTAT3 levels were reduced by 43.9%, 66.5% and 81.1%, in Huh7, PLC/PRF/5 and HepaG2 cells respectively (Figure 1C).
Figure 1. C188-9 treatment reduced viability of hepatoma cells.
(A) Lysates prepared from asynchronous cultures of Huh7, PLC/PRF/5, HepG2 and THLE3 cells were examined for levels of pSTAT3 and GAPDH by luminex assay. GAPDH-normalized pSTAT3 levels of replicate experiments (×100) are shown in the Y-axis. (B) Huh7, PLC/PRF/5 and HepG2 cell lines were treated with C188-9 at the concentrations indicated for 48 h under anchorage-dependent conditions. Cell viability was calculated as a percentage of the DMSO-treated control wells. The half-maximal inhibitory concentration (IC50) values were derived after plotting viability values on a logarithmic curve. (C) Lysates prepared from Huh7, PLC/PRF/5 and HepG2 cells untreated or treated with 10 and 30 μM of C188-9 were subjected to luminex-based detection of pSTAT3 and GAPDH levels. The GAPDH-normalized pSTAT3 levels of each treatment were expressed as a percent of untreated cells. Data shown are representative results of triplicate experiments for each cell line.
C188-9 Treatment Resulted in Tumor Growth Arrest in mice with hepatic deletion of Pten
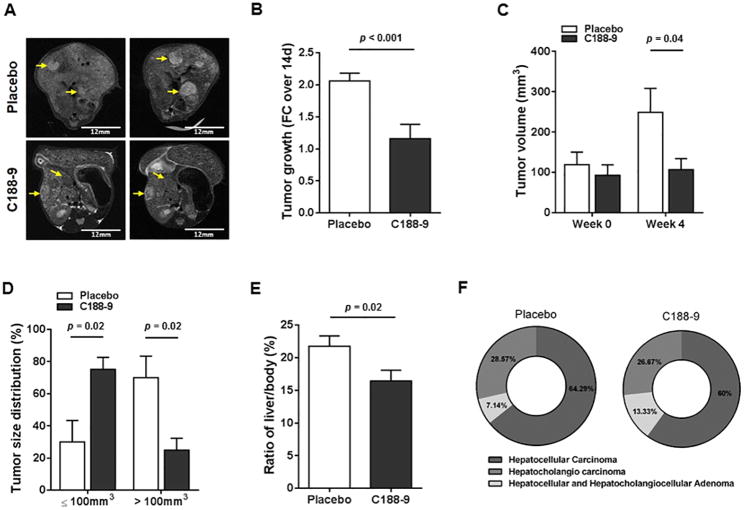
We then evaluated the effect of targeting STAT3 in vivo, using the hepatocyte-specific Pten deletion (HepPten-) mouse model. We treated 11 month-old HepPten- male mice for 4 weeks, with daily intraperitoneal injections of C188-9 or vehicle. All treated mice had developed tumors prior to treatment as confirmed by MRI. MRI imaging showed continuous tumor growth increase in vehicle-treated mice, but arrest of tumor growth in C188-9-treated mice (Figure 2A). Indeed, while the average tumor growth rate in the vehicle group was 2.06 fold over 14 days, the average tumor growth rate was 1.16-fold in the C188-9 treated group (p<0.001) (Figure 2B). The tumor growth arrest induced by C188-9 treatment was further confirmed by comparing the average tumor volume determined by MRI in both groups prior to treatment (119.02mm3 and 92.64mm3 for vehicle- and C188-9-treated mice, respectively; p=ns) and at the end of treatment (249.02mm3 and 106.48mm3 for vehicle- and C188-9-treated mice, respectively; p=0.04) (Figure 2C). Necropsy results showed that while 70% of the tumors collected at time of necropsy from the vehicle group were >100mm3, only 24.85% of the tumors in C188-9 treated mice were >100mm3 (p=0.02) (Figure 2D). Necropsy results also showed a significant reduction of liver to body weight ratio in C188-9-treated group compared to vehicle-treated mice (p=0.02), further supporting the reduction of tumor growth upon C188-9 treatment (Figure 2E). Finally, blinded analysis of all tumors by a pathologist showed that C188-9 treatment resulted in decreased percentage of carcinomas upon C188-9 treatment (from 92.87% to 86.67%) and concomitant increased percentage of adenomas (from 7.13% to 13.33%), indicating that tumor malignancy progression was inhibited upon C188-9 treatment (Figure 2F).
Figure 2. Effect of C188-9 treatment on tumor growth and tumor phenotype.
(A) Liver of mice from vehicle- and C188-9-treated groups were imaged with MRI to measure tumor growth during treatment. Representative tumors detected by MRI before treatment and at the end of treatment, are indicated by yellow arrows. (B) Tumor growth expressed as fold change over 2 weeks of treatment, in vehicle-treated and C188-9-treated groups as determined by MRI. (C) Tumor volume before and after treatment as determined by MRI. (D) Tumor size distribution in each treatment group. The tumors are separated into ≤ 100 mm3 or > 100 mm3. Average percentages of tumors are represented on the Y-axis and tumor size categories on the X-axis. (E) Liver-to-body weight ratio at the end of treatment. For panels B to E, data are presented as means ± SEM (Mann-Whitney test). (F) Pie charts representing the overall tumor histology distribution within the two treatment groups.
C188-9 Treatment Improves NASH and Liver Function
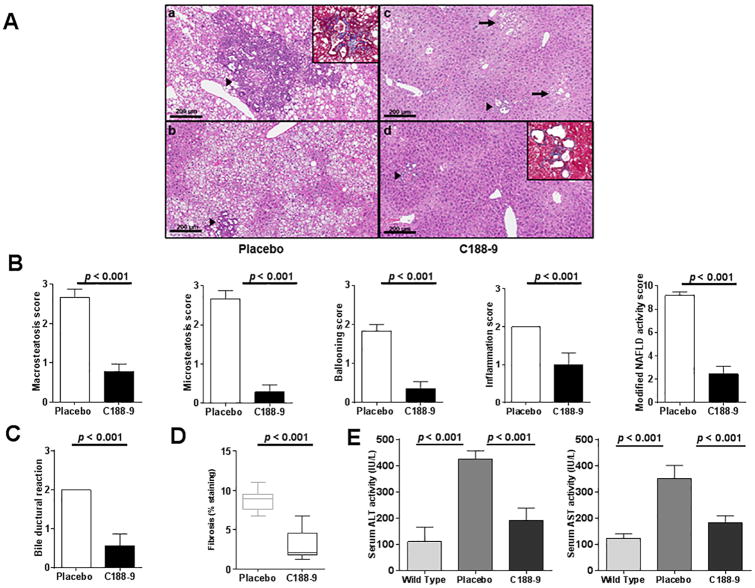
Since HepPten- mice develop NASH prior to HCC, we also evaluated the effect of C188-9 treatment on this underlying liver pathology. The liver parenchyma in the vehicle-treated mice showed diffuse mixed macro- and microsteatosis extending from zones 1 to 3 (Figure 3A, panels a-b). In contrast, the C188-9-treated mice showed minimal steatosis, with only a few small foci of steatosis observed in the centrilobular area (Figure 3A, arrows in panel c). As shown in Figure 3B, steatosis scoring further confirmed that C188-9 treatment resulted in decrease in hepatic steatosis for both macrovesicular and microvesicular steatosis (2.7±0.2 vs.0.8±0.2 and 2.7±0.2 vs. 0.3±0.2, respectively, p<0.001 for both). In line with the effect of C188-9 onreducing the steatosis, ballooning of hepatocytes and inflammation were also significantly decreased in mice treated with C188-9 compared to vehicle-treated mice (1.8±0.2 vs. 0.4±0.2; p=0.002 and 2 vs.1±0.3; p=0.016). Using a modified NAFLD activity score (mNAS) to include both macrovesicular and microvesicular steatosis grades, we observed an overall inhibition of NASH and reversion to a phenotype with milder pathology upon C188-9 treatment (9.2±0.3 vs. 2.4±0.7; p<0.001) (Figure 3B). The vehicle-treated mice also showed frequent areas of prominent bile ductular reaction with peribiliary neutrophilic inflammation (Figure 3A, panels a-b). In contrast, most of the C188-9-treated mice appeared similar to untreated control mice, with the exception of rare focal bile ductular reactions, which had less associated inflammation and were significantly smaller in size when compared to those observed in the vehicle group (Figure 3A, panels c-d). The bile ductular reaction scores were significantly reduced upon C188-9 treatment (2 vs. 0.6±0.8; p=0.004) (Figure 3C). In addition, C188-9 treatment significantly reduced fibrosis in these areas, measured by Masson's trichrome staining, from 8.77% to 3.07% (p<0.001) (Figure 3D) (Figure 3A, panels a-d). Most remarkably and in agreement with NASH improvement, serum biochemical analysis showed a significant reduction of hepatocellular injury upon C188-9 treatment as ALT and AST levels decreased upon C188-9 treatment, from 434IU/L to 165.43IU/L (p=0.003) and from 350 IU/L to 182.57IU/L (p=0.004), respectively (Figure 3E), reaching near to normal values.
Figure 3. Effect of C188-9 on hepatic histopathology and liver function in experimental mice.
(A) Representative images of H&E stained liver sections from vehicle- and C188-9-treated mice are shown (scale Bars, 200 μm). The liver parenchyma in the vehicle-treated mice showed nearly diffuse mixed macrovesicular and microvesicular steatosis affecting all zones of hepatic lobules (from zones 1 to 3) (a-b). In contrast, the livers from C188-9-treated mice showed minimal steatosis, with only a few small foci of perivenular steatosis in the centrilobular zone (c, arrows). The livers of vehicle-treated mice also showed frequent areas of prominent bile ductular reaction with peribiliary neutrophilic inflammation (a-b, arrow heads). Most livers of C188-9-treated mice appeared similar to untreated control mice (data not shown), with the exception of rare focal bile ductular reactions, which had less associated inflammation and were much smaller in size when compared to those observed in the vehicle group (c-d, arrowheads). Vehicle-treated mouse livers also had increased peribiliary fibrosis (a, inset) when compared to those from C188-9-treated mice (d, inset), which was confirmed by Masson's trichrome stains. (B) Steatosis was individually scored by macrosteatosis and microsteatosis (0 for <5%, 1 for 5–33%, 2 for 33–66%, and 3 for more than 66%). Ballooning score was 0 for no ballooned cells per field, 1 for few, and 2 for many. Inflammation was graded by overall assessment of all inflammatory foci (0 for no foci, 1 for <2, 2 for 2–4 foci, and 3 for >4 foci). Modified NAS is the sum of marcrosteatosis, microsteatosis, inflammation, and ballooning scores. (C) Bile ductural reaction was assessed on a 1 to 4 scale. (D) Liver fibrosis was measured by Masson's Trichrome staining. The fibrosis was quantified by determining the percentage of the positive staining area (blue staining) out of the whole liver tissue area. (E) Serum alanine transaminase (ALT) and aspartate aminotransferase (AST) levels were measured in the serum of vehicle-treated and C188-9-treated mice. For all panels, values represent mean ± SEM (Mann Whitney test).
C188-9 Treatment Reduces Tumor Development in vivo
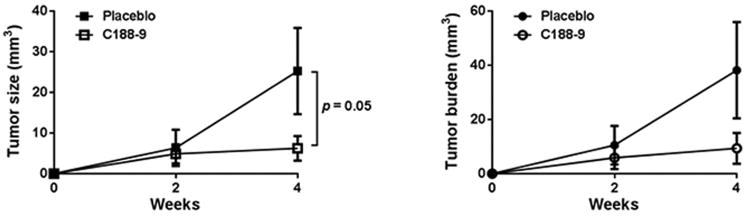
Because of the effects of C188-9 on NASH severity, we evaluated the effect of C188-9 on tumor development in the HepPten-mice. Mice without tumors as determined by MRI, were treated with C188-9 or vehicle and tumor development was monitored over a 4-week period by MRI. At the end of treatment, the average tumor size (6.24±3.02mm3 vs. 25.25±10.6mm3; p=0.05) and average tumor burden (9.36±5.67mm3 vs. 38.18±17.76mm3) were lower in C188.9 treated mice compared to the vehicle group (Figure 4), demonstrating that treatment of C188-9 reduces tumor development in HepPten- mice.
Figure 4. Effects of C188-9 on tumor prevention in vivo.
(A) Average tumor volume and (B) average tumor burden in vehicle- and C188-9 treated HepPten-mice, over the 4-week treatment. Tumor volume was determined using MRI. Data are represented as mean ± SEM (Mann Whitney test).
Gene Expression Changes upon C188-9 treatment in Tumors and Adjacent Liver in HepPten- mice
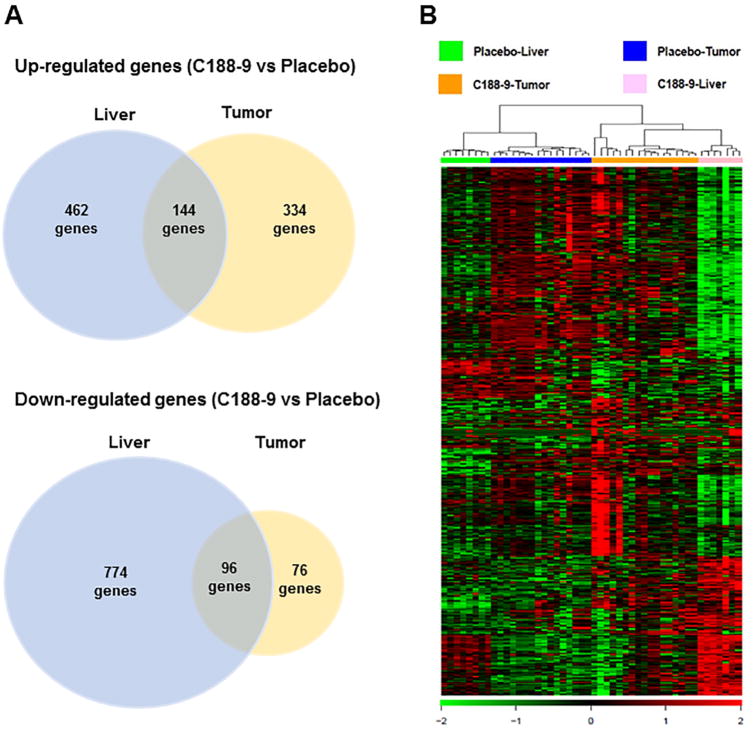
To identify the mechanism of C188-9 effect in vivo, we measured mRNA expression using RNA-sequencing in both liver and tumors from C188-9 treated group and vehicle group. Using p value<0.05 and fold change >1.5, we identified 606 up-regulated and 870 down-regulated genes in liver and 478 up-regulated and 172 down-regulated genes in tumors, upon C188-9 treatment. Among these genes, 144 genes were commonly up-regulated and 96 genes were commonly down-regulated in both liver and tumor (Figure 5A). Hierarchical clustering based on these differentially expressed genes (DEGs) distinguished four main clusters corresponding to vehicle liver, vehicle tumor, C188-9 tumor and C188-9 liver, with the widest separation being between vehicle liver and C188-9 liver. The clustering analysis demonstrated that all mice responded similarly to C188-9 treatment and that the effects on gene expression were largely specific to the liver or the tumor, with the largest effect being observed in liver (Figure 5B).
Figure 5. Clustering of differentially expression genes.
(A) Venn diagrams showing up-regulated and down-regulated genes inC188-9 vs. vehicle-treated liver and in C188-9 vs. vehicle-treated tumors. The diagram shows the number of genes identified in each group following the criteria described in the Methods section. (B) Hierarchical clustering and heat map of 1,868 differentially expressed genes in samples from four groups (vehicle livers, vehicle tumors, C188-9 livers, and C188-9 tumors). Green color indicates strength of C188-9-induced down-regulation and red indicates up-regulation.
We performed Ingenuity Pathway Analysis (IPA) for all DEGs significantly down-regulated or up-regulated or in liver, in tumor, or in both. The top biological functions and canonical pathways that were down-regulated by C188-9 were: TREM1 signaling, Role of pattern recognition receptors in recognition of bacteria and viruses in liver; Cellular growth and Cell death and survival in tumor; Mitotic role of polo-like kinase, and interferon signaling in both liver and tumor (Supplementary Table S2). IPA analysis identified STAT3 as the main upstream regulator of the downregulated DEGs. Inhibition of STAT3 activity upon C188-9 treatment was confirmed by measuring pY-STAT3 levels, pY-STAT3 levels were increased in tumors compared to liver in the vehicle group (p=0.02) confirming the activation of STAT3 in HCC in HepPten- mice. C188-9 treatment resulted in a reduction of pY-STAT3 levels in tumors to levels similar to those observed in vehicle-treated tumors (p=0.014) (Supplementary Figure S1). TGFβ1 and IFNγ were also identified as upstream regulators in liver. In agreement, IFNγ-inducible genes and toll like receptors were overrepresented among the downregulated DEGs. A large number of pro-fibrotic and tumor promoter genes were also found downregulated in liver and tumor (Supplementary Table S2). The effect on IFN-inducible genes suggested that STAT1 activity could also be inhibited upon C188-9 treatment in HepPten- tumors. Indeed, pY-STAT1 levels were decreased upon C188-9 treatment in tumors in HepPten-mice (p=0.002) (Supplementary Figure S1). The top biological functions and canonical pathways that were up-regulated by C188-9 were: FXR/RXR activation, Estrogen biosynthesis and Cholesterol biosynthesis in liver; Granulocyte adhesion in tumor; LXR/RXR activation in both liver and tumor (Supplementary Table S3). Interestingly, Hnf4a, a major hepatocytic differentiation transcription factor, was identified as the main upstream regulator in liver, and other liver specification and differentiation genes were also upregulated such as Acox2 in liver and Hnf3a and aldolase C in tumor. A large number of cytochrome P450 genes were upregulated in both liver and tumor. Other genes associated with inhibition of fibrosis and tumor suppression in liver were also identified (Supplementary Table 3). We also randomly selected 5 genes from the list of DEGs, for validation by qRT-PCR. RNA sequencing data were validated by qRT-PCR for the downregulated genes CENPF, CD34, and HMMR and the upregulated genesCD163 and CYP1A1 (Supplementary FigureS2).
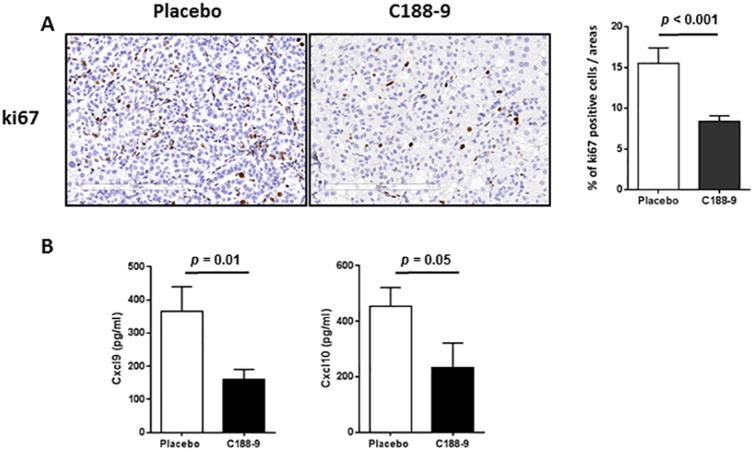
C188-9 Treatment Inhibits Cell Proliferation in vivo and is Associated with Reduction in Circulating Inflammatory Chemokines
Gene expression analysis identified ki67 as a gene commonly downregulated in both liver and tumor upon C188-9 treatment in HepPten-mice. Sections from vehicle and C188-9 treated tumor were stained with ki67 antibody, and the percentages of ki67-positive cells were calculated. As shown in Fig. 6A, C188-9 treated group exhibited less staining for ki67, when compared with the vehicle treated group (15.5 ± 1.9 vs. 8.4 ± 0.7; p<0.001). Because inflammation was reduced based on histology analysis and because several inflammatory cytokines and chemokines were found modulated by C188-9 in gene expression analysis, we measured a panel of cytokines/chemokines in serum collected from the mice treated with either vehicle or C188-9. CXCL9 and CXCL10 levels were found to be dramatically decreased in C188-9-treated mice (364.6 ± 75.2 pg/ml vs. 159.9± 30.6 pg/ml, p=0.01 and455.1± 66.7 pg/ml vs. 234.0± 87.7 pg/ml, p=0.05, respectively) (Figure6B).
Figure 6. Effects of C188-9 treatment on tumor cell proliferation and chemokine levels in serum.
In both vehicle and C188-9-treated mice, ki67 immunohistochemical (IHC) staining was used to evaluate the amount of tumor cell proliferation. (A) Representative sections are shown. Scale Bars, 200 μm. (B) Quantitation of IHC-positive staining for ki67 expressed as percentage of positive tumor cells. (C) Serum levels of CXCL9 and CXCL10 in vehicle- and C188-9-treated HepPten-mice. Values represent mean ± SEM (Mann-Whitney test).
Discussion
While causal roles for STAT3 in hepatic tumorigenesis have been proposed (32), the importance of STAT3 in HCC preconditions, such as liver fibrosis or NASH, has received far less attention, a side from two studies suggesting that STAT3 inhibition mediates the antifibrotic effect of sorafenib (33) and that STAT3 signaling is activated in NAFLD (34). Herein, we used a clinically relevant model of NASH-associated HCC to evaluate the therapeutic potential of C188-9, a small molecule inhibitor of STAT3. C188-9 binds to the SH2 domain of STAT3 and blocks its recruitment to the kinase-containing receptor complexes, its tyrosine phosphorylation, and its homodimerization. STAT3 activation can lead to tumorigenesis in diverse type of cancers (18-21). We demonstrated that C188-9 had beneficial effects on both hepatic tumor and NASH in liver, with blockage of tumor growth and progression, prevention of tumor development, reduction of steatosis, inflammation and hepatocellular ballooning, resulting in NASH reversion, and reduction of bile ductular reaction and associated fibrosis. Evaluation of C188-9 treatment on additional models of HCC and NASH will be needed to demonstrate that the effects reported in this study are independent of hepatic PTEN loss. Future studies aimed at further characterizing STAT3 activation in human NASH-HCCs are also warranted.
Genomic analysis identified a main inhibitory effect on immune response including downregulation of TREM-1 signaling and reduced expression of Toll-like receptors (TLRs) and interferon-inducible genes. It was previously reported that the proinflammatory myeloid cell receptor TREM-1 controls development of HCC (35) and that TREM-1 expression in hepatic stellate cells is a prognostic marker for hepatitis B-related HCC (36). Toll-like receptors are pattern recognition receptors that sense and respond to microbial pathogens and damage-associated molecular patterns by facilitating inflammation (37). TLR are present on host immune cells, especially macrophages and dendritic cells, but also on epithelial cells (38). STAT3 is an important regulator of TLR2 and TLR4 activity and a mediator of TLR4 and TLR9 signaling (39-42). TLR3/4/9-mediated inflammatory responses are an important contributor of chronic inflammation-associated HCC (43). The reduction of TLRs in C188-9 treated HepPten- mice may contribute to the concomitant reduction in inflammation measured by histology and in reduced levels of inflammatory cytokines in serum of these mice and in the overall reduction of NASH severity. TLR2, TLR4 and TLR9 has been implicated in the development of ASH, NASH, liver fibrosis, and HCC and increased TLRs is involved in increased inflammation in these chronic liver disease (44-47. Whether the observed reduction in TLRs in liver and tumor upon C188-9 treatment corresponds to a reduction in immune cell infiltration or a reduced expression on epithelial cells or both requires further evaluation. A comprehensive immune profiling follow-up study aimed at identifying the immune cell populations affected by C188-9 treatment in the HepPten- model would be highly relevant. Such populations include Th2 and Th17 cells: STAT3 drives development of Th17 cells and cytokine production by Th2 and Th17 cells and we have recently reported that C188-9 prevents the accumulation of Th2 and Th17 cells in a murine asthma model (48).
In contrast, the Farnesoid X Receptor (FXR) signaling pathway was activated upon C188-9 treatment. FXR is highly expressed in liver and has an important role in protecting against HCC by inhibiting cell growth and inducing cell cycle arrest at G1 phase (49). Genomic analysis also identified HNF4a, a major hepatocytic transcription factor, as the main upstream regulator in liver and other liver specification genes upregulated in liver and tumor, suggesting that C188-9 induced hepatocytic differentiation in both liver and tumor. We recently showed that induction of hepatocytic differentiation in the HepPten-model inhibits tumor growth, reduced liver fibrosis and prevented tumor development (7). Therefore, the improved differentiation status of hepatic cells may contribute to the therapeutic effect of C188-9 treatment on tumor and liver and the observed improvement in liver injury tests.
C188-9 has been shown to have inhibitory effects on STAT1 in prior studies involving head and neck squamous cell carcinoma (20). While we identified STAT3 as the main upstream regulator targeted by C188-9 treatment in both liver and tumor, the significant overrepresentation of interferon-inducible genes among the C188-9-dependent downregulated genes, suggested that STAT1 activity was also inhibited. C188-9-induced inhibition of STAT1 activity was indeed confirmed in both liver and hepatic tumors. The differential contribution of inhibition of STAT1vs. STAT3 to the C188-9 treatment effects in HepPten-model will be further investigated in follow-up studies.
In conclusion, this study identified the novel small-molecule C188-9 as a highly promising therapeutic drug for treatment and prevention of HCC. C188-9 showed not only preventive and antitumor activity but also significant reduced pathologic lesions of NASH and hepatocellular injury, with an overall improvement of liver functions. This dual effect of C188-9 may be highly beneficial to HCC patients with NASH or liver cirrhosis. A Phase 1 trial will be initiated later this year to examine the potential of C188-9 treatment in HCC patients.
Supplementary Material
Translational Relevance.
The incidence of hepatocellular carcinoma (HCC) is increasing in the United States and is the second leading cause of cancer-related mortality worldwide. Non-alcoholic steatohepatitis (NASH) is becoming an important risk for HCC and most patients with HCC have underlying liver cirrhosis and compromised liver function, which limit treatment options. Thus, novel therapeutic strategies to prevent or treat HCC in the context of NASH and cirrhosis are urgently needed. This study identified the novel small-molecule C188-9 as a highly promising therapeutic drug for treatment and prevention of HCC. C188-9 showed not only preventive and antitumor activity but also significant reduced pathologic lesions of NASH and hepatocellular injury, with an overall improvement of liver functions. This dual effect of C188-9 may be highly beneficial to HCC patients with NASH or liver cirrhosis. A Phase 1 trial will be initiated later this year to examine the potential of C188-9 treatment in HCC patients.
Acknowledgments
Financial Support: This work was supported in part by the National Institutes of Health through Cancer Center Support grant (P30CA016672 to L. Beretta), by the Sequencing and Microarray Facility at the University of Texas MD Anderson Cancer Center CA016672, and by Cancer Prevention and Research Institute of Texas grant (DP150069 to D. Tweardy).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011 Mar-Apr;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004 Nov;127(5 Suppl 1):S27–34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, et al. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016 May 1;122(9):1312–37. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka S, Arii S. Molecular targeted therapies in hepatocellular carcinoma. Seminars in oncology. 2012 Aug;39(4):486–92. doi: 10.1053/j.seminoncol.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Cervello M, McCubrey JA, Cusimano A, Lampiasi N, Azzolina A, Montalto G. Targeted therapy for hepatocellular carcinoma: novel agents on the horizon. Oncotarget. 2012 Mar;3(3):236–60. doi: 10.18632/oncotarget.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muntane J, De la Rosa AJ, Docobo F, Garcia-Carbonero R, Padillo FJ. Targeting tyrosine kinase receptors in hepatocellular carcinoma. Current cancer drug targets. 2013 Mar;13(3):300–12. doi: 10.2174/15680096113139990075. [DOI] [PubMed] [Google Scholar]
- 7.Bruix J, Sherman M American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology. 2011 Mar;53(3):1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Park YN. Hepatocellular carcinomas expressing ‘stemness’-related markers: clinicopathological characteristics. Digestive diseases. 2014;32(6):778–85. doi: 10.1159/000368021. [DOI] [PubMed] [Google Scholar]
- 9.Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anti-cancer drugs. 2005 Jul;16(6):601–7. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Furtek SL, Backos DS, Matheson CJ, Reigan P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS chemical biology. 2016 Feb 19;11(2):308–18. doi: 10.1021/acschembio.5b00945. [DOI] [PubMed] [Google Scholar]
- 11.Bharadwaj UK, M M, Tweardy DJ. STAT3 inhibitors in Cancer: A Comprehenive Update. In: Ward AC, editor. STAT Inhibitors in Cancer. Switzerland: Springer International Publishing; 2016. [Google Scholar]
- 12.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006 Apr;130(4):1117–28. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 13.He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, et al. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer cell. 2010 Mar 16;17(3):286–97. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Won C, Kim BH, Yi EH, Choi KJ, Kim EK, Jeong JM, et al. Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology. 2015 Oct;62(4):1160–73. doi: 10.1002/hep.27968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell research. 2011 Jan;21(1):159–68. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Lafdil F, Kong X, Gao B. Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. International journal of biological sciences. 2011;7(5):536–50. doi: 10.7150/ijbs.7.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, Hsu SH, Frankel W, Ghoshal K, Jacob ST. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology. 2012 Jul;56(1):186–97. doi: 10.1002/hep.25632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Kasembeli MM, Jiang X, Tweardy BJ, Tweardy DJ. Chemical probes that competitively and selectively inhibit Stat3 activation. PloS one. 2009;4(3):e4783. doi: 10.1371/journal.pone.0004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood. 2011 May 26;117(21):5701–9. doi: 10.1182/blood-2010-04-280123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bharadwaj U, Eckols TK, Xu X, Kasembeli MM, Chen Y, Adachi M, et al. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget. 2016 Mar;:25. doi: 10.18632/oncotarget.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis KM, Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Fridley C, et al. Small-molecule targeting of signal transducer and activator of transcription (STAT) 3 to treat non-small cell lung cancer. Lung cancer. 2015 Nov;90(2):182–90. doi: 10.1016/j.lungcan.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. The Journal of clinical investigation. 2004 Jun;113(12):1774–83. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim YJ, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected] Proceedings of the National Academy of Sciences of the United States of America. 2004 Feb 17;101(7):2082–7. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai KK, Shang S, Lohia N, Booth GC, Masse DJ, Fausto N, et al. Extracellular matrix dynamics in hepatocarcinogenesis: a comparative proteomics study of PDGFC transgenic and Pten null mouse models. PLoS genetics. 2011 Jun;7(6):e1002147. doi: 10.1371/journal.pgen.1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muir K, Hazim A, He Y, Peyressatre M, Kim DY, Song X, et al. Proteomic and lipidomic signatures of lipid metabolism in NASH-associated hepatocellular carcinoma. Cancer research. 2013 Aug 1;73(15):4722–31. doi: 10.1158/0008-5472.CAN-12-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Jiao J, Cermelli S, Muir K, Jung KH, Zou R, et al. miR-21 Inhibition Reduces Liver Fibrosis and Prevents Tumor Development by Inducing Apoptosis of CD24+ Progenitor Cells. Cancer research. 2015 May 1;75(9):1859–67. doi: 10.1158/0008-5472.CAN-14-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung KH, Zhang J, Zhou C, Shen H, Gagea M, Rodriguez-Aguayo C, et al. Differentiation therapy for hepatocellular carcinoma: Multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology. 2016 Mar;63(3):864–79. doi: 10.1002/hep.28367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Nonalcoholic Steatohepatitis Clinical Research Network. Hepatology. 2005 Jun;41(6):1313–21. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 29.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009 May 1;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast cancer research : BCR. 2009;11(4):R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim BK, Lee JW, Park PJ, Shin YS, Lee WY, Lee KA, et al. The multiplex bead array approach to identifying serum biomarkers associated with breast cancer. Breast cancer research : BCR. 2009;11(2):R22. doi: 10.1186/bcr2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong AL, Soo RA, Tan DS, Lee SC, Lim JS, Marban PC, et al. Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015 May;26(5):998–1005. doi: 10.1093/annonc/mdv026. [DOI] [PubMed] [Google Scholar]
- 33.Su TH, Shiau CW, Jao P, Liu CH, Liu CJ, Tai WT, et al. Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2015 Jun 9;112(23):7243–8. doi: 10.1073/pnas.1507499112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min HK, Mirshahi F, Verdianelli A, Pacana T, Patel V, Park CG, et al. Activation of the GP130-STAT3 axis and its potential implications in nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol. 2015 Mar;308:6. G794–G803. doi: 10.1152/ajpgi.00390.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer research. 2012 Aug 15;72(16):3977–86. doi: 10.1158/0008-5472.CAN-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao R, Sun TW, Yi Y, Wu H, Li YW, Wang JX, et al. Expression of TREM-1 in hepatic stellate cells and prognostic value in hepatitis B-related hepatocellular carcinoma. Cancer science. 2012 Jun;103(6):984–92. doi: 10.1111/j.1349-7006.2012.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011 May 27;34(5):637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Shaykhiev R, Behr J, Bals R. Microbial patterns signaling via Toll-like receptors 2 and 5 contribute to epithelial repair, growth and survival. PloS one. 2008;3(1):e1393. doi: 10.1371/journal.pone.0001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tye H, Kennedy CL, Najdovska M, McLeod L, McCormack W, Hughes N, et al. STAT3-driven upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer cell. 2012 Oct 16;22(4):466–78. doi: 10.1016/j.ccr.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Greenhill CJ, Rose-John S, Lissilaa R, Ferlin W, Ernst M, Hertzog PJ, et al. IL-6 trans-signaling modulates TLR4-dependent inflammatory responses via STAT3. Journal of immunology. 2011 Jan 15;186(2):1199–208. doi: 10.4049/jimmunol.1002971. [DOI] [PubMed] [Google Scholar]
- 41.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nature reviews Cancer. 2014 Nov;14(11):736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 42.Eyking A, Ey B, Runzi M, Roig AI, Reis H, Schmid KW, et al. Toll-like receptor 4 variant D299G induces features of neoplastic progression in Caco-2 intestinal cells and is associated with advanced human colon cancer. Gastroenterology. 2011 Dec;141(6):2154–65. doi: 10.1053/j.gastro.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Xiao J, Zhou X, Xu M, Hu C, Xu X, et al. STK4 regulates TLR pathways and protects against chronic inflammation-related hepatocellular carcinoma. The Journal of clinical investigation. 2015 Nov 2;125(11):4239–54. doi: 10.1172/JCI81203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roh YS, Seki E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. Journal of gastroenterology and hepatology. 2013 Aug;28(Suppl 1):38–42. doi: 10.1111/jgh.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawada K, Ohtake T, Hasebe T, Abe M, Tanaka H, Ikuta K, et al. Augmented hepatic Toll-like receptors by fatty acids trigger the pro-inflammatory state of non-alcoholic fatty liver disease in mice. Hepatology research : the official journal of the Japan Society of Hepatology. 2014 Aug;44(8):920–34. doi: 10.1111/hepr.12199. [DOI] [PubMed] [Google Scholar]
- 46.Miura K, Ohnishi H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World journal of gastroenterology : WJG. 2014 Jun 21;20(23):7381–91. doi: 10.3748/wjg.v20.i23.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapil S, Duseja A, Sharma BK, Singla B, Chakraborti A, Das A, et al. Small intestinal bacterial overgrowth and toll-like receptor signaling in patients with non-alcoholic fatty liver disease. Journal of gastroenterology and hepatology. 2016 Jan;31(1):213–21. doi: 10.1111/jgh.13058. [DOI] [PubMed] [Google Scholar]
- 48.Gavino AC, Nahmod K, Bharadwaj U, Makedonas G, Tweardy DJ. STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy. 2016;71:1684–1692. doi: 10.1111/all.12937. [DOI] [PubMed] [Google Scholar]
- 49.Guo F, Xu Z, Zhang Y, Jiang P, Huang G, Chen S, et al. FXR induces SOCS3 and suppresses hepatocellular carcinoma. Oncotarget. 2015 Oct 27;6(33):34606–16. doi: 10.18632/oncotarget.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.