Abstract
It is becoming increasingly clear that genomics is beginning to have a major impact in guiding diagnoses and treatment of many disorders. As the cost of DNA sequencing continues to drop and more patient genomes are sequenced, the challenge is to deliver this knowledge to the clinic, particularly in cancer. DNA sequencing of patients with rare disease is revealing novel druggable pathways for more common disorders. Genetic tools for treating disease are also advancing rapidly. Genetic approaches, thought to be pipedreams five years ago for diseases such as Duchenne muscular dystrophy, are now showing promise in clinical trials and many of these methodologies are being applied more widely for other diseases. The era of genomic medicine has arrived.
Introduction
The pace of change in human genetics research over the last few decades has been rapid and major scientific discoveries have proceeded in tandem with breakthroughs in the development of enabling technologies, particularly genome sequencing. Together, these advances are transforming our understanding of how genes underpin biological processes in health and disease and have the potential to generate major health benefits in the coming decades. The challenge will be to use personal genomic information to provide care plans at every stage of disease. The concept of ‘personalized medicine’ is not new and was anticipated in the late 1800s by a Canadian physician, Sir William Osler, who gave the Harveian Oration in 1906 and noted ‘the great variability among individuals'. However, the modern definition has evolved to incorporate personal genomic information into a patient's clinical assessment and family history to guide medical management.1
The first draft of the human genome sequence was completed in year 2000 and the current whole-genome and exome sequencing projects are rapidly becoming tools in the diagnosis of rare mendelian disorders. Sequencing the human genome took a decade to complete and cost $3 billion. Since then, the price of sequencing has dropped rapidly to where it stands now at approximately $1,000 per genome and whole genomes can be sequenced in a day.
Even before the human genome sequence was complete, it was declared to be a landmark in medical research. Now there are many published articles describing how the genomics revolution is long overdue and has not lived up to its promise. However, clinical development does take a long time. Going from the germ theory of disease to antibiotics took 60 years. As I shall review in this oration, genomic medicine might beat that.
Genomic medicine for Duchenne muscular dystrophy
The pace of technological change and its impact on the treatment of genetic disorders through personalised medicine is dramatically illustrated by looking at progress in the treatment of Duchenne muscular dystrophy (DMD). Five years ago, I used to stand up in front of patient groups and tell parents that effective treatments might be on the horizon. Today, I am talking of treatments that have almost reached the clinic. There is now real hope for the sufferers of this terrible disease.
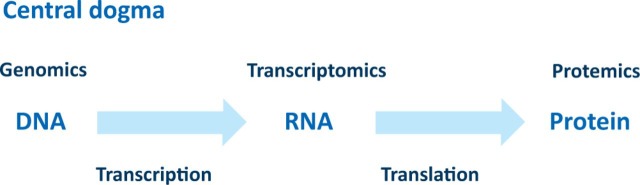
Therapies for DMD build on the central dogma in genetics which defines the flow of information from DNA to RNA to protein as proposed initially by Watson and elaborated by Crick (Fig 1).2 The DNA (the genome) contains the genetic information of the individual which is transcribed into RNA (the transcriptome). This is then processed via splicing into coding mRNA and then translated into protein (the proteome). Any pathology resulting from germ line or somatic mutations in the genome can be studied at the transcriptome or proteome level, thus providing the necessary mechanistic insights into the disease process which is the first step in the development of effective treatments. As William Harvey knew only too well, an understanding of the basic structure of the circulatory system and centrality of the heart led to a physiological understanding which was essential for the development of therapies. As I will illustrate, these same principles of studying the detailed mechanisms apply to the understanding and design of therapeutic strategies for DMD.
Fig 1.
The central dogma of genetics.
DMD is a relentless, X-linked recessive muscle-wasting disease with one of the highest new mutation rates known, meaning that many cases in the clinic have no previous family history.3 Affected DMD boys are usually diagnosed at 3–5 years of age and go into a wheelchair at about 12 years of age. All show abnormal ECGs by their late teens. Patients usually die in their twenties from respiratory or heart failure and a minority of patients may also show mental impairment. Becker had described a milder similar form of muscular dystrophy (Becker muscular dystrophy (BMD)) but it was unknown whether this disease was caused by mutations in the same gene. In BMD, loss of ambulation usually occurs in the late teens or early twenties but in some cases the only manifestation is a cardiomyopathy and patients can live into their 80th decade. Prenatal diagnosis of DMD did not exist before DNA markers were identified and no effective treatment is currently available.
DNA markers specific to the X chromosome established the location of the mutated gene and the first prenatal diagnosis and reliable carrier detection of DMD.4 Moreover, the milder form of the disease, BMD, was found to be allelic.5 The gene was finally identified using DNA from a patient with a large deletion covering the region who suffered from DMD, retinitis pigmentosa and chronic granulomatous disease.6 The protein product was named dystrophin as the lack of dystrophin causes dystrophy.
The DMD gene is remarkable in that it is spread over more than two million base pairs of DNA and is made up of 79 individual exons, which in part explains the high new mutation rate. Sixty five per cent of patient mutations are out-of-frame deletions.7 Detailed analysis of these deletions provided an explanation for the severe DMD and mild BMD phenotypes. Deletions in DMD patients are out of frame and result in the absence of protein product whereas BMD deletions result from in-frame deletions of the gene which lead to the production of truncated, partially functional dystrophin protein.8
Dystrophin is a large cytoskeleton structural protein which is absent or present at very low levels, in DMD patients. Dystrophin is essential for muscle-membrane stability as it provides an important link between the dystrophin-associated protein complex (DAPC) at the muscle membrane and the actin cytoskeleton.9
There are many pharmacological strategies which tackle the secondary effects of DMD but many are only partially effective because they only treat one aspect of the pathogenesis and may be toxic in the longer term.10 Thus there has been great interest in developing genetic approaches to the disease which tackle the primary defect. The challenge is that any effective therapy will need to replace about 20% of normal levels of dystrophin in both skeletal muscle and heart and the treatment would need to be life-long.
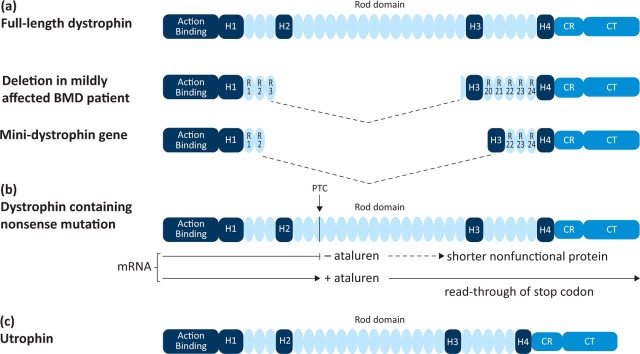
The pioneering approaches now entering the clinic for DMD are also applicable to many other diseases. The genetic strategies to DMD therapy include direct replacement of the protein and the manipulation of the cellular machinery at the level of mRNA processing, translation or transcriptional modulation of a related gene (Fig 2).
Fig 2.
Genetic strategies for Duchenne muscular dystrophy therapy. Reproduced with permission from Fairclough et al (2013).65
Viral gene therapy
The use of viruses for the delivery of functional genes has had a long history over at least two decades but is now coming into clinical use. Viral vectors are rendered harmless through the removal of genes that cause pathology and this generates enough space in the viral genome for the insertion of the coding region of the therapeutic gene. Early applications of gene therapy, however, have been hampered by adverse events in clinical trials due to a poor understanding of the integration of the virus into the host genome and the immune reactions to viral proteins.11 More recently, novel safer vector designs generally based on adenovirus or lentivirus vectors virus have been developed.12 In 2012, the long-awaited market approval of gene therapy in the Western world was granted for the viral delivery of the lipoprotein lipase gene.13
For DMD, the challenge is the large size of the mRNA (14 kb) and the need to target all muscles. The most commonly used virus to infect muscle is adeno-associated virus (AAV), which cannot accommodate more than about 5 kb of foreign DNA. This problem was solved by the remarkable observation that a very mildly affected BMD patient possessed a deletion encompassing almost 50% of the coding sequence of the gene.14 Other patients with similar large deletions were later reported. Thus dystrophin mini- or micro-genes (approximately 5 kb or less) could be designed based on the deleted genes in these patients which fit into AAV vectors even when muscle-specific promoters are incorporated (Fig 2a).15
Four out of six patients involved in a recent clinical trial using AAV delivery of a mini-dystrophin gene locally to the arm showed an immune response directed at the transgene product.16 However, this may have been due to the lack of muscle-specific expression as a trial delivering alpha-sarcoglycan transgene to limb girdle muscular dystrophy (LGMD) patients did not invoke an immune response.17 More needs to be understood about why certain patients have T cells to dystrophin epitopes and the choice of promoter will be important. Recent studies have shown that revertant fibres produced through restoring the reading frame by exon skipping or second site mutation, can express an immunogenic novel epitope never encountered previously by the patient.18 It will be important to pre-screen patients for immunity to dystrophin before admission into trials. Even though systemic delivery of dystrophin to all muscle remains a challenge because of the immune response, transient immunosuppression may minimise this problem.19
Expression of transgenes from AAV vectors can be long lasting. For example, transgene expression has been reported in a haemophilia B patient 10 years after gene transfer into muscle.20 However, if repeated administration is required, it may be necessary to use different AAV serotypes in successive treatments.
Gene therapy is now coming of age for a whole range of different disorders as different types of viruses are being developed some of which integrate into the host genome, such as lentiviruses. Perhaps the most spectacular recent application of gene therapy is for eye disorders. For example results of clinical trials for choroideremia are looking very promising. Gene therapy is rapidly moving into mainstream ophthalmology.21
Exon skipping
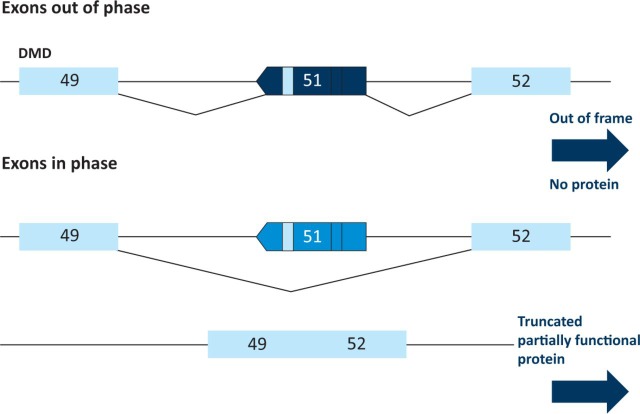
The application of exon skipping for the therapy of DMD depends on the identification of the precise mutation in the genome of the patient and the manipulation of the transcriptome. As discussed earlier, approximately 65% of DMD mutations are deletions which disrupt the open reading frame resulting in the absence of functional dystrophin protein at the sarcolemma.7 More mildly affected BMD patients have deletions which result in in-frame mutations that allow the production of shorter, partially functional dystrophin. Thus the correction of the reading frame in DMD patients should in many cases result in a BMD phenotype and some patients should be amenable to correction via the skipping of specific exons.22 Analysis of the deletions in DMD patients suggested that the skipping of exon 51 would restore the reading frame in the largest number of patients and thus this has been the focus of the initial studies and clinical trials (Fig 3).
Fig 3.
Skipping of exon 51 restores the reading frame in the largest number of Duchenne muscular dystrophy patients.
Antisense oligonucleotides (AONs) hybridise to complementary sequences around the target exon, modulate pre-mRNA splicing and promote the restoration of the reading frame. Proof-of-principle of this approach was established in phase-I/II clinical trials led by groups in the Netherlands (Leiden University Medical Centre / Prosensa-GlaxoSmithKline (GSK)) and the UK (MDEX Consortium / Sarepta Therapeutics). Different AON chemistries, namely 2′O methyl phosphorothioate (2′OMe; drisapersen) and morpholino phosphorodiamidate (PMO; eteplirsen) respectively, were used for exon 51 skipping in these trials. Both were well tolerated and restored dystrophin protein via either intramuscular23,24 or systemic routes,25,26 although only one muscle could be biopsied and there was no targeting to the heart. ProSensa/GSK is due to complete a phase-III trial of drisapersen at the end of this year. Drisapersen has been awarded ‘breakthrough therapy’ status in the USA, based on the results of a 53-patient phase-II clinical trial which were reported in April 2013. The accepted clinical endpoint for these trials was the Six Minute Walk Test (6MWT)27,28 – a standardised test of ambulation. The results showed that after 24 weeks, boys with DMD who were given drisapersen were able to walk 35 metres further in the 6MWT than those given a placebo. The results of a phase-IIb clinical trial of eteplirsen also show promise although only 12 boys were included in this initial trial. After DMD patients received the drug for 48 weeks, production of the dystrophin protein was restored in up to 50% of the muscle fibres that were examined. Boys who took eteplirsen for 48 weeks were able to walk, on average, 67.3 metres further in the 6MWT than those who took a placebo for 24 weeks followed by eteplirsen for 24 weeks.
Thus progress for exon skipping is very promising and other exons are now being targeted, although the efficacy of skipping is very dependent on the exon needing to be skipped.29 Only limited analysis is available so far and only one muscle has been analysed. Another important consideration is that the restored dystrophin protein is truncated and semi-functional, and therefore at best, the clinical outcome is the conversion to the corresponding BMD phenotype. The efficacy of exon-skipping AONs remains limited by poor cellular uptake and rapid clearance, resulting in relatively low dystrophin protein restoration in skeletal muscle and little or no restoration in heart. These problems may be solved by the development of a second generation of AONs with improved skeletal and cardiac muscle penetration.30
This mutation-specific requirement for DMD therapy has direct cost and regulatory implications given that each AON in development is currently regarded by the Food and Drug Administration (FDA) and other regulatory agencies as a new drug. Hopefully this will change as the success and safety of the current trials becomes evident.
The general approach of exon skipping is applicable to many other disorders, notably spinal muscular atrophy (SMA) and myotonic dystrophy.22 AONs are also being used to modulate pre-mRNA splicing in certain forms of cancer where disrupted alternative splicing commonly occurs, and where splicing reprogramming of, for example, signal transducer and activator of transcription 3 (STAT3) can lead to a favourable anti-oncogenic outcome.31
Termination codon read-through
Another approach to the therapy of DMD depends on the precise definition of the genomic mutation and the manipulation of the translational machinery in the cell. Strategies designed to promote ribosomal read-through of premature termination codon (PTC) mutations can lead to the expression of full-length protein (Fig 2b). Such stop codon mutations occur in approximately 15% of DMD patients. Aminoglycosides restore protein translation and these have shown some efficacy but their clinical use is limited because of long-term toxic effects.27,32 PTC Therapeutics used a high throughput screen to identify drugs which promote read-through of stop codons both for DMD and cystic fibrosis. The first drug in this class, ataluren, showed convincing proof-of-concept in the mdx mouse model of the disease but initial human trials have been disappointing, with low levels of dystrophin expression and variability between patients.28,33 Results from the 48-week phase-IIb study showed that treated patients walked on average 30 metres further than patients on placebo in the 6MWT. The use of ataluren for DMD has thus demonstrated proof-of-principle and other compounds which may show greater efficacy in read-through of stop codons have been reported.34
Ten per cent of inherited diseases are caused by premature termination codon mutations and thus this mutation precision approach has much wider applicability beyond DMD.32 Stop codon read-through is being applied to other genetic disorders such as cystic fibrosis and SMA. This approach is now also being used for some hereditary cancer syndromes. For example, it is potentially applicable to 24% of cases for the treatment of adenomatous polyposis coli-associated colorectal cancers.35
Increasing levels of utrophin
Another strategy for the therapy of DMD comes from the analysis of the genome coupled with the study of the transcriptional control and localisation of dystrophin during early human development. The dystrophin-related protein, utrophin, shows sequence and structural similarity to dystrophin and can functionally compensate for the lack of dystrophin in the mdx mouse model of the disease (Fig 2c).10,36,37 The utrophin gene is located on chromosome 6 and genomic sequence analysis shows it to be related to the dystrophin gene by a genomic duplication event. The major differences in the two genes lie in their promoter regions. In early fetal life, utrophin is found at the sarcolemma with dystrophin, but in adult muscle it is restricted to the neuromuscular and myotendinous junction. This localisation is defined by DNA sequences in the promoter region of utrophin not present in the equivalent genomic region of dystrophin. In the absence of dystrophin, small amounts of utrophin can be found at the sarcolemma due to its stabilisation by the dystrophin-associated complex. Encouragingly, the amount of naturally occurring utrophin protein present in DMD patients positively correlates with the age of confinement to a wheelchair, demonstrating that even small increases might be beneficial.38
Utrophin levels can be increased by direct delivery of the protein,39 stabilisation of the protein40 or RNA,41,42 or by transcriptional modulation.43 A high through-put screen based on the promoter region of utrophin has led to the development of a drug which increases utrophin levels in the mdx mice twofold. Summit plc have reported positive data in a phase-I trial for SMT C1100.43 Phase-II trials are planned and other drugs are being developed based on the same principle.
Increasing utrophin should be effective in all DMD patients irrespective of their dystrophin mutation and thus avoids the need for genome sequence information on each patient. This approach also circumvents the immunological challenges faced by dystrophin-based therapies.18
The change of expression of a related or similar protein was first used for the treatment of beta thalassaemia where increased levels of fetal haemoglobin were used to compensate for the lack of the adult beta globin gene.44 Such an approach could be applicable to many other disorders as, even if there is not a protein similar to the defective one, it may be possible to modulate a compensating pathway.
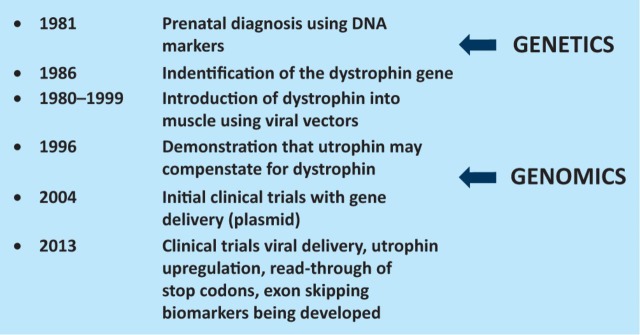
In summary, the delivery of therapy for DMD relies on the identification of the precise mutation in the patient and the understanding of the genome, transcriptome and proteome. The gene was identified in 1986 and only now are therapies entering the clinic, but the rate of progress is increasing (Fig 4).
Fig 4.
Milestones in DMD research. DMD = Duchenne muscular dystrophy.
More generally, genomic technologies continue to be developed which shorten the timeline between understanding the disease mechanism and the delivery of an effective treatment. This is very elegantly demonstrated in the case of cancer where delivery of personalised medicine has been revolutionary for clinical trials and treatment.
Genomic medicine for cancer
Much was already known about mechanism of cancer when Sir Walter Bodmer delivered his Harveian Oration45 where he described the various genes which were known to be involved in the somatic evolution of cancer. Since then, sequencing either of whole genomes (DNA) or transcriptomes (RNA) has increased our understanding of the disease and is becoming a valuable tool in diagnosis, risk stratification and the development of individualised treatment of patients.
BRCA1 and BRCA2 are the best studied genes and are found associated with a high risk of breast cancer, although the function of these genes is not fully understood.46 Routine screening for mutations in these genes is now commonplace for those most at risk because of family history. Managing the risk remains difficult but treatment is improving as is survival.
Genomic information is allowing the division of cancers into subgroups, which is already having an impact on patient management. Knowledge of the expression profiles of particular tumours is used to determine which treatment is likely to be most effective. This stratification of patient tumour groups in turn results in more informative clinic trials. Breast cancer has seen the greatest progress in targeting treatment to specific molecular abnormalities, particularly in the 20% of women whose tumours are driven by overexpression of human epidermal growth factor receptor 2 (HER2).47 Indeed, so much is now known about this pathway that even when drug resistance appears, there are opportunities to block other pathways at different levels. However, even 15 years after approval of the ground-breaking HER2 monoclonal antibody trastuzumab, clinicians still grapple with the problem of heterogeneity within subgroups of patients.
We are also realising that cancer is more complex the more we learn about it. Sequencing studies have revealed that tumours usually have thousands to tens of thousands of somatic mutations and yet only 1–10 of these in each cancer is recurrent and likely to be involved in ‘driving’ the cancer. Most of the rest are likely to be ‘passengers', although defining the passenger/driver status of a mutation remains difficult. The sequencing of a small-cell lung-cancer cell line revealed that the mutational signature is typical of that expected from tobacco smoking including defects in transcription-coupled repair pathways.48 This reveals the potential for next-generation sequencing of tumours to reveal insights into the mutational processes associated with cancer which might inform prevention, early diagnosis and eventually treatment.49
There are now many examples of where targeted therapies for different cancers have been developed. The collation of data through international cancer consortia is being very effective in bringing together the genomic information on various tumours so that the latest information on pathways can be used by the oncologist to inform patient management.50
One important aspect for the development of drugs from the understanding of the biology is that the development times can be drastically reduced. A good example of this is the recognition in 2007 that approximately 5% of non-small-cell lung cancer (NSCLC) is caused by the rearrangement of the anaplastic lymphoma kinase (ALK). Crizotinib, a mesenchymal-epithelial transition/ALK multi-targeted receptor tyrosine kinase inhibitor went into phase-I trial the same year and was approved in 2011.51 Thus a drug for NSCLC was developed in an unprecedented four years after the discovery of the causal rearrangement because the treatment could be targeted to the patient subgroup. More recently, the characterisation of ALK mutations is guiding the development of second-generation inhibitors.
Another example is the rapid development of selective BRAF (serine/threonine-protein kinase B-Raf) inhibitors which are now established as a standard of care option for patients diagnosed with metastatic melanoma whose tumours carry a BRAF mutation.52 Since its discovery 10 years ago, BRAF mutations have been reported in a number of malignancies, including melanoma, colorectal cancer, papillary thyroid cancer, and most recently hairy-cell leukaemia; in all accounting for 7–8% of all cancers. Knowledge of the pathway again is leading to strategies to deal with those tumours that become resistant to treatment. This is a great success story for cancer genomic medicine and more will follow.
Rare variants and their relevance to more common diseases
Whole-genome sequencing of patients is now leading to the identification of many novel causative genes for a variety of disorders, which is useful for diagnosis and may in some cases lead to novel treatment. A particularly interesting example of how rare variants can provide insights into new treatments for disease is illustrated by neonatal diabetes (ND). This is a rare but potentially devastating monogenic form of diabetes that usually develops within the first six months of life. ND sufferers were previously assumed to have type 1 diabetes and thus were treated with insulin injections. This is unpleasant (especially for babies), and impacts significantly on patients' lifestyles at all ages. Mutations were found in the ATP-sensitive K+ channel, Kir6.2, and functional studies showed that the mutations impair the ability of adenosine triphosphate to shut the channel.53 Because channel closure is required for insulin secretion, this leads to impaired insulin release and thus to ND. Most of the mutant channels can be closed by sulphonylurea drugs, which were already in routine clinical use for type 2 diabetes. Clinical studies showed that oral sulphonylureas can be successfully used to treat these ND patients.
There are now several examples of where the identification of the gene responsible for a rare mendelian disorder is leading to new opportunities for the treatment of more common disorders. For example, the identification of mutations in sclerostin in three families suffering from a progressive sclerosing bone dysplasia identified this gene as being key in the suppression of bone formation. This observation suggested that inhibiting this pathway may therefore be more broadly applicable to osteoporosis, and indeed antibodies to sclerostin are currently moving into phase-III clinical trials for this condition.54,55 A form of hypercholesterolemia was found to result from a mutation in the gene for an enzyme called pro-protein convertase subtilisn Kexin type 9 (PCSK9).56 Loss-of-function mutations lead to degradation of hepatic low-density lipoprotein (LDL) receptors and a reduction in LDL concentrations. Monoclonal antibodies raised against PCSK9 are currently in phase-III clinical trials for coronary heart disease.
Biomarkers to ensure precision care
As illustrated above, the development of an effective treatment for DMD relied on an assessment of the ability to walk which is difficult to measure because of variability, meaning that lengthy clinical trials have to be undertaken. This is the challenge in many disorders and the development of disease-specific biomarkers is becoming increasingly important for the design of clinical trials. The methodology here can depend on profiling gene expression (transcriptome analysis) in the blood sample of patients and those at risk of developing the disease. A recent paper documented a signature for Alzheimer's disease based on the profile of expression of certain miRNAs in the blood of patients, which differentiated it from other neurodegenerative disorders and also identified those at the early stages of the disease.57
Measurement of expression patterns can be performed at the RNA, protein and metabolite level (transcriptomics, proteomics and metabolomics respectively). Precision diagnosis of the mutation in a patient coupled with reliable biomarkers for the assessment of the efficacy of the personalised treatment will be essential in future clinical trials. The development of these approaches is such that individual biotechnology companies are now routinely offering such services.
The epigenome
Garrod was one of first individuals to recognise the importance of environmental factors in his essay, The inborn factors in disease.58 It is now well established that the genome is constantly being modified by the environmental influences such as ageing, diet, stress and toxins which manifest themselves as changes in DNA methylation or chromatin modifications.59 Some cancers can be caused by methylation changes alone. The importance of aberrant methylation not only in tumorigenesis but also in other disorders is now recognised and the technology to monitor these changes is in place.60 Such studies will provide the vital link needed between the genome and environmental influences, which in due course should lead to strategies for prevention. However, as David Weatherall noted in his Harveian Oration in 1992:
…with some notable exceptions, we are uncertain about the extent to which environment is responsible for cancer, and even if it turns out that it is, how far we can modify it. For many other diseases of western society, diabetes, arthritis, the major psychoses, dementia and the rest, the relative role of nature and nurture are even less clear.61
Thus we will need an understanding of disease mechanisms through genomics as well as minimisation of risk through manipulation of the environment, where possible, for future medical care.
Conclusions
There are many examples today of where genomics is leading to more effective treatments. Five years ago, therapy for DMD seemed an impossible dream and few pharmaceutical companies were interested in a relatively rare orphan disease. However, that has all changed as DMD is now seen as a model for the development of treatment based on an individual's mutation (personalised/precision medicine) and advances in technology have made many of the problems more tractable. The combination of better biomarkers, more defined phenotyping and genomic/transcriptomic/proteomic analyses should eventually allow real progress in the therapy for currently intractable neurological disorders, such as Alzheimer's disease.
Personalised medicine will not only lead to novel therapies, but will also allow more rapid development because of better designed clinical trials. We are entering an era where individuals are able to monitor their health status on their mobile phones and integrate these data with blood-sample testing of biomarkers. This information can be coupled with genome information which defines individual disease risk, as emphasised by John Bell in his Harveian Oration in 2010.62 This will transform preventative medicine. There will be further benefits if the human genomic information can be integrated with monitoring of physiological states through genomic analysis of the bacteria of the gut and viral infections.63 There are significant challenges in the protection of patient privacy, the education of tomorrow's doctors, and in changing regulation as indicated in the House of Lords Select Committee report on genomic medicine.64 Nevertheless, the genomic era has never looked brighter.
William Harvey really started modern physiology and the concepts of form and function which lead to modern medicine. The continued excitement which is moving from form and function of genomes, transcriptomes and proteomes and their relation to disease is perhaps a restatement of Harvey.
Acknowledgements
I would like to thank Alastair Buchan, Donald Chambers, Mike Stratton, Matthew Wood and John Mattick for discussions on genomic medicine. I am grateful for the DMD patients and their families who have kept me focused on the development of genomic medicine for DMD and for the many colleagues who have helped us move to clinical trials. I am also indebted to the Medical Research Council, the Muscular Dystrophy Campaign (UK) and the Muscular Dystrophy Association (USA) for realising that from bench to bedside is a long game.
References
- 1.Chan IS, Ginsburg GS. Personalized medicine: progress and promise. Annu Rev Genomics Hum Genet. 2011;12:217–44. doi: 10.1146/annurev-genom-082410-101446. doi: 10.1146/annurev-genom-082410-101446. PubMed PMID: 21721939. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD. The annotated and illustrated double helix. New York: Simon & Schuster; 2012. p. 163. [Google Scholar]
- 3.Emery AEH. Muscular dystrophy (the facts) Third edition. Oxford: Oxford University Press; 2008. [Google Scholar]
- 4.Davies KE, Pearson PL, Harper PS, et al. Linkage analysis of two cloned DNA sequences flanking the Duchenne muscular dystrophy locus on the short arm of the human X chromosome. Nucleic Acids Res. 1983;11(8):2303–12. doi: 10.1093/nar/11.8.2303. doi: 10.1093/nar/11.8.2303. PubMed PMID: 6304647. Pubmed Central PMCID: 325885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kingston HM, Sarfarazi M, Thomas NS, Harper PS. Localisation of the Becker muscular dystrophy gene on the short arm of the X chromosome by linkage to cloned DNA sequences. Hum Genet. 1984;67(1):6–17. doi: 10.1007/BF00270551. doi: 10.1007/BF00270551. PubMed PMID: 6086495. [DOI] [PubMed] [Google Scholar]
- 6.Monaco AP, Neve RL, Colletti-Feener C, et al. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323(6089):646–50. doi: 10.1038/323646a0. doi: 10.1038/323646a0. PubMed PMID: 3773991. [DOI] [PubMed] [Google Scholar]
- 7.Aartsma-Rus A, Fokkema I, Verschuuren J, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30(3):293–9. doi: 10.1002/humu.20918. doi: 10.1002/humu.20918. PubMed PMID: 19156838. Epub 2009/01/22. eng. [DOI] [PubMed] [Google Scholar]
- 8.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2(1):90–5. doi: 10.1016/0888-7543(88)90113-9. doi: 10.1016/0888-7543(88)90113-9. PubMed PMID: 3384440. [DOI] [PubMed] [Google Scholar]
- 9.Cohn RD, Campbell KP. Molecular basis of muscular dystrophies. Muscle Nerve. 2000;23(10):1456–71. doi: 10.1002/1097-4598(200010)23:10<1456::aid-mus2>3.0.co;2-t. PubMed PMID: 11003781. Epub 2000/09/26. eng. [DOI] [PubMed] [Google Scholar]
- 10.Fairclough RJ, Perkins KJ, Davies KE. Pharmacologically targeting the primary defect and downstream pathology in Duchenne muscular dystrophy. Curr Gene Ther. 2012;12(3):206–44. doi: 10.2174/156652312800840595. PubMed PMID: 22571500. Epub 2012/05/11.eng. [DOI] [PubMed] [Google Scholar]
- 11.Cavazzana-Calvo M, Fischer A. Gene therapy for severe combined immunodeficiency: are we there yet? J Clin Invest. 2007;117(6):1456–65. doi: 10.1172/JCI30953. doi: 10.1172/JCI30953. PubMed PMID: 17549248. Pubmed Central PMCID: 1878528. Epub 2007/06/06.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson JM. Bulls, bubbles, and biotech. Hum Gene Ther. 2013;24(8):715–6. doi: 10.1089/hum.2013.2509. doi: 10.1089/hum.2013.2509. PubMed PMID: 23944718. [DOI] [PubMed] [Google Scholar]
- 13.Kastelein JJ, Ross CJ, Hayden MR. From mutation identification to therapy: discovery and origins of the first approved gene therapy in the Western world. Hum Gene Ther. 2013;24(5):472–8. doi: 10.1089/hum.2013.063. doi: 10.1089/hum.2013.063. PubMed PMID: 23578007. [DOI] [PubMed] [Google Scholar]
- 14.England SB, Nicholson LV, Johnson MA, et al. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343(6254):180–2. doi: 10.1038/343180a0. PubMed PMID: 2404210. [DOI] [PubMed] [Google Scholar]
- 15.Pichavant C, Aartsma-Rus A, Clemens PR, et al. Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol Ther (the journal of the American Society of Gene Therapy) 2011;19(5):830–40. doi: 10.1038/mt.2011.59. doi: 10.1038/mt.2011.59. PubMed PMID: 21468001. Pubmed Central PMCID: 3098643. Epub 2011/04/07.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendell JR, Rodino-Klapac L, Sahenk Z, et al. Gene therapy for muscular dystrophy: lessons learned and path forward. Neurosci Lett. 2012;527(2):90–9. doi: 10.1016/j.neulet.2012.04.078. doi: 10.1016/j.neulet.2012.04.078. PubMed PMID: 22609847. Pubmed Central PMCID: 3492936. Epub 2012/05/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell JR, Rodino-Klapac LR, Rosales XQ, et al. Sustained alpha-sarcoglycan gene expression after gene transfer in limb-girdle muscular dystrophy, type 2D. Ann Neurol. 2010;68(5):629–38. doi: 10.1002/ana.22251. doi: 10.1002/ana.22251. PubMed PMID: 21031578. Pubmed Central PMCID: 2970162. Epub 2010/10/30. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendell JR, Campbell K, Rodino-Klapac L, et al. Dystrophin immunity in Duchenne's muscular dystrophy. N Engl J Med. 2010;363(15):1429–37. doi: 10.1056/NEJMoa1000228. PubMed PMID: 20925545. Pubmed Central PMCID: 3014106. Epub 2010/10/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Storb R, Halbert CL, et al. Successful regional delivery and long-term expression of a dystrophin gene in canine muscular dystrophy: a preclinical model for human therapies. Mol Ther (the journal of the American Society of Gene Therapy) 2012;20(8):1501–7. doi: 10.1038/mt.2012.111. doi: 10.1038/mt.2012.111. PubMed PMID: 22692496. Pubmed Central PMCID: 3412492. Epub 2012/06/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchlis G, Podsakoff GM, Radu A, et al. Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood. 2012;119(13):3038–41. doi: 10.1182/blood-2011-09-382317. doi: 10.1182/blood-2011-09-382317. PubMed PMID: 22271447. Pubmed Central PMCID: 3321866. Epub 2012/01/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipinski DM, Thake M, MacLaren RE. Clinical applications of retinal gene therapy. Prog Retin Eye Res. 2013;32:22–47. doi: 10.1016/j.preteyeres.2012.09.001. doi: 10.1016/j.preteyeres.2012.09.001. PubMed PMID: 22995954. [DOI] [PubMed] [Google Scholar]
- 22.Muntoni F, Wood MJ. Targeting RNA to treat neuromuscular disease. Nat Rev Drug Discov. 2011;10(8):621–37. doi: 10.1038/nrd3459. doi: 10.1038/nrd3459. PubMed PMID: 21804598. Epub 2011/08/02. eng. [DOI] [PubMed] [Google Scholar]
- 23.Kinali M, Arechavala-Gomeza V, Feng L, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8(10):918–28. doi: 10.1016/S1474-4422(09)70211-X. doi: 10.1016/S1474-4422(09)70211-X. PubMed PMID: 19713152. Pubmed Central PMCID: 2755039. Epub 2009/08/29. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357(26):2677–86. doi: 10.1056/NEJMoa073108. doi: 10.1056/NEJMoa073108. PubMed PMID: 18160687. [DOI] [PubMed] [Google Scholar]
- 25.Cirak S, Arechavala-Gomeza V, Guglieri M, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378(9791):595–605. doi: 10.1016/S0140-6736(11)60756-3. doi: 10.1016/S0140-6736(11)60756-3. PubMed PMID: 21784508. Pubmed Central PMCID: 3156980. Epub 2011/07/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goemans NM, Tulinius M, van den Akker JT, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011;364(16):1513–22. doi: 10.1056/NEJMoa1011367. PubMed PMID: 21428760. Epub 2011/03/25. eng. [DOI] [PubMed] [Google Scholar]
- 27.Finkel RS. Read-through strategies for suppression of nonsense mutations in Duchenne/Becker muscular dystrophy: aminoglycosides and ataluren (PTC124) J Child Neurol. 2010;25(9):1158–64. doi: 10.1177/0883073810371129. doi: 10.1177/0883073810371129. PubMed PMID: 20519671. Epub 2010/06/04. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirawat S, Welch EM, Elfring GL, et al. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J Clin Pharmacol. 2007;47(4):430–44. doi: 10.1177/0091270006297140. doi: 10.1177/0091270006297140. PubMed PMID: 17389552. Epub 2007/03/29. eng. [DOI] [PubMed] [Google Scholar]
- 29.Mitrpant C, Adams AM, Meloni PL, et al. Rational design of antisense oligomers to induce dystrophin exon skipping. Mol Ther (the journal of the American Society of Gene Therapy) 2009;17(8):1418–26. doi: 10.1038/mt.2009.49. doi: 10.1038/mt.2009.49. PubMed PMID: 19293776. Pubmed Central PMCID: 2835229. Epub 2009/03/19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betts CA, Hammond SM, Yin HF, Wood MJ. Optimizing tissue-specific antisense oligonucleotide-peptide conjugates. Methods Mol Biol. 2012;867:415–35. doi: 10.1007/978-1-61779-767-5_27. doi: 10.1007/978-1-61779-767-5_27. PubMed PMID: 22454077. Epub 2012/03/29. eng. [DOI] [PubMed] [Google Scholar]
- 31.Zammarchi F, de Stanchina E, Bournazou E, et al. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci USA. 2011;108(43):17779–84. doi: 10.1073/pnas.1108482108. doi: 10.1073/pnas.1108482108. PubMed PMID: 22006329. Pubmed Central PMCID: 3203802. Epub 2011/10/19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bidou L, Allamand V, Rousset JP, Namy O. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol Med. 2012;18(11):679–88. doi: 10.1016/j.molmed.2012.09.008. doi: 10.1016/j.molmed.2012.09.008. PubMed PMID: 23083810. [DOI] [PubMed] [Google Scholar]
- 33.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447(7140):87–91. doi: 10.1038/nature05756. doi: 10.1038/nature05756. PubMed PMID: 17450125. Epub 2007/04/24. eng. [DOI] [PubMed] [Google Scholar]
- 34.Kayali R, Ku JM, Khitrov G, et al. Read-through compound 13 restores dystrophin expression and improves muscle function in the mdx mouse model for Duchenne muscular dystrophy. Hum Mol Genet. 2012;21(18):4007–20. doi: 10.1093/hmg/dds223. doi: 10.1093/hmg/dds223. PubMed PMID: 22692682. Epub 2012/06/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bordeira-Carrico R, Pego AP, Santos M, Oliveira C. Cancer syndromes and therapy by stop-codon readthrough. Trends Mol Med. 2012;18(11):667–78. doi: 10.1016/j.molmed.2012.09.004. doi: 10.1016/j.molmed.2012.09.004. PubMed PMID: 23044248. [DOI] [PubMed] [Google Scholar]
- 36.Miura P, Jasmin BJ. Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: how close are we? Trends Mol Med. 2006;12(3):122–9. doi: 10.1016/j.molmed.2006.01.002. doi: 10.1016/j.molmed.2006.01.002. PubMed PMID: 16443393. Epub 2006/01/31. eng. [DOI] [PubMed] [Google Scholar]
- 37.Khurana TS, Davies KE. Pharmacological strategies for muscular dystrophy. Nat Rev Drug Discov. 2003;2(5):379–90. doi: 10.1038/nrd1085. doi: 10.1038/nrd1085. PubMed PMID: 12750741. [DOI] [PubMed] [Google Scholar]
- 38.Kleopa KA, Drousiotou A, Mavrikiou E, Ormiston A, Kyriakides T. Naturally occurring utrophin correlates with disease severity in Duchenne muscular dystrophy. Hum Mol Genet. 2006;15(10):1623–8. doi: 10.1093/hmg/ddl083. doi: 10.1093/hmg/ddl083. PubMed PMID: 16595608. Epub 2006/04/06. eng. [DOI] [PubMed] [Google Scholar]
- 39.Sonnemann KJ, Heun-Johnson H, Turner AJ, et al. Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med. 2009;6(5):e1000083. doi: 10.1371/journal.pmed.1000083. doi: 10.1371/journal.pmed.1000083. PubMed PMID: 19478831. Pubmed Central PMCID: 2680620. Epub 2009/05/30. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amenta AR, Yilmaz A, Bogdanovich S, et al. Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice. Proc Natl Acad Sci USA. 2011;108(2):762–7. doi: 10.1073/pnas.1013067108. doi: 10.1073/pnas.1013067108. PubMed PMID: 21187385. Epub 2010/12/29. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moorwood C, Soni N, Patel G, Wilton SD, Khurana TS. A Cell-Based High-Throughput Screening Assay for Posttranscriptional Utrophin Upregulation. J Biomol Screen. 2013;18(4):400–6. doi: 10.1177/1087057112465648. doi: 10.1177/1087057112465648. PubMed PMID: 23112083. Epub 2012/11/01. eng. [DOI] [PubMed] [Google Scholar]
- 42.Chakkalakal JV, Miura P, Belanger G, Michel RN, Jasmin BJ. Modulation of utrophin A mRNA stability in fast versus slow muscles via an AU-rich element and calcineurin signaling. Nucleic Acids Res. 2008;36(3):826–38. doi: 10.1093/nar/gkm1107. doi: 10.1093/nar/gkm1107. PubMed PMID: 18084024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tinsley JM, Fairclough RJ, Storer R, et al. Daily treatment with SMTC1100, a novel small molecule utrophin upregulator, dramatically reduces the dystrophic symptoms in the mdx mouse. PloS One. 2011;(6)(5):e19189. doi: 10.1371/journal.pone.0019189. doi: 10.1371/journal.pone.0019189. PubMed PMID: 21573153. Pubmed Central PMCID: 3089598. Epub 2011/05/17. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gambari R, Fibach E. Medicinal chemistry of fetal hemoglobin inducers for treatment of beta-thalassemia. Curr Med Chem. 2007;14(2):199–212. doi: 10.2174/092986707779313318. PubMed PMID: 17266579. Epub 2007/02/03. eng. [DOI] [PubMed] [Google Scholar]
- 45.Bodmer W. Harveain Oration. London: Royal College of Physicians; 1996. The somatic evolution of cancer. [PMC free article] [PubMed] [Google Scholar]
- 46.King MC, Marks JH, Mandell JB, New York Breast Cancer Study G Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–6. doi: 10.1126/science.1088759. doi: 10.1126/science.1088759. PubMed PMID: 14576434. [DOI] [PubMed] [Google Scholar]
- 47.Piccart M. Personalised cancer management: closer, but not here yet. Ann Oncol – official journal of the European Society for Medical Oncology (ESMO) 2013;24(8):1951–5. doi: 10.1093/annonc/mdt260. doi: 10.1093/annonc/mdt260. PubMed PMID: 23878113. [DOI] [PubMed] [Google Scholar]
- 48.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–90. doi: 10.1038/nature08629. doi: 10.1038/nature08629. PubMed PMID: 20016488. Pubmed Central PMCID: 2880489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. doi: 10.1038/nature12477. PubMed PMID: 23945592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. doi: 10.1038/nature11252. PubMed PMID: 22810696. Pubmed Central PMCID: 3401966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ou SH. Crizotinib: a novel and first-in-class multitargeted tyrosine kinase inhibitor for the treatment of anaplastic lymphoma kinase rearranged non-small cell lung cancer and beyond. Drug Des Devel Ther. 2011;5:471–85. doi: 10.2147/DDDT.S19045. PubMed PMID: 22162641. Pubmed Central PMCID: 3232174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salama AK, Flaherty KT. BRAF in Melanoma: Current strategies and future directions. Clin Cancer Res (an official journal of the American Association for Cancer Research) 2013;19(16):4326–34. doi: 10.1158/1078-0432.CCR-13-0779. doi: 10.1158/1078-0432.CCR-13-0779. PubMed PMID: 23770823. [DOI] [PubMed] [Google Scholar]
- 53.Hattersley AT, Ashcroft FM. Activating mutations in Kir6.2 and neonatal diabetes: new clinical syndromes, new scientific insights, and new therapy. Diabetes. 2005;54(9):2503–13. doi: 10.2337/diabetes.54.9.2503. doi: 10.2337/diabetes.54.9.2503. PubMed PMID: 16123337. [DOI] [PubMed] [Google Scholar]
- 54.Papapoulos SE. Targeting sclerostin as potential treatment of osteoporosis. Ann Rheum Dis. 2011;70(Suppl 1):i119–22. doi: 10.1136/ard.2010.141150. doi: 10.1136/ard.2010.141150. PubMed PMID: 21339215. [DOI] [PubMed] [Google Scholar]
- 55.Lewiecki EM. Monoclonal antibodies for the treatment of osteoporosis. Expert Opin Biol Ther. 2013;13(2):183–96. doi: 10.1517/14712598.2012.740006. doi: 10.1517/14712598.2012.740006. PubMed PMID: 23253281. [DOI] [PubMed] [Google Scholar]
- 56.Lambert G, Sjouke B, Choque B, Kastelein JJ, Hovingh GK. The PCSK9 decade. J Lipid Res. 2012;53(12):2515–24. doi: 10.1194/jlr.R026658. doi: 10.1194/jlr.R026658. PubMed PMID: 22811413. Pubmed Central PMCID: 3494258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leidinger P, Backes C, Deutscher S, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14(7):R78. doi: 10.1186/gb-2013-14-7-r78. PubMed PMID: 23895045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garrod AE. The inborn factors in disease: an essay. Oxford Clarendon Press; 1931. [Google Scholar]
- 59.Bell CG, Beck S. The epigenomic interface between genome and environment in common complex diseases. Brief Funct Genomics. 2010;9(5–6):477–85. doi: 10.1093/bfgp/elq026. doi: 10.1093/bfgp/elq026. PubMed PMID: 21062751. Pubmed Central PMCID: 3080746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brena RM, Costello JF. Genome-epigenome interactions in cancer. Hum Mol Genet. 2007;16(Spec No 1):R96–105. doi: 10.1093/hmg/ddm073. doi: 10.1093/hmg/ddm073. PubMed PMID: 17613554. [DOI] [PubMed] [Google Scholar]
- 61.Weatherall DJ. Harveain Oration. London: Royal College of Physicians; 1992. The role of nature and nurture in common diseases: Garrod's legacy. [Google Scholar]
- 62.Bell JI. Harverian Oration. London: Royal College of Physicians; 2010. Redefining disease. [Google Scholar]
- 63.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–307. doi: 10.1016/j.cell.2012.02.009. doi: 10.1016/j.cell.2012.02.009. PubMed PMID: 22424236. Pubmed Central PMCID: 3341616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Committee HoLSaT. London: the Stationery Office; 2009. Genomic medicine (2nd report of session 2008–09) [Google Scholar]
- 65.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14:373–8. doi: 10.1038/nrg3460. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]