Abstract
The evolutionary relationships of gnathostomes (jawed vertebrates), which comprise chondrichthyans (cartilaginous fishes), lobe-finned fishes (coelacanths and lungfishes), tetrapods, and actinopterygians (ray-finned fishes), have been debated for almost a century. Phylogenetic analyses based on fossils, morphology, and molecular sequences have generated different models of relationships that remain unresolved. We identified 13 derived shared molecular markers (synapomorphies) that define clades in the vertebrate lineage and used them to resolve the phylogenetic relationships of extant jawed vertebrates. Our markers include the presence or absence of insertions and deletions in coding sequences, nuclear introns, and alternatively spliced transcripts. The synapomorphies identified by us are congruent with each other and give rise to a single phylogenetic tree. This tree confirms that chondrichthyans are basal to all living gnathostomes, that lungfishes (Dipnoi) are the closest living relatives of tetrapods, and that bichirs (Cladistia) are the living members of the most ancient family of ray-finned fishes. Our study also provides molecular evidence to support the monophyly of living tetrapods and teleosts.
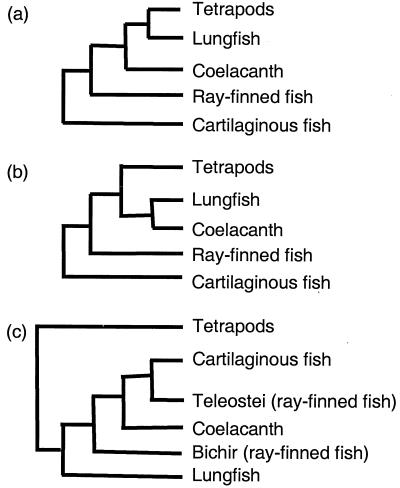
The jawed vertebrates (gnathostomes) fall into two major taxa, the Chondrichthyes (cartilaginous fishes) and Osteichthyes (bony fishes). The latter taxon includes sarcopterygians (coelacanths, lungfishes, and tetrapods) and actinopterygians (ray-finned fishes). The evolutionary relationships of gnathostomes have been the subject of study for almost a century. Fossils, morphological characters, and molecular sequences have been used to infer their phylogenetic relationships. However, these studies have proposed different models of relationships (Fig. 1) that have not been resolved with confidence (1–6). Phylogenetic analyses of mitochondrial and some nuclear gene sequences have generated conflicting models, indicating that molecular sequences may not be adequate for resolving the deep branches of gnathostomes (3, 4, 6). Thus there is a need to explore novel markers that can reliably resolve the deep branches of gnathostomes.
Figure 1.
Alternative models of evolutionary relationships of gnathostomes proposed by earlier studies (4–5). Model a proposes a lungfish + tetrapod clade, whereas model b supports a lungfish + coelacanth clade (4). Model c places cartilaginous fishes as the sister group of teleost fishes and the lungfish at the base of all fishes (5). This study supports model a.
The presence or absence of insertions or deletions within well-conserved coding sequences has been shown to be a useful molecular marker for defining clades in evolution (7, 8). We searched for such markers in vertebrates by comparing sequences of orthologous genes from distantly related vertebrates such as humans and fugu and identified several insertions and deletions in the coding sequences of the recombinase-activating proteins 1 and 2 (RAG1 and RAG2) and proopiomelanocortin (POMC) genes. We validated them as phylogenetic markers by cloning from representatives of major taxa of jawed vertebrates. We also cloned the gene for DM20, which shows alternative splicing in different phylogenetic groups of vertebrates. DM20 produces a single transcript in cartilaginous fishes, lobe-finned fishes, zebrafish, and trout, whereas in mammals and birds a second transcript (proteolipid protein, termed PLP) is produced with the use of an alternative splice site 105 bp downstream of the 5′ splice site of intron three. In frogs only PLP is produced (9–12).
We have shown that the presence or absence of nuclear introns serves as a marker to define clades in the ray-finned fish lineage (13). We have now extended our study by cloning RAG1 and rhodopsin genes from other major taxa of jawed vertebrates and used the data, together with the data on the presence or absence of indels in the coding sequences of the RAG1, RAG2, and POMC genes, and the alternative transcripts of DM20, to resolve the phylogenetic relationships of extant gnathostomes.
Materials and Methods
DNA and cDNA Preparation.
The genomic DNA of coelacanth was extracted from gill tissue that was preserved in ethanol by standard protocols. Genomic DNA from other vertebrates was extracted from fresh or frozen tissues. Total RNA was extracted from fresh tissues with the use of TRIzol reagent (GIBCO/BRL). 5′ Rapid amplification of cDNA ends (5′-RACE)-Ready and 3′-RACE-Ready cDNAs were synthesized from total RNA with a SMART RACE cDNA Amplification Kit (CLONTECH) according to the manufacturer's protocols.
PCR Amplification.
Gene fragments were amplified by PCR from cDNA or genomic DNA. The genomic structure of genes cloned from cDNA was determined by amplifying genomic DNA with the use of primers specific for the coding sequences. PCR cycles were as reported (13). The PCR products were cloned into a T-vector and sequenced with an Applied Biosystems 377 automated DNA sequencer.
PCR Primers.
The RAG1 fragment (corresponding to human RAG1 sequence encoding residues 482 to 804) was amplified with the use of the primers 5′-AGYCARTAYCAYAARATGTA-3′ and 5′-GCRTTNCCDATRTCRCARTG-3′. The RAG2 fragment (corresponding to human RAG2 sequence encoding residues 31 to 460) was amplified with the use of the primers 5′-TTYGGNCARAARGGNTGGCC-3′ and 5′-TCCATRCAYTGNGCRTGNACCCA-3′. The 3′-end coding sequence of RAG2 (corresponding to human RAG2 sequence encoding amino acids 460 to 527) was cloned by 3′-RACE or by PCR with the use of a forward primer specific for the upstream RAG2 sequence and a redundant reverse primer (5′-CTYCKNARRAANGAYTTYTT-3′). The POMC gene fragment (corresponding to the human POMC sequence encoding residues 141 to 260) was amplified with the use of the primers 5′-ATGGARCAYTTYCGNTGGGG-3′ and 5′-TTNACYATNACRTTYTTRAA-3′. The PLP/DM20 sequences (corresponding to human PLP sequence encoding residues 33 to 218) were cloned from brain cDNA with the use of the primers 5′-TGYGGNTGYGGNCAYGARGC-3′ and 5′-TTNCCNGGNRTNGCNKTCCA-3′.
The coding sequences of RAG1 that span the position of introns 1a and 1b present in the RAG1 gene from fugu and some other teleosts (13) were cloned from coelacanth, lungfish, salamander, and caecilian with the use of the primers reported (13). The RAG1 sequence of the banded shark was cloned by 5′-RACE, and its genomic structure was confirmed by the amplification of genomic DNA with the use of specific primers. The rhodopsin gene was amplified with the use of primers reported (13).
Vertebrates Used in This Study.
The following vertebrates were used: leopard shark, Triakis sp.; banded shark, Chiloscyllium punctatum; electric ray, Torpedo californica; bichir, Polypterus sp.; paddlefish, Polyodon spathula; sturgeon, Acipenser sp.; gar, Lepisosteus osseus; bowfin, Amia calva; arowana, Osteoglossum sp.; elephantfish, Gnathonemus sp.; notopterus, Notopterus notopterus; butterflyfish, Pantodon buchholzi; cod, Gadus morhua; mullet, Mugil cephalus; sea bass, Lates calcarifer; flounder, Pleuronectes americanus; fugu, Fugu rubripes; lungfish, Protopterus sp.; coelacanth, Latimeria menadoensis; frog, Xenopus tropicalis; salamander, Pachytriton sp.; caecilian, Typhlonectes natans; python, Python reticulatus; turtle, Pelodiscus sinensis.
Results
Insertions and Deletions in Coding Sequences.
The insertions and deletions were identified by aligning the amino acid sequences with the use of clustal x. Altogether we identified eight single-amino-acid deletions in RAG1 and RAG2 that are specific to distinct groups of vertebrates (Fig. 2–4), and a 2-aa deletion in the β-endorphin moiety of POMC of tetrapods (Fig. 5).
Figure 2.
Deletions and insertions in the coding sequence of RAG1. The positions of amino acids correspond to the human RAG1 sequence, and deletions (▴) and insertions (▾) are indicated in relation to the human sequence. GenBank accession numbers: human, M29474; rabbit, M77666; chicken, M58530; sparrow, AF143738; penguin, AF143734; ostrich, AF143727; alligator, AF143724; gharial, AF143725; frog, L19324; fugu, AF108420; trout, U15663; zebrafish, U71093; shark, U62645.
Figure 4.
Deletions and insertions in the amino-terminal region of RAG2 sequences. The positions of amino acids correspond to the human RAG2 sequence, and deletions (▴) and insertions (▾) are indicated in relation to the human sequence. GenBank accession numbers: human, AAC35287; rabbit, M77667; mouse, NM009020; chicken, M58531; frog, L19325; fugu, AF108420; trout, U31670; zebrafish, U71094.
Figure 5.
A 2-aa deletion in the POMC sequence from tetrapods that is absent in other vertebrates. The positions of amino acids correspond to the human POMC sequence, and deletions (▴) are indicated in relation to the human sequence. GenBank accession numbers: human, V01510; macaque, CTMKP; bovine, P01190; guinea pig, S78260; rat, J00759; mouse, P01193; chicken, AB019555; Xenopus, X59370 and X59369; toad, AF115251; bullfrog, AF194966; African lungfish, AF100164; Australian lungfish, AF141926; flounder, AF184066 and AF191593; trout, Q04617 and Q04618; carp, Y14617 and Y14618; eel, AF194969; gar, U59910; sturgeon, AF092937 and AF092936; paddlefish, AF117302 and AF117303; stingray, AB020972; dogfish, AB017198; lamprey, U30886.
Alternative Transcripts of DM20.
We cloned the cDNA for PLP/DM20 from bichir, sturgeon, fugu, salamander, caecilian, and turtle. Our data indicate that only DM20 transcripts are produced in the ray-finned fishes similar to the cartilaginous fishes and lobe-finned fishes, whereas the three living groups of amphibians generate only PLP transcripts. Reptiles generate both PLP and DM20 similar to those of birds and mammals (Fig. 6). We also cloned the genomic sequence for the fugu DM20 and found that it lacks a 105-bp ORF in intron 3 that gives rise to PLP in amphibians and tetrapods (data not shown). These results suggest that DM20 is the ancestral transcript, and the longer PLP transcript was generated in a common ancestor of tetrapods after it diverged from the lungfish lineage. Subsequently the ability to generate the original DM20 transcript was lost in the lineage that gave rise to amphibians. We tried to identify mutations that potentially inhibit splicing of the DM20 exon in amphibians by comparing the sequences flanking the 5′ donor splice site from tetrapods and amphibians but found no bases that are unique to amphibians (data not shown). In fact, the first nine bases of intron 3 of DM20 in mammals and the corresponding bases in salamander are identical (/GTAACAGGG). It appears that the generation of DM20 is suppressed in amphibians because of mutations in other elements or factors.
Figure 6.
Alternatively spliced DM20 and PLP. The positions of amino acids correspond to the human sequences. Arrows indicate splice sites. GenBank accession numbers: human, M27110; mouse, X07215; dog, X55317; chicken, X61661; zebrafinch, S75729; frog, Z19522 and Z19523; lungfish, AB038774; coelacanth, AB025933; trout, U21686; zebrafish, AW281594; shark, U02973; electric ray, AAB32823.
Presence or Absence of Nuclear Introns.
We have shown that the RAG1 gene in some teleosts contains two introns, in contrast to the intronless RAG1 gene in reptiles, birds, and mammals (13). We cloned the RAG1 gene from a shark, coelacanth, lungfish, salamander, and caecilian. Our data, together with our previous findings, show that in contrast to the intronless RAG1 genes in cartilaginous fishes, lobe-finned fishes, and tetrapods, RAG1 genes in bichir, paddlefish, sturgeon, gar, and bowfin contain a single intron (RAG1b), whereas RAG1 genes in teleosts contain two introns (RAG1a and -1b).
Our previous study had shown that the rhodopsin gene from some ray-finned fishes is intronless, unlike the rhodopsin gene in lampreys, skates (a cartilaginous fish), bichir, coelacanths, lizards, chickens, and mammals, which contains introns (13). We have now cloned the rhodopsin gene from the paddlefish, lungfish, frog, salamander, and caecilian and show that whereas rhodopsin gene from cartilaginous fishes, lobe-finned fishes and tetrapods contains introns, rhodopsin from ray-finned fishes, with the exception of bichirs, contains no introns.
Discussion
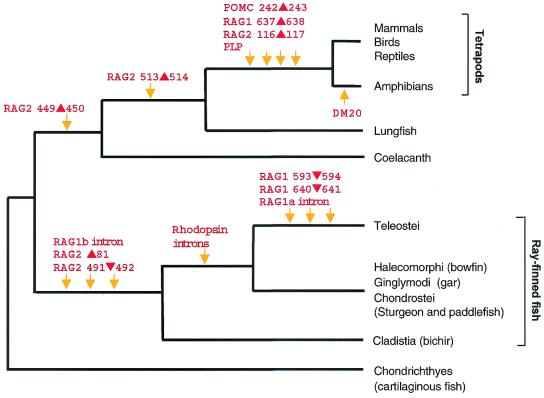
The markers cloned by us identify several clades in the vertebrate lineage and, when combined, give rise to a single phylogenetic tree of extant jawed vertebrates (Fig. 7). This tree resolves several controversies in the models of phylogenetic relationships of jawed vertebrates proposed by previous studies. First, it confirms that cartilaginous fishes are ancestral to bony vertebrates. Although it is traditionally accepted that cartilaginous fishes are ancestral to bony vertebrates, based on their cartilaginous endoskeleton and other morphological characters (2, 14), some recent studies based on the mitochondrial sequence have challenged this assumption and proposed that cartilaginous fishes lie within the bony-fish tree, in which lungfishes are basal to all gnathostomes (Fig. 1c) (5, 15, 16). Our study provides robust molecular evidence to refute this unorthodox view.
Figure 7.
Phylogenetic relationships of gnathostomes inferred from the molecular markers identified in this study. Arrows indicate the timing of deletions and insertions of amino acids, alternative splicing, and “gain” or “loss” of introns. DM20 transcripts are found in all taxa except amphibians. The rhodopsin gene from ray-finned fishes, with the exception of bichir, is intronless.
Second, our markers demonstrate that the extant tetrapods are monophyletic and lungfishes are the closest living relatives of tetrapods as proposed in phylogenetic model 1a (Fig. 1). The extant tetrapods can be divided into two groups: amphibians and amniotes, with amphibians represented by three living orders, Anura (frogs), Caudata (salamanders), and Gymnophiona (caecilians), and the amniotes, including reptiles, birds, and mammals. Although a large number of morphological characters supports a monophyletic status of these tetrapods (1, 2), some systematists have argued a diphyletic origin of tetrapods, suggesting that salamanders and newts arose from a lobe-finned fish lineage different from that which led to frogs and other tetrapods (17, 18). Furthermore, it has not been possible so far to resolve the phylogenetic position of tetrapods in relation to the extant lobe-finned fishes, the lungfish and the coelacanth (3–6). Our synapomorphies confirm the monophyletic status of the living tetrapods and provide evidence for a sister-group relationship between the living lungfishes and tetrapods.
Third, the markers cloned by us resolve the phylogenetic position of bichirs, a group of freshwater fishes endemic to Africa. The classification of bichirs has been problematic, as they display a mixture of ancestral and derived characters and share many characters with both lobe-finned fishes and ray-finned fishes (2). For example, they possess paired fins with fleshy bases and lungs that arise from the ventral side of the digestive tract, similar to the anatomy of lobe-finned fishes, but ganoid rhombic scales and a single elongated dorsal fin that is directly inserted into the body, as in ray-finned fishes (2). Phylogenetic analysis of the complete mitochondrial sequence of bichirs was also unable to determine whether they are ray-finned fishes or lobe-finned fishes (19). Based on the phylogenetic distribution of some nuclear introns, we had shown that bichirs lie outside the clade comprising (sturgeon + gar + bowfin + Teleostei) but did not resolve whether bichirs are within the ray-finned fish lineage or lobe-finned fish lineage (13). The additional data presented here clearly demonstrate that bichirs are in the ray-finned fish lineage basal to all other living ray-finned fishes. Thus bichirs occupy an important position in the ray-finned fish lineage and are a useful model for understanding the evolution, development, and physiology of the diverse ray-finned fishes.
Fourth, our analysis confirms the monophyly of Teleostei, the largest group of vertebrates, which includes more than 23,000 living species. Although several morphological characters support the monophyly of teleosts, molecular data that corroborate them are lacking (20). We provide here a set of diverse molecular markers that clearly indicate the monophyletic origin of living teleosts. Interestingly, the RAG2 sequence from paddlefish contains a 2-aa insertion between residues 84 and 85, whereas sturgeon, gar, and bowfin contain a 3-aa insertion at the same position, suggesting that the insertion occurred in a common ancestor of these groups (Fig. 3). This hypothesis of a common ancestry implies that the taxa Chondrostei (sturgeon and paddlefish), Ginglymodi (gar), and Halecomorphi (bowfin) constitute a single clade (Fig. 7). But this clade is inconsistent with the morphology-based phylogeny that groups sturgeon and paddlefish together as a clade and bowfin (represented by a single species) as a sister group of Teleostei (2). Sampling of more species from these groups should help to confirm the grouping suggested by the insertion in RAG2.
Figure 3.
Deletions and insertions in the carboxy-terminal region of RAG2 sequences. The positions of amino acids correspond to the human RAG2 sequence, and deletions (▴) and insertions (▾) are indicated in relation to the human sequence. GenBank accession numbers: human, AAC35287; rabbit, M77667; mouse, NM009020; cat, AF203771; bat, AF203770; chicken, M58531; frog, L19325; fugu, AF108420; needlefish, AF306489; trout, U31670; zebrafish, U71094; electric fish, AF201659; chitala, AF201626; butterflyfish, AF201647.
Discovering the deep branches in the evolution of vertebrates is a major challenge for evolutionary biologists. Analyses of molecular sequences have given conflicting models even when large data sets were used (3–6, 19). The pitfalls of molecular sequences include variations in the rate of evolution of different genes and the large amounts of phylogenetic “noise” that have accumulated from reversible changes over long evolutionary times. In the case of gnathostomes, these problems with sequences are further compounded by the fact that there was a rapid radiation of vertebrates during a very short window of time in the Devonian period (21). On the other hand, molecular events that change gene structure such as those identified here are not affected by such events. Although homoplasy due to parallel changes or reversal of a change is possible in these markers, it involves multiple events and is uncommon. Furthermore, homoplasy in multiple independent markers such as those identified here is highly unlikely. More significantly, all of the markers identified by us are congruent with each other and are compatible with only one phylogenetic tree. The phylogenetic relationships inferred from this tree are also consistent with proposed phylogenies based on morphological and/or molecular sequence data. Thus we believe these markers are robust and define the true phylogenetic relationships of living gnathostomes.
Acknowledgments
We thank K. P. Lim and N. Sivasothi for help in obtaining and identifying specimens and B. H. Tay and D. Tan for technical help in cloning and sequencing. This work was funded by the National Science and Technology Board of Singapore.
Abbreviations
- POMC
proopiomelanocortin
- RAG1 and RAG2
recombinase-activating proteins 1 and 2
- PLP
proteolipid protein
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Cloutier R, Ahlberg P E. In: Interrelationships of Fishes. Stiassy M L, Parenti L R, Johnson G D, editors. New York: Academic; 1996. pp. 445–480. [Google Scholar]
- 2.Janvier P. Early Vertebrates. Oxford: Clarendon; 1996. [Google Scholar]
- 3.Cao Y, Waddell P J, Okada N, Hasegawa M. Mol Biol Evol. 1998;15:1637–1646. doi: 10.1093/oxfordjournals.molbev.a025891. [DOI] [PubMed] [Google Scholar]
- 4.Zardoya R, Cao Y, Hasegawa M, Meyer A. Mol Biol Evol. 1998;15:506–517. doi: 10.1093/oxfordjournals.molbev.a025950. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen A S, Arnason U. Proc Natl Acad Sci USA. 1999;96:2177–2182. doi: 10.1073/pnas.96.5.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zardoya R, Meyer A. Genetics. 2000;155:765–775. doi: 10.1093/genetics/155.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera M C, Lake J A. Science. 1992;257:74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- 8.Keeling P J, Palmer J D. Nature (London) 2000;405:635–637. doi: 10.1038/35015167. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida M, Colman D R. Neuron. 1996;16:1115–1126. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]
- 10.Tohyama Y, Kasama-Yoshida H, Sakuma M, Kobayashi Y, Cao Y, Hasegawa M, Kojima H, Tamai Y, Tanokura M, Kurihara T. Neurochem Res. 1999;24:867–873. doi: 10.1023/a:1020958014398. [DOI] [PubMed] [Google Scholar]
- 11.Tohyama Y, Ichimiya T, Kasama-Yoshida H, Cao Y, Hasegawa M, Kojima H, Tamai Y, Kurihara T. Mol Brain Res. 2000;80:256–259. doi: 10.1016/s0169-328x(00)00143-1. [DOI] [PubMed] [Google Scholar]
- 12.Tang S, Panno J P, McKeown B A. Mol Brain Res. 1996;41:134–139. doi: 10.1016/0169-328x(96)00082-4. [DOI] [PubMed] [Google Scholar]
- 13.Venkatesh B, Ning Y, Brenner S. Proc Natl Acad Sci USA. 1999;96:10267–10271. doi: 10.1073/pnas.96.18.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maisey J G. J Vertebrate Palaeontol. 1984;4:359–371. [Google Scholar]
- 15.Rasmussen A S, Janke A, Arnason U. J Mol Evol. 1998;46:382–388. doi: 10.1007/pl00006317. [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen A S, Arnason U. J Mol Evol. 1999;48:118–123. doi: 10.1007/pl00006439. [DOI] [PubMed] [Google Scholar]
- 17.Jarvik E. Basic Structure and Evolution of Vertebrates. Vol. 1. London: Academic; 1980. [Google Scholar]
- 18.Bjerring H. Palaeontographica. 1991;A219:89–95. [Google Scholar]
- 19.Noack K, Zardoya R, Meyer A. Genetics. 1996;144:1165–1180. doi: 10.1093/genetics/144.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Pinna M C C. In: Interrelationships of Fishes. Stiassy M L, Parenti L R, Johnson G D, editors. New York: Academic; 1996. pp. 147–162. [Google Scholar]
- 21.Meyer A. Trends Ecol Evol. 1995;10:111–116. doi: 10.1016/s0169-5347(00)89004-7. [DOI] [PubMed] [Google Scholar]