The major forms of chronic inflammatory bowel disease (IBD), Crohn's disease (CD) and ulcerative colitis (UC) are characterised by inflammation of the intestinal tract. These complex, lifelong conditions can present at any age with peak incidences in the second and third decades. The natural history of IBD varies worldwide,1 with gradients in incidence, phenotype and recurrence having been demonstrated in Europe and North America.2,3 The exact aetiological factors involved in IBD pathogenesis remain elusive, however it is becoming clear that genetic predisposition, environmental influences and a dysregulated immune response to the intestinal microflora are involved.4–6
Since the first description of Crohn's disease in the early 20th century there have been several lines of epidemiological evidence implicating genetic susceptibility in the pathogenesis of IBD.7,8 The most notable of these include the familial clustering of IBD cases, ethic differences in disease susceptibility and high concordance rates in twins.9–11 The observation of increased twin concordance was demonstrated in the late 1980s from large studies carried out in Scandinavia and the UK.12 These studies showed that the risk of CD in monozygotic twins is 36% and for UC 16%, in contrast to 4% for both diseases in dizygotic twins.13
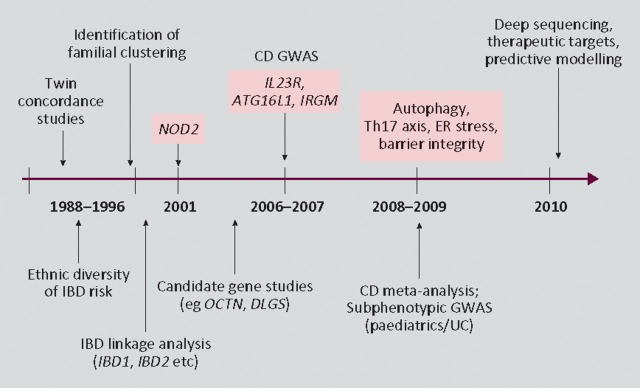
Following this strong evidence for heritable factors non-parametric linkage analysis was employed to further delineate possible genes causing IBD susceptibility in the 1990s (Fig 1).13 Linkage analysis involves the study of a limited set of genetic markers across the entire genome and does not explicitly specify the mode of inheritance, looking instead for excessive allele sharing in affected relatives. This method of gene identification has come under scrutiny in recent years, mainly due to newer techniques failing to replicate findings in these early analyses. However, it should be noted that the gene identified in these studies (NOD2/CARD15) remains the strongest individual signal using genome-wide association scanning (GWAS).4
Fig 1.
Timeline of epidemiological and genetic discoveries in inflammatory bowel disease. CD = Crohn's disease; GWAS = genome-wide association scan; IBD = inflammatory bowel disease; UC = ulcerative colitis.
Without a doubt the major breakthrough in not only IBD genetics but complex polygenic disease as a whole was the discovery of NOD2/CARD15 in 1996. This gene was discovered by the French paediatric gastroenterologist Hugot following fine mapping of the IBD1 locus.15 Hugot's group later identified variants in the leucine-rich repeat region (LRR) of the gene as conferring susceptibility to IBD, something which has been widely replicated in both adult and early-onset disease.16,17 The gene encoded by NOD2/CARD15 has gut expression limited to anti-microbial Paneth cells, with the LRR domain recognising muramyl dipeptide (MDP), a product of bacterial cell wall degradation.18,19 Since these initial studies, however, further work has demonstrated a wide range of functions for this pattern recognition receptor including the regulation of commensal bacteria, the control of certain immune cell populations and viral recognition.20–22
In more recent years the introduction of the Human Genome Project and other international collaborations has allowed the detailed examination of the entire genetic code.23 This information, coupled with the creation of high-throughput genotyping platforms, has led to the ability to analyse up to 500,000 single nucleotide polymorphisms (SNPs) using hypothesis-free GWAS. To date there have been a total of 14 GWAS performed in IBD patients leading to an explosion of multiple candidate genes implicated in IBD pathogenesis.17,24 Before the first major CD meta-analysis published in 2008 by Barrett and colleagues, only 16 loci had been identified, with the meta-analysis confirming many of these and providing evidence for 21 additional SNPs.25,26 This has been expanded further still through recent work by the International IBD Genetics Consortium with the confirmed loci now numbering 69 in CD alone.24 Although these identified loci will undoubtedly generate decades of future research, it is postulated that they still only represent approximately 25% of the genetic variance, with only the option of deep sequencing and functional studies likely to increase understanding of these susceptibility loci. What has become very clear, however, is that the genetic architecture exposed by these scans has led to a clearer understanding of the disease pathways and mechanisms involved in IBD.
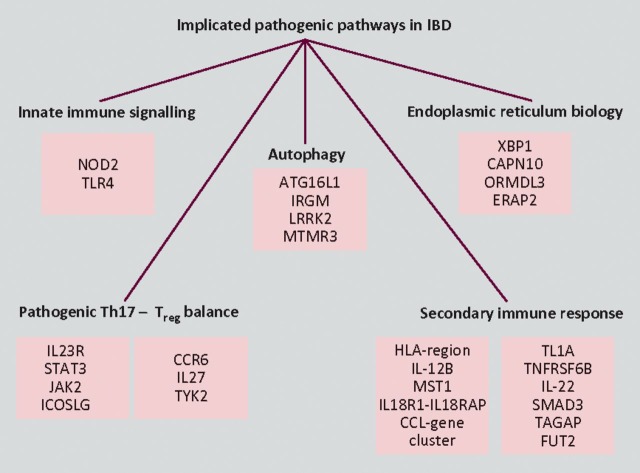
This insight provided by GWAS has uncovered candidate genes involved in pathways broadly involving the control of intestinal barrier function, the mucosal response to luminal bacteria and the secondary downstream immune response (Fig 2). Besides NOD2/CARD15, the two pathways generating the most interest at present are autophagy (the degradation of unwanted cell organelles and the elimination of pathogenic microorganisms) and the IL12–IL23 pathway (involved in pathogenic T cell differentiation).11 SNPs in the regions of the autophagy genes ATG16L1 and IRGM have been widely replicated as conferring susceptibility to CD, with functional work beginning to unravel the functional implications of their encoded proteins.27 The association with CD of a germline variation of the IL23R (interleukin-23 receptor) gene in 2006 provided the initial stimulus to look more closely at the IL17-producing subset of CD4 + Th17 cells.28 Since then risk and protective haplotypes of IL23R have been described with other genes in the Th17 pathway, such as STAT3, JAK2 and ICOSLG also implicated. In addition, the exciting recent discovery that bacterial handling and MHC class-II presentation are regulated by NOD2 autophagy induction through ATG16L1 has ignited the drive to establish a cohesive model of disease pathogenesis.29,30
Fig 2.
Broad overview of the genetic architecture of inflammatory bowel disease giving examples of confirmed and candidate genes implicated in each pathway. T® = regulatory T cell.
Not only has the study of genetics led to advances in the understanding of IBD aetiopathogenesis it has also enabled researchers to focus on key areas of potential therapeutic targets. Although gene therapy is unlikely to play a part in IBD management in the near future, several lines of pharmaceutical intervention have already been highlighted from gene discovery. The targeting of the IL12-IL23 pathway has already shown promise with the use of anti-p40 monoclonal antibody for induction of remission in CD.31 Additionally, the use of Sirolimus (which alters cellular autophagy) has been shown to be of use in refractory IBD and in the treatment of experimentally-induced murine colitis.32,33 Although specific drug treatments are still under development, genotype analysis has already been shown to inform clinicians regarding the natural history of IBD and in predicting treatment response to newer biological drugs.34,35 It is hoped that in the coming years the combination of genotyping, biomarkers and detailed phenotyping will allow a tailored approach to clinical management using a validated predictive model.
It can be seen that IBD genetics has come a long way in a short space of time, from twin studies to a clinical tool in less than 30 years. Although there is still a great deal to be learned, gaining unprecedented insight into complex polygenic disease pathophysiology will no doubt lead to rapid discoveries not only in IBD but other similar diseases. Within IBD research there is now a pressing need to explore the gene-environmental interactions especially with relation to CD-associated adherent-invasive Escherichia coli, mycobacterium and cigarette smoke.36–38 These interactions, in addition to the deep sequencing of the candidate genes to unlock the disease-causing mutations, will hopefully lead us even further towards a panacea for this debilitating disease.
Sources of support
PH is funded by a Medical Research Council project grant (No. G0800675).
References
- 1.Vatn MH. Natural history and complications of IBD. Curr Gastroenterol Rep. 2009;11:481–7. doi: 10.1007/s11894-009-0073-8. [DOI] [PubMed] [Google Scholar]
- 2.Shivananda S, Lennard-Jones J, Logan R, et al. Incidence of inflammatory bowel disease across Europe: is there a difference between north and south? Results of the European Collaborative Study on Inflammatory Bowel Disease (EC-IBD) Gut. 1996;39:690–7. doi: 10.1136/gut.39.5.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein CN, Wajda A, Svenson LW, et al. The epidemiology of inflammatory bowel disease in Canada: a population-based study. Am J Gastroenterol. 2006;101:1559–68. doi: 10.1111/j.1572-0241.2006.00603.x. [DOI] [PubMed] [Google Scholar]
- 4.Van Limbergen J, Wilson D, Satsangi J. The genetics of Crohn's disease. Annu Rev Genomics Hum Genet. 2009;10:89–116. doi: 10.1146/annurev-genom-082908-150013. [DOI] [PubMed] [Google Scholar]
- 5.Armitage EL, Aldhous MC, Anderson N, et al. Incidence of juvenile-onset Crohn's disease in Scotland: association with northern latitude and affluence. Gastroenterology. 2004;127:1051–7. doi: 10.1053/j.gastro.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol. 2010;26:327–31. doi: 10.1097/MOG.0b013e328339536b. [DOI] [PubMed] [Google Scholar]
- 7.Dalziel TK. Chronic interstitial enteritis. BMJ. 1913;(ii):1068–70. [Google Scholar]
- 8.Crohn BB, Ginsburg L, Oppenheimer GD. Regional ileitis: a clinical and pathologic entity. JAMA. 1932;99:1323–9. [Google Scholar]
- 9.Peeters M, Nevens H, Baert F, et al. Familial aggregation in Crohn's disease: increased age-adjusted risk and concordance in clinical characteristics. Gastroenterology. 1996;111:597–603. doi: 10.1053/gast.1996.v111.pm8780562. [DOI] [PubMed] [Google Scholar]
- 10.Roth MP, Petersen GM, McElree C, et al. Familial empiric risk estimates of inflammatory bowel disease in Ashkenazi Jews. Gastroenterology. 1989;96:1016–20. doi: 10.1016/0016-5085(89)91618-1. [DOI] [PubMed] [Google Scholar]
- 11.Lees CW, Satsangi J. Genetics of inflammatory bowel disease: implications for disease pathogenesis and natural history. Expert Rev Gastroenterol Hepatol. 2009;3:513–34. doi: 10.1586/egh.09.45. [DOI] [PubMed] [Google Scholar]
- 12.Tysk C, Lindberg E, Jarnerot G, et al. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–6. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell RK, Satsangi J. IBD: a family affair. Best Pract Res Clin Gastroenterol. 2004;18:525–39. doi: 10.1016/j.bpg.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Noble C, Nimmo E, Gaya D, et al. Novel susceptibility genes in inflammatory bowel disease. World J Gastroenterol. 2006;12:1991–9. doi: 10.3748/wjg.v12.i13.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugot JP, Laurent-Puig P, Gower-Rousseau C, et al. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821–3. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 16.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 17.Henderson P, Van Limbergen J, Wilson DC, et al. Genetics of childhood-onset inflammatory bowel disease. Inflamm Bowel Dis. 2010;17:346–61. doi: 10.1002/ibd.21283. [DOI] [PubMed] [Google Scholar]
- 18.Lala S, Ogura Y, Osborne C, et al. Crohn's disease and the NOD2 gene: a role for paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/S0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 19.Kelsall B. Getting to the guts of NOD2. Nat Med. 2005;11:383–4. doi: 10.1038/nm0405-383. [DOI] [PubMed] [Google Scholar]
- 20.Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106:15813–8. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman MK, Midtling EH, Svingen PA, et al. The Pathogen Recognition Receptor NOD2 Regulates Human FOXP3+ T Cell Survival. J Immunol. 2010;184:7247–56. doi: 10.4049/jimmunol.0901479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabbah A, Chang TH, Harnack R, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–80. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. International HapMap Project. Database of genes associated with human disease and response to pharmaceuticals [HapMap web site]. www.hapmap.org.
- 24.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathew CG. New links to the pathogenesis of Crohn's disease provided by genome-wide association scans. Nat Rev Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 30.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 31.Mannon PJ, Fuss IJ, Mayer L, et al. Anti-interleukin-12 antibody for active Crohn's disease. N Engl J Med. 2004;351:2069–79. doi: 10.1056/NEJMoa033402. [DOI] [PubMed] [Google Scholar]
- 32.Massey DC, Bredin F, Parkes M. Use of sirolimus (rapamycin) to treat refractory Crohn's disease. Gut. 2008;57:1294–6. doi: 10.1136/gut.2008.157297. [DOI] [PubMed] [Google Scholar]
- 33.Farkas S, Hornung M, Sattler C, et al. Rapamycin decreases leukocyte migration in vivo and effectively reduces experimentally induced chronic colitis. Int J Colorectal Dis. 2006;21:747–53. doi: 10.1007/s00384-005-0793-7. [DOI] [PubMed] [Google Scholar]
- 34.Latiano A, Palmieri O, Corritore G, et al. Variants at the 3p21 locus influence susceptibility and phenotype both in adults and early-onset patients with inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1108–17. doi: 10.1002/ibd.21176. [DOI] [PubMed] [Google Scholar]
- 35.Jurgens M, Laubender RP, Hartl F, et al. Disease activity, ANCA, and IL23R Genotype status determine early response to infliximab in patients with ulcerative colitis. Am J Gastroenterol. 2010;105:1811–9. doi: 10.1038/ajg.2010.95. [DOI] [PubMed] [Google Scholar]
- 36.Peeters H, Bogaert S, Laukens D, et al. CARD15 variants determine a disturbed early response of monocytes to adherent-invasive Escherichia coli strain LF82 in Crohn's disease. Int J Immunogenet. 2007;34:181–91. doi: 10.1111/j.1744-313X.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- 37.Kirkwood CD, Wagner J, Boniface K, et al. Mycobacterium avium subspecies paratuberculosis in children with early-onset Crohn's disease. Inflamm Bowel Dis. 2009;15:1643–55. doi: 10.1002/ibd.20967. [DOI] [PubMed] [Google Scholar]
- 38.Aldhous MC, Prescott RJ, Roberts S, et al. Does nicotine influence cytokine profile and subsequent cell cycling/apoptotic responses in inflammatory bowel disease? Inflamm Bowel Dis. 2008;14:1469–82. doi: 10.1002/ibd.20523. [DOI] [PubMed] [Google Scholar]