Abstract
During epithelial-mesenchymal transition (EMT) epithelial cancer cells transdifferentiate into highly motile, invasive, mesenchymal-like cells, giving rise to disseminating tumor cells. Few of these disseminated cells successfully metastasize. Immune cells and inflammation in the tumor microenvironment were shown to drive EMT, but few studies investigated the consequences of EMT for tumor immunosurveillance. In addition to initiating metastasis, we demonstrate that EMT confers increased susceptibility to natural killer (NK) cells and contributes, in part, to the inefficiency of the metastatic process. Depletion of NK cells allowed spontaneous metastasis without affecting primary tumor growth. EMT-induced modulation of E-cadherin and cell adhesion molecule 1 (CADM1) mediated increased susceptibility to NK cytotoxicity. Higher CADM1 expression correlates with improved patient survival in 2 lung and 1 breast adenocarcinoma patient cohorts and decreased metastasis. Our observations reveal a novel NK-mediated, metastasis-specific immunosurveillance in lung cancer and present a window of opportunity for preventing metastasis by boosting NK cell activity.
Keywords: Immunology, Oncology
Keywords: Immunotherapy, Lung cancer, NK cells
Introduction
Significant progress has been made in determining and defining the key regulators of immunosurveillance in primary tumors, wherein the balance of protumor and antitumor responses dictates tumor progression. Targeting these key regulators led to the successful development of checkpoint inhibitors that boost T cell–mediated immunity against multiple tumor types, including non–small cell lung cancer (NSCLC) (1, 2). It is now increasingly clear that the microenvironment and the conditions tumor cells encounter in the primary tumor are disparate from what they encounter during metastasis. This suggests that mechanisms of immunosurveillance in the primary tumor may also be different from those that regulate metastatic spread (3), indicating the existence of metastasis-specific immunosurveillance (3–8).
Epithelial-mesenchymal transition (EMT) involves dissolution of cell-cell adhesions, downregulation of epithelial markers, induction of mesenchymal markers, and breach of the basement membrane to migrate and invade (9, 10). In addition, EMT also endows cancer cells with stem cell–like properties, resistance to targeted therapies, and the ability to evade host immunosurveillance (11, 12). The multifunctional cytokine transforming growth factor-β (TGF-β) is a potent inducer of EMT and promotes tumor progression in late-stage tumors (13). Consistently, expression of TGF-β is frequently upregulated in cancers, including NSCLC (14), and is correlated with enhanced invasion, metastasis, and poor prognosis for patients with lung cancer (15). In NSCLC, we have shown that TGF-β–induced EMT confers a migratory and invasive phenotype promoting metastasis (16, 17); inhibition of EMT blocked tumor metastasis (18, 19), and an EMT-associated secretory phenotype was predictive of outcomes in NSCLC patients (20), demonstrating the biological and clinical significance of EMT in lung tumor progression.
Given the extensive molecular and cellular changes that occur during EMT, it is likely that tumor cell interactions with both innate and adaptive immune cells in the tumor microenvironment will be altered. Several studies implicated both innate and adaptive immune cells as drivers of EMT (21). However, few studies have investigated the immunomodulatory consequences of EMT (22). Studies to date have shown evasion of T cell responses and suppression of dendritic cell functions by cells undergoing EMT, suggesting an important role for EMT in immune editing (23, 24). Consistent with this, we demonstrated an immune-evasive consequence of EMT by which tumor cells escape complement-dependent cytotoxicity by modulating the complement regulatory protein CD59 (25). In contrast, here we report a novel and unexpected consequence of EMT that confers enhanced susceptibility to NK cell–mediated killing.
Natural killer (NK) cells are innate lymphoid cells known for their ability to recognize and rapidly eliminate infected or transformed cells (26–28). In humans, there is a correlation between low NK cell cytotoxicity in peripheral blood and increased cancer risk (29). Conversely, NK cell infiltration into tumor tissue is associated with better patient prognosis in multiple malignancies, including NSCLC (29). However, studies investigating the role of NK cells in tumor progression and metastasis are very limited. NK cells express an array of germline-encoded receptors that enable them to detect and respond to their targets while sparing normal cells. Depending on the signal they transmit, these receptors are classified into inhibitory and activating receptors (30). Integration of signaling from these receptors determines whether or not an NK cell becomes activated. NK cells use their inhibitory receptors to detect the presence of MHC class I as self-molecules on potential target cells. NK cells also express other non–MHC-dependent inhibitory receptors, including killer lectin-like receptor G1 (KLRG1) (31). The type I, epithelial cadherin (E-cad) has been identified as an inhibitory ligand that engages KLRG1 in both mice and humans (32). Cell adhesion molecule 1 (CADM1) has been identified as an activating NK ligand and binds to the cytotoxic and regulatory T cell–associated molecule (CRTAM) receptor on NK cells (33). CADM1 is identified as a tumor suppressor and is frequently downregulated in multiple different malignancies (34, 35). Here, we demonstrate that EMT-induced CADM1 expression, along with the downregulation of E-cad, regulates NK-mediated, metastasis-specific immunosurveillance.
Results
EMT differentially regulates NK ligands and confers enhanced susceptibility to NK cytotoxicity.
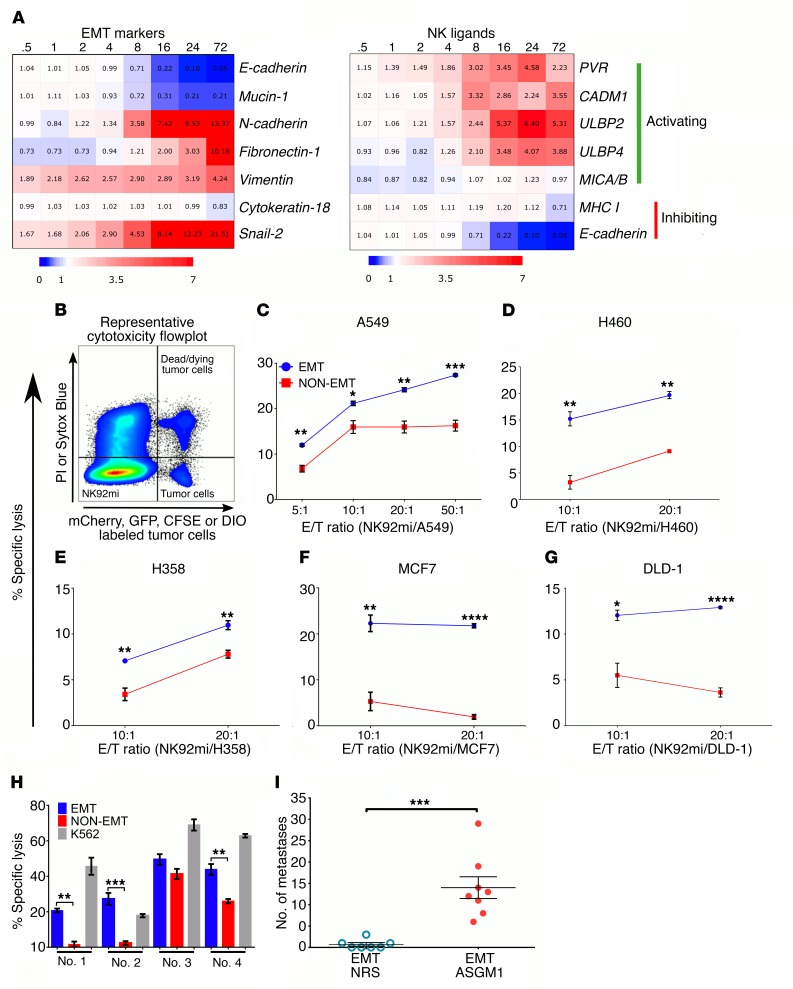
NK cell reactivity is regulated by the balance of activating and inhibitory receptor engagement by ligands expressed on the target cells. Analysis of a previously obtained time course gene expression profile during TGF-β–induced EMT (17, 36) identified differential modulation of several NK-activating ligands, in addition to the modulation of epithelial and mesenchymal markers. We observed a significant time-dependent induction of mRNA for the NK-activating ligands PVR, CADM1, ULBP2, and ULBP4 (Figure 1A). Similarly, among the known inhibitory ligands, E-cad expression was significantly suppressed, but there was no significant change in the expression of MHC I in response to EMT. Collectively, the increased expression of activating ligands and decreased expression of inhibitory ligands in tumor cells undergoing EMT suggest a potential increased susceptibility to NK-mediated cytotoxicity. To test this, we assessed the susceptibility of a panel of human cancer cell lines to NK-mediated cytotoxicity before and after EMT. All cell lines were stimulated with TGF-β to induce EMT and then cocultured with the human NK cell line NK92mi to assess tumor cell–specific killing by flow cytometry (Figure 1B). Consistent with the ligand expression profile, we observed a significantly increased susceptibility to NK-mediated cytotoxicity in EMT cells compared with the controls at all tumor/NK cell ratios tested (Figure 1, C–G). The EMT-induced susceptibility to NK cells was not specific to lung cancer cells but was also observed in breast (Figure 1F) and colon (Figure 1G) cancer cells, suggesting that it may be a more general phenomenon in response to EMT. Similarly, murine lung cancer cells also demonstrated increased susceptibility to NK-mediated cytotoxicity after EMT (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI97611DS1). In this case, CD45+, NK1.1+, CD3e– NK cells isolated from total splenic cells after overnight culture were used as effector cells against 344SQ murine lung cancer cells as targets.
Figure 1. EMT differentially regulates NK ligands and promotes susceptibility to NK-mediated cytotoxicity.
(A) Heatmap (blue: downregulation; red: upregulation) representing fold changes, from 0 hours to 72 hours, time course of differentially expressed EMT markers and NK ligand genes during TGF-β–induced EMT, from a previously published gene expression profile data set (GEO GSE17708) (36). (B) Representative flow cytometric plot of cytotoxicity assay showing locations of effector, NK92mi (fluorophore-null cells), and target cells (fluorophore positive) and their exclusion DNA-binding dye status (viability indicator). PI, propidium iodide. (C–G) NK92mi-mediated cytotoxicity plots after 4 hours of coculture at indicated E/T ratios per cell type and treatment. Cell lines were treated with TGF-β (5 ng/ml) for 3, 6, 12, 6, and 6 days to induce optimum EMT, as assessed by complete E-cad downregulation and induction of vimentin or N-cadherin, in A549 (C), H460 (D), H358 (E), MCF7 (F), and DLD-1 (G), respectively. Data represent triplicate mean ± SEM, and 2-tailed unpaired t tests were performed. All experiments were repeated at least twice. (H) Freshly isolated human peripheral blood–derived NK cells were used as effector cells (E/T, 10:1) against A549 cells. K562 cells were used as a positive control for cytotoxicity. Data represent mean ± SEM, and 2-tailed, unpaired, t tests were performed. (I) To assess experimental metastasis, A549 cells were treated with TGF-β (5 ng/ml) in vitro and injected through the tail vein into RAG–/– mice. After 8 weeks, lungs were harvested to assess tumor burden. NRS, normal rabbit serum. Data represent 2 independent experiments, n = 3–4 for each group, and pooled results are shown. Error bars are SEM; Mann-Whitney U test was performed, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To demonstrate that this increased cytotoxicity is not specific to the NK92mi cell line, untouched human NK cells were isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors as effectors against EMT and control A549 cells as targets. The differential susceptibility between EMT and control A549 cells was also observed when primary PBMC-derived NK cells were used as effectors (Figure 1H). K562 cells were used as positive controls to ensure that cytotoxic-capable NK cells were isolated. Next, to demonstrate the direct link between EMT and NK-mediated immunosurveillance in vivo, we induced EMT in A549-GFP cells as described earlier (19) and injected them into the tail vein of 2 groups of RAG–/– mice. In one group, NK cells were depleted with weekly anti–asialo GM1 (anti-ASGM1) treatment, and mice were sacrificed after 6–8 weeks to assess lung metastasis. We observed metastatic lung nodules only in the mice treated with ASGM1 and not in the control group (Figure 1I), demonstrating EMT-induced susceptibility to NK-mediated immunosurveillance, in vivo.
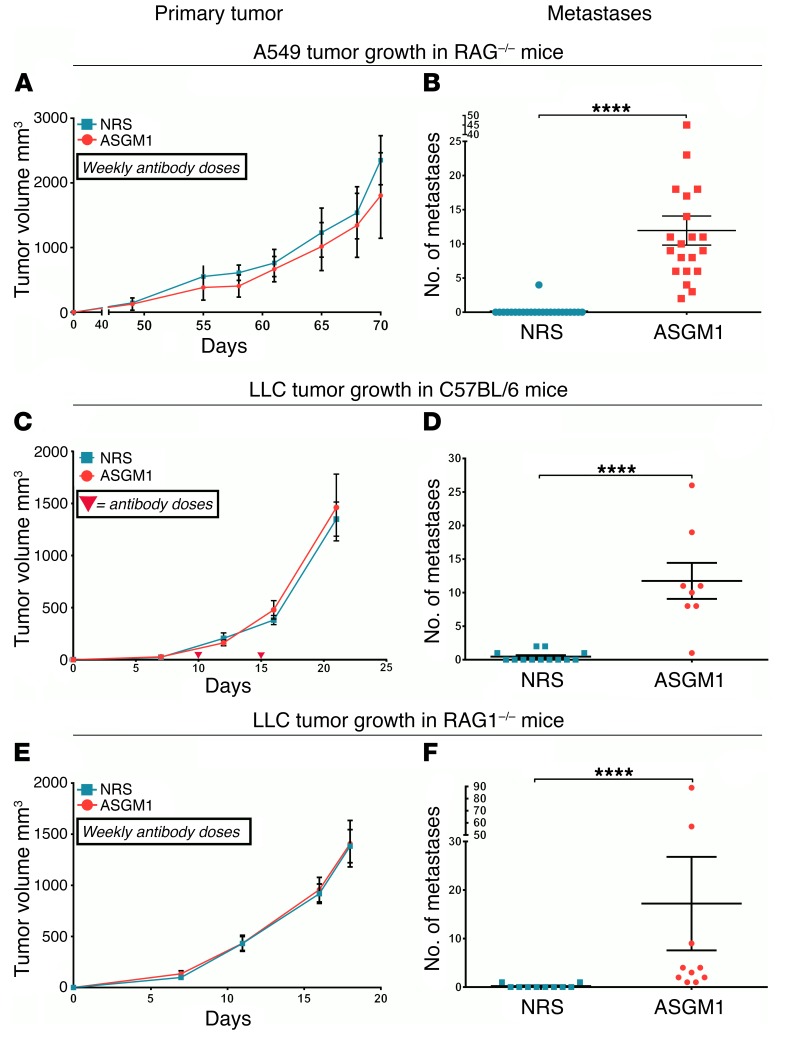
NK cell depletion allows spontaneous metastasis without affecting primary tumor growth.
In earlier studies we demonstrated that TGF-β–induced EMT in A549 cells triggers a migratory and invasive phenotype in vitro and promotes metastasis in vivo (16, 19). Inhibition of TGF-β signaling, which prevents EMT, blocked metastasis of A549 cells (19), indicating an EMT-dependent metastasis model. To assess the role of NK cells in the above model, we implanted 106 A549 cells in 2 groups of RAG1–/– mice on either side of the dorsal flank. RAG1–/– mice do not have T and B cells but have functional NK cells. One group of mice were treated weekly with ASGM1 antibody to deplete NK cells. Interestingly, depletion of NK cells had no effect on the primary tumor growth (Figure 2A). In contrast, we observed spontaneous lung metastasis only in the mice that were depleted of NK cells (Figure 2B), suggesting the presence of an NK-mediated metastasis-specific immunosurveillance. Together with the observations in Figure 1, this also suggests an EMT-dependent mechanism. To assess the extent of depletion, splenic cells were isolated and analyzed by flow cytometry. We observed nearly a 90% reduction in the number of NK cells (CD45+NK1.1+CD3e– cells) with ASGM1 treatment (Supplemental Figure 2B).
Figure 2. NK cell depletion allows spontaneous metastatic spread without affecting primary tumor growth.
(A–F) To assess the effect of NK cell depletion on primary tumor growth and metastasis, indicated cell lines were implanted subcutaneously under the dorsal flanks of RAG1–/– or C57BL/6 mice. Mice were treated weekly with anti–asialo GM1 antibody (ASGM1) to deplete NK cells or with normal rabbit serum (NRS) as control. (A, C, and E) Primary tumor growth was monitored, and mean tumor volumes are plotted with error bars as SEM. Representative data from a single experiment of at least duplicates. (B, D, and F) Overt lung nodules were counted on the excised lungs to assess spontaneous metastasis. Mouse strains and tumor cell implants are designated. Error bars are SEM; Mann-Whitney U test was performed, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data represent at least 2 experiments, n = 4–5 per group, and pooled data are shown.
To assess the role of NK cells in metastasis in an immunocompetent host, we used a syngeneic murine model of Lewis lung carcinoma (LLC) cells. We subcutaneously implanted LLC cells in C57BL/6 hosts with and without NK cell depletion using ASGM1 antibody. LLC cells grow very aggressively in the syngeneic host but do not metastasize. As with the xenograft model above, depletion of NK cells had no effect on the primary tumor growth (Figure 2C), but we observed spontaneous lung metastasis only in mice that were treated with ASGM1 antibody (Figure 2D). In a parallel experiment, we used NK1.1 antibody instead of ASGM1 to deplete NK cells and observed similar results (Supplemental Figure 3, A and B). Since there is no single method that can exclusively deplete only NK cells, our data with ASGM1 (which is also known to affect basophil populations) and NK1.1 antibody (which is also known to deplete NK-T cells) showing similar effects demonstrate an NK cell–dependent phenomena.
The potential contribution of T and B cells to the observed metastasis-specific immunosurveillance was assessed by implanting of LLC cells in T and B cell–deficient RAG–/– mice. Notably, we observed similar spontaneous metastasis only upon NK depletion (Figure 2F), and there was no effect on the growth of the primary tumor with and without NK cell depletion (Figure 2E). We also observed similar results when 344SQ cells were implanted in C57BL/6 mice with and without ASGM1 NK depletion (Supplemental Figure 1, C and D). Collectively, similar observations in 4 different models above demonstrate the presence of an NK cell–dependent, metastasis-specific immunosurveillance mechanism.
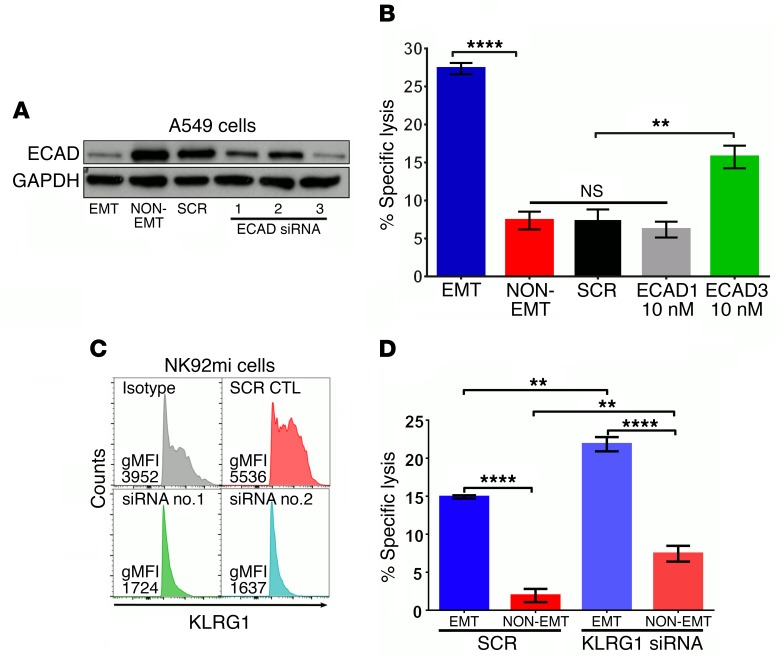
Loss of E-cad expression sensitizes tumor cells to NK cytotoxicity through KLRG1.
Classically, inhibitory signals on NK cells are mediated by receptors that recognize MHC I molecules. However, recent studies demonstrate that NK cells also express other receptors like KLRG1 that can recognize MHC-independent inhibitory signals such as E-cad (32). Since E-cad is downregulated during EMT with a concomitant increase in the susceptibility to NK cell–mediated cytotoxicity, we reasoned that E-cad may be an important inhibitory signal that protects epithelial cells from NK cell–mediated cytotoxicity. To test this, we inhibited E-cad expression by 3 different siRNA molecules in A549 cells (Figure 3A), before assessing their susceptibility to NK cell cytotoxicity using NK92mi cells as described for Figure 3. We observed that E-cad inhibition increased susceptibility of A549 cells to NK-mediated cytotoxicity (Figure 3B). However, the magnitude of cytotoxicity did not reach the levels observed after EMT. Next, we determined whether E-cad–induced inhibitory signaling is indeed mediated through KLRG1 on NK cells. siRNA-mediated inhibition of KLRG1 expression in NK92mi cells (Figure 3C) enhanced their cytotoxicity against the non-EMT control as well as EMT-induced tumor cells (Figure 3D), demonstrating that KLRG1 mediates E-cad–induced inhibitory signaling. Inhibiting the E-cad/KLRG1 axis did not fully recapitulate the enhanced cytotoxicity against A549 cells undergoing EMT, suggesting that it may also require the induction of an activating ligand.
Figure 3. Loss of E-cad expression sensitizes tumor cells to NK-mediated cytotoxicity through KLRG1.
(A) A549 cells were transfected with 10 nM of scrambled (SCR) or 3 different E-cad–specific siRNA molecules. After 24 hours, cells were treated with (EMT) or without (NON-EMT) TGF-β (5 ng/ml) for 72 hours. E-cad and GAPDH expressions were assessed by Western immunoblotting. (B) Susceptibility to NK cytotoxicity was assessed using NK92mi cells as effectors, as described for Figure 1. Mean ± SEM is shown; 1-way ANOVA with Tukey’s post hoc analysis was performed, **P < 0.01, ****P < 0.0001. (C) NK92mi cells were transfected with 10 nM of SCR or KLRG1-specific siRNA. After 72 hours, KLRG1 expression was assessed by flow cytometry. gMFI, geometric mean fluorescence intensity. (D) NK92mi cells from C were used as effectors against EMT or non-EMT A549 cells in the NK cytotoxicity assay. Mean ± SEM is shown; 2-way ANOVA with Tukey’s post hoc analysis was performed, **P < 0.01, ****P < 0.0001. All experiments were repeated twice, and data are representative of 1 experiment.
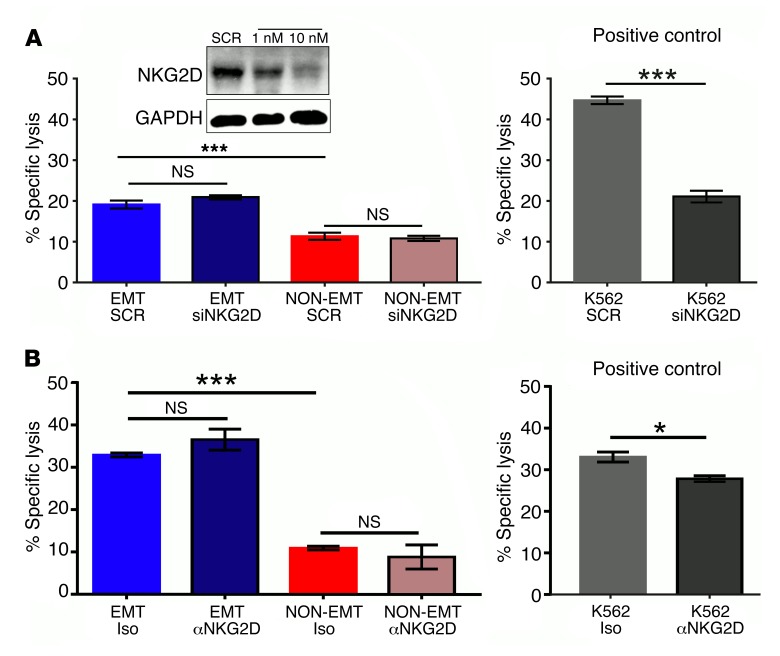
NKG2D receptor is not involved in the EMT-induced susceptibility to NK-mediated cytotoxicity.
Since we observed a robust induction of the NKG2D ligands ULBP2 and ULBP4 during EMT, we tested the potential involvement of NKG2D as an activating NK cell receptor mediating tumor cell killing after EMT. We assessed the involvement of NKG2D by independently blocking its expression, using siRNA, and function, using neutralizing antibodies, in NK92mi cells. K562 cells, whose NK-mediated cytotoxicity is partly NKG2D dependent, were used as a positive control (37). Interestingly, inhibition of expression or function of NKG2D receptors had no effect on A549 tumor cell killing before or after EMT (Figure 4, A and B). This suggests that alternative activating receptors may be critical for NK cell recognition of A549 cells after EMT.
Figure 4. NKG2D receptor is not involved in EMT-induced susceptibility to NK-mediated cytotoxicity.
(A) NK92mi cells were transfected with 10 nM of scrambled (SCR) or NKG2D-specific siRNA. After 72 hours, NKG2D expression was assessed by Western immunoblotting (inset), and these cells were used as effectors against A549 cells that were treated with (EMT) or without (NON-EMT) TGF-β (5 ng/ml) for 72 hours in the NK cytotoxicity assay. (B) NKG2D receptors were neutralized by treatment of NK92mi cells with anti-NKG2D receptor antibody (50 μg/ml) 45 minutes before coculturing with EMT or non-EMT A549 cells in the NK cytotoxicity assay. Iso, isotype control. All experiments were repeated twice; representative data of 1 experiment are shown. Mean ± SEM is shown; 2-way ANOVA with Tukey’s post hoc analysis was performed, *P < 0.05, ***P < 0.001. For positive control K562 cytotoxicity, mean ± SEM is shown; 2-tailed, unpaired t tests were performed, *P < 0.05, ***P < 0.001.
CADM1 expression is modulated by EMT-MET cycling and mediates tumor cell susceptibility to NK cytotoxicity.
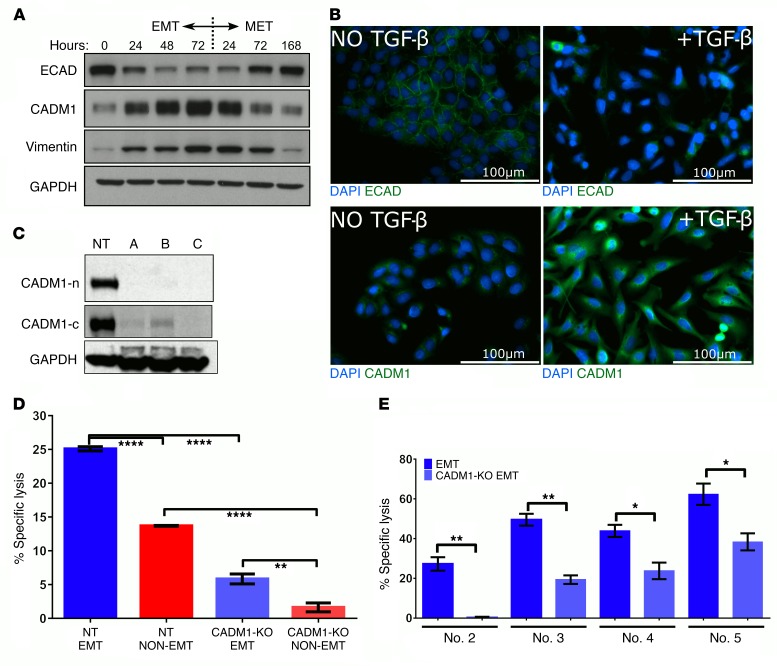
Given that neutralizing NKG2D receptors had no effect, we tested the role of CADM1, the next most abundant NK ligand induced during EMT, as a potential activating ligand mediating EMT-induced susceptibility to NK cells. CADM1, also known as TSLC1, is identified as a tumor suppressor in lung cancer, and its expression is frequently lost in 40% of NSCLC (34, 38, 39). Interestingly, CADM1 was also shown to form heterophilic interactions with an immunoglobulin family receptor known as CRTAM that serves as its cognate receptor and is expressed on activated NK cells, suggesting a role for CADM1 in immunosurveillance (33). We observed a robust modulation of CADM1 protein in response to TGF-β–induced EMT and mesenchymal-epithelial transition (MET) cycling, along with the epithelial marker E-cad and the mesenchymal marker Vim, in a time-dependent fashion (Figure 5A). We further validated TGF-β–induced CADM1 expression with concomitant downregulation of E-cad by immunofluorescence (Figure 5B).
Figure 5. CADM1 expression is modulated by EMT-MET cycling and mediates tumor cell susceptibility to NK cytotoxicity.
(A) A549 cells were treated with TGF-β (5 ng/ml) to induce EMT, and total proteins were extracted at the indicated times. After 72 hours, cells were washed 3 times and replaced with fresh media to induce mesenchymal-epithelial transition (MET), and total proteins were extracted at the indicated times. Protein expression of E-cad, CADM1, vimentin, and GAPDH was assessed by Western immunoblotting. (B) A549 cells treated with and without TGF-β for 72 hours were fixed and assessed for E-cad and CADM1 expression by immunofluorescence staining. Scale bars: 100 μm. (C) To stably knock out CADM1, Cas9-expressing A549 cells were transduced with lentiviruses expressing 3 different CADM1-specific CRISPR sgRNAs and a nontargeting (NT) control sgRNA. CADM1 knockout was assessed by Western immunoblotting using 2 different CADM1 antibodies raised against C-terminal (CADM1-c) and N-terminal (CADM1-n) portions. (D and E) Susceptibility of CADM1-KO A549 cells to NK cytotoxicity was assessed by coculturing with either NK92mi cells or primary human blood–derived NK cells from 4 different donors. In D, mean ± SEM is shown; 2-way ANOVA with Tukey’s post hoc analysis was performed, **P < 0.01, ****P < 0.0001. In E, EMT controls are from Figure 1H, as these experiments were performed simultaneously. Mean ± SEM is shown; 2-tailed, unpaired t tests were performed, *P < 0.05, **P < 0.01.
To determine the role of CADM1 in NK-mediated cytotoxicity, we developed stable A549 cells with CADM1 knockout by genome editing using CRISPR/Cas9 technology along with constitutive mCherry expression (A549–CADM1-KO) together with a corresponding nontargeting control cell line (A549-NT) (Figure 5C) and assessed the susceptibility, before and after EMT, to NK cell cytotoxicity. We observed that CADM1 inhibition abrogated EMT-induced susceptibility of A549 cells to NK92mi cytotoxicity (Figure 5D) as well as to the primary donor-derived NK cell cytotoxicity (Figure 5E). Together with E-cad data (Figure 3), this demonstrates that modulation of both E-cad and CADM1 expression can regulate tumor cell susceptibility to NK-mediated cytotoxicity. Analysis of E-cad and CADM1 expression in primary A549 tumors from Figure 2A (Supplemental Figure 4, A and B) showed no difference with or without NK cell depletion, which is consistent with lack of effect on primary tumor growth kinetics. The analysis of E-cad and CADM1 expression in susceptibility cell lines (Supplemental Figure 5) further suggests that potentially it is the ratio between E-cad and CADM1 that dictates susceptibility to NK-mediated cytotoxicity. Interestingly, CADM1 knockout had no effect on TGF-β–induced EMT (Supplemental Figure 6).
Inhibition of CADM1 in tumor cells enables immune evasion and promotes metastasis.
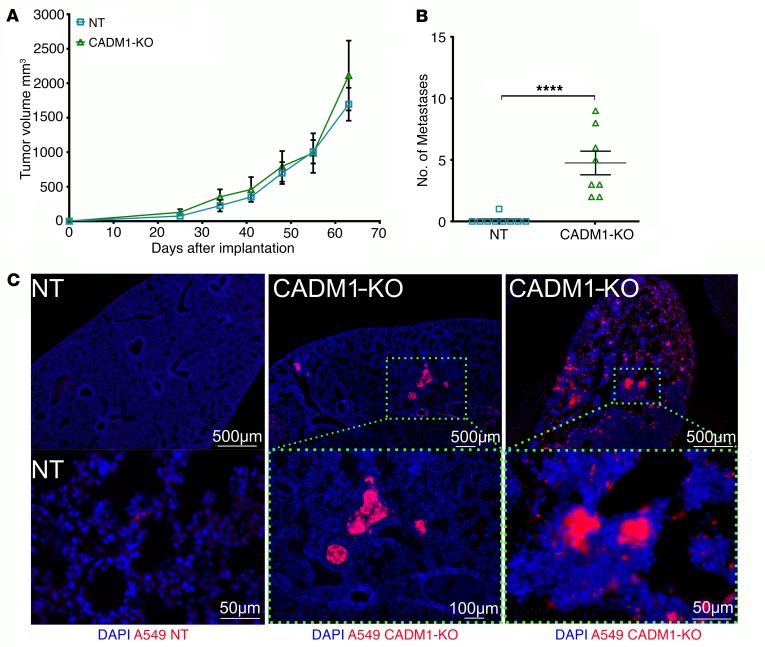
To assess the impact of CADM1 inhibition on tumor metastasis, we implanted A549–CADM1-KO and A549-NT cells into the dorsal flanks of RAG1–/– mice and assessed primary tumor growth and lung metastasis. Notably, there was no difference in the kinetics of primary tumor growth between A549–CADM1-KO and A549-NT due to CADM1 inhibition (Figure 6A). As expected, the tumors from the control A549-NT cells did not metastasize. However, there was a striking increase in overt lung metastasis from A549–CADM1-KO cells even without NK cell depletion, as assessed by gross counting (Figure 6B) and visualizing of mCherry-positive tumor cell colonies in the lung (Figure 6C). This indicates that CADM1 inhibition alone is sufficient to allow metastasis that is otherwise blocked by NK cell immunosurveillance. Putatively, loss of CADM1 observed in various cancers may be an immune-evasive strategy used by tumors.
Figure 6. Inhibition of CADM1 in tumor cells allows spontaneous metastasis without affecting primary tumor growth.
(A) mCherry-expressing CADM1 KO and control A549 cells were subcutaneously implanted into the dorsal flanks of RAG1–/– mice. Primary tumor growth was monitored, and mean tumor volumes are plotted; mean ± SEM shown. Data are representative of 1 experiment (n = 4–5 mice per group). (B) Overt lung nodules were counted on the excised lungs to assess spontaneous metastasis. Data represent 2 independent experiments, and pooled data are shown; error bars are SEM, and Mann-Whitney U test was performed, ****P < 0.0001. (C) Presence, or lack thereof, of metastatic spread was further confirmed by visualization of mCherry-positive tumor cells in the cross sections of the lungs by immunofluorescence. Top row scale bars: 500 μm; lower row: 50 μm, 100 μm, and 50μm, respectively.
Restoring CADM1 expression in tumor cells is sufficient to confer susceptibility to NK cytotoxicity.
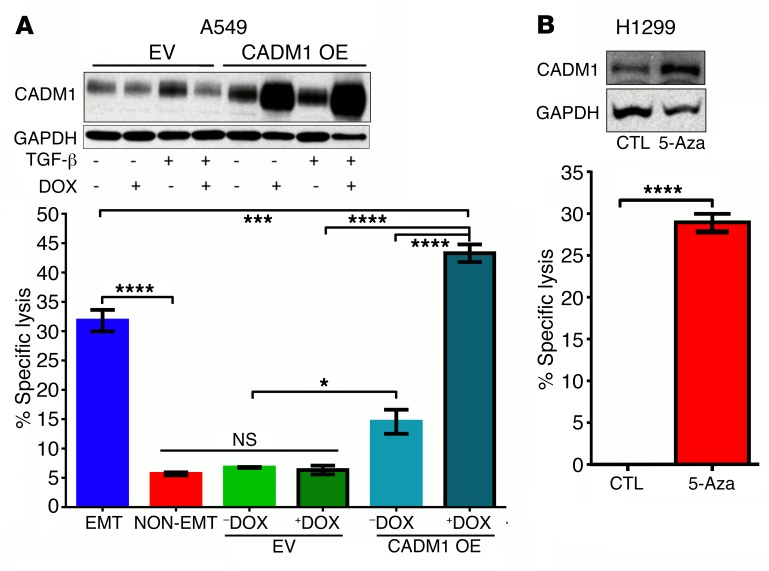
It is well documented that CADM1 expression is frequently lost in 40% of lung cancers as a result of either promoter hypermethylation or loss of heterozygosity (LOH) (35). Here we tested the efficacy of 2 approaches of restoring CADM1 expression on NK-mediated cytotoxicity. First, in an A549 cell line with LOH for CADM1, we generated a stable cell line expressing a doxycycline-inducible CADM1 (A549–CADM1-OE). Doxycycline-induced overexpression of CADM1 had no effect on growth or on TGF-β–induced EMT in A549–CADM1-OE cells (Supplemental Figure 7). However, doxycycline-induced CADM1 overexpression alone was sufficient to confer susceptibility to NK-mediated cytotoxicity, without the induction of EMT (Figure 7A). Similarly, in a cell line with CADM1 promoter hypermethylation (H1299) (40), we were able to restore CADM1 expression by culturing H1299 cells in the presence of 5′-azadeoxycytidine (5-aza), a pan-demethylating agent, for 6 days (Figure 7B). Consistent with the restored CADM1 expression, 5-aza treatment also rendered H1299 cells susceptible to NK-mediated cytotoxicity (Figure 7B). The efficacy of the above 2 methods demonstrates that restoring CADM1 expression can be a potential immunotherapeutic strategy for lung cancer.
Figure 7. Restoring CADM1 expression in tumor cells is sufficient to confer susceptibility to NK cytotoxicity.
(A) Stable A549 cell lines expressing empty vector (EV) or vector with doxycycline-inducible (DOX-inducible) human CADM1 overexpression (CADM1 OE) were developed. Expression of TGF-β–induced CADM1 was assessed in the presence and absence of doxycycline by Western immunoblotting, and susceptibility to NK cytotoxicity was assessed using NK92mi cells as effectors, as described for Figure 1. Mean ± SEM is shown; 1-way ANOVA with Tukey’s post hoc analysis was performed, *P < 0.05, ***P < 0.001, ****P < 0.0001. (B) H1299 cells harboring CADM1 promoter hypermethylation were cultured in the presence and absence of a pan–DNA methylase inhibitor, 5-azadeoxycytidine (5-Aza). CADM1 expression and NK cytotoxicity were assessed as described above. Mean ± SEM is shown; 2-tailed, unpaired t tests were performed, ****P < 0.0001.
CADM1 expression in primary tumor correlates with improved patient survival and decreased metastasis.
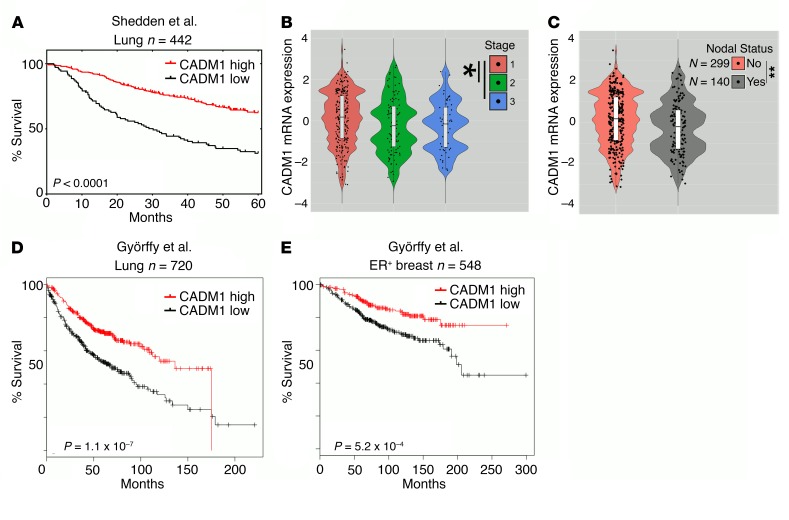
To further demonstrate clinical significance, gene expression of CADM1 was assessed in a primary human lung adenocarcinoma data set (n = 442) (41). Univariate Cox overall survival analysis revealed that increased expression of CADM1 strongly correlated with prolonged patient survival (Figure 8A). CADM1 expression inversely correlated with higher tumor stage and positive nodal status (Figure 8, B and C). We further validated the survival analysis in an independent lung adenocarcinoma data set (n = 720) (Figure 8D) (42), and also in an estrogen receptor–positive breast cancer data set (n = 548) (Figure 8E) (43), as we saw an enhanced EMT-induced NK cytotoxicity in MCF7, an estrogen receptor–positive breast cancer cell line (Figure 1F). In all cases we observed similar survival benefit with increasing CADM1 expression. Additionally, we investigated the correlation between E-cad expression and patient survival in the same cohorts and found no effect on patient outcome (Supplemental Figure 8). This observation is consistent with the modest effect of E-cad/KLRG1 inhibition (Figure 3) on NK cytotoxicity, indicating the need of an activating ligand for optimum NK activation.
Figure 8. Reduced CADM1 expression is correlated to worse patient survival and metastasis.
(A) The lung adenocarcinoma patient cohort of Shedden et al. (41) (Lung n = 442) was stratified into groups expressing low and high CADM1 and was assessed for overall survival. (B and C) The Shedden et al. data set is further classified into subgroups based on tumor stage and nodal status recorded at the time of diagnosis. Mean CADM1 expression and its distribution are depicted in the violin plots with box-and-whisker overlays in white. Two-tailed, unpaired t tests were performed, *P < 0.05; **P < 0.01. (D) The lung adenocarcinoma patient cohort of Györffy et al. (42) (Lung n = 720) was stratified into groups expressing low and high CADM1 and was assessed for overall survival. (E) The breast carcinoma patient cohort of Györffy et al. (43) [estrogen receptor–positive (ER+) breast n = 548] was stratified into groups expressing low and high CADM1 and was assessed for overall survival. (A, D, and E) Data sets shown here are Kaplan-Meier survival curves with log-rank P values comparing the groups.
Discussion
Studies thus far have focused on understanding how a small proportion of disseminating cells escape host surveillance and metastasize. Unfortunately, very little attention is paid to the understanding of the mechanisms that successfully eradicate more than 99% of tumor cells. It is now increasingly appreciated that the immune system plays an important role in the surveillance against metastasis (3–7). Since EMT is critical for metastasis, exclusive focus on evasive or resistance mechanisms that cells acquire after EMT may have promoted an unintended bias: that cells undergoing EMT must be resistant to host antitumor responses. On the contrary, it is equally feasible that metastatic cells after EMT are also vulnerable to host immunosurveillance. In other words, when cancer cells exit the immunosuppressive primary tumor microenvironment it is possible that they may pay a toll to metastasize by becoming more susceptible to host immunosurveillance. Strongly supporting the above notion, our studies show that in addition to promoting an invasive phenotype (18, 19, 44), EMT renders cancer cells more susceptible to NK-mediated killing, in vitro, by modulating activating and inhibitory ligands. This is consistent with a singular in vitro study that showed increased susceptibility of a colon cancer cell line to NK cells (45). More importantly, for the first time to our knowledge, we have demonstrated the effect of this enhanced susceptibility on tumor metastasis in vivo in multiple mouse models. In addition, we also observed this phenomenon with breast cancer cell lines, indicating the broader significance of this study. Together, this suggests that the NK cell–mediated immunosurveillance mechanism may contribute in part to the inefficiency of the metastatic process.
To date, a significant number of studies in mouse models have implicated NK cells in the control of metastasis (46) and also showed that evasion of NK-mediated immunosurveillance plays a critical role in maintaining metastatic latency (47). Supporting this, clinical studies in multiple solid tumors demonstrated an inverse correlation between the number of circulating or tumor-infiltrating NK cells and the presence of metastasis at the time of diagnosis (46). Similarly, higher expression of NK cell–activating receptors was shown to correlate with better prognosis in patients with or at risk of metastasis. However, mechanisms involved in this metastasis-specific control are unknown. Our current observations are not only consistent with the above notion but also provide a mechanistic basis by determining the activating and inhibitory axis involved. Moreover, we did not observe any effect of NK cell depletion on the growth of the primary tumors. TGF-β is known to inhibit NK cell function (48, 49), and the TGF-β–rich tumor microenvironment may explain why NK cell depletion has no effect on the primary tumor growth. However, TGF-β levels in circulation or at the site of metastasis are substantially lower than those in the primary tumor microenvironment, enabling metastasis-specific immunosurveillance. An earlier study demonstrated that EMT allows cancer cells to escape T cell immunosurveillance (23) and enables them to leave the primary tumor — whereas our data suggest that concomitant modulation of NK ligands due to EMT will make them vulnerable to NK-mediated cytotoxicity as they abandon the immunosuppressive primary tumor microenvironment, thereby allowing metastasis-specific control.
The direct interaction between KLRG1 and E-cad has been established. It is implicated in the recognition of non-MHC ligand-mediated inhibitory signaling in NK cells (32). Even though the role of this interaction has been proposed in tumor progression, to our knowledge it has never been demonstrated in a physiologically relevant context, until this study. Our data suggest that the ratio between E-cad and CADM1 expression may dictate the susceptibility of tumor cells to NK-mediated cytotoxicity. In the case of CADM1, its tumor suppressor activity has been attributed to its cell adhesion functions through homophilic interactions, mediated by its C-terminal intracellular domain and downstream membrane-associated guanylate kinases (MaGUKs) (50, 51). The role of CADM1 in the evasion of immunosurveillance has never been considered as a potential mechanism for tumor suppression. Earlier studies that demonstrated the inhibition of tumor growth by CADM1 overexpression were carried out in nude mice or mice that carry functional NK cells and overlooked the role of NK cells in the observed tumor suppression (34, 52–54). Our data clearly demonstrate that CADM1 overexpression is sufficient to make cancer cells sensitive to NK cells, in vitro. Therefore, assessment of the growth of CADM1-overexpressing cells in the presence and absence of NK cells may clarify the mechanism by which CADM1 exerts its tumor-suppressive activity.
One limitation of our study is that we still do not know at which step(s) of the metastatic cascade NK cells operate to control metastases. To address this, it is important to investigate the effect of NK cell depletion on various steps of metastatic cascade, including local invasion, intravasation, persistence of circulating tumor cells, extravasation, and survival at the distant site. Animal models that allow for the precise monitoring of each of the above steps are essential to adequately address the above question. Although NK cells were the focus of this study, it is important to investigate the role of other immune cells in the control of metastasis, either in conjunction or independent of NK cells. The role of cytotoxic T cells might be particularly important, as subpopulations as these cells also express KLRG1 and CRTAM receptors that recognize E-cad and CADM1 (55, 56).
In summary, contrasting with the prevailing notion that EMT confers only tumor-promoting functions in cancer cells, this study strongly suggests that EMT also renders cancer cells more susceptible to NK cell cytotoxicity and contributes to the inefficiency of metastasis. This novel metastasis-specific immunosurveillance mechanism presents a potential window of opportunity to prevent metastasis by boosting NK cell functions. Multiple NK-based therapeutic strategies, at both the preclinical and the clinical stage, are under development for cancer immunotherapy with promising outcomes (57, 58). Approaches that can induce or restore activating ligands or that can neutralize inhibitory ligands or receptors may boost NK cell functions. Analogous to chimeric antigen receptor T cells, these approaches can also aid in the design of “super-NK” cells for cancer immunotherapy.
Methods
Cell lines and culture conditions
Murine Lewis lung carcinoma (LLC) and 344SQ and human A549, NK92mi, H1299, H460, H358, K562, and MCF7 cells were grown in 95% air/5% CO2 at 37°C. Culture medium for LLC cells was DMEM supplemented with 10% heat-inactivated FBS, 0.3 mg/ml l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. Human cell lines were cultured according to the guidelines from the American Type Culture Collection. For EMT experiments, cells were grown in 6-well plates to 30%–40% confluence (50,000 cells per well) in complete medium. They were serum-starved for 24 hours and then treated with TGF-β (5 ng/ml) for 72 hours.
Mice
Eight-week-old male or female C57BL/6J, B6.129S7-Rag1tm1Mom/J (RAG1−/−) mice were purchased from The Jackson Laboratory and housed in specific pathogen–free conditions. All animal experiments were conducted in accordance with procedures approved by the University Committee on Use and Care of Animals and conformed to the policies and procedures of the Unit for Laboratory Animal Medicine at the University of Michigan.
Tumor models
Cancer cells were grown in their respective media. Cells were lifted using EDTA and removed with complete media. Cells were spun at 250 g in a tabletop centrifuge and washed 2 times with plain serum-free media. Cells were then resuspended in plain media at 10 × 106 cells per milliliter of media and stored on ice until injection.
Subcutaneous.
Cells were loaded into a 1-ml syringe fitted with a 26-gauge needle, and 100 μl of this cell suspension was injected subcutaneously in the right and left dorsal flanks of mice, resulting in 2 injections of 1 × 106 cells each per mouse. Tumors were measured twice weekly with manual calipers, and a modified ellipsoidal volume equation, (L × W2)/2 = volume, was used to determine tumor sizes. Mice were euthanized upon reaching a 2,500-mm3 tumor volume limit.
Experimental metastasis.
Cells were grown and lifted via EDTA, washed twice in Dulbecco’s PBS (DPBS), and resuspended at 5 × 106 cells per milliliter of DPBS. Two hundred microliters of this cell suspension was injected via lateral tail vein of mice. Mice were monitored, and after 8 weeks mice were euthanized and lungs excised for lung module counts.
In all models, lungs and primary tumors were excised and fixed with ice-cold 4% paraformaldehyde, and overt lung metastases were counted under a magnifying glass. Additionally, lungs were cryoprotected in 30% sucrose solution, embedded in OCT compound, frozen, and sectioned at 10-μm thickness on a cryostat (Leica). These sections were then mounted with ProLong Gold Antifade with DAPI (Thermo Fisher Scientific, catalog P36931) and observed on an epifluorescent microscope.
siRNA transfection
CDH1, KLRG1, NKG2D, and CADM1 siRNA molecules were purchased from Integrated DNA Technologies. A scrambled sequence from the same company was used as control. Cells at 30%–40% confluence (50,000 cells per well) were transfected with siRNA using Lipofectamine 2000 and Opti-MEM medium (Thermo Fisher Scientific). After 6 hours of transfection, cells were allowed to recover overnight from transfection in RPMI-1640 medium with 10% FBS before induction of EMT or further assessments as indicated. One million NK92mi cells were transfected in 5-ml round-bottom tubes.
Doxycycline-inducible CADM1 gene expression
CADM1 cDNA, approximately 1,362 bp, was created using the primers 5′-GGGCGGCCGCCAGGTGCCCGACATGGC-3′, NotI containing N-terminal, and 5′-AAGGAAAAAAGAATTCCAGTTGGACACCTCATTGAA-3′, EcoRI containing C-terminal, at an annealing temperature of 65°C for 35 cycles with a 40-second annealing time and 90-second extensions, using Promega GoTaq Green Master Mix (catalog M7122); primer concentrations were 1 μM each. Lentiviral pLVX-TRE3g and pLVX-TET3g plasmids (catalog 631187, Clontech) were used to create stable doxycycline-inducible cell lines. pLVX-TRE3g was cut with EcoRI-HF (catalog R3101S) and NotI-HF (catalog R3189S, New England Biolabs Inc.) restriction enzymes. These same enzymes were used to cut the PCR CADM1 product overnight and were subsequently heat-inactivated and gel-purified in 2% agarose and visualized with ethidium bromide for 1 hour at 110 V. Bands were excised and purified using Qiagen Gel Extraction Kit (catalog 28704). Vector and insert were combined at a 1:3 ratio and ligated overnight at approximately 16°C using T4 DNA ligase (catalog M0202T, New England Biolabs Inc.).
Ligation reactions were transformed into One Shot STBL3 E. coli cells (catalog C737303, Thermo Fisher Scientific) via heat shock for 42 seconds and plated on 100 μg/ml ampicillin agar plates overnight. Colonies were isolated in 20 μl LB media, and PCR was performed to initially screen for CADM1 cDNA expression. Colonies that tested positive for CADM1 expression were transferred to 5 ml LB media containing 100 μg/ml ampicillin and grown overnight. Plasmids were purified from overnight cultures with the QIAprep Spin Miniprep Kit) (catalog 27104, Qiagen).
Purified plasmids were digested with EcoRI and NotI, and gel-electrophoresed plasmids containing inserts were sequenced using MSCV reverse primer 5′-CAGCGGGGCTGCTAAAGCGCATGC-3′. Sequencing was performed at the University of Michigan Sequencing Core. Sequence-validated plasmids were then used in lentiviral production. Lentiviral particles were made at the University of Michigan Vector Core Cancer Center (Cancer Center Support Grant P30 CA046592).
CRISPR/Cas9–mediated deletion
A549 cells expressing Cas9 were purchased from Genecopiea (catalog SL504). Single-guide RNAs (sgRNAs) were also purchased from Genecopiea. CADM1 (catalog HCP206321-LvSG03-3-B) and NT (catalog CCPCTR01-LvSG03-B) bacterial stocks were plated, and a single colony was selected for plasmid growth and subsequent virus production. Lentiviral particles containing the specific targeting constructs were then used to infect A549-Cas9 cells. These new cell lines were verified by puromycin selection (1 μg/ml) and mCherry expression validated by flow cytometry.
Western blot analysis
Cells were washed with DPBS, pH 7.4, after treatment and lysed in radioimmunoprecipitation assay buffer (RIPA) containing NaF, Na3VO4, and protease inhibitor. Samples containing 10–20 μg of total protein were electrophoresed on SDS-polyacrylamide gels and transferred onto a PVDF membrane by electroblotting. Membranes were probed with the following primary antibodies: anti-CADM1 (3E1; catalog CM004-3, MBL International; 1:1,000), anti-CADM1 (polyclonal; catalog ABT6, EMD Millipore; 1:500), anti-SynCAM (polyclonal; catalog S4945, Sigma-Aldrich; 1:1,000), anti–E-cadherin (36/E-Cadherin; catalog 610182, BD Transduction Laboratories; 1:1,000), anti–N-cadherin (32/E-Cadherin; catalog 610921, BD Transduction Laboratories; 1:1,000), anti-GAPDH–HRP (GAPDH 71.1; catalog G9295, Sigma-Aldrich; 1:10,000), and anti-vimentin (13.2; catalog V5255, Sigma-Aldrich; 1:1,000). Secondary antibodies, containing HRP, included anti-mouse–HRP (polyclonal; catalog A9044, Sigma-Aldrich; 1:80,000) and anti-chicken–HRP (polyclonal; catalog 31401, Thermo Fisher Scientific; 1:10,000). See complete unedited blots in the supplemental material.
Immunodepletion
The following antibodies were administered i.p. to deplete NK cells: 25 μl per mouse of stock rabbit polyclonal anti–asialo GM1 (anti-ASGM1) (catalog 986-10001, Wako) diluted to a final volume of 100 μl in DPBS; and 200 μg per mouse of murine monoclonal anti-NK1.1 (clone PK136; catalog BP0036, BioXcell) diluted to a final volume of 100 μl in double-distilled H2O (ddH2O). One hundred microliters of undiluted normal rabbit serum (catalog 16120, Life Technologies) was used as the control for ASGM1. Two hundred micrograms murine monoclonal anti-IgG2A (clone C1.18.4; catalog BP0085, BioXcell) diluted to a final volume of 100 μl in ddH2O was used as the control for anti-NK1.1.
Flow cytometry
Cells were collected and spun at 250 g and resuspended in FACS buffer (DPBS with 1% BSA). Nonspecific antibody binding was blocked with either CD16/32 for mouse or 5% human serum for 10 minutes at room temperature. Cells were spun and washed in FACS buffer. Primary antibody incubations were performed at the following dilutions for 30 minutes on ice: CD3e (145-2C11), 1:200; NK1.1 (PK136), 1:100; CD45 (30-F11), 1:200 (BD Biosciences); KLRG1 (13F12F2), 1:100 (eBioscience). Cells were analyzed with an Attune Acoustic Flow cytometer (Applied Biosystems). Sorting was performed on a FACSAria IIIu at the University of Michigan Flow Cytometry Core. Analysis was performed with FlowJo version 10 (FlowJo LLC).
Human NK cell isolation
Human NK cells were isolated in an untouched manner using a commercially available MACS NK isolation kit (Miltenyi Biotec, catalog 130-092-657). This was performed to the manufacturer’s specifications. Briefly, donated peripheral blood samples from healthy donors were subjected to ACK red blood lysis, and white blood cells were collected by centrifugation. Cells were counted and appropriate amounts of antibody cocktail were added from the NK isolation kit.
Cytotoxicity assay
Target cells were cultured and treated with TGF-β (5 ng/ml) in 1% serum media for 3–12 days, depending on cancer cell type, as previously described. These cells were then lifted with EDTA, washed in DPBS, and labeled with CFSE at 0.5 μM concentration for 10 minutes. The staining was stopped with ice-cold complete media for 5 minutes, spun at 250 g, and washed twice. Cells that were already GFP or mCherry positive were not stained and were immediately used. These cells were counted and cocultured for 4 hours with NK92mi killer cells in complete media corresponding to the cancer cells. These reaction tubes were then placed on ice after 4 hours, and propidium iodide or Sytox Blue (Thermo Fisher Scientific, catalog S34857) was added and incubated for 5 minutes and then analyzed via flow cytometry. Cells that were labeling dye or fluorophore positive and propidium iodide or Sytox Blue positive were considered dead, and to determine the NK-specific lysis, the spontaneous death from cultures lacking NK cells was subtracted.
Immunocytochemistry/immunohistochemistry
Cells were grown on glass coverslips; after treatments, cells were fixed with ice-cold methanol for 15 minutes at –20°C. Cells were labeled with anti–E-cadherin (36/E-cadherin) conjugated with FITC (BD Transduction Laboratories) or with CADM1 (ABT66; EMD Millipore) with subsequent anti-rabbit–Alexa Fluor 488 (R37116) secondary antibody. Cells were mounted with ProLong Gold Antifade with DAPI and observed under a Nikon Eclipse Ti epifluorescent microscope. Primary tumor sections were immunolabeled with anti–E-cadherin (36/E-cadherin) conjugated with FITC (BD Transduction Laboratories) or with anti-SynCAM (S4945; Sigma-Aldrich) with subsequent anti-rabbit–Alexa Fluor 488 (R37116) secondary antibody.
Statistics
All statistical analysis was performed in GraphPad Prism 6 (GraphPad Software). Violin plots were created in R with ggplot2 package version 1.0.143 (RStudio Inc.). Heatmaps were generated with Morpheus (https://software.broadinstitute.org/morpheus/). Metastatic quantifications were analyzed with a Mann-Whitney U test. All survival analyses are Kaplan-Meier survival curves with Mantel-Cox log rank. Multiple group analyses were performed with a 1-way or 2-way ANOVA with Tukey’s post hoc analysis. All other statistics are unpaired, 2-tailed Student’s t tests. Significance is delineated by *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and mean ± SEM is shown.
Study approval
All animal experiments were approved by the IACUC of the University of Michigan and performed according to NIH guidelines. Blood samples were obtained with written and informed consent under IRBMED HUM00075841 from the University of Michigan.
Author contributions
PJC and JC performed experiments. PJC and VGK designed experiments, analyzed data, and wrote the manuscript. VGK supervised the study. DGB, GC, and TJS gave reagents, analyzed data, and reviewed the manuscript. GC helped with in silico analysis.
Supplementary Material
Acknowledgments
We thank S. Kunkel, C. Bonifant, R. Vittal, and C. Lumeng for reagents; B. Moore and M. Schaller for helpful comments and critiques; and M. Newstead for laboratory support. We thank J. Kurie (MD Anderson Cancer Center) for generously gifting the 344SQ cell line. This research is funded by NIH/National Cancer Institute grant CA132571-01 and the Elizabeth A. Crary Fund (to VGK); and by the Rackham Graduate Experimental Research Grant, the Rackham Graduate Predoctoral Fellowship, the Miller Award for Innovative Immunology Research, and NIH grant T32 AI007413 (to PJC). Research reported in this article was also supported by the NIH/National Cancer Institute (P30 CA046592) to University of Michigan Comprehensive Cancer Center.
Version 1. 01/11/2018
In-Press Preview
Version 2. 02/26/2018
Electronic publication
Version 3. 04/02/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(4):1384–1396.https://doi.org/10.1172/JCI97611.
Contributor Information
Peter J. Chockley, Email: ramshorn@umich.edu.
Jun Chen, Email: junchen@umich.edu.
Guoan Chen, Email: guoanche@umich.edu.
David G. Beer, Email: dgbeer@umich.edu.
Theodore J. Standiford, Email: tstandif@med.umich.edu.
Venkateshwar G. Keshamouni, Email: vkeshamo@umich.edu.
References
- 1.Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res. 2013;19(19):5300–5309. doi: 10.1158/1078-0432.CCR-13-0143. [DOI] [PubMed] [Google Scholar]
- 2.Romano E, Romero P. The therapeutic promise of disrupting the PD-1/PD-L1 immune checkpoint in cancer: unleashing the CD8 T cell mediated anti-tumor activity results in significant, unprecedented clinical efficacy in various solid tumors. J Immunother Cancer. 2015;3:15. doi: 10.1186/s40425-015-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slaney CY, Rautela J, Parker BS. The emerging role of immunosurveillance in dictating metastatic spread in breast cancer. Cancer Res. 2013;73(19):5852–5857. doi: 10.1158/0008-5472.CAN-13-1642. [DOI] [PubMed] [Google Scholar]
- 4.Hong H, et al. Depletion of CD4+CD25+ regulatory T cells enhances natural killer T cell-mediated anti-tumour immunity in a murine mammary breast cancer model. Clin Exp Immunol. 2010;159(1):93–99. doi: 10.1111/j.1365-2249.2009.04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olkhanud PB, et al. Breast cancer lung metastasis requires expression of chemokine receptor CCR4 and regulatory T cells. Cancer Res. 2009;69(14):5996–6004. doi: 10.1158/0008-5472.CAN-08-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paolino M, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507(7493):508–512. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidwell BN, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18(8):1224–1231. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- 9.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 10.Tarin D, Thompson EW, Newgreen DF. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005;65(14):5996–6000; discussion 6000. doi: 10.1158/0008-5472.CAN-05-0699. [DOI] [PubMed] [Google Scholar]
- 11.Zavadil J, Haley J, Kalluri R, Muthuswamy SK, Thompson E. Epithelial-mesenchymal transition. Cancer Res. 2008;68(23):9574–9577. doi: 10.1158/0008-5472.CAN-08-2316. [DOI] [PubMed] [Google Scholar]
- 12.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zavadil J, Bottinger EP. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 14.Kim WS, et al. Reduced transforming growth factor-beta type II receptor (TGF-β RII) expression in adenocarcinoma of the lung. Anticancer Res. 1999;19(1A):301–306. [PubMed] [Google Scholar]
- 15.Kong F, Jirtle RL, Huang DH, Clough RW, Anscher MS. Plasma transforming growth factor-beta1 level before radiotherapy correlates with long term outcome of patients with lung carcinoma. Cancer. 1999;86(9):1712–1719. doi: 10.1002/(SICI)1097-0142(19991101)86:9<1712::AID-CNCR12>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Keshamouni VG, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res. 2006;5(5):1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 17.Keshamouni VG, et al. Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-β-induced epithelial-mesenchymal transition. J Proteome Res. 2009;8(1):35–47. doi: 10.1021/pr8006478. [DOI] [PubMed] [Google Scholar]
- 18.Reka AK, Kuick R, Kurapati H, Standiford TJ, Omenn GS, Keshamouni VG. Identifying inhibitors of epithelial-mesenchymal transition by connectivity map-based systems approach. J Thorac Oncol. 2011;6(11):1784–1792. doi: 10.1097/JTO.0b013e31822adfb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reka AK, et al. Peroxisome proliferator-activated receptor-γ activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol Cancer Ther. 2010;9(12):3221–3232. doi: 10.1158/1535-7163.MCT-10-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reka AK, et al. Epithelial-mesenchymal transition-associated secretory phenotype predicts survival in lung cancer patients. Carcinogenesis. 2014;35(6):1292–1300. doi: 10.1093/carcin/bgu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chockley PJ, Keshamouni VG. Immunological consequences of epithelial-mesenchymal transition in tumor progression. J Immunol. 2016;197(3):691–698. doi: 10.4049/jimmunol.1600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akalay I, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73(8):2418–2427. doi: 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
- 24.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 25.Goswami MT, et al. Regulation of complement-dependent cytotoxicity by TGF-β-induced epithelial-mesenchymal transition. Oncogene. 2016;35(15):1888–1898. doi: 10.1038/onc.2015.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5(2):117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 27.West WH, Cannon GB, Kay HD, Bonnard GD, Herberman RB. Natural cytotoxic reactivity of human lymphocytes against a myeloid cell line: characterization of effector cells. J Immunol. 1977;118(1):355–361. [PubMed] [Google Scholar]
- 28.Spits H, Bernink JH, Lanier L. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol. 2016;17(7):758–764. doi: 10.1038/ni.3482. [DOI] [PubMed] [Google Scholar]
- 29.Imai K, Matsuyama S, Miyake S, Suga K, Nakachi K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. Lancet. 2000;356(9244):1795–1799. doi: 10.1016/S0140-6736(00)03231-1. [DOI] [PubMed] [Google Scholar]
- 30.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15(4):243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 32.Schwartzkopff S, et al. Tumor-associated E-cadherin mutations affect binding to the killer cell lectin-like receptor G1 in humans. J Immunol. 2007;179(2):1022–1029. doi: 10.4049/jimmunol.179.2.1022. [DOI] [PubMed] [Google Scholar]
- 33.Boles KS, Barchet W, Diacovo T, Cella M, Colonna M. The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood. 2005;106(3):779–786. doi: 10.1182/blood-2005-02-0817. [DOI] [PubMed] [Google Scholar]
- 34.Kuramochi M, et al. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27(4):427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96(9):543–552. doi: 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sartor MA, et al. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics. 2010;26(4):456–463. doi: 10.1093/bioinformatics/btp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C, et al. JNK MAP kinase activation is required for MTOC and granule polarization in NKG2D-mediated NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105(8):3017–3022. doi: 10.1073/pnas.0712310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi S, et al. Hypermethylation of the TSLC1/IGSF4 promoter is associated with tobacco smoking and a poor prognosis in primary nonsmall cell lung carcinoma. Cancer. 2006;106(8):1751–1758. doi: 10.1002/cncr.21800. [DOI] [PubMed] [Google Scholar]
- 39.Murakami Y. Functional cloning of a tumor suppressor gene, TSLC1, in human non-small cell lung cancer. Oncogene. 2002;21(45):6936–6948. doi: 10.1038/sj.onc.1205825. [DOI] [PubMed] [Google Scholar]
- 40.Heller G, et al. Expression and methylation pattern of TSLC1 cascade genes in lung carcinomas. Oncogene. 2006;25(6):959–968. doi: 10.1038/sj.onc.1209115. [DOI] [PubMed] [Google Scholar]
- 41.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Györffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 44.Keshamouni VG, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res. 2006;5(5):1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 45.López-Soto A, et al. Epithelial-mesenchymal transition induces an antitumor immune response mediated by NKG2D receptor. J Immunol. 2013;190(8):4408–4419. doi: 10.4049/jimmunol.1202950. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of metastasis by NK Cells. Cancer Cell. 2017;32(2):135–154. doi: 10.1016/j.ccell.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 47.Malladi S, et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;165(1):45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghiringhelli F, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J Exp Med. 2005;202(8):1075–1085. doi: 10.1084/jem.20051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viel S, et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal. 2016;9(415):ra19. doi: 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 50.Murakami S, Sakurai-Yageta M, Maruyama T, Murakami Y. Trans-homophilic interaction of CADM1 activates PI3K by forming a complex with MAGuK-family proteins MPP3 and Dlg. PLoS One. 2014;9(2):e82894. doi: 10.1371/journal.pone.0082894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masuda M, et al. The tumor suppressor protein TSLC1 is involved in cell-cell adhesion. J Biol Chem. 2002;277(34):31014–31019. doi: 10.1074/jbc.M203620200. [DOI] [PubMed] [Google Scholar]
- 52.Faraji F, Pang Y, Walker RC, Nieves Borges R, Yang L, Hunter KW. Cadm1 is a metastasis susceptibility gene that suppresses metastasis by modifying tumor interaction with the cell-mediated immunity. PLoS Genet. 2012;8(9):e1002926. doi: 10.1371/journal.pgen.1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seong BK, et al. A metastatic mouse model identifies genes that regulate neuroblastoma metastasis. Cancer Res. 2017;77(3):696–706. doi: 10.1158/0008-5472.CAN-16-1502. [DOI] [PubMed] [Google Scholar]
- 54.Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re-expression of TSLC1 in a non-small-cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene. 2004;23(33):5632–5642. doi: 10.1038/sj.onc.1207756. [DOI] [PubMed] [Google Scholar]
- 55.Henson SM, Akbar AN. KLRG1 — more than a marker for T cell senescence. Age (Dordr) 2009;31(4):285–291. doi: 10.1007/s11357-009-9100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leavy O. T-cell activation: polarity and CRTAM: a matter of timing. Nat Rev Immunol. 2008;8(4):246. doi: 10.1038/nri2308. [DOI] [Google Scholar]
- 57.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17(9):1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 58.Gulley JL, Repasky EA, Wood LS, Butterfield LH. Highlights of the 31st annual meeting of the Society for Immunotherapy of Cancer (SITC), 2016. J Immunother Cancer. 2017;5(1):55. doi: 10.1186/s40425-017-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.