Abstract
Bromodomain containing 4 (BRD4), a member of the bromodomain and extra-terminal family, has become a promising drug target for numerous types of cancer. BRD4 has been reported to be deregulated in gliomas; however, the precise molecular pathways regulated by BRD4 remained elusive. In the present study, BRD4 expression was silenced in the glioma cell line U251 and the results demonstrated that BRD4 knockdown attenuated cell proliferation and promoted cell apoptosis. A genome-wide analysis of BRD4-regulated transcripts in U251 cells was performed using microarray to reveal the possible molecular mechanism. A total of 3,529 differentially expressed genes were identified; 1,648 of these genes were upregulated and 1,881 were downregulated. The results of the gene ontology analysis revealed that these genes were mainly involved in membrane organization, mitotic cell cycle, cell division and DNA replication. Pathway analysis revealed that the pathways altered following BRD4 knockdown included multiple cellular processes, such as cell cycle and apoptosis. Candidate genes were identified through global signal transduction network analysis and were validated using reverse transcription-quantitative polymerase chain reaction and western blot analyses. The results demonstrated that BRD4 knockdown decreased the expression of KRAS proto-oncogene GTPase (KRAS). Downregulated KRAS expression in U251 cells restrained cell proliferation and promoted cell apoptosis, suggesting that the effect of BRD4 on glioma cells might occur through the Ras pathway. In conclusion, the present results confirmed the role of BRD4 in glioma and provided information for further exploration of the molecular mechanism of BRD4 in glioma development and progression.
Keywords: bromodomain containing 4, human glioma, proliferation, apoptosis, transcriptional analysis, KRAS proto-oncogene GTPase
Introduction
Glioma is the most common and aggressive malignant adult brain tumour with an overall 5-year survival rate of 10%. Current treatments for patients with glioma include surgical resection, radiotherapy and chemotherapy. Despite recent advances in cancer diagnosis and treatment, the survival of patients with glioma is only ~14 months (1). Therefore, developing novel treatments to increase the survival of patients with glioma remains a great challenge.
Epigenetic regulators have recently emerged as a new class of therapeutic targets in cancer treatment. Members of the bromodomain and extra-terminal (BET) family are epigenetic modulators that bind to acetylated histone H3 and H4 and influence gene regulation (2). Andrieu et al (3) firstly established a link between the expression and function of a BET protein and human cancer. These proteins have become promising drug targets for numerous cancer pathways characterized by changes in the epigenetic cell signature (4,5). Among the bromodomain proteins, bromodomain containing 4 (BRD4) has been extensively studied. Similar to other BET family members, BRD4 has two bromodomains by which it recognizes acetylated histones, followed by recruitment of Mediator, a transcription initiation cofactor, present on the promoter regions (6–8). These events lead to the phosphorylation of RNA polymerase II, which is necessary for transcriptional activation.
In addition to its role in gene regulation, BRD4 has an important role in human diseases. Several recent studies have reported BRD4 upregulation in multiple types of cancer, including melanoma, hepatocellular carcinoma, lung cancer, acute myeloid leukaemia, and glioblastoma (2,9–12). BET bromodomain inhibitors, such as JQ1 and I-BETs, effectively inhibit cell proliferation and tumour growth, mainly through the downregulation of MYC proto-oncogene bHLH transcription factor (c-MYC), a factor that regulates transcriptional pause release and is important for tumour cell viability (11,13). However, c-MYC is not always the main transcriptional target for BET proteins (14,15). This suggests the existence of additional, unidentified downstream targets of BET proteins in human cancers. Therefore, it is essential to elucidate the transcriptome and the molecular pathways controlled by BRD4 in glioma.
In the present study, BRD4 expression was silenced by using a lentivirus-mediated short hairpin RNA (shRNA) approach in the glioma cell line U251 and the impact of BRD4 knockdown on cell proliferation and apoptosis was examined. Furthermore, genome-wide screening was performed using an Affymetrix microarray to identify the gene expression profiles mediated by BRD4. The data were subjected to gene ontology (GO), pathway and global signal transduction network analyses, and the important or core differentially expressed genes regulated by BRD4 were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot assays.
Materials and methods
Patients and specimens
All specimens were obtained from patients with primary glioma during surgical operation between October 2014 and December 2016 at the Department of Neurosurgery, Qilu Hospital of Shandong University (Jinan, China). Tumour specimens were verified by pathological analysis, and pathological classification was based on the WHO classification standard. A total of 77 patients with glioma included 16 low-grade samples with a median age of 38 years (WHO grade II; age range, 21–62 years old; 10 males and 6 females), and 61 high-grade tumours with a median age of 52 years (WHO grade III and IV; age range, 15–78 years old; 40 males and 21 females). Ten samples of normal adult brain tissue were obtained from surgical resections of trauma patients with a median age of 40 years (age range, 22–62 years old; 7 males and 3 females). The study was approved by the Ethics Committee of Qilu Hospital (Jinan, China), and informed written consent was obtained from each patient.
Cell culture and transfection
The human glioma cell line U251 was obtained from the Shanghai Institutes for Biological Sciences Cell Resource Center (Shanghai, China) and maintained in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). Primary normal human astrocytes (HA) were obtained from ScienCell Research Laboratories (Carlsbad, CA, USA) and were cultured in astrocyte media (ScienCell Research Laboratories).
To knockdown BRD4 in U251 cells, two different shRNA sequences (BRD4-shRNA1 and BRD4-shRNA2) were designed, as described previously (16). shRNA lentiviruses co-expressing enhanced green fluorescent protein (EGFP) and shRNA against human BRD4 were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). Scrambled shRNA (Scr-shRNA) that targeted a non-specific sequence (5′-TTCTCCGAACGTGTCACGT-3′) was used as the control.
siRNA targeting KRAS proto-oncogene GTPase (KRAS) and the negative control siRNA (cat. nos. sc-35731 and sc-37007, respectively) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Cells were plated in 6-well plates at 1.5×105 cells per well, grown for 24 h, then transfected with 50 pmol siRNA for 6 h using RNAiMAX transfection reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Cells were harvested for analysis three days post-transfection.
Immunohistochemistry
Immunostaining was performed as previously described (17). KRAS was assessed using a rabbit polyclonal antibody (cat. no. 12063-1-AP; ProteinTech Group, Inc., Chicago, IL, USA) at a 1:200 dilution.
The sections were deparaffinised with xylene and dehydrated in a graded concentration of ethanol. Then, antigen retrieval was performed using heat treatment in a microwave oven. Tissue sections (5 µm thick) were treated with peroxidase blocking solution and incubated with primary antibodies at 4°C overnight, followed by staining with an ABC kit (Santa Cruz Biotechnology, Inc.). Hematoxylin was used as a counterstain.
The staining score was assessed using the H-score system, as described previously (18,19). Briefly, a staining intensity score of 0 to 3 was assigned for the intensity of tumour cells (0, negative; 1+, weak positive; 2+, positive; and 3+, strong positive). A proportional score is the % of positive cells over total. The H-score was calculated using the formula: H-score = ∑ (I × Pi), where I = intensity of staining and Pi = percentage of stained tumour cells, producing a score ranging from 0 to 300. Five vital tumour fields were evaluated (at ×400 magnification) and a final mean score for each tumour was calculated.
Cell proliferation assay
Cell proliferation was assessed using the cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) and the Cell-Light EdU assays (RiboBio Co., Ltd., Guangzhou, China) according to the manufacturer's instructions. Each experiment was performed in triplicate.
Flow cytometry analysis for cell apoptosis
Cells were cultured in 6-well plates and harvested when they reached ~85% confluence. The Muse Annexin V and Dead Cell kit (Millipore; Merck KGaA, Darmstadt, Germany) was used to detect cell apoptosis, following the manufacturer's protocol. The analysis of apoptosis was performed using a Millipore Muse Cell Analyzer.
RT-qPCR
Total RNA was isolated from cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) and then transcribed to cDNA using a PrimeScript II 1st Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China), according to manufacturer's instructions. qPCR was performed in technical triplicates using SYBR Green reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The thermocycling conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. This was followed by a dissociation stage of 95°C for 15 sec, 60°C for 30 sec, and 95°C for 15 sec. The primers used for qPCR are listed in Table I. Expression levels were calculated using the 2-ΔΔCq method (20), with the Cq values normalized using GAPDH as an internal control.
Table I.
Primers used in quantitative polymerase chain reaction analysis.
| Gene | Sequence (5′–3′) |
|---|---|
| KRAS | F: CAGTAGACACAAAACAGGCTCAG |
| R: TGTCGGATCTCCCTCACCAATG | |
| BRAF | F: AACGAGACCGATCCTCATCAGC |
| R: GGTAGCAGACAAACCTGTGGTTG | |
| CALM2 | F: AGTGCTGCAGAACTTCGCCATG |
| R: CAAGGTCTTCACTTTGCTGTCATC | |
| ARAF | F: CCCACATTCCAAGTCACCAGCA |
| R: CCTCCCAGTAATAGCCTGAGTC | |
| RHOA | F: TCTGTCCCAACGTGCCCATCAT |
| R: CTGCCTTCTTCAGGTTTCACCG | |
| MAPK8 (JNK1) | F: GACGCCTTATGTAGTGACTCGC |
| R: TCCTGGAAAGAGGATTTTGTGGC | |
| PLCB3 | F: GGTGAATGCCACTGAGGAGATG |
| R: CTTGGTCTCCACAAAGGACGAC | |
| MAPK10 (JNK3) | F: GTGTGGAAGTGGGAGACTCAAC |
| R: GTCAAGGACAGCATCATACGCG | |
| GNAI1 | F: AGCACTGAGTGACTACGACCTG |
| R: GGATGTATCTGTAAACCACTTGTTG | |
| ADCY6 | F: CACATAGCACCGCAGTTGGCAT |
| R: AGGCAGTGATGTCAGCAGGTGT | |
| GAPDH | F: CCGGGAAACTGTGGCGTGATGG |
| R: AGGTGGAGGAGTGGGTGTCGCTGTT |
KRAS, KRAS proto-oncogene GTPase; BRAF, B-Raf proto-oncogene serine/threonine kinase; CALM2, calmodulin 2; ARAF, A-Raf proto-oncogene serine/threonine kinase; RHOA, ras homolog family member A; MAPK, mitogen-activated protein kinase; JNK, c-Jun N-terminal kinase; PLCB3, phospholipase C β3; GNAI1, G protein subunit α i1; ADCY6, adenylate cyclase 6; F, forward; R, reverse.
Western blotting
For western blot analysis, proteins were extracted from cells using RIPA buffer containing protein inhibitors (Beyotime Institute of Biotechnology, Haimen, China). Protein concentrations were measured by the BCA protein assay (Beyotime Institute of Biotechnology) using bovine serum albumin as standard. Equal amounts (20 µg/lane) of protein samples were loaded on 12% SDS-PAGE gel for separation and then transferred to polyvinylidene fluoride membrane (Millipore; Merck KGaA). Membranes were blocked with 5% skim milk at room temperature for 2 h and then incubated with primary antibodies at 4°C overnight. Primary antibodies against the following proteins were used: BRD4 (1:1,000; cat. no. ab128874), B-Raf proto-oncogene serine/threonine kinase (BRAF; 1:1,000; cat. no. ab33899), ras homolog family member A (RHOA; 1:1,000; cat. no. ab54835), mitogen-activated protein kinase 8 (MAPK8, also known as JNK1; 1:2,000; cat. no. ab54835), MAPK10 (also known as JNK3; 1:1,000; cat. no. ab87404) (all from Abcam, Cambridge, UK), KRAS (1:1,000; cat. no. 12063-1-AP; ProteinTech) and GAPDH (1;10,000; cat. no. SAB2100894; Sigma-Aldrich; Merck KGaA). Following this, the membranes were washed and incubated with the appropriate secondary antibody (horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG; cat. nos. SA00001-1 and SA00001-2, respectively; 1:10,000; ProteinTech) for 2 h at room temperature. An enhanced chemiluminescence kit (Millipore; Merck KGaA) was used to detect the signal on the membrane. The ratios of target proteins to internal control were calculated using Tanon GIS 1D analysis software version 4.2 (Tanon, Shanghai, China).
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
TUNEL assay was performed to assess apoptosis using the In Situ Death Detection kit (Roche Diagnostics GmbH, Mannheim, Germany). Briefly, cells were fixed with paraformaldehyde, permeabilized with Triton X-100, and labelled with labelling solutions provided in the kit. Nuclei were stained with DAPI. Apoptosis was assessed by the ratio of the number of TUNEL-positive cells divided by the number of DAPI-positive cells.
Microarray analysis
Total RNA was extracted from scrambled and BRD4-shRNA U251 cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) and purified with an RNeasy mini kit (Qiagen GmbH, Hilden, Germany). cDNA was synthesized with One-Cycle Target Labeling and Control Reagents, and cRNA was created using a GeneChip IVT Labeling kit (both from Affymetrix; Thermo Fisher Scientific, Inc.). cRNA was fragmented, and then hybridized to an Affymetrix Human Clariom D Array (Affymetrix; Thermo Fisher Scientific, Inc). GeneChips were washed and stained in the Affymetrix Fluidics Station 450. All arrays were scanned using the Affymetrix GeneChip Command Console, which was installed in the GeneChip Scanner 3000 7G. The data were analysed with the Robust Multichip Analysis (RMA) algorithm using the Affymetrix default analysis settings and global scaling as a normalization method. Values presented are log2 RMA signal intensities.
The microarray data discussed in this article are publicly available at the NCBI Gene Expression Omnibus (GEO) under the accession number GSE 97791.
Data analysis
Differentially expressed genes were identified using the Student's t-test for comparison of the two groups. Genes were considered differentially regulated between the two groups when there was >2-fold difference in expression with P<0.05. Differentially expressed genes with at least a 2-fold change in either the positive or negative direction were considered up- or downregulated, respectively. Hierarchical clustering was performed on the differentially expressed genes using Cluster_Treeview software from Stanford University (Palo Alto, CA, USA) (21).
GO analysis was performed to assess the primary function of the differential expression of mRNAs regulated by BRD4. GO analysis organizes genes into hierarchical categories and uncovers the gene regulatory network based on biological process and molecular function. Fisher's exact test was performed to select the significant GO categories, and the threshold of significance was P<0.05.
Pathway analysis was used to determine the significant pathways that the differentially expressed genes participated in, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Fisher's exact test was used to select the significant pathways, and the threshold of significance was defined as P<0.05.
A gene-gene interaction network (global signal transduction network) was constructed based on the KEGG database regarding the interactions between differentially expressed genes to examine the core genes that would be predicted to have an important role in this network. Networks are displayed as graphs, where the nodes represent genes, and the lines represent relation types between the nodes, such as activation or dephosphorylation. The nodes were connected when their corresponding encoded gene products were connected directly or indirectly by a linker gene in the interaction network. The link number of one node with upstream genes, downstream genes or all binding genes is reported as the in-degree, out-degree, or degree, respectively. A higher degree indicates that the gene has a strong correlation with other genes, implying a more important role in the signalling network.
Statistical analysis
Experimental data are presented as the mean ± standard deviation of at least three experiments. Statistical analyses were performed with SPSS software (version 22.0; IBM SPSS, Armonk, NY, USA). Significant differences were assessed by Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
BRD4 is highly expressed in U251 glioma cells
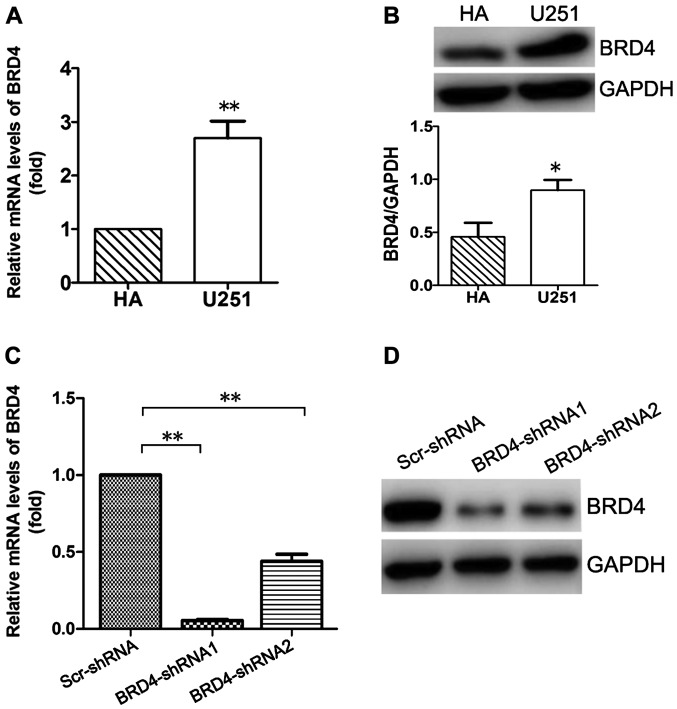
The expression of BRD4 was investigated in glioma U251 and normal HA cells by qPCR and western blotting. As illustrated in Fig. 1A, the mRNA expression levels of BRD4 were significantly higher in U251 cells compared with HA cells (P<0.01). The protein expression levels of BRD4 were consistent with the mRNA levels, as evidenced by the western blot analysis results (P<0.05; Fig. 1B).
Figure 1.
Evaluation of BRD4 expression in HA and glioma U251 cells. (A) mRNA expression levels of BRD4 in glioma U251 cells and normal HA cells. (B) Western blot analysis of the protein expression levels of BRD4 in U251 cells and normal HA cells. (C) mRNA levels and (D) protein expression levels of BRD4 were effectively inhibited by BRD4-shRNAs. GAPDH was used as an internal control. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01, with comparisons indicated by lines. BRD4, bromodomain containing 4; HA, human astrocytes; sh, short hairpin; Scr, scrambled control.
BRD4-shRNAs effectively downregulated BRD4 expression at both the mRNA and protein levels (Fig. 1C and D). Compared with BRD4-shRNA2, BRD4-shRNA1 was more effective at silencing BRD4 expression at the mRNA and protein levels (Fig. 1C and D). Therefore, the BRD4-shRNA1 was used in subsequent experiments in order to knockdown BRD4 expression in U251 cells.
Effect of BRD4 knockdown on proliferation and apoptosis of U251 cells
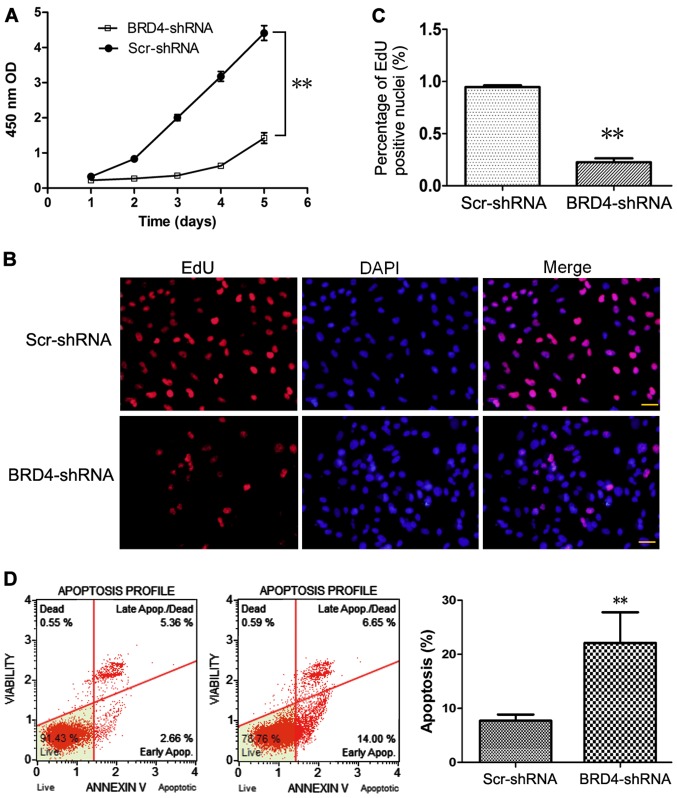
As presented in Fig. 2A, CCK-8 assays were performed at different time-points to determine the effect of BRD4 knockdown on U251 cell proliferation. Compared with control Scr-shRNA, BRD4-shRNA significantly decreased the proliferation of U251 cells (Fig. 2A), suggesting that BRD4 promoted the proliferation of glioma cells. Consistent with this result, the EdU incorporation assay indicated that the % of proliferating cells was decreased in the BRD4-shRNA group compared with the Scr-shRNA group (Fig. 2B and C).
Figure 2.
BRD4 knockdown attenuates the proliferation and promotes the apoptosis of U251 cells. (A) A Cell counting kit-8 assay was performed to detect the proliferation of U251 cells transduced with BRD4-shRNA or Scr-shRNA. Each experiment was performed in triplicate. (B) An EdU assay was performed to determine cell proliferation rates following BRD4 knockdown. Nuclei were counterstained blue with DAPI. Red indicates the cells undergoing proliferation. Scale bar, 20 µm. (C) Quantification of the EdU assay results demonstrated that U251 cell proliferation was reduced following BRD4 knockdown. (D) Representative plots and quantification of flow cytometry analysis of apoptosis following BRD4 knockdown in U251 cells. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01. BRD4, bromodomain containing 4; sh, short hairpin; Scr, scrambled control; OD, optical density.
Cell apoptosis was evaluated by flow cytometry using a Muse Annexin V Dead Cell kit
Apoptosis was detected in only 7.7±1.2% of control Scr-shRNA-transduced U251 cells, while 22.1±5.7% of cells were apoptotic following BRD4-shRNA transduction (Fig. 2D).
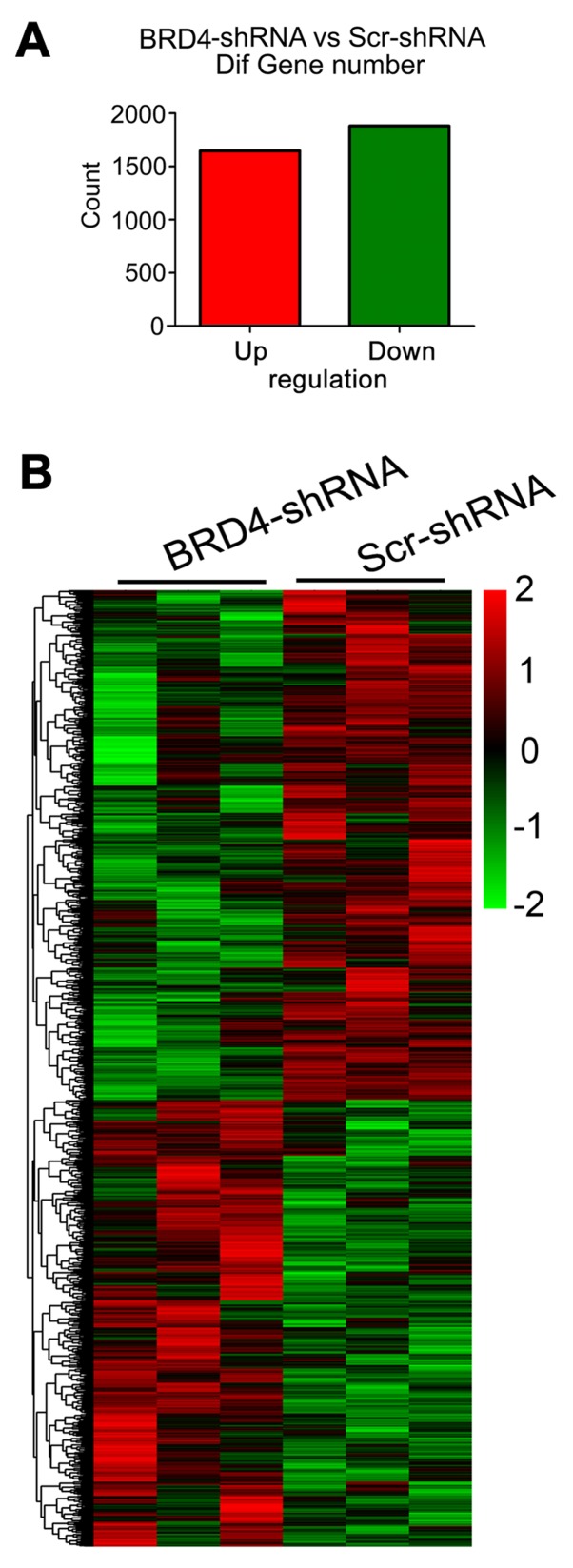
Analysis of differentially expressed mRNA between BRD4-shRNA and Scr-shRNA-transduced U251 cells
In total, 3,529 separately and differentially expressed mRNAs were identified between the BRD4-shRNA and Scr-shRNA groups by microarray analysis; 1,648 of these genes were upregulated and 1,881 were downregulated (Fig. 3A). The data regarding the differentially expressed genes between the two groups were subjected to unsupervised hierarchical clustering and TreeView analysis. The heatmap demonstrated distinguishable mRNA expression profiles between the two groups (Fig. 3B).
Figure 3.
Differentially expressed genes induced by BRD4 knockdown. (A) There were 3,529 differentially expressed genes identified in U251 cells following BRD4 knockdown compared with the control (filtering criteria, fold change >2 and P<0.05), of which 1,648 were upregulated and 1,881 were downregulated. (B) Heatmap of the 3,529 differentially expressed genes resulting from the microarray analysis. Genes and samples are listed in rows and columns, respectively. The colour scale at the top right of the heatmap represents the normalized expression data (red indicates upregulation and green indicates downregulation). BRD4, bromodomain containing 4; sh, short hairpin; Scr, scrambled control.
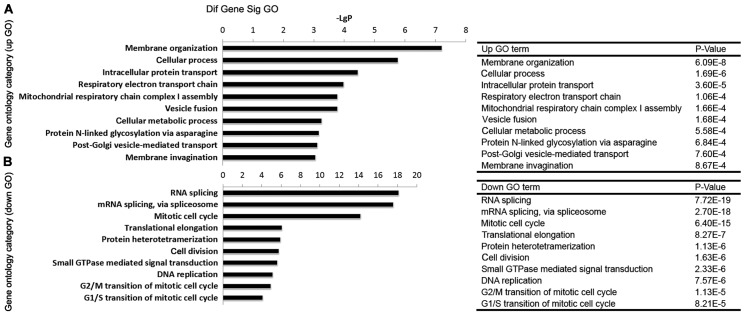
GO analysis of BRD4-regulated genes
The differentially expressed genes were subjected to GO analysis, in order to explore the main cell functions affected by BRD4. As presented in Fig. 4A, the top five GOs involving the upregulated genes (upGOs) following BRD4 knockdown were membrane organization, cellular process, intracellular protein transport, respiratory electron transport chain, and mitochondrial respiratory chain complex I assembly. The relevant GOs involving the downregulated genes (downGOs) following BRD4 knockdown included mitotic cell cycle, cell division, small GTPase-mediated signal transduction, DNA replication, G2/M transition of mitotic cell cycle, and G1/S transition of mitotic cell cycle (Fig. 4B).
Figure 4.
Significantly changed GOs induced by BRD4 knockdown in U251 cells. GO categories involving (A) upregulated genes (upGOs) and (B) downregulated genes (downGOs) are shown. GOs with a P<0.05 were identified as significantly changed. The x-axis represents the value of -LogP. The P-values of GOs are presented in the table at the right-hand side of the figure. The y-axis represents the category of GO biological processes. GO, gene ontology; BRD4, bromodomain containing 4.
Analysis of pathways affected by BRD4 knockdown
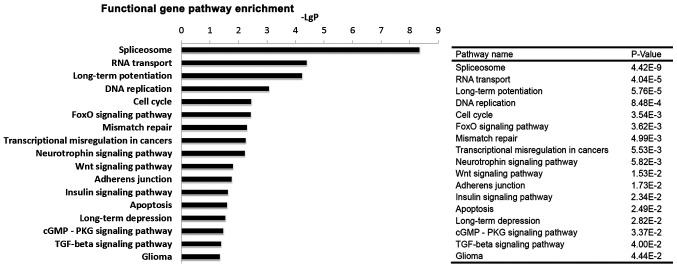
Based on the KEGG database, the pathways in which the differentially expressed genes were involved were analysed. As presented in Table II, 53 pathways were identified and classified into 19 categories, of which 8 pathways were involved in the nervous system, 6 pathways in cancer, and 3 pathways in cell growth and death. Among the 53 pathways, some important pathways, including spliceosome, DNA replication, cell cycle, the forkhead box O (FoxO) signalling pathway, transcriptional misregulation in cancer, the Wnt signalling pathway, the insulin signalling pathway, apoptosis, the transforming growth factor (TGF)-β signalling pathway, and glioma were significantly altered (Fig. 5).
Table II.
Category distribution of the 53 pathways enriched with the differentially expressed genes.
| Category | No. of pathways |
|---|---|
| Nervous system | 8 |
| Cancer | 6 |
| Endocrine system | 5 |
| Replication and repair | 5 |
| Signal transduction | 5 |
| Cell growth and death | 3 |
| Infectious diseases | 3 |
| Neurodegenerative diseases | 3 |
| Translation | 3 |
| Transport and catabolism | 3 |
| Carbohydrate metabolism | 1 |
| Cellular immunity | 1 |
| Development | 1 |
| Endocrine and metabolic diseases | 1 |
| Energy metabolism | 1 |
| Folding, sorting and degradation | 1 |
| Immune diseases | 1 |
| Substance dependence | 1 |
| Transcription | 1 |
Figure 5.
Significantly changed pathways induced by BRD4 knockdown in U251 cells. The y-axis represents the significantly modulated pathways, and the x-axis represents the value of -LogP. A larger -LgP indicates a smaller P-value. The P-values of the enriched pathways are presented in the table at the right-hand side of the figure. BRD4, bromodomain containing 4.
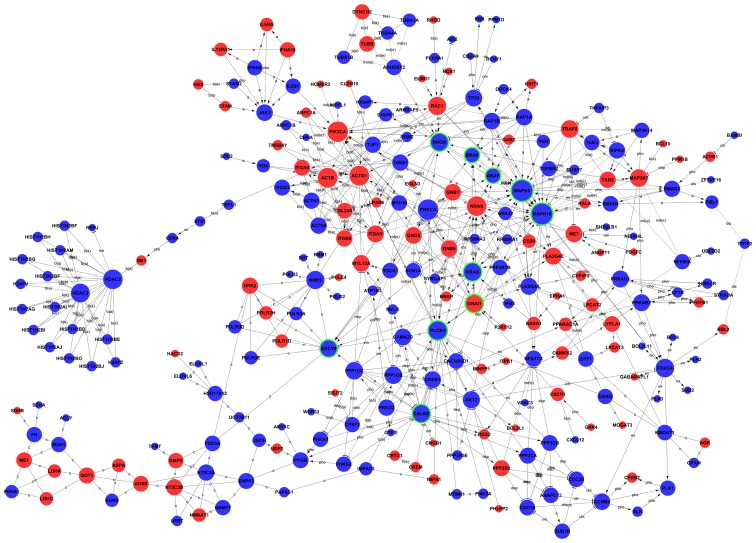
Analysis of the global signal transduction network predicts the key candidate genes regulated by BRD4
The global signal transduction network was analysed to screen for the key candidate genes regulated by BRD4 in U251 cells. As illustrated in Fig. 6, the results revealed that the high degree genes were KRAS, BRAF, calmodulin 2 (CALM2), A-Raf proto-oncogene (ARAF), RHOA, MAPK8, phospholipase C β3 (PLCB3), MAPK10, G protein subunit α i1 (GNAI1) and adenylate cyclase 6 (ADCY6). Among these ten genes, GNAI1 was upregulated, and the others were downregulated (Table III).
Figure 6.
Global signal transduction network of regulated genes following BRD4 knockdown. Red circles represent upregulated genes, and blue circles represent downregulated genes. Circles with green rings represent the genes that were selected to be experimentally validated. The area of the circle represents the degree, which is defined as the number of binding genes. A higher degree indicated that the gene had a strong correlation with other genes. The line exhibits the interaction between the genes. BRD4, bromodomain containing 4; a, activation; dep, dephosphorylation; pho, phosphorylation; ex, expression; b (a), binding-association; inh, inhibition; ind (e), indirect effect; c, compound; ubi, ubiquitination; dis, dissociation.
Table III.
Ten key genes identified by global signal transduction network analysis.
| Gene symbol | Official full name | Degree | Indegree | Outdegree | Style |
|---|---|---|---|---|---|
| KRAS | KRAS proto-oncogene GTPase | 74 | 59 | 15 | Down |
| BRAF | B-Raf proto-oncogene serine/threonine kinase | 64 | 2 | 62 | Down |
| CALM2 | Calmodulin 2 | 59 | 54 | 5 | Down |
| ARAF | A-Raf proto-oncogene serine/threonine kinase | 44 | 2 | 42 | Down |
| RHOA | Ras homolog family member A | 34 | 25 | 9 | Down |
| MAPK8 | Mitogen-activated protein kinase 8 | 33 | 8 | 25 | Down |
| PLCB3 | Phospholipase C β3 | 33 | 13 | 20 | Down |
| MAPK10 | Mitogen-activated protein kinase 10 | 33 | 7 | 26 | Down |
| GNAI1 | G protein subunit α i1 | 25 | 22 | 3 | Up |
| ADCY6 | Adenylate cyclase 6 | 19 | 7 | 12 | Down |
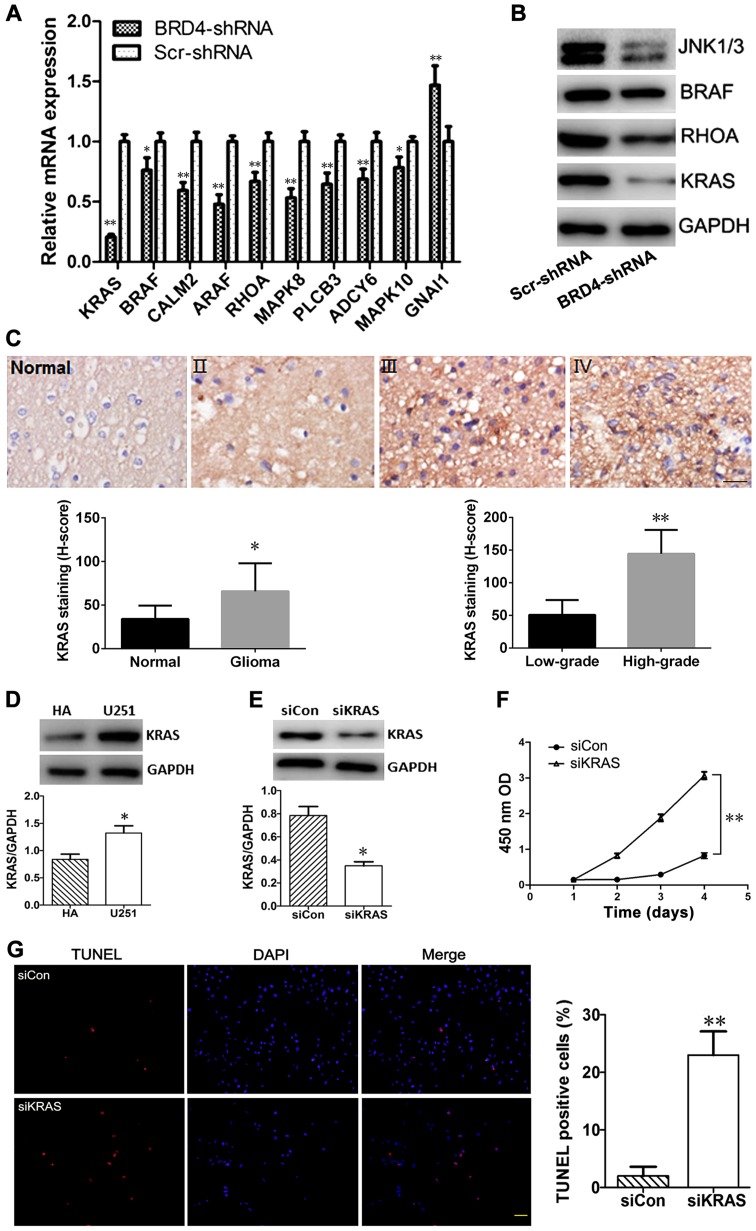
The expression levels of these ten differentially expressed mRNAs were validated by RT-qPCR. The relative change in mRNA expression as detected by RT-qPCR was consistent with the results obtained from the microarray experiment; GNAI1 was upregulated, while the rest of the genes were downregulated following BRD4 knockdown (Fig. 7A). In addition, the results of western blot analysis revealed that the protein expression levels of KRAS, BRAF, RHOA, MAPK8 (also known as JNK1) and MAPK10 (also known as JNK3) were markedly decreased in BRD4-shRNA cells compared with Scr-shRNA cells (Fig. 7B), further confirming the results from the microarray analysis.
Figure 7.
Experimental validation of microarray results. (A) Reverse transcription-quantitative polymerase chain reaction results for the mRNA expression levels of the ten key genes identified by global signal transduction network analysis. (B) Western blotting validation of the protein expression changes of key genes in the BRD4-shRNA and the Scr-shRNA groups. GAPDH was used as an internal control. (C) Representative photographs and quantification of KRAS immunostaining in normal brain tissue and glioma tissues of grades II, III and IV. Scale bar, 20 µm. (D) Western blot analysis of KRAS levels in HA and U251 cells. (E) KRAS silencing following siRNA transfection in U251 cells was confirmed by western blotting (at 72 h post-transfection). (F) A cell counting kit-8 assay was performed to detect the proliferation rates of siKRAS and or siCon-transfected U251 cells. Each experiment was performed in triplicate. (G) The apoptosis rates of siKRAS and siCon-transfected U251 cells were determined by TUNEL staining (red). Nuclei were counterstained with DAPI (blue). Scar bar, 50 µm. Experimental data are presented as the mean ± standard deviation of at least three experiments. *P<0.05 and **P<0.01. BRD4, bromodomain containing 4; sh, short hairpin; Scr, scrambled control; KRAS, KRAS proto-oncogene GTPase; HA, human astrocytes; si, small interfering; Con, control; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labelling; OD, optical density.
siRNA-mediated knockdown of KRAS suppressed the growth of U251 cells
The expression of KRAS in glioma tissues was detected by immunohistochemistry. The results demonstrated that KRAS staining was obviously stronger in glioma tissues compared with normal tissues (H-score, 65.88±32.22 and 34.13±15.34, respectively; P<0.05), and the protein expression of KRAS in the high-grade group (grade III and IV) was higher compared with the low-grade group (144.38±36.34 and 50.83±22.80, respectively; P<0.01). Representative images from the immunostaining of KRAS in glioma and normal brain tissues are presented in Fig. 7C. The results of the western blot analysis indicated that the protein expression levels of KRAS were significantly increased in U251 cells compared with HA cells (P<0.05; Fig. 7D).
KRAS-siRNA transfection effectively downregulated the protein expression of KRAS in U251 cells compared with cells transfected with control siRNA (P<0.05; Fig. 7E). Cell proliferation was measured using the CCK-8 kit. Compared with control siRNA, KRAS-siRNA inhibited the proliferation of U251 cells (P<0.01; Fig. 7F). Results from a TUNEL assay demonstrated that the % of TUNEL-positive cells was significantly higher in the KRAS-siRNA group compared with the control siRNA group (P<0.01; Fig. 7G).
Discussion
BRD4 is a therapeutic target for multiple cancers. Inhibition of the bromodomains in BRD4 by small molecules or by targeting shRNAs has been extensively evaluated and proven effective in alleviating several types of cancer (2,9–12,22–25). Therefore, it is important to study the transcriptome and molecular pathways controlled by BRD4 in cancer. In the present study, microarray analysis was performed in order to explore the molecular pathways regulated by BRD4 in glioma cells.
In the current study, BRD4 was demonstrated to be over-expressed in U251 glioma cells compared with normal HA cells. Then, BRD4 expression was silenced in U251 cells, and efficiency was confirmed by RT-qPCR and western blot analyses. BRD4 knockdown by shRNA resulted in a reduction of proliferation and induction of apoptosis in the U251 cells.
These results implied that BRD4 may have a pro-proliferation effect on glioma cells. Pastori et al (26) have reported that BET bromodomain proteins were elevated in glioblastomas and were required for glioblastoma cell proliferation; these results are similar with the present study. However, the present study focused on the molecular pathways controlled by BRD4 in glioma and further addressed BRD4-regulated mRNAs and pathways in U251 cells. Collectively, the present results confirmed the role of BRD4 in promoting glioma cell proliferation.
Using genome-wide transcriptional analysis, 3,529 differentially expressed genes were identified between BRD4-shRNA and Scr-shRNA-transduced U251 cells. Among these genes, 1,648 genes were upregulated, and 1,881 genes were downregulated, suggesting that BRD4 acts primarily as a transcriptional activator.
GO classification and pathway analysis were performed to understand the functional significance of the differentially expressed genes. GO analysis organized the differentially expressed genes into hierarchical categories according to their pertinent biological processes. BRD4 is known to bind acetylated lysine residues on histone tails and has been identified as a chromatin reader that regulates many key biological processes, including the G2/M transition, DNA replication, chromatin remodelling, and transcriptional regulation (27–31). The present data exhibited that BRD4 was involved in regulating 'gene expression', 'mitotic cell cycle', 'membrane organization', 'cell division', 'DNA replication', and 'G2/M transition of mitotic cell cycle'. These results demonstrated that the genes regulated by BRD4 in glioma had biological functions relevant to glioma growth and the maintenance of its malignant phenotype. The tight overlap between the existing literature and the present enrichment analysis highlights the strength of the data from the micro-array analysis.
Pathway analysis identified 53 pathways, and the highest enrichment was in categories related to 'nervous system', 'cancer', 'endocrine system', and 'replication and repair'. Among the 53 significant pathways, spliceosome, the most deregulated pathway following BRD4 knockdown, is critical for tumorigenesis (32–34). Other enriched pathways altered in the analysis were DNA replication, cell cycle, FoxO signalling pathway, transcriptional misregulation, Wnt signalling pathway, insulin signalling pathway, apoptosis and TGF-β signalling pathway, all of which are closely associated with cancer development (35–39). For instance, the FoxO signalling pathway contributes to the maintenance of leukemia-initiating cells in acute and chronic myeloid leukemia (35). The Wnt signalling pathway is frequently dysregulated in multiple tumour types and has critical roles in tumour progression (40–44). Emerging evidence has suggested that glioma formation and progression are associated with dysregulation of many Wnt signalling pathway components involved in the regulation of migration, invasion, and proliferation of malignant glioma cells (45–47). The pathway analysis of BRD4-regulated genes revealed the key role of BRD4 in the occurrence and development of glioma.
To screen for the key genes regulated by BRD4 in glioma development, a global signal transduction network analysis was performed by overlapping the significantly regulated GOs and pathways in the present study. KRAS, BRAF, CALM2, ARAF, RHOA, MAPK8, PLCB3, MAPK10, GNAI1 and ADCY6 were selected as candidate genes. Among the ten genes, KRAS, having the highest degrees, is one of the most widely known oncogenes. KRAS is a member of the RAS family and is reportedly involved in the progression of numerous tumours, such as pilocytic astrocytoma (48) and glioblastoma (49). The present results suggested that KRAS was upregulated in glioma compared with the normal controls, and its expression was decreased in U251 cells transduced with BRD4-shRNA. Downregulation of KRAS expression by KRAS-specific siRNA attenuated the proliferation and promoted the apoptosis of U251 cells. This effect was similar to the effect of BRD4 knockdown on U251 cells. Therefore, it can be speculated that the BRD4-mediated proliferation of glioma cells might occur through regulating KRAS and subsequently the Ras pathway. The inhibition of BRD4 by siRNA or inhibitors has exhibited potent anticancer effects in several types of cancer, including osteosarcoma, leukemia, lymphoma, lung cancer and prostate cancer (10,13,50,51). However, the precise molecular mechanisms mediating the actions of BET inhibition in glioblastoma remain elusive.
c-MYC is a well-known and important transcription factor gene directly regulated by BRD4, and inhibiting BET bromodomains effectively reduces cell proliferation and tumour growth, mainly through the downregulation of c-MYC (11,52). However, several studies have proposed that c-MYC may not be the only transcriptional target of BET proteins and demonstrated that other genes undergoing expressional changes simultaneously contributed to the decrease of cell proliferation (10,15,22). Cheng et al (14) reported that downregulation of c-MYC upon BRD4 inhibition was rather modest and appeared to only serve a minor role in mediating the effects of JQ1 in glioblastoma of different genetic background. This indicates the existence of additional, unidentified targets of BRD4 in glioma. The main purpose of the present study was to screen for novel potential targets of BRD4 in glioma through microarray analysis. Consistent with the observations of Cheng et al, the present microarray results demonstrated that the mRNA expression of c-MYC was not significantly changed in the BRD4-shRNA group compared with the control group (expression levels, 6.14 vs. 6.93, respectively; P=0.08), and c-MYC was not in a core position of the global gene-gene interaction network. These results indicated that the effects of BRD4 in carcinogenesis are mediated by complex mechanisms including, but not limited, to MYC-mediated transcriptional regulation.
The discovery that KRAS might be a downstream mediator of BRD4 in glioma will help us better understand the potential activity of BRD4 inhibitors and the central BET-dependent transcriptional program. In addition, the ability of BRD4 inhibition to reduce expression of KRAS highlights the potential of this novel therapeutic strategy to improve treatment for cancers that require KRAS activity. However, it remains to be determined whether BRD4 activates KRAS directly or indirectly and whether the effect of BRD4 inhibition in glioma is KRAS-dependent. Although several BRD4 chromatin immunoprecipitation (ChIP)-sequencing data have been previously published (accession nos. GSM2635249, GSM823382 and GSM823378), those ChIP assays were performed in K562 cells (53), human embryonic stem cells (54), and human CD4+ T cells (54), respectively. The target genes of BRD4 might be different in different tissues. For example, FoxM1 is a target of BRD4 in ovarian cancer (15), and interleukin 7 receptor is a target of BRD4 in lymphoblastic leukemia (10). Therefore, ChIP experiments will be necessary to determine whether KRAS is a direct target of BRD4 in glioma in further studies.
In conclusion, a comprehensive analysis of the gene expression profiles of U251 cells with or without BRD4 knockdown was conducted. The strong overlap between the existing literature and the present enrichment analysis highlights the robustness of the results obtained in the current study. This integrated analysis provides information for further research on the molecular mechanisms of BRD4 in glioma tumorigenesis.
Acknowledgments
We thank Dr Jing Li (Cnkingbio Company, Ltd., Beijing, China) for technical support.
Footnotes
Funding
This study was supported by the Natural Science Foundation of Shandong Province (grant no. ZR2013HQ057), the Medical Science and Technology Development Plan of Shandong Province (grant nos. 2014WS0147 and 2014WS0416), the Shandong Province outstanding Young Scientists Research Award Fund (grant no. BS2015YY026), the China Postdoctoral Science Foundation (grant nos. 2015M572055, 2016M590642 and 2016M600543), and the Shandong Postdoctoral Innovation Fund (grant no. 20160213).
Author's contributions
ZD performed experiments, contributed to analysis and interpretation of the data, and was a major contributor in writing the manuscript. XS and FY performed experiments, and contributed in drafting and revising the manuscript. JW collected the clinical data, helped to perform the cell experiments, and contributed in the revision of the manuscript. YZ and SL contributed to the conception and design of the study, the data analysis and interpretation, and the writing and revision of the manuscript, as well as were responsible for acquisition of funding. All authors read and approved the final manuscript.
Availability of data and materials
The microarray data discussed in this article are publicly available at the NCBI Gene Expression Omnibus (GEO) under the accession number GSE 97791.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Qilu Hospital (Jinan, China), and informed written consent was obtained from each patient.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrieu G, Tran AH, Strissel KJ, Denis GV. BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 2016;76:6555–6567. doi: 10.1158/0008-5472.CAN-16-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papavassiliou KA, Papavassiliou AG. Bromodomains: Pockets with therapeutic potential. Trends Mol Med. 2014;20:477–478. doi: 10.1016/j.molmed.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 6.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 7.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 8.Zeng L, Zhou MM. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/S0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 9.Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, González-Gomez P, Morante M, Jubierre L, Zhang W, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73:6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Ma P, Jing Y, Yan Y, Cai MC, Zhang M, Zhang S, Peng H, Ji ZL, Di W, et al. BET Bromodomain inhibition as a therapeutic strategy in ovarian cancer by downregulating FoxM1. Theranostics. 2016;6:219–230. doi: 10.7150/thno.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao YF, Wu YB, Long X, Zhu SQ, Jin C, Xu JJ, Ding JY. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget. 2016;7:9491–9500. doi: 10.18632/oncotarget.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling EA, Hao A. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–733. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- 18.Azim HA, Jr, Peccatori FA, Brohée S, Branstetter D, Loi S, Viale G, Piccart M, Dougall WC, Pruneri G, Sotiriou C. RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res. 2015;17:24. doi: 10.1186/s13058-015-0538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo W, Chan SL, Mo FK, Chu CM, Hui JW, Tong JH, Chan AW, Koh J, Hui EP, Loong H, et al. Phase I/II study of temsirolimus for patients with unresectable Hepatocellular Carcinoma (HCC)-a correlative study to explore potential biomarkers for response. BMC Cancer. 2015;15:395. doi: 10.1186/s12885-015-1334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Saldanha AJ. Java Treeview - extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 22.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, Bolin S, Schumacher SE, Zeid R, Masoud S, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastori C, Daniel M, Penas C, Volmar CH, Johnstone AL, Brothers SP, Graham RM, Allen B, Sarkaria JN, Komotar RJ, et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics. 2014;9:611–620. doi: 10.4161/epi.27906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson MA, Kouzarides T, Huntly BJP. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 28.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/MCB.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn CN, Venugopal P, Scott HS, Hiwase DK. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol Rev. 2015;263:257–278. doi: 10.1111/imr.12241. [DOI] [PubMed] [Google Scholar]
- 34.Wojtuszkiewicz A, Assaraf YG, Maas MJ, Kaspers GJ, Jansen G, Cloos J. Pre-mRNA splicing in cancer: The relevance in oncogenesis, treatment and drug resistance. Expert Opin Drug Metab Toxicol. 2015;11:673–689. doi: 10.1517/17425255.2015.993316. [DOI] [PubMed] [Google Scholar]
- 35.Coomans de Brachène A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73:1159–1172. doi: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powathil GG, Gordon KE, Hill LA, Chaplain MA. Modelling the effects of cell-cycle heterogeneity on the response of a solid tumour to chemotherapy: Biological insights from a hybrid multiscale cellular automaton model. J Theor Biol. 2012;308:1–19. doi: 10.1016/j.jtbi.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Hallett RM, Huang C, Motazedian A, Auf der Mauer S, Pond GR, Hassell JA, Nordon RE, Draper JS. Treatment-induced cell cycle kinetics dictate tumor response to chemotherapy. Oncotarget. 2015;6:7040–7052. doi: 10.18632/oncotarget.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koval A, Ahmed K, Katanaev VL. Inhibition of Wnt signalling and breast tumour growth by the multi-purpose drug suramin through suppression of heterotrimeric G proteins and Wnt endocytosis. Biochem J. 2016;473:371–381. doi: 10.1042/BJ20150913. [DOI] [PubMed] [Google Scholar]
- 39.Leek RD, Stratford I, Harris AL. The role of hypoxia-inducible factor-1 in three-dimensional tumor growth, apoptosis, and regulation by the insulin-signaling pathway. Cancer Res. 2005;65:4147–4152. doi: 10.1158/0008-5472.CAN-04-2184. [DOI] [PubMed] [Google Scholar]
- 40.Sawa M, Masuda M, Yamada T. Targeting the Wnt signaling pathway in colorectal cancer. Expert Opin Ther Targets. 2016;20:419–429. doi: 10.1517/14728222.2016.1098619. [DOI] [PubMed] [Google Scholar]
- 41.Ramos-Solano M, Álvarez-Zavala M, García-Castro B, Jave-Suárez LF, Aguilar-Lemarroy A. Wnt signalling pathway and cervical cancer. Rev Med Inst Mex Seguro Soc. 2015;53(Suppl 2):S218–S224. In Spanish. [PubMed] [Google Scholar]
- 42.Yong X, Tang B, Xiao YF, Xie R, Qin Y, Luo G, Hu CJ, Dong H, Yang SM. Helicobacter pylori upregulates Nanog and Oct4 via Wnt/β-catenin signaling pathway to promote cancer stem cell-like properties in human gastric cancer. Cancer Lett. 2016;374:292–303. doi: 10.1016/j.canlet.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 43.Yang L, Tang H, Kong Y, Xie X, Chen J, Song C, Liu X, Ye F, Li N, Wang N, et al. LGR5 Promotes Breast Cancer Progression and Maintains Stem-Like Cells Through Activation of Wnt/β-Catenin Signaling. Stem Cells. 2015;33:2913–2924. doi: 10.1002/stem.2083. [DOI] [PubMed] [Google Scholar]
- 44.Zhi X, Lin L, Yang S, Bhuvaneshwar K, Wang H, Gusev Y, Lee MH, Kallakury B, Shivapurkar N, Cahn K, et al. βII-Spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology. 2015;61:598–612. doi: 10.1002/hep.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, Xue J, Liu M, Wang Y, et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al. LAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang X, Yu Y, Yang HW, Agar NY, Frado L, Johnson MD. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J Biol Chem. 2010;285:8472–8480. doi: 10.1074/jbc.M109.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryu MJ, Liu Y, Zhong X, Du J, Peterson N, Kong G, Li H, Wang J, Salamat S, Chang Q, et al. Oncogenic Kras expression in post-mitotic neurons leads to S100A8-S100A9 protein overexpression and gliosis. J Biol Chem. 2012;287:22948–22958. doi: 10.1074/jbc.M112.357772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milinkovic VP, Skender Gazibara MK, Manojlovic Gacic EM, Gazibara TM, Tanic NT. The impact of TP53 and RAS mutations on cerebellar glioblastomas. Exp Mol Pathol. 2014;97:202–207. doi: 10.1016/j.yexmp.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Lee DH, Qi J, Bradner JE, Said JW, Doan NB, Forscher C, Yang H, Koeffler HP. Synergistic effect of JQ1 and rapamycin for treatment of human osteosarcoma. Int J Cancer. 2015;136:2055–2064. doi: 10.1002/ijc.29269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., III Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170:1028–1043.e19. doi: 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]