Abstract
Obesity is a major risk factor for insulin resistance and type 2 diabetes. In adipose tissue, obesity-mediated insulin resistance correlates with the accumulation of proinflammatory macrophages and inflammation. However, the causal relationship of these events is unclear. Here, we report that obesity-induced insulin resistance in mice precedes macrophage accumulation and inflammation in adipose tissue. Using a mouse model that combines genetically induced, adipose-specific insulin resistance (mTORC2-knockout) and diet-induced obesity, we found that insulin resistance causes local accumulation of proinflammatory macrophages. Mechanistically, insulin resistance in adipocytes results in production of the chemokine monocyte chemoattractant protein 1 (MCP1), which recruits monocytes and activates proinflammatory macrophages. Finally, insulin resistance (high homeostatic model assessment of insulin resistance [HOMA-IR]) correlated with reduced insulin/mTORC2 signaling and elevated MCP1 production in visceral adipose tissue from obese human subjects. Our findings suggest that insulin resistance in adipose tissue leads to inflammation rather than vice versa.
Keywords: Inflammation, Metabolism
Keywords: Adipose tissue, Macrophages, Obesity
Introduction
Obesity-induced insulin resistance is a major risk factor for type 2 diabetes, hyperlipidemia, cardiovascular disease, and some types of cancer (1–3). Although the mechanism by which obesity causes insulin resistance is unclear, inflammation has been linked to the development of local and systemic insulin resistance, especially when the inflammation occurs in white adipose tissue (WAT) (4, 5).
WAT consists of adipocytes and stromal vascular cells (SVCs) including endothelial cells, preadipocytes, and immune cells (6). Among the immune cells, macrophages play a particularly important role in obesity-induced adipose tissue inflammation. Macrophages are classified into 2 types: proinflammatory M1 and antiinflammatory M2 macrophages. M1 macrophages express the surface marker CD11c (7, 8) and produce proinflammatory cytokines such as TNF-α (9, 10). M2 macrophages express the surface marker CD301 and produce antiinflammatory cytokines such as IL-10 (7). The relative and absolute number of M1 macrophages increases in WAT upon obesity, thereby promoting adipose tissue inflammation (7–11). Studies in rodents have suggested that adipose tissue inflammation causes local and systemic insulin resistance (9, 11–13). However, it has been demonstrated that immunocompromised mice are not protected from systemic insulin resistance induced by a short-term high-fat diet (HFD) (14). Furthermore, Tian et al. have shown that adipose tissue inflammation is dispensable for local and systemic insulin resistance (15). Another study has shown that inhibition of adipose tissue inflammation results in glucose intolerance, suggesting that inflammation may even be a mechanism to counter insulin resistance (16). In humans, expression of the macrophage markers CD68 and TNF-α in WAT correlates with BMI, suggesting that obesity may induce the accumulation of adipose tissue macrophages and inflammation in humans (9, 17). However, clinical trials targeting TNF-α have shown little or no beneficial effect on systemic insulin sensitivity (18–21). Thus, the causal relationship between adipose tissue inflammation and insulin resistance is unclear.
Two models have been proposed to explain the increase in the number of M1 macrophages in WAT upon obesity. The first is that circulating monocytes are recruited to WAT, where they differentiate into M1 macrophages (7, 22). The second is that obesity induces the proliferation of resident macrophages in WAT (23). The monocyte chemoattractant protein 1/C-C chemokine ligand 2 (MCP1/CCL2), presumably produced by adipocytes, is required to increase the number of macrophages in WAT (12, 23, 24). However, the mechanism(s) controlling MCP1 expression in adipocytes upon obesity are poorly understood.
The target of rapamycin complex 2 (TORC2) is an evolutionarily conserved serine/threonine protein kinase complex that controls growth and metabolism (reviewed in ref. 25). In mammals, mTORC2 consists of mTOR, rapamycin-insensitive companion of mTOR (RICTOR), mammalian stress-activated protein kinase–interacting protein 1 (mSIN1), and mammalian lethal with SEC thirteen 8 (mLST8) (26–31). Insulin stimulates mTORC2 to promote glucose uptake in adipose tissue (32–34), liver (35–37), and skeletal muscle (38, 39). Previously, we and others have shown that adipose-specific Rictor knockout (AdRiKO) exacerbates obesity-related complications in mice, such as systemic insulin resistance and hepatic steatosis (32–34).
Here, we used mTORC2-deficient and therefore insulin-resistant AdRiKO mice to investigate the causal relationship between insulin resistance and inflammation. We found that inhibition of the insulin/mTORC2 pathway resulted in enhanced Mcp1 transcription in mouse and human adipocytes and thereby promoted inflammation in visceral WAT. Furthermore, obesity-induced insulin resistance developed before the accumulation of proinflammatory M1 macrophages in visceral WAT of WT mice. Thus, insulin resistance precedes and causes inflammation in adipose tissue.
Results
Insulin/mTORC2 signaling in WAT negatively controls inflammation.
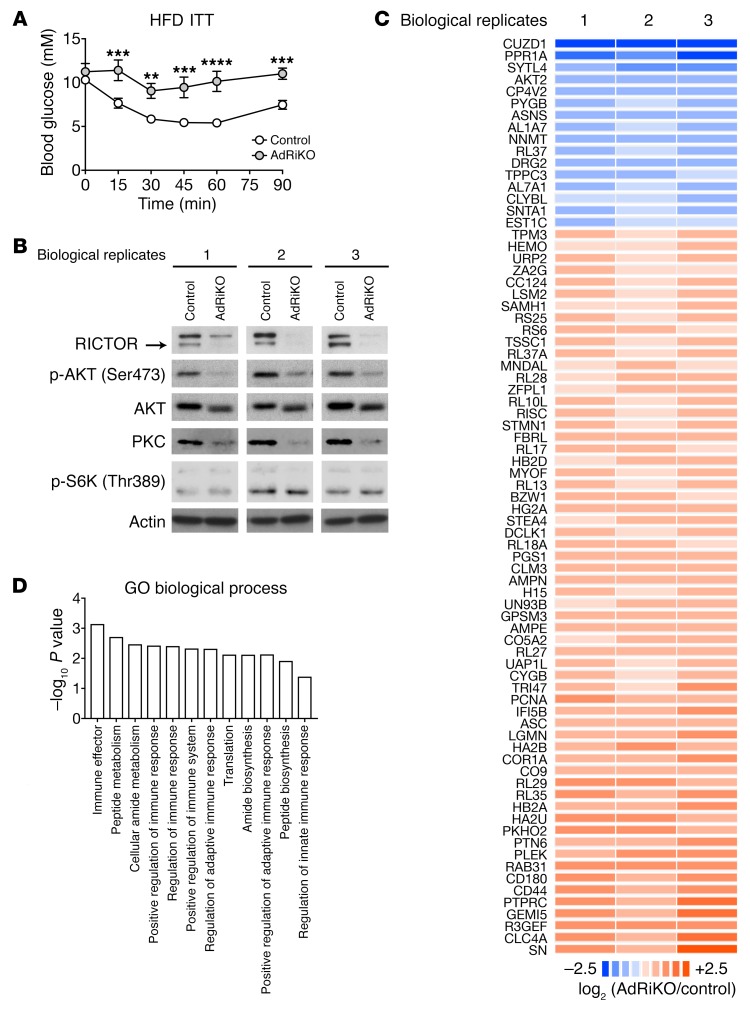
AdRiKO exacerbates systemic insulin resistance upon obesity, as evidenced by impaired glucose clearance in response to insulin treatment (Figure 1A and refs. 32–34). Thus, the AdRiKO mouse is a good model to investigate the causal relationship between insulin resistance and inflammation upon obesity. To determine whether insulin resistance due to impaired insulin/mTORC2 signaling in WAT affects adipose inflammation, we performed quantitative proteomics on epididymal WAT (eWAT) from HFD-fed (for 10 weeks) AdRiKO (aP2-Cre, Rictorfl/fl) and control (Rictorfl/fl) mice. As previously reported (32–34), HFD-fed AdRiKO mice had an increased body size with no change in adiposity (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI96139DS1). Furthermore, we confirmed that RICTOR expression, AKT (Ser473) phosphorylation, and PKC expression, readouts for mTORC2 signaling, were decreased, while S6K (Thr389) phosphorylation, a readout for mTORC1 signaling, was not affected in eWAT (Figure 1B; see complete unedited blots in the supplemental material). Among approximately 3,000 proteins identified in the proteome, 61 and 16 were up- and downregulated, respectively, in AdRiKO mice compared with controls (Figure 1C and Supplemental Table 1). Gene Ontology (GO) analysis of the regulated proteins revealed enrichment of immune response–related biological processes (Figure 1D), suggesting that insulin resistance due to loss of mTORC2 signaling may cause inflammation upon obesity.
Figure 1. Quantitative proteome analysis reveals insulin/mTORC2 signaling functions in adipose tissue inflammation.
(A) ITT for AdRiKO and control mice fed a HFD for 10 weeks. Mice were fasted for 5 hours prior to the ITT. Data are presented as the mean ± SEM. **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-way ANOVA. n = 10 (control) and n = 5 (AdRiKO). (B) Immunoblots of eWAT from HFD-fed AdRiKO and control mice. eWAT samples were collected from ad libitum–fed mice. The same lysates were used for proteome analysis. (C) Regulated proteome with 3 biological replicates. See also Supplemental Table 1. (D) GO term analysis of the regulated proteome. Data are presented as the mean ± SEM.
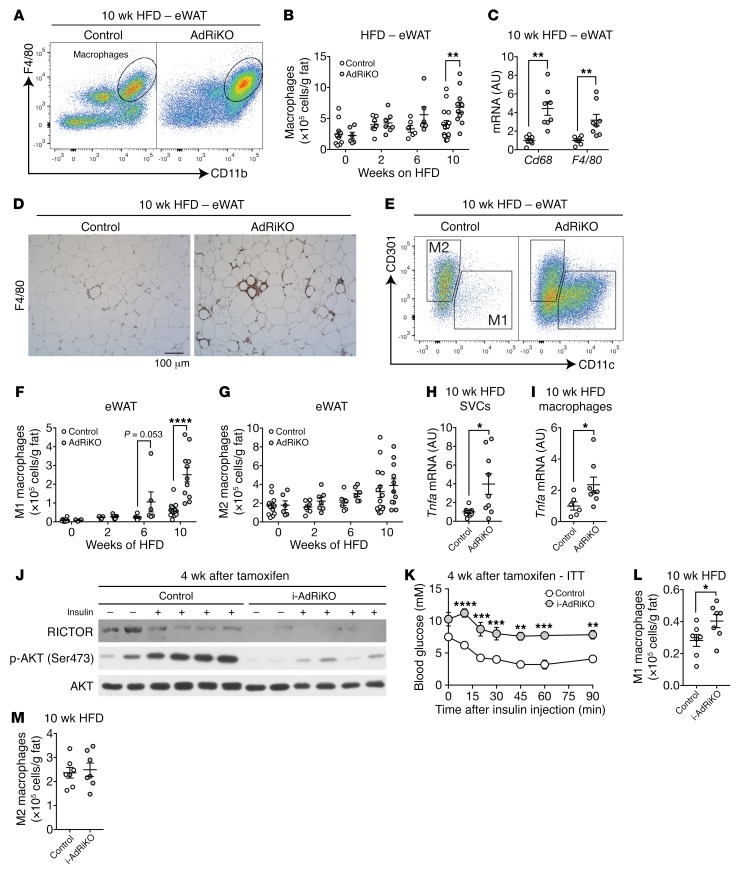
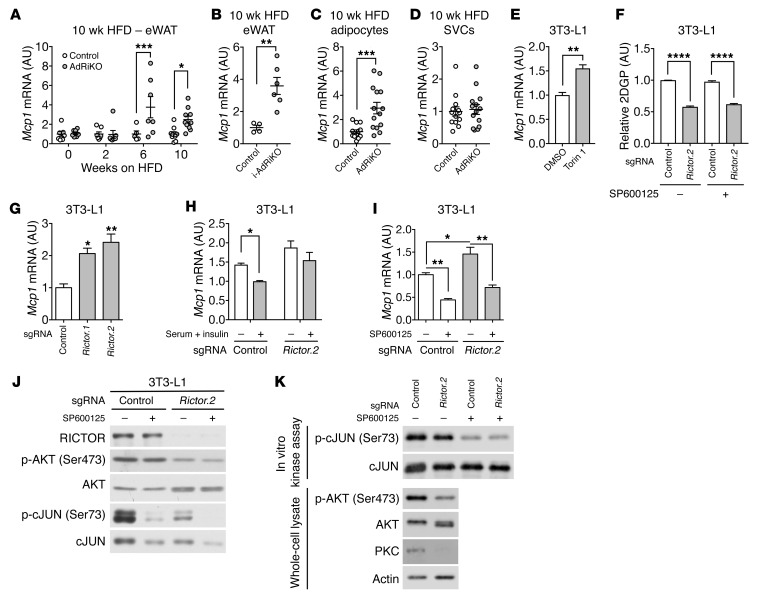
To examine further whether mTORC2 in WAT controls inflammation, we quantified immune cells in eWAT of HFD-fed AdRiKO and control mice by flow cytometry. While the numbers of B and T cells did not differ (Supplemental Figure 1D), the number of macrophages (F4/80+CD11b+) increased in AdRiKO eWAT at 10 weeks of HFD feeding (Figure 2, A and B, and Supplemental Figure 2A). The increase in macrophages correlated with increased macrophage gene expression (Cd68 and F4/80) and F4/80 staining (Figure 2, C and D). Next, we determined whether the increase in the number of macrophages in AdRiKO eWAT was due to an increase in proinflammatory M1 (F4/80+CD11b+CD11c+) and/or antiinflammatory M2 (F4/80+CD11b+CD301+) macrophages. The numbers of both M1 and M2 macrophages increased in both AdRiKO and control mice during the HFD time course (Figure 2, E–G). AdRiKO eWAT showed a disproportionately large increase in M1 macrophages, starting at 6 weeks of HFD feeding (Figure 2, E and F). Quantification of proinflammatory cytokine TNF-α mRNA in SVCs and macrophages isolated from HFD-fed AdRiKO and control mice confirmed the disproportionate increase in M1 macrophages in AdRiKO eWAT (Figure 2, H and I, and Supplemental Figure 2, B and C). These observations indicate that AdRiKO leads to the accumulation of M1 macrophages, confirming that genetically induced insulin resistance due to loss of mTORC2 signaling in WAT promotes inflammation. We note that there was no difference in macrophage numbers between AdRiKO and control mice on a normal diet (ND) (Supplemental Figure 3, A–E), indicating that AdRiKO potentiates inflammation only in response to obesity.
Figure 2. AdRiKO eWAT accumulates M1 macrophages.
(A and B) Numbers of macrophages (CD45+F4/80+CD11b+) in SVCs isolated from eWAT of HFD-fed AdRiKO and control mice. Representative FACS profiles are shown in A, and quantification is shown in B. **P < 0.01, by multiple Student’s t test. n = 6–15. (C) Gene expression of macrophage markers in eWAT from HFD-fed AdRiKO and control mice. **P < 0.01, by multiple Student’s t test. n = 7–8. (D) Representative F4/80 staining of eWAT from HFD-fed AdRiKO and control mice. n = 4. Scale bar: 100 μm. (E–G) Numbers of M1 macrophages (CD45+F4/80+CD11b+CD11c+) and M2 macrophages (CD45+F4/80+CD11b+CD301+) in SVCs from eWAT of HFD-fed AdRiKO and control mice. Representative FACS profiles are shown in E, and quantification is shown in F and G. ****P < 0.0001 and P = 0.053, by multiple Student’s t test. n = 6–15. (H and I) Tnfa gene expression in SVCs (H) (n = 9) and isolated macrophages (I) (n = 6–8) from eWAT of HFD-fed AdRiKO and control mice. *P < 0.05, by unpaired Student’s t test. (J) Immunoblots of eWAT from i-AdRiKO and control mice. Mice were treated with tamoxifen for 5 days. After 4 weeks, mice were fasted for 5 hours and then treated with PBS or insulin. (K) ITT for i-AdRiKO and control mice 4 weeks after induction of Rictor knockout. Mice were fasted for 5 hours prior to the ITT. **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-way ANOVA. n = 5 (control) and n = 6 (i-AdRiKO). (L and M) Numbers of M1 macrophages (CD45+F4/80+CD11b+CD11c+) (L) and M2 macrophages (CD45+F4/80+CD11b+CD301+) (M) in SVCs from eWAT of HFD-fed i-AdRiKO and control mice. *P < 0.05, by unpaired Student’s t test. n = 7. Data are presented as the mean ± SEM.
Our AdRiKO model relies on the adipose-specific promoter aP2 to drive Cre expression and thereby knock out Rictor. However, aP2-Cre can be expressed in cell types other than adipocytes including macrophages (40, 41). Three lines of evidence suggest that our findings are not due to confounding effects of ectopic knockout of Rictor in macrophages. First, Rictor expression was unchanged in macrophages isolated from the HFD-fed AdRiKO mice compared with those from control mice (Supplemental Figure 4, A and B). Second, adipose-specific deletion of Rictor via expression of Cre from the Adipoq promoter (i-AdRiKO: Adipoq-CreERT2, Rictorfl/fl) (40, 42) also showed an increase in M1 macrophages in eWAT upon obesity, as observed in aP2-Cre AdRiKO mice (Figure 2, J–M, and Supplemental Figure 4, C and D). Third, macrophage-specific knockout of Rictor, due to expression of Cre from the LysM promoter (LysM-Cre Rictorfl/fl), had no effect on macrophages in WAT of mice fed a HFD (43).
HFD-induced insulin resistance precedes the accumulation of adipose tissue M1 macrophages.
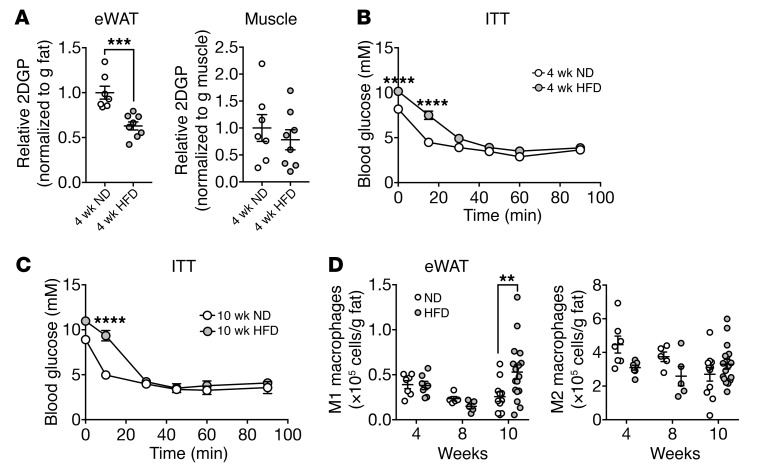
Our findings indicate that genetically induced local insulin resistance causes the accumulation of M1 macrophages and thus inflammation in WAT upon obesity. This predicts that HFD-induced insulin resistance precedes inflammation in WT mice. To test this prediction, we performed a longitudinal study with HFD-fed WT mice (Supplemental Figure 5A). WT mice developed eWAT and systemic insulin resistance by week 4 of HFD feeding, as measured by insulin-stimulated glucose uptake and an insulin tolerance test (ITT) (Figure 3, A and B, and Supplemental Figure 5B), respectively. Mice fed a HFD for 10 weeks remained insulin resistant compared with mice on a ND (Figure 3C and Supplemental Figure 5C). The M1 macrophage population in eWAT mildly increased in mice by week 10 of a HFD, but not by 4 or 8 weeks (Figure 3D). Tnfa expression did not increase in mice at 4 or 10 weeks of a HFD (Supplemental Figure 5D). These findings are consistent with previous reports showing that mice develop adipose and systemic insulin resistance within several days to 4 weeks of a HFD (44, 45), whereas the number of M1 macrophages in WAT increases only within 8 to 10 weeks of a HFD (10, 15, 45, 46). The finding that HFD-induced insulin resistance precedes the accumulation of M1 macrophages is consistent with our above conclusion that insulin resistance leads to inflammation.
Figure 3. HFD-induced insulin resistance precedes the accumulation of M1 macrophages.
(A) Insulin-stimulated 2DGP accumulation in eWAT and muscle from WT mice fed a ND or HFD for 4 weeks. Mice were fasted for 5 hours, injected with insulin at 0 minutes and 2DG at 10 minutes, and sacrificed at 30 minutes. ***P < 0.001, by unpaired Student’s t test. n = 7–8. (B and C) ITT for WT mice fed a ND or HFD for 4 weeks (B) or 10 weeks (C). Mice were fasted for 5 hours prior to the ITT. ****P < 0.0001, by 2-way ANOVA. n = 15 (4 wk ND), n = 17 (4 wk HFD), n = 3 (10 wk ND), and n = 4 (10 wk HFD). (D) Numbers of M1 macrophages (CD45+F4/80+CD11b+CD11c+) and M2 macrophages (CD45+F4/80+CD11b+CD301+) in eWAT of WT mice fed a HFD for 4, 8, or 10 weeks. **P < 0.01, by multiple Student’s t test. n = 5–17. Rictorfl/fl mice were used as WT controls. Data are presented as the mean ± SEM.
Insulin resistance–induced inflammation is specific to visceral WAT.
Adipose-specific loss of mTORC2 signaling directly causes insulin resistance in all WAT depots and indirectly leads to systemic insulin resistance (32–34). To test whether AdRiKO causes inflammation in liver or in fat depots other than eWAT (see above), we examined macrophage numbers in peri-renal WAT (prWAT), subcutaneous WAT (sWAT), and liver of HFD-fed AdRiKO and control mice. AdRiKO prWAT, but not sWAT or liver, had increased numbers of M1 macrophages, (Supplemental Figure 6, A–C). Thus, AdRiKO promotes inflammation specifically in visceral WAT (eWAT and prWAT).
To test whether local insulin resistance causes inflammation in the liver, we examined hepatic macrophages in liver-specific Rictor-knockout mice (LiRiKO: Alb-Cre, Rictorfl/fl), which have hepatic insulin resistance due to loss of insulin/mTORC2 signaling in liver (35–37). HFD-fed LiRiKO mice had a moderate but nonsignificant increase in the number of hepatic macrophages compared with HFD-fed control mice (Supplemental Figure 6D). Furthermore, the numbers of M1 and M2 macrophages in eWAT were identical in LiRiKO and control mice (Supplemental Figure 6E). Thus, the ability of local insulin resistance to promote inflammation is specific to visceral WAT.
Rictor knockout in adipocytes increases expression of the chemokine MCP1.
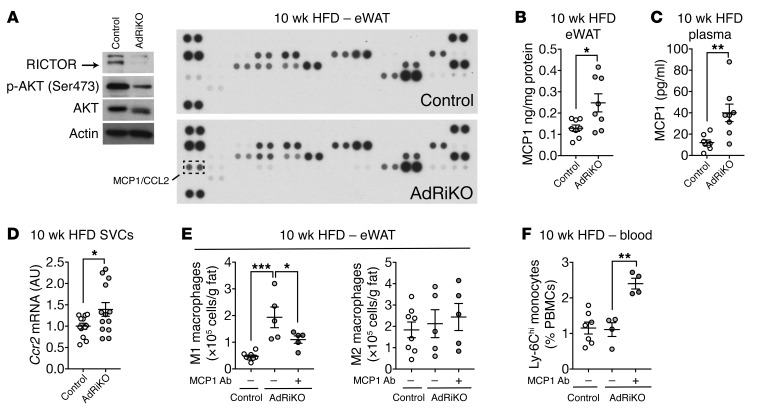
How does insulin resistance in visceral WAT cause local accumulation of M1 macrophages and inflammation? Amano et al. have suggested that a HFD induces local proliferation of macrophages (23). Alternatively, others have proposed that WAT recruits circulating monocytes, which then differentiate into M1 macrophages (7, 22). To distinguish between these 2 models, we used flow cytometry to measure the proliferation marker Ki-67 in macrophages in eWAT of HFD-fed mice. The percentage of Ki-67+ M1 macrophages in AdRiKO mice was similar to that in control mice (Supplemental Figure 7A), suggesting that the increase in M1 macrophages in AdRiKO eWAT is not due to local proliferation. To investigate the possibility that WAT recruits monocytes, we examined the expression of WAT-derived chemokines in eWAT from HFD-fed AdRiKO and control mice. A chemokine array and ELISA revealed increased expression of monocyte chemoattractant protein 1 (MCP1, also known as C-C motif ligand 2 [CCL2]) in AdRiKO eWAT (Figure 4, A and B). Increased levels of MCP1 were also detected in the plasma of HFD-fed AdRiKO mice (Figure 4C). Furthermore, in SVCs isolated from AdRiKO eWAT, we observed significantly increased expression of C-C chemokine receptor type 2 (Ccr2), encoding an MCP1 receptor, as a result of increased numbers of Ccr2-expressing cells such as monocytes and macrophages (Figure 4D). These findings suggest that insulin-resistant visceral WAT, via MCP1 expression, recruits monocytes, which then differentiate into M1 macrophages.
Figure 4. Insulin/mTORC2 signaling inhibits Mcp1 transcription and M1 macrophage accumulation in vivo.
(A) Adipokine array of eWAT from HFD-fed AdRiKO and control mice. Immunoblots show the reduction of RICTOR expression and mTORC2 signaling. n = 8 (data from 8 mice were pooled). (B) MCP1 protein levels in eWAT from HFD-fed AdRiKO and control mice. *P < 0.05, by unpaired Student’s t test. n = 8. (C) MCP1 protein levels in plasma from HFD-fed AdRiKO and control mice. **P < 0.01, by unpaired Student’s t test. n = 8. (D) Ccr2 mRNA levels in SVCs isolated from eWAT of HFD-fed AdRiKO and control mice. *P < 0.05, by unpaired Student’s t test. n = 12. (E) Numbers of M1 macrophages (CD45+F4/80+CD11b+CD11c+) and M2 macrophages (CD45+F4/80+CD11b+CD301+) in eWAT. Mice were fed a HFD for 8 weeks and treated with a control or MCP1-neutralizing antibody for 2 weeks with ongoing HFD feeding. *P < 0.05 and ***P < 0.001, by 1-way ANOVA. n = 5–8. (F) Percentage of inflammatory monocytes in peripheral blood mononuclear cells (PBMCs). Mice were treated as in E. **P < 0.01, by 1-way ANOVA. n = 4–7. Data are presented as the mean ± SEM.
We next tested whether the increase in MCP1 is responsible for M1 macrophage accumulation in AdRiKO eWAT. Mice were fed a HFD for 8 weeks and then treated with an MCP1-neutralizing or control antibody for 2 weeks along with ongoing HFD feeding. The antibody treatments had no impact on body weight (Supplemental Figure 7B). The MCP1-neutralizing antibody inhibited the accumulation of M1 macrophages in AdRiKO eWAT by 50%, with no impact on M2 macrophages (Figure 4E). The MCP1-neutralizing antibody also caused a 2-fold increase in monocytes (Ly-6ChiCD11b+) in the blood of AdRiKO mice (Figure 4F). Thus, MCP1 appears to mediate the increase in M1 macrophages in AdRiKO eWAT. Altogether, our data suggest that mTORC2 inhibition in WAT results in Mcp1 expression, followed by infiltration of monocytes in an MCP1-CCR2–dependent manner.
Insulin/mTORC2 signaling inhibits Mcp1 transcription in adipocytes.
Expression of the Mcp1 gene was increased in the eWAT of HFD-fed AdRiKO and i-AdRiKO mice compared with expression levels in control eWAT (Figure 5, A and B), suggesting that MCP1 is upregulated in AdRiKO WAT at the transcriptional level. Furthermore, we note that the increase in Mcp1 expression (Figure 5A) coincided with an increase in the number of M1 macrophages in AdRiKO eWAT (Figure 2F). The number of macrophages and expression levels of Mcp1 were unchanged in AdRiKO and control eWAT in ND-fed mice (Supplemental Figure 3 and Supplemental Figure 8A). To identify the cells in which Mcp1 expression was induced, we measured Mcp1 mRNA levels in adipocytes and SVCs isolated from eWAT of HFD-fed AdRiKO and control mice (Supplemental Figure 2B). AdRiKO adipocytes, but not SVCs, showed increased Mcp1 expression (Figure 5, C and D). To determine whether the regulation of Mcp1 transcription by mTORC2 is cell autonomous, we first treated 3T3-L1 adipocytes with the mTOR inhibitor torin 1 (47). 3T3-L1 adipocytes treated with torin 1 had increased Mcp1 expression (Figure 5E). Next, we generated 2 Rictor-knockout 3T3-L1 adipocyte cell lines (Figure 5F and Supplemental Figure 8B) using the genome-editing CRISPR-Cas9 system (48). Rictor-knockout 3T3-L1 adipocytes were able to differentiate, albeit at a slower rate compared with control cells (Supplemental Figure 8C). Consistent with our in vivo data, Mcp1 expression was increased in the Rictor-knockout 3T3-L1 adipocytes (Figure 5G and Supplemental Figure 8B). Serum and insulin treatment suppressed Mcp1 expression in control but not Rictor-knockout 3T3-L1 adipocytes (Figure 5H and Supplemental Figure 8D). In WT mice, Mcp1 expression increased by 10 weeks, but not 4 weeks, of HFD feeding (Supplemental Figure 8E). These data support the notion that insulin resistance precedes and promotes Mcp1 transcription in adipocytes. We note that Rictor knockout in liver (LiRiKO) did not result in hepatic Mcp1 expression (Supplemental Figure 8F), consistent with our above finding that LiRiKO failed to stimulate inflammation in liver.
Figure 5. Insulin/mTORC2 signaling inhibits Mcp1 transcription in adipocytes.
(A) Mcp1 mRNA levels in eWAT from AdRiKO and control mice during the HFD time course. *P < 0.05 and ***P < 0.001, by multiple Student’s t test. n = 5–10. (B) Mcp1 mRNA levels in eWAT from HFD-fed i-AdRiKO and control mice. **P < 0.01, by unpaired Student’s t test. n = 4–6. (C and D) Mcp1 mRNA levels in adipocytes (C) and SVCs (D) isolated from eWAT of HFD-fed AdRiKO and control mice. ***P < 0.001, by unpaired Student’s t test. n = 13–14. (E) Mcp1 mRNA levels in 3T3-L1 adipocytes treated with DMSO or 250 nM torin 1 for 6 hours. **P < 0.01, by unpaired Student’s t test. N >3. (F) 2DGP accumulation in insulin-stimulated Rictor-knockout or control 3T3-L1 adipocytes treated with DMSO or the JNK inhibitor SP600125 (20 μM). N = 3. (G) Mcp1 mRNA levels in Rictor-knockout and control 3T3-L1 adipocytes. *P < 0.05 and **P < 0.01, by unpaired Student’s t test. N = 3. (H) Mcp1 mRNA levels in Rictor-knockout and control 3T3-L1 adipocytes treated with or without serum and insulin. *P < 0.05, by unpaired Student’s t test. N = 3. (I) Mcp1 mRNA levels in Rictor-knockout and control 3T3-L1 cells treated with DMSO or the JNK inhibitor SP600125 (20 μM) for 6 hours. *P < 0.05 and **P < 0.01, by 1-way ANOVA. N = 3. (J) Immunoblots of Rictor-knockout and control 3T3-L1 adipocytes treated with DMSO or the JNK inhibitor SP600125 (20 μM) for 6 hours. N = 3. (K) In vitro JNK kinase assay. Active JNK was immunoprecipitated from Rictor-knockout or control 3T3-L1 adipocytes, and JNK activity was assessed toward its substrate cJUN. SP600125 treatment served as a negative control. N = 3. Data are presented as the mean ± SEM.
How does mTORC2 loss lead to Mcp1 transcription? It has been suggested that JNK is required for MCP1 expression and secretion in cultured 3T3-L1 adipocytes (49). Consistent with that report, treatment with the JNK inhibitor SP600125 reduced Mcp1 expression in Rictor-knockout 3T3-L1 cells (Figure 5I). Inhibition of JNK was confirmed by loss of cJUN (Ser73) phosphorylation (Figure 5J). SP600125 did not restore AKT (Ser 473) phosphorylation (Figure 5J) or insulin-stimulated glucose uptake (Figure 5F) in the Rictor-knockout 3T3-L1 cells, indicating that the effect of the drug was independent of mTORC2 and insulin resistance. Furthermore, JNK activity was unaffected by Rictor knockout (Figure 5, J and K). Thus, mTORC2 and JNK control Mcp1 expression independently.
Impaired insulin/mTORC2 signaling and increased MCP1 expression in omental WAT of obese patients.
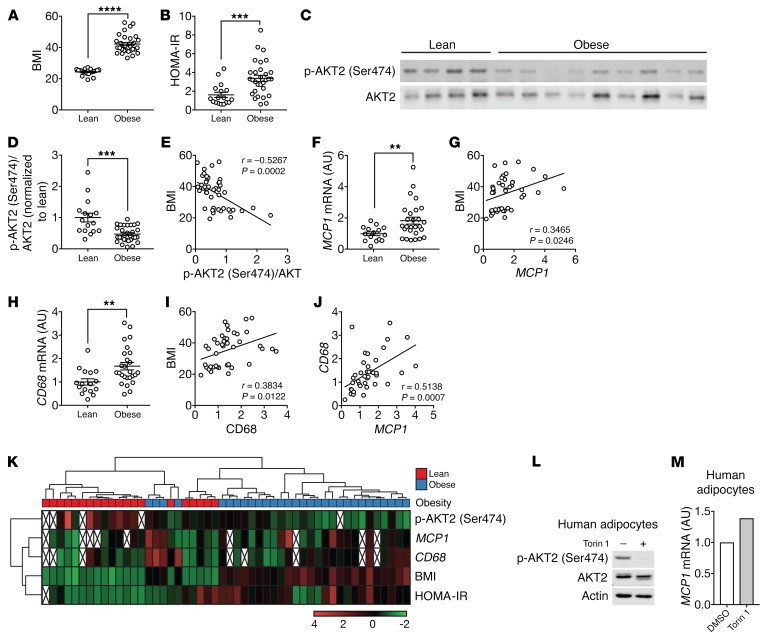
MCP1 mRNA levels in omental WAT (oWAT) correlate with BMI in obese human subjects (9, 50). However, how MCP1 transcription is regulated in human adipose tissue is unknown. Our finding that insulin/mTORC2 signaling negatively controls Mcp1 transcription and adipose tissue inflammation in mice prompted us to examine insulin/mTORC2 signaling, MCP1 expression, and macrophage accumulation in human visceral WAT, i.e., oWAT. To this end, oWAT samples were collected from 20 lean and 30 obese human patients who were under general anesthesia (Figure 6A and Supplemental Table 2). The obese patients were insulin resistant as determined by high homeostatic model assessment of insulin resistance (HOMA-IR) (Figure 6B and Supplemental Figure 9A). We note that oWAT was collected from patients who, because of standard procedure before undergoing general anesthesia, had fasted overnight, a condition that might not be optimal for the evaluation of insulin/mTORC2 signaling. Nevertheless, in oWAT, AKT2 (Ser474) phosphorylation, a readout for mTORC2 signaling, was lower in obese patients than in lean patients (Figure 6, C and D). AKT2 (Ser474) phosphorylation negatively correlated with BMI (Figure 6E). These data suggest that insulin/mTORC2 signaling is impaired in oWAT of obese and insulin-resistant patients and validate AdRiKO mice as a model for human insulin resistance. MCP1 expression was higher in obese subjects and positively correlated with BMI (Figure 6, F and G, and Supplemental Figure 9B). CD68 expression was also higher in the obese subjects and positively correlated with BMI (Figure 6, H and I, and Supplemental Figure 9C). MCP1 and CD68 expression levels also correlated (Figure 6J). AKT2 (Ser474) phosphorylation did not correlate with MCP1 and CD68 expression (Supplemental Figure 9, D and E) when comparing the entire cohort of 50 patients, consistent with our above observation in mice that inhibition of insulin/mTORC2 signaling alone was not sufficient to promote adipose tissue inflammation (Supplemental Figure 3). However, approximately one-third of the obese patients (9 of 30) had low AKT (Ser474) phosphorylation and high HOMA-IR, MCP1, and CD68 (Figure 6K), suggesting that AdRiKO mice may phenocopy this subgroup of patients. Finally, torin 1 treatment led to an increase in MCP1 expression in human primary adipocytes, suggesting that insulin/mTOR signaling inhibits MCP1 expression also in humans (Figure 6, L and M).
Figure 6. Impaired insulin/mTORC2 signaling and increased MCP1 expression in oWAT of insulin-resistant obese patients.
(A and B) BMI and HOMA-IR of lean and obese patients. ***P < 0.001 and ****P < 0.0001, by Mann-Whitney U test. See also Supplemental Table 2. (C) Representative immunoblots for p-AKT2 (Ser474) and AKT2 in human oWAT. (D) Quantification of p-AKT2 (Ser474) normalized to total AKT2. ***P < 0.001, by Mann-Whitney U test. (E) p-AKT2 (Ser474)/AKT2 negatively correlated with BMI. Significance was determined by Pearson’s correlation analysis. (F) MCP1 mRNA levels in human oWAT. **P < 0.01, by Mann-Whitney U test. (G) MCP1 positively correlated with BMI. Significance was determined by Pearson’s correlation analysis. (H) CD68 mRNA levels in human oWAT. **P < 0.01, by Mann-Whitney U test. (I and J) CD68 positively correlated with BMI (I) and MCP1 levels (J). Significance in I and J was determined by Pearson’s correlation analysis. (K) Cluster analysis of BMI, HOMA-IR, p-AKT2 (Ser474)/AKT2, MCP1, and CD68 levels. (L and M) Effect of torin 1 on insulin/mTORC2 signaling (L) and MCP1 mRNA levels (M) in human primary adipocytes. Differentiated human primary adipocytes were treated with DMSO or 250 nM torin 1 for 6 hours.
Discussion
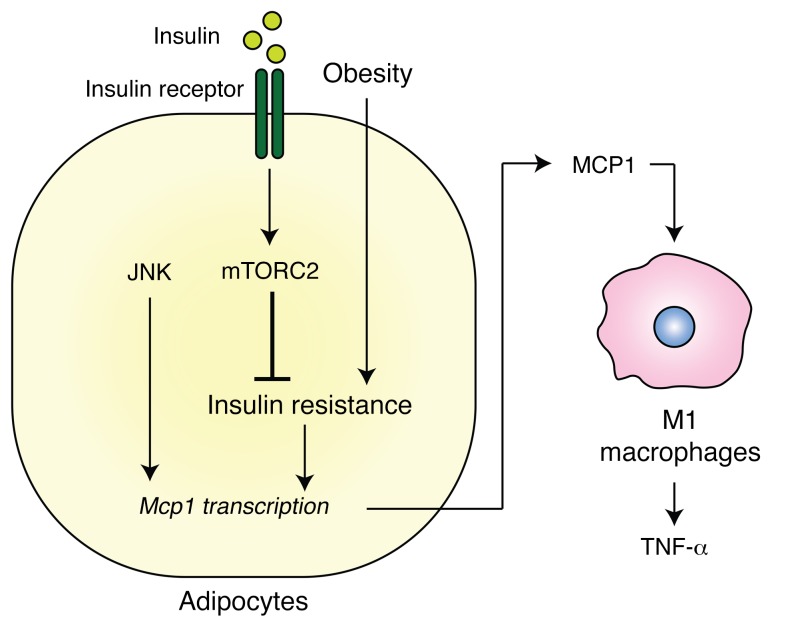
We provide 2 lines of evidence that insulin resistance promotes the accumulation of M1 macrophages and thereby fosters inflammation (Figure 7). First, we show that knockout of mTORC2, i.e., genetically induced insulin resistance, in mouse adipocytes derepressed Mcp1 expression. As a consequence, monocytes were recruited to visceral WAT, where they differentiated into M1 macrophages and caused inflammation. Second, HFD-induced insulin resistance in WT mice preceded the accumulation of proinflammatory M1 macrophages. We also show that oWAT from obese, insulin-resistant patients had low mTORC2 signaling, high MCP1 expression, and high macrophage content, suggesting that our findings in mice have human relevance (Figure 6).
Figure 7. Insulin resistance causes inflammation in adipose tissue.
Insulin resistance, due to obesity and loss of insulin/mTORC2 signaling, results in enhanced production of MCP1 in adipocytes. MCP1 in turn recruits monocytes and activates proinflammatory M1 macrophages.
Our findings are consistent with observations made in mice genetically modified in other components of the insulin signaling pathway. Two studies demonstrated that deletion of PTEN or PIK3R1, negative regulators of insulin signaling, causes enhanced insulin sensitivity and a reduced number of macrophages in adipose tissue (51, 52). More recently, Shearin et al. (53) showed that adipose-specific Akt1- and Akt2-knockout mice exhibit insulin resistance and increased macrophage infiltration in adipose tissue.
The above studies altogether may disentangle the “chicken-and-egg” relationship (see Introduction) of insulin resistance and inflammation, at least in adipose tissue. Obesity induces insulin resistance, via a yet-to-be defined mechanism, which in turn promotes inflammation. As suggested previously (16), this inflammation may contribute to adipose tissue remodeling and expansion to maintain glucose hemostasis. It has been suggested that activated M1 macrophages undergo metabolic reprogramming from oxidative phosphorylation to glycolysis (54, 55). Thus, a physiological role of M1 macrophages could be to clear excess local glucose. Our finding that insulin resistance precedes inflammation may account for the observation that inhibition of TNF-α is ineffective in the treatment of obesity-induced insulin resistance (18–21).
HFD-fed AdRiKO mice showed reduced AKT (Ser473) phosphorylation (Figure 1B), decreased glucose uptake (Figure 1A), and extensive inflammation (Figure 2) in eWAT. HFD-fed WT mice also displayed decreased glucose uptake (Figure 3, A–C), which was followed by mild inflammation (Figure 3D). Unexpectedly, AKT (Ser473) phosphorylation was not reduced in WT mice fed a HFD for 4 or 10 weeks (Supplemental Figure 5, E and F) (56), although we still observed mild inflammation in eWAT by week 10 of HFD feeding. We note that the number of M1 macrophages in AdRiKO eWAT was much higher than that in control eWAT by week 10 of the HFD (Figure 2F). These findings suggest that obesity-induced insulin resistance promotes mild inflammation downstream or independently of mTORC2, whereas chronic insulin resistance leads to mTORC2 inhibition and therefore extensive inflammation, as observed in AdRiKO mice and obese patients.
Our experiments reveal that loss of mTORC2 leads to increased Mcp1 expression in adipocytes (Figure 5). What is the downstream effector through which mTORC2 controls Mcp1 transcription? One candidate is the phosphatidic acid phosphatase LIPIN1, whose knockdown results in Mcp1 expression in 3T3-L1 adipocytes (57). LIPIN1, independently of its phosphatase activity, also functions as a transcriptional repressor (58). We found that Lipin1 expression was reduced in Rictor-knockout eWAT and adipocytes (Supplemental Figure 10, A and B), suggesting that mTORC2 may negatively control Mcp1 transcription via LIPIN1. An adipose-specific transcription factor is another candidate through which mTORC2 may control Mcp1 expression. Rictor knockout increased Mcp1 expression in adipocytes, but not in liver or fibroblasts (Supplemental Figure 8F and Supplemental Figure 10C), indicating that the regulation of Mcp1 transcription by mTORC2 is specific to adipocytes.
Why does AdRiKO eWAT accumulate a disproportionately high number of M1 macrophages only in response to obesity? The increase in Mcp1 transcription in mTORC2-knockout adipocytes required JNK activity (Figure 5I), which is high only in WAT from obese mice (59, 60). Thus, obesity might be a prerequisite for JNK activation, which in turn stimulates Mcp1 expression in conjunction with loss of mTORC2. We note that mTORC2 did not control JNK (Figure 5, J and K).
In summary, we propose that obesity-mediated insulin resistance is a cause of inflammation in visceral WAT. Although our findings do not rule out the possibility that inflammation promotes insulin resistance in other tissues or conditions, they bring into question whether antiinflammation therapy in adipose tissue is an effective strategy in the prevention of type 2 diabetes.
Methods
Mice.
Adipose tissue–specific Rictor-knockout (AdRiKO) and liver-specific Rictor-knockout (LiRiKO) mice were described previously (32, 35, 61). Since AdRiKO mice are infertile, we bred Rictorfl/+ aP2-Cre mice with Rictorfl/fl mice to generate mice for our experiments. For experiments with AdRiKO mice, age-matched Cre-negative males were used as controls. To generate LiRiKO mice for experiments, Rictorfl/fl Alb-Cre mice were crossed with Rictorfl/fl mice. To generate an i-AdRiKO mouse line, Rictorfl/fl mice were bred with adipoq-CreERT2 mice, provided by Stefan Offermanns (Max Planck Institute for Heart and Lung Research [MPI-HLR], Bad Nauheim, Germany) (42). Once the i-AdRiKO mouse line (Rictorfl/fl adipoq-CreERT2) was generated, these mice were bred with Rictorfl/fl mice to produce mice for experiments. For experiments with LiRiKO and i-AdRiKO mice, Cre-negative littermate male mice were used as controls. All mice used in this study were on a C57BL/6 background. For i-AdRiKO mice, Rictor knockout was induced by i.p. injection of 1 mg/mouse tamoxifen (Sigma-Aldrich) resuspended in corn oil daily for 5 days. Mice were housed at 22°C in a conventional facility under a 12-hour light/12-hour dark cycle with unlimited access to water and were fed a ND or a HFD (60% kcal from fat NAFAG 2127, KLIBA). The HFD-feeding experiment was conducted for 10 weeks unless otherwise specified. Cre-negative animals were used as a control. For i-AdRiKO mouse experiments, control mice were also treated with tamoxifen. Mice were randomly assigned for each experiment. Only male, 6- to 16-week-old mice were used for experiments.
Cell culture.
3T3-L1 cells were cultured and differentiated as described previously (62). In brief, 3T3-L1 preadipocyte cells were maintained in M1 medium (DMEM supplemented with 4 mM glutamine, 1 mM sodium pyruvate, 1× penicillin and streptomycin, and 10% FBS) at 37°C in an incubator with 5% CO2. For differentiation, cells were maintained in M1 medium for 2 days after reaching confluence. The medium was replaced with M2 medium (M1 medium supplemented with 1.5 μg/ml insulin, 0.5 mM IBMX, 1 μM dexamethasone, and 2 μM rosiglitazone), defined as day 0 of differentiation. After 2 days, the medium was replaced with M3 medium (M1 with 1.5 μg/ml insulin). The medium was replaced with M2 on day 4 of differentiation. From day 6, cells were maintained in M3 with a medium change every 2 days. For all experiments, cells differentiated for 10 to 14 days were used. Torin 1 was purchased from Tocris Bioscience and dissolved in DMSO.
Human oWAT biopsies.
oWAT biopsies were obtained from lean subjects with a BMI below 27 kg/m2 and from obese subjects with a BMI above 35 kg/m2 (Supplemental Table 2). Patients were fasted overnight and then underwent general anesthesia. All oWAT samples were obtained between 8:30 and 12:00, snap-frozen in liquid nitrogen, and stored at –80°C.
Human primary adipocytes.
Human primary visceral preadipocytes (from a 56-year-oldwoman with a BMI of 23) were obtained from Lonza. Cells were cultured and differentiated according to the manufacturer’s instructions. After 14 days of differentiation, cells were treated with DMSO or 250 nM torin1 for 6 hours.
Generation of Rictor-knockout cells by CRISPR/Cas9-mediated genome editing.
Rictor-knockout 3T3-L1 cells (Hall laboratory stock) were generated using the CRISPR/Cas9 system. Oligonucleotides containing single-guide RNAs (sgRNAs) (Rictor.1, forward: CACCGGAAGATACTGATCCCGCTTG, sgRNA Rictor.1, reverse: AAACCAAGCGGGATCAGTATCTTCC; sgRNA Rictor.2, forward: CACCGTGCCTCCTAGGGCTTGGTCG, sgRNA Rictor.1, reverse: AAACCGACCAAGCCCTAGGAGGCAC) were cloned into lentiCRISPRv2 (Addgene plasmid 52961) (48), a gift of Feng Zhang (MIT, Cambridge Massachusetts, USA). Plasmids were amplified by bacterial transformation and isolated by Miniprep (Zymo Research). LentiCRISPRv2 plasmids were cotransfected with psPAX2 (Addgene plasmid 12260; a gift of Didier Trono, EPFL, Lausanne, Switzerland) and pCMV-VSV-G (63) (Addgene plasmid 8454; a gift of Robert Weinberg, MIT, Cambridge, Massachusetts, USA) into HEK293T cells (Hall laboratory stock). Supernatants containing lentiviruses were collected 1 day after transfection and used to transduce undifferentiated 3T3-L1 cells. Transduced cells were selected by puromycin.
2-Deoxyglucose uptake assay.
Mice were fed a ND or HFD for 4 weeks, fasted for 5 hours, and given Humalog insulin i.p. (Lilly; 0.75 U/kg body weight). After 10 minutes, 2-deoxyglucose (2DG) (Sigma-Aldrich) was given i.p. (32.8 μg/g body weight), and tissues (eWAT, muscle) were collected 20 minutes later. For in vitro 2DG uptake, differentiated adipocytes were cultured in serum-free M1 medium, washed 3 times with HKRP buffer (1.2 mM KH2PO4, 1.2 mM MgSO4, 1.3 mM CaCl2, 118 mM NaCl, 5 mM KCl, and 30 mM HEPES, pH7.5), and cultured in HKRP buffer with 1% BSA for 30 minutes. Cells were stimulated with 100 nM insulin for 20 minutes and subsequently cultured with 1 mM 2DG for 20 minutes. Tissues or cells were lysed in 10 mM Tris-HCL, pH 8.0, by boiling for 15 minutes. 2-Deoxyglucose-6-phosphate (2DGP) was measured using a Glucose Uptake-Glo Assay Kit (Promega) following the manufacturer’s instructions.
MCP1 ELISA and adipokine array.
Mice were fed a HFD for 10 weeks, and MCP1 levels in eWAT and plasma were measured using a commercial ELISA kit (R&D Systems) following the manufacturer’s instructions. Adipokine array (R&D Systems) was performed according to the manufacturer’s instructions.
MCP1-neutralizing antibody treatment.
Mice were fed a HFD for 8 weeks. Mice were given i.p. 1 mg/kg body weight isotype control IgG (BD Biosciences; catalog 553968, lot 4113848) or anti–MCP1 IgG (BD Biosciences; catalog 554440, lot 5203535) every 2 days for 2 weeks.
ITT.
Mice were fasted for 5 hours, Humalog insulin (Lilly) was given i.p. (0.75U/kg body weight), and blood glucose levels were measured with an Accu-Chek blood glucose meter.
In vivo insulin stimulation.
Mice were fasted for 5 hours, Humalog insulin (Lilly) was administered i.p. (0.75U/kg body weight), and tissues were collected.
Isolation of adipocytes and SVCs and flow cytometric analysis.
SVCs were isolated and stained with antibody as previously described (64). In brief, fat pads were excised and minced in HBSS++ supplemented with 0.5% BSA and digested with 1 mg/ml type II collagenase (Sigma-Aldrich) at 37°C for 40 minutes with vigorous shaking. After digestion, final 10 mM EDTA was added and incubated for 10 minutes to dissociate SVCs. The resulting suspension was filtered through a 100-μm cell strainer (Corning) and centrifuged at 500 g for 10 minutes. After centrifugation, floating adipocytes were collected, and SVC-containing pellets were subjected to red blood cell lysis in 1× Red Blood Cell Lysis Buffer (eBioscience). SVCs were blocked with Fc-block (BD Biosciences) and stained with the following antibodies and reagents: F4/80-PE (eBioscience; catalog 12-4801-80); CD11b-APC eFluor 780 (eBioscience; catalog 47-0112-80); CD11c-PE-Cy7 (eBioscience; catalog 25-0114-81); CD45.2-PerCP (eBioscience; catalog 45-0454-80); CD301–Alexa Fluor 647 (AbD Serotec; catalog MCA2392A647T); CD4-FITC (eBioscience; catalog 11-0041-81); CD8a-PE (eBioscience; catalog 12-0081-81); CD3-APC (eBioscience; catalog 17-0032-80); CD45R (B220) PerCP-Cyanine 5.5 (eBioscience; catalog 45-0452-80); CD25 APC-eFluor 780 (eBioscience; catalog 47-0251-80); and Ki-67–Alexa Fluor 488 (BD Pharmingen; catalog 561165), along with a FOXP3 Flow Kit (BioLegend, catalog 320021) and a Live/Dead Fixable Dead Cell Kit (Thermo Fisher; catalog L34955). Stained SVCs were analyzed using the FACSCanto II (BD Biosciences) or sorted with a FACSAria IIIu (BD Biosciences).
Immunohistochemistry.
WATs were fixed overnight in 4% formalin at room temperature, dehydrated, embedded in paraffin, and cut into 5-μm-thick sections. Adipose tissue macrophages were stained with anti-F4/80 antibody (Abcam; catalog ab6640) and a secondary antibody conjugated with HRP (VECTOR Laboratories), followed by incubation in ImmPACT DAB peroxidase substrate solution (VECTOR Laboratories) and subsequent counterstaining with hematoxylin (VECTOR Laboratories). Images were obtained using DM600B (Leica) and analyzed with Fiji software (ImageJ; NIH) (65).
RNA isolation and quantitative real-time PCR.
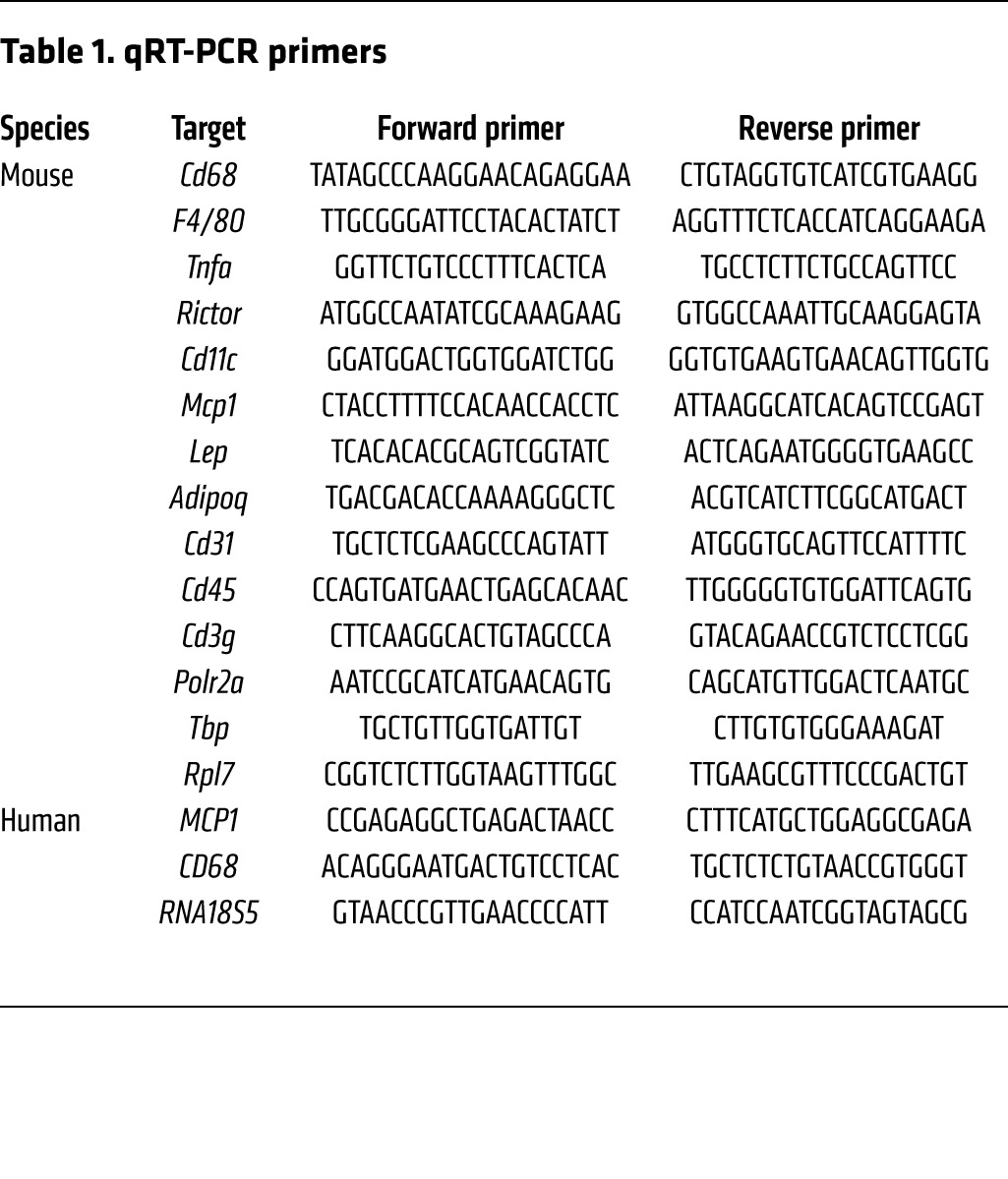
Total RNA was isolated with TRIzol Reagent (Sigma-Aldrich) and an RNeasy Kit (QIAGEN). For RNA isolation from sorted macrophages, an RNeasy Micro Kit (QIAGEN) was used. RNA was reverse transcribed to cDNA using an iScript cDNA Synthesis Kit (Bio-Rad). Semiquantitative real-time PCR analysis was performed using Fast SYBR Green (Applied Biosystems). Relative expression levels were determined by normalizing each Ct value to either Polr2a, Tbp, or Rpl7 expression for mice and RNA18S5 for human samples using the ΔΔCt method. The primer sequences used in this study are shown in Table 1.
Table 1. qRT-PCR primers.
Protein isolation and immunoblots.
Tissues were homogenized in lysis buffer containing 100 mM Tris-HCl, pH7.5, 2 mM EDTA, 2 mM EGTA, 150 mM NaCl, 1% Triton X-100, cOmplete Inhibitor Cocktail (Roche), and PhosSTOP (Roche). The protein concentration was determined by the Bradford assay, and equal amounts of protein were separated by SDS-PAGE and transferred onto nitrocellulose membranes (GE Healthcare). The following antibodies were used in this study and were purchased from Cell Signaling Technology: AKT (catalog 4685); phosphorylated AKT (p-AKT) (Ser473) (catalog 4060), RICTOR (catalog 2140), PKC (catalog 2056), p-S6K (Thr389) (catalog 9234), S6K (catalog 2708), p-JNK (Thr183/Tyr185) (catalog 4668), JNK (catalog 9252), p-cJUN (Ser73) (catalog 3270), cJUN (catalog 9165). Actin monoclonal antibody was purchased from Sigma-Aldrich (catalog MAB1501).
JNK kinase assay.
JNK activity was measured using a SAPK/JNK Kinase Assay Kit (Nonradioactive) (Cell Signaling Technology; catalog 8794).
Proteome analysis.
eWAT was homogenized in urea lysis buffer containing 50 mM Tris-HCl, pH 8.2, 8 M urea, 75 mM NaCl, cOmplete inhibitor cocktail (Roche), and PhosSTOP (Roche). The extracts were incubated for 30 minutes at 4°C and centrifuged for 15 minutes at 14,000 g. Protein concentration was measured with the Bradford assay. Proteins were reduced with 2.5 mM DTT for 40 minutes at 56°C and alkylated with 7.5 mM iodoacetamide for 40 minutes at room temperature in the dark with gentle shaking. The urea concentration was lowered to 4 M with 25 mM Tris-HCl, pH 8. The lysates were digested with 2 rounds of endoproteinase LysC (Wako) at an enzyme/protein ratio of 1:100 (w/w) at 37°C for 2 hours. After the LysC digestion, the urea concentration was lowered to 1 M with 25 mM Tris-HCl, pH 8.0. The lysates were digested with 2 rounds of trypsin (Worthington) at an enzyme/protein ratio of 1:100 (w/w) for 2 hours, followed by overnight digestion at 37°C. Digestion was stopped by adding trifluoroacetic acid (TFA) to reach a final concentration of 0.4%. The digests were centrifuged for 5 minutes at 3,000 g and desalted on a C18 SepPak cartridge (Waters) that had been equilibrated with 0.1% TFA. The peptides were applied onto the cartridge, washed with 0.1% TFA, and subsequently eluted with 0.5% AcOH/80% AcCN. The peptide concentration was estimated by measuring the UV absorbance at 280 nm (66). The desalted peptides were dried in a SpeedVac (Thermo Fisher Scientific). For strong cation separation, the dried peptides were dissolved in 1.5 ml 7 mM KH2PO4, pH 2.65, and 30% AcCN (v/v) (SCX buffer A) and centrifuged at 10,600 × g. The peptides were applied onto a HiTrap SP cartridge (GE Healthcare) that had been equilibrated with 3 ml SCX buffer A, and the cartridge was washed with 3 ml SCX buffer A. Bound peptides were stepwise eluted with 1.5 ml each of SCX buffer A containing 50 mM, 100 mM, 150 mM, 250 mM, and 350 mM KCl, and each fraction was collected manually. The peptide concentration was estimated by absorbance at 280 nm. The fractions were dried in a SpeedVac and desalted on either micro or macro spin C18 columns (The Nest Group). The peptides were desalted with 0.1% TFA and subsequently eluted with 0.5% AcOH/80% AcCN. The dried peptides (20 μg) were dissolved in 20 μl of 0.1% formic acid and 0.005% TFA and analyzed on an Orbitrap Elite FT Hybrid Instrument (Thermo Fisher Scientific).
The peptides from the SCX step-off fractions were analyzed by capillary liquid chromatography tandem MS (LC-MS/MS) using a home-packed separating column (0.075 mm × 15 cm) packed with Reprosil C18 reverse-phase material (2.4 m particle size; provided by Albin Maisch, Ammerbuch-Entringen, Germany). The column was connected online to an Orbitrap Elite FT Hybrid Instrument. The solvents used for peptide separation were 0.1% formic acid in water/0.005% TFA (solvent A) and 0.1% formic acid/0.005% TFA and 80% acetonitrile in water (solvent B). Peptide digest (2 μl) was injected with a Proxeon nLC Capillary Pump (Thermo Fisher Scientific) set to 0.3 μl/min. A linear gradient from 0% to 40% of solvent B in solvent A in 95 minutes was delivered with the nano pump at a flow rate of 0.3 μl/min. After 95 minutes, the percentage of solvent B was increased to 75% in 10 minutes. The eluting peptides were ionized at 2.5 kV. The mass spectrometer was operated in data-dependent mode. The precursor scan was done in the Orbitrap, set to 60,000 resolution, while the fragment ions were mass analyzed in the LTQ Orbitrap Instrument. A top-10 method was run so that the 10 most intense precursors were selected for fragmentation. Each biological replicate was measured in 3 technical replicates.
The LC-MS/MS data were searched with Proteome Discoverer 1.4 (Thermo Fisher Scientific), set to Mascot and Sequest HT against a mouse UniProtKB databank (67). The precursor tolerance was set to 10 ppm, while the fragment ion tolerance was set to 0.5 Da. The following variable modifications were used during the search: carbamidomethyl-cystein, oxidized methionine, and N-terminal protein acetylation. The peptide search matches were set at “high confidence” (1% FDR).
All LC-MS/MS runs were aligned with Progenesis software (Waters). For identification of the aligned features, the corresponding Proteome Discoverer 1.4 results files were imported into the aligned Progenesis data file. The data aligned in Progenesis were exported as .txt files to the R-based Perseus program (68). Volcano plots and ANOVA 2-sample t tests were performed with a FDR of 5%. Proteins were considered regulated when the ANOVA 2-sample t test was below 0.05 in each technical replicate and the Student’s t test P value was below 0.05 in at least 2 of 3 biological replicates. GO process analysis was performed according to the framework provided by the Gene Ontology Consortium (69, 70).
Statistics.
Immunoblots for p-AKT2 (Ser474) and AKT2 on human oWAT were quantified in Fiji (65). Samples for which we failed to detect AKT2 were excluded from further analysis. Sample size was chosen according to our previous studies and published reports in which similar experimental procedures were described. The investigators were not blinded to the treatment groups. All data are shown as the mean ± SEM. Sample numbers are indicated in each figure legend. For mouse experiments, n represents the number of animals, and for cell culture experiments, N indicates the number of independent experiments. To determine the statistical significance between 2 groups, an unpaired 2-tailed Student’s t test was performed. For more than 3 groups, 1-way ANOVA was performed. For ITT data, 2-way ANOVA was performed. For human samples, statistical outliers were excluded in Figure 6 according to the robust regression and outlier removal test (ROUT) (Q = 1%) but are included in Supplemental Figure 9. In all cases, a Mann-Whitney U test was performed to determine statistical significance. All statistical analysis was performed using GraphPad Prism 7 (GraphPad Software). Cluster analysis was performed using ClustVis (71). A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal experiments were performed in accordance with federal guidelines for animal experimentation and approved by the Kantonales Veterinäramt of the Kanton Basel-Stadt (Basel, Switzerland). For human biopsies, the study protocol was approved by the Ethikkomission Nordwest-und Zentralschweiz (EKNZ) (Basel, Switzerland). All patients provided informed consent to provide adipose tissue samples.
Author contributions
MS and MNH designed and directed the research. MS, VA, RP, ICF, DW, ACMG, NC, SM, MC, JAM, MMS, and PJ performed experiments and analyzed data. MS, RP, BW, and CB designed the protocol for the collection of human adipose tissue biopsies. MS and MNH wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Didier Trono (EPFL, Switzerland), Robert Weinberg (MIT, USA), and Feng Zhang (MIT, USA), Stefan Offermanns (MPI-HLR, Germany), the Imaging Core Facility (Biozentrum), and the FACS Core Facility (Biozentrum) for reagents, a mouse strain, equipment, or technical support. We acknowledge support from the Swiss National Science Foundation (to MS and MNH), the Louis Jeantet Foundation (to MNH), and the Canton of Basel (to MNH).
Version 1. 03/12/2018
Electronic publication
Version 2. 04/02/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(4):1538–1550.https://doi.org/10.1172/JCI96139.
Contributor Information
Mitsugu Shimobayashi, Email: mitsugu.shimobayashi@unibas.ch.
Verena Albert, Email: albeve@scripps.edu.
Diana Weissenberger, Email: diana.weissenberger@unibas.ch.
Anne Christin Meyer-Gerspach, Email: annechristin.meyergerspach@unibas.ch.
Nicolas Clement, Email: nicolas.clement@claraspital.ch.
Suzette Moes, Email: suzette.moes@unibas.ch.
Marco Colombi, Email: marco.colombi@unibas.ch.
Paul Jenö, Email: paul.jenoe@unibas.ch.
Christoph Beglinger, Email: christoph.beglinger@unibas.ch.
Ralph Peterli, Email: Ralph.Peterli@claraspital.ch.
References
- 1.Rocha VZ, Libby P. Obesity, inflammation, and atherosclerosis. Nat Rev Cardiol. 2009;6(6):399–409. doi: 10.1038/nrcardio.2009.55. [DOI] [PubMed] [Google Scholar]
- 2.Odegaard JI, Chawla A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013;339(6116):172–177. doi: 10.1126/science.1230721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J, Morley TS, Kim M, Clegg DJ, Scherer PE. Obesity and cancer — mechanisms underlying tumour progression and recurrence. Nat Rev Endocrinol. 2014;10(8):455–465. doi: 10.1038/nrendo.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lackey DE, Olefsky JM. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. 2016;12(1):15–28. doi: 10.1038/nrendo.2015.189. [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin T, Ackerman SE, Shen L, Engleman E. Role of innate and adaptive immunity in obesity-associated metabolic disease. J Clin Invest. 2017;127(1):5–13. doi: 10.1172/JCI88876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen MT, et al. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem. 2007;282(48):35279–35292. doi: 10.1074/jbc.M706762200. [DOI] [PubMed] [Google Scholar]
- 9.Weisberg SP, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 12.Kanda H, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest. 2006;116(6):1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patsouris D, Li PP, Thapar D, Chapman J, Olefsky JM, Neels JG. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 2008;8(4):301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YS, et al. Inflammation is necessary for long-term but not short-term high-fat diet-induced insulin resistance. Diabetes. 2011;60(10):2474–2483. doi: 10.2337/db11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian XY, et al. Thermoneutral housing accelerates metabolic inflammation to potentiate atherosclerosis but not insulin resistance. Cell Metab. 2016;23(1):165–178. doi: 10.1016/j.cmet.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wernstedt Asterholm I, et al. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 2014;20(1):103–118. doi: 10.1016/j.cmet.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kern PA, Saghizadeh M, Ong JM, Bosch RJ, Deem R, Simsolo RB. The expression of tumor necrosis factor in human adipose tissue. Regulation by obesity, weight loss, and relationship to lipoprotein lipase. J Clin Invest. 1995;95(5):2111–2119. doi: 10.1172/JCI117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ofei F, Hurel S, Newkirk J, Sopwith M, Taylor R. Effects of an engineered human anti-TNF-α antibody (CDP571) on insulin sensitivity and glycemic control in patients with NIDDM. Diabetes. 1996;45(7):881–885. doi: 10.2337/diab.45.7.881. [DOI] [PubMed] [Google Scholar]
- 19.Paquot N, Castillo MJ, Lefebvre PJ, Scheen AJ. No increased insulin sensitivity after a single intravenous administration of a recombinant human tumor necrosis factor receptor: Fc fusion protein in obese insulin-resistant patients. J Clin Endocrinol Metab. 2000;85(3):1316–1319. doi: 10.1210/jcem.85.3.6417. [DOI] [PubMed] [Google Scholar]
- 20.Stanley TL, et al. TNF-α antagonism with etanercept decreases glucose and increases the proportion of high molecular weight adiponectin in obese subjects with features of the metabolic syndrome. J Clin Endocrinol Metab. 2011;96(1):E146–E150. doi: 10.1210/jc.2010-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wascher TC, Lindeman JH, Sourij H, Kooistra T, Pacini G, Roden M. Chronic TNF-alpha neutralization does not improve insulin resistance or endothelial function in “healthy” men with metabolic syndrome. Mol Med. 2011;17(3-4):189–193. doi: 10.2119/molmed.2010.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagareddy PR, et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 2014;19(5):821–835. doi: 10.1016/j.cmet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amano SU, et al. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab. 2014;19(1):162–171. doi: 10.1016/j.cmet.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamei N, et al. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J Biol Chem. 2006;281(36):26602–26614. doi: 10.1074/jbc.M601284200. [DOI] [PubMed] [Google Scholar]
- 25.Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 27.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 29.Frias MA, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16(18):1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127(1):125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20(20):2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci U S A. 2009;106(24):9902–9907. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, et al. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59(6):1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y, et al. Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nat Commun. 2016;7:11365. doi: 10.1038/ncomms11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15(5):725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan M, Pino E, Wu L, Kacergis M, Soukas AA. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem. 2012;287(35):29579–29588. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bentzinger CF, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8(5):411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, et al. Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances Basal glycogen synthase activity. Mol Cell Biol. 2008;28(1):61–70. doi: 10.1128/MCB.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KY, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makowski L, et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med. 2001;7(6):699–705. doi: 10.1038/89076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sassmann A, Offermanns S, Wettschureck N. Tamoxifen-inducible Cre-mediated recombination in adipocytes. Genesis. 2010;48(10):618–625. doi: 10.1002/dvg.20665. [DOI] [PubMed] [Google Scholar]
- 43.Festuccia WT, Pouliot P, Bakan I, Sabatini DM, Laplante M. Myeloid-specific Rictor deletion induces M1 macrophage polarization and potentiates in vivo pro-inflammatory response to lipopolysaccharide. PLoS One. 2014;9(4):e95432. doi: 10.1371/journal.pone.0095432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SY, et al. Unraveling the temporal pattern of diet-induced insulin resistance in individual organs and cardiac dysfunction in C57BL/6 mice. Diabetes. 2005;54(12):3530–3540. doi: 10.2337/diabetes.54.12.3530. [DOI] [PubMed] [Google Scholar]
- 45.Turner N, et al. Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia. 2013;56(7):1638–1648. doi: 10.1007/s00125-013-2913-1. [DOI] [PubMed] [Google Scholar]
- 46.Kawano Y, et al. Colonic Pro-inflammatory macrophages cause insulin resistance in an intestinal Ccl2/Ccr2-dependent manner. Cell Metab. 2016;24(2):295–310. doi: 10.1016/j.cmet.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi K, et al. JNK- and IκB-dependent pathways regulate MCP-1 but not adiponectin release from artificially hypertrophied 3T3-L1 adipocytes preloaded with palmitate in vitro. Am J Physiol Endocrinol Metab. 2008;294(5):E898–909. doi: 10.1152/ajpendo.00131.2007. [DOI] [PubMed] [Google Scholar]
- 50.Christiansen T, Richelsen B, Bruun JM. Monocyte chemoattractant protein-1 is produced in isolated adipocytes, associated with adiposity and reduced after weight loss in morbid obese subjects. Int J Obes (Lond) 2005;29(1):146–150. doi: 10.1038/sj.ijo.0802839. [DOI] [PubMed] [Google Scholar]
- 51.Morley TS, Xia JY, Scherer PE. Selective enhancement of insulin sensitivity in the mature adipocyte is sufficient for systemic metabolic improvements. Nat Commun. 2015;6:7906. doi: 10.1038/ncomms8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCurdy CE, et al. Attenuated Pik3r1 expression prevents insulin resistance and adipose tissue macrophage accumulation in diet-induced obese mice. Diabetes. 2012;61(10):2495–2505. doi: 10.2337/db11-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shearin AL, Monks BR, Seale P, Birnbaum MJ. Lack of AKT in adipocytes causes severe lipodystrophy. Mol Metab. 2016;5(7):472–479. doi: 10.1016/j.molmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang SC, et al. Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity. 2016;45(4):817–830. doi: 10.1016/j.immuni.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213(1):15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan SX, et al. Selective insulin resistance in adipocytes. J Biol Chem. 2015;290(18):11337–11348. doi: 10.1074/jbc.M114.623686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi N, et al. Suppression of lipin-1 expression increases monocyte chemoattractant protein-1 expression in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 2011;415(1):200–205. doi: 10.1016/j.bbrc.2011.10.060. [DOI] [PubMed] [Google Scholar]
- 58.Kim HB, et al. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol Cell Biol. 2010;30(12):3126–3139. doi: 10.1128/MCB.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirosumi J, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420(6913):333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 60.Sabio G, et al. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science. 2008;322(5907):1539–1543. doi: 10.1126/science.1160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albert V, et al. mTORC2 sustains thermogenesis via Akt-induced glucose uptake and glycolysis in brown adipose tissue. EMBO Mol Med. 2016;8(3):232–246. doi: 10.15252/emmm.201505610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zebisch K, Voigt V, Wabitsch M, Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal Biochem. 2012;425(1):88–90. doi: 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 63.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho KW, Morris DL, Lumeng CN. Flow cytometry analyses of adipose tissue macrophages. Methods Enzymol. 2014;537:297–314. doi: 10.1016/B978-0-12-411619-1.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 67.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 69.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.