Abstract
Cellular senescence is a highly stable cell cycle arrest that is elicited in response to different stresses. By imposing a growth arrest, senescence limits the replication of old or damaged cells. Besides exiting the cell cycle, senescent cells undergo many other phenotypic alterations such as metabolic reprogramming, chromatin rearrangement, or autophagy modulation. In addition, senescent cells produce and secrete a complex combination of factors, collectively referred as the senescence-associated secretory phenotype, that mediate most of their non–cell-autonomous effects. Because senescent cells influence the outcome of a variety of physiological and pathological processes, including cancer and age-related diseases, pro-senescent and anti-senescent therapies are actively being explored. In this Review, we discuss the mechanisms regulating different aspects of the senescence phenotype and their functional implications. This knowledge is essential to improve the identification and characterization of senescent cells in vivo and will help to develop rational strategies to modulate the senescence program for therapeutic benefit.
Introduction
Cellular senescence was originally identified as a stable exit from the cell cycle caused by the finite proliferative capacity of cultured human fibroblasts (1, 2). Currently, senescence is considered a stress response that can be induced by a wide range of intrinsic and extrinsic insults, including oncogenic activation, oxidative and genotoxic stress, mitochondrial dysfunction, irradiation, or chemotherapeutic agents (3).
While the defining characteristic of senescence is the establishment of a stable growth arrest that limits the replication of damaged and old cells, many other phenotypic alterations associated with the senescent program are relevant to understanding the pathophysiological functions of senescent cells (4). For example, senescent cells undergo morphology changes, chromatin remodeling, and metabolic reprogramming, and secrete a complex mix of mostly proinflammatory factors termed the senescence-associated secretory phenotype (SASP) (Figure 1). Here, we review the molecular mechanisms controlling cellular senescence with a special focus on their translational relevance and suitability for identifying and characterizing senescent cells in vivo.
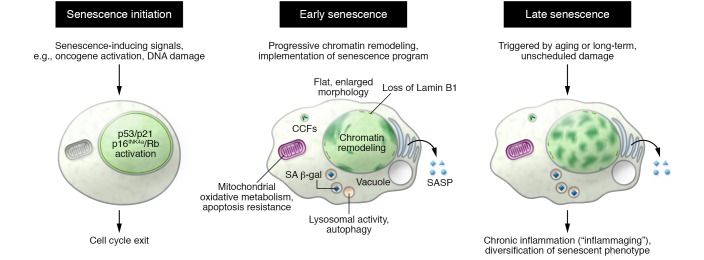
Figure 1. Phenotypic characteristics of senescent cells.
Diagram depicting some of the phenotypic alterations associated with senescence initiation, early senescence, and late phases of senescence.
Physiological roles of senescence
Cellular senescence was initially dismissed as a tissue culture artifact. However, a wealth of data has demonstrated that senescent cells can influence disease and aging, as well as normal tissue homeostasis (5). Indeed, senescence can be engaged during development (6, 7) and is also necessary for tissue remodeling. For instance, transient induction of senescent cells is observed during wound healing and contributes to wound resolution (8, 9). Senescence can also be a protective stress response. In fact, senescence is best known as a potent anticancer mechanism that prevents malignancies by limiting the replication of preneoplastic cells (10).
However, the accumulation of senescent cells also drives aging and age-related diseases (11, 12). The connection between senescence and aging was initially grounded on observations of the accumulation of senescent cells in aged tissues (13). It was suggested that, during aging, senescence of stem and progenitor cells could hinder tissue homeostasis by interfering with the capacity of tissues to repair and regenerate. In the last 10 years, our understanding of senescence’s detrimental consequences in aging and age-related pathologies has significantly expanded. Two lines of research have facilitated this awareness. First, the use of transgenic models that allow for the detection of senescent cells has enabled a systematic identification of these cells in many age-related pathologies (5). Second, the development of genetic and drug strategies to selectively eliminate senescent cells, spearheaded by the van Deursen laboratory, has demonstrated that senescent cells can indeed play a causal role in aging and related pathologies (11).
The confirmation that selectively killing senescent cells significantly improves the health span of mice in the context of normal aging and ameliorates the consequences of age-related disease or cancer therapy (14–19) has ignited interest in the identification of compounds that can clear senescent cells. These so-called senolytic therapies, however, still face important caveats. In addition to their potential side effects, the evaluation of senolytic compounds is compromised by limitations such as the lack of universal senescence biomarkers and the heterogeneity of senescent phenotypes in vivo (20). Ongoing research into the pathways that initiate and maintain senescence will provide insights to identify biomarkers and potential therapies to target senescent cells.
Senescence as a dynamic program
Senescence has been traditionally considered as a defined, static cell fate. However, it is now recognized that senescence is a dynamic multistep process (11). A simplified model (Figure 1) suggests that although the initial senescence-inducing signals are sufficient to initiate cell cycle exit, this merely constitutes an early step in the senescence process. Senescent cells progressively remodel their chromatin and start to sequentially implement other aspects of the senescence program, such as the SASP, to enter into a second step of “full senescence.” If these senescent cells persist for extended periods of time, they continue evolving and can be categorized as entering into a third step of “late senescence,” which can involve adaptation and diversification of the senescent phenotype. It is tempting to suggest that the concept of senescence progression may help account for the heterogeneity of senescent cells and their associated phenotypes in vivo. Indeed, the senescent responses occurring in vivo can be categorized into two types. Acute senescence seems to be a programmed process that is triggered in response to discrete stressors, is established with fast kinetics, and normally contributes to tissue homeostasis. In contrast, chronic senescence may result from long-term unscheduled damage, and it is often associated with detrimental processes such as aging. Only chronic senescence is associated with the presence of cells in a state of late, deep senescence. Nevertheless, there is no conclusive evidence proving an irrefutable relationship between the different stages of senescence identified in cultured cells and the physiopathological functions associated with senescent cells. In fact, recent evidence suggests that the functionality of senescent cells in vivo strongly relies on the SASP’s effect on the surrounding environment and the associated immune responses. However, the effects of the SASP may be difficult to predict. On one hand, the senescent secretome is a heterogeneous mix of proteins whose composition depends not only on the stage of senescence progression but also on the affected cell type and the nature of the inducing stressor(s). The efficiency and kinetics of the clearance of senescent cells may vary depending on the organ in which they accumulate or the general ability to mount an effective immune response. For example, while cells undergoing oncogene-induced senescence (OIS) in the liver are efficiently cleared by the immune system (21), senescent cells in melanocytic nevi often manage to evade immune clearance and persist (22).
Overall, therapeutic approaches aiming to modulate senescent phenotypes will benefit from a better understanding of the steps driving and defining the evolution of senescent cells in vitro.
Cell cycle arrest
One of the defining features of senescent cells is their stable cell cycle arrest. This cell cycle exit is controlled by activation of the p53/p21CIP1 and p16INK4a/Rb tumor suppressor pathways (Figure 2). Unlike quiescent cells, senescent cells are nonresponsive to mitogenic or growth factor stimuli; thus, they are unable to reenter the cell cycle even in advantageous growth conditions. Senescent cells are also distinct from terminally differentiated cells, which are also irreversibly withdrawn from cell cycle. While terminal differentiation is the result of a defined developmental program, which turns undifferentiated precursors into specialized effector cells, senescence is mainly implemented as a cellular stress response. However, terminally differentiated cells such as neurons, adipocytes, and hepatocytes can also undergo senescence, or at least show senescence-like features, during aging or in response to oncogenic activation or DNA damage (21, 23–25). This indicates that the onset of senescence can occur independently of an active cell cycle arrest.
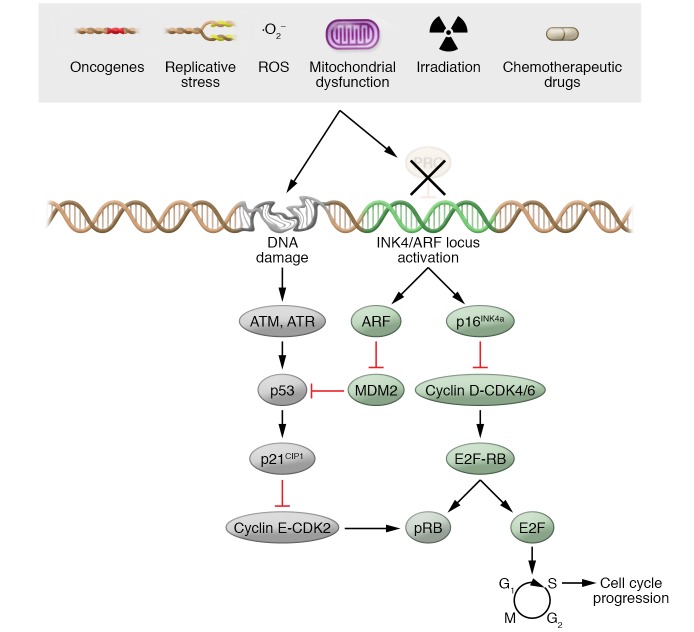
Figure 2. Molecular pathways controlling growth arrest during senescence.
A variety of stressors induce senescence-associated growth arrest. Cell cycle exit is regulated by induction of the p16INK4a/Rb and p53/p21CIP1 pathways. Figure reproduced with permission from McHugh and Gil (126).
DNA damage response
The senescence growth arrest is often triggered by a persistent DNA damage response (DDR) caused by either intrinsic (oxidative damage, telomere attrition, hyperproliferation) or external insults (ultraviolet, γ-irradiation, chemotherapeutic drugs) (26). For instance, during replicative senescence of human fibroblasts, progressive telomere shortening ultimately exposes an uncapped, double-stranded chromosome free end, which is sensed as a double-strand break by the DDR machinery (27). In cells undergoing OIS, the initial hyperproliferative phase that follows oncogene activation induces this DDR. In response to mitotic signals, an increase in usage of DNA replication origins leads to accumulation of genomic damage and activation of a DDR because of stalled replication forks (9, 28, 29). Senescence is associated with a persistent DDR that results in irreparable DNA damage (30). The DDR associated with replicative senescence is telomere-dependent: it correlates with telomere uncapping and an overall loss of telomeric length (27). During OIS, DDR occurs independently of telomeric length, but it is still associated with telomeric dysfunction (31). Regardless of which mechanism drives the damage, the DDR is characterized by increased deposition of γ-H2Ax (the histone H2AX phosphorylated at Ser139) and p53-binding protein 1 (53BP1) in the chromatin as well as activation of a kinase cascade involving first the serine/threonine-nonspecific kinases ATM and ATR and later the checkpoint serine/threonine kinases CHK1 and CHK2, which eventually leads to activation of the p53/p21CIP1 axis (26, 27).
Despite DDR’s role in initiating senescence, DDR markers have limited utility for identifying senescence in vivo. In fact, activation of p53 and/or p21CIP1 during senescence can occur in a DDR-independent manner (6, 7, 32, 33). Moreover, the majority of cells expressing markers of DNA damage in vivo, especially in a nonpathological setting, are not in fact senescent; rather, they are responding to a transient reparable damage.
Induction of the p53/p21CIP1 pathway
The senescent growth arrest is implemented by activation of the p53 and p16INK4a/Rb tumor suppressor networks (34). Once activated, p53 regulates a complex antiproliferative transcriptional program (35). The most relevant function of p53 in senescence is to induce the transcription of the cyclin-dependent kinase inhibitor (CDKi) p21CIP1, which in turn blocks CDK2 activity, resulting in hypophosphorylated Rb and cell cycle exit (26). In agreement with this, inactivation of p53 signaling by different means interferes with the onset of cellular senescence (36–39).
Importantly, if the stress that triggers senescence is transient, p53 induction can enact a quiescent state and activate DNA repair processes. Upon resolution of the stress, cells can resume cycling (12). Persistent stress or additional signals can activate p16INK4a, an inhibitor of CDK4 and CDK6, contributing to a long-lasting arrest (40). It has been suggested that the role of p21CIP1 may be limited to the onset of senescence, whereas p16INK4a maintains a durable growth arrest, possibly signifying the existence of differently regulated phases of senescence. Indeed, although the induction of p21CIP1 is important for senescence initiation, its expression does not necessarily persist in senescent cells (20). In agreement with this, upregulation of p21CIP1 (but not p53 or p16INK4a) is the driver of developmental senescence: a short-term “programmed” type of cellular senescence that occurs during mammalian embryonic development (6, 7). Although p21CIP1 can be a valuable marker of senescence in some settings, it is also induced during transient cell cycle arrest or in response to DNA damage, and it should be used as a senescence marker only in combination with others.
Activation of the INK4/ARF locus
The INK4/ARF locus encodes for three tumor suppressors: ARF (p14ARF in human and p19Arf in mouse) and p16INK4a — each encoded by the CDKN2A gene — and p15INK4b, encoded by CDKN2B (41, 42). Both p16INK4a and p19ARF have key roles in maintaining the senescent growth arrest. p16INK4a selectively inhibits CDK4 and CDK6, and p19ARF prevents p53 degradation. Indeed, loss-of-function mutations in p16INK4a are among the most frequent in human malignant cancers (43), suggesting that loss of p16INK4a enables senescence bypass and tumor progression.
The functional importance of the INK4/ARF locus has driven strong efforts to understand its regulation. In normal cells, the INK4/ARF locus is silenced by the Polycomb repressive complexes PRC1 and PRC2. Thus, disruption of PRC1 or PRC2 components, such as CBX7, BMI1, or EZH2, and the consequent loss of the trimethyl histone H3K27 repressive mark and/or PRC1 from the INK4/ARF locus are sufficient to activate p16INK4a and induce senescence (44–47). Epigenetic regulation of the INK4/ARF locus, however, goes beyond Polycomb proteins. Other epigenetic regulators such as MLL1, JMJD3, or ZRF1 are involved in the regulation of the locus (41, 48–51). How the recruitment and displacement of PRCs and other epigenetic components to the INK4/ARF locus are controlled is still not completely understood. The suggested model is that interaction with transcription factors (e.g., TWIST1 or transcription factors of the homeobox family) and long intergenic noncoding RNAs (e.g., ANRIL) can dictate the recruitment specificity (52–55). On the other hand, senescence-dependent transcriptional regulation of some of these chromatin modifiers, such as JMJD3 or EZH2, may drive the activation of the locus.
Further highlighting its functional relevance, p16INK4a stands out as one of most specific markers of senescence in vivo (13). While p16INK4a expression is almost undetectable in young healthy organisms, it markedly increases during tumorigenesis and aging. This evidence is the combined result of expression studies and the use of mouse models reporting for p16INK4 (56, 57). For instance, the Sharpless group (57) demonstrated an exponential increase in p16INK4a expression during aging. In these studies, while p16INK4a activity did not correlate with mortality, it did predict cancer initiation with higher sensitivity than glucose uptake (measured by fluorodeoxyglucose-PET).
More recently, mouse models have been generated in which p16INK4a-positive cells can be selectively eliminated based on the expression of inducible suicide genes under the control of p16INK4a regulatory elements (9, 15). These models have served to unequivocally show the causal roles of senescent cells in aging, age-related diseases, wound healing, and cancer therapy. The findings reported in these studies will be described in more detail in the other Reviews of this series.
Despite the clear benefits of exploiting p16INK4a activation as a tool to understand the role of senescence in pathophysiology, its use as an in vivo biomarker of senescence has limitations. First, forms of p16INK4a-independent senescence can occur in vitro. More importantly, p16INK4a can be expressed in nonsenescent cells (e.g., cancer cells with inactivated Rb), and it is unclear whether p16INK4a expression in macrophages or lymphocytes reflects a senescence phenotype or cell fates such as differentiation or exhaustion (20). Finally, the currently available antibodies are rather poor at detecting p16INK4a in murine tissues.
Non–cell-autonomous effects of senescence
Cellular senescence was initially considered to be a cell-intrinsic program. Increasing evidence, however, has shown that senescent cells have the ability to signal and influence their surrounding environment. Senescent cells produce a complex mixture of soluble and insoluble factors that are collectively termed senescence-associated secretory phenotype (SASP) or senescence-messaging secretome (58, 59). SASP is the general term given to the combination of cytokines, chemokines, extracellular matrix proteases, growth factors, and other signaling molecules secreted by senescent cells. Importantly, its specific composition varies depending on the cell type and the senescence inducer. Likewise, the functions attributed to the SASP, or at least some of its members, are also very diverse and depend not only on the nature of the SASP, but on the surrounding environment and the genetic context of the cells being exposed to the senescent secretome. The SASP is the best-studied mechanism by which senescent cells influence their neighbors, but is not the only one. For example, senescent cells can signal and influence adjacent cells through juxtacrine NOTCH/JAG1 signaling (60) or ROS production (61), or by cargo transfer, which occurs via formation of cytoplasmic bridges (62) or release of exosomes (63).
Functions of the SASP.
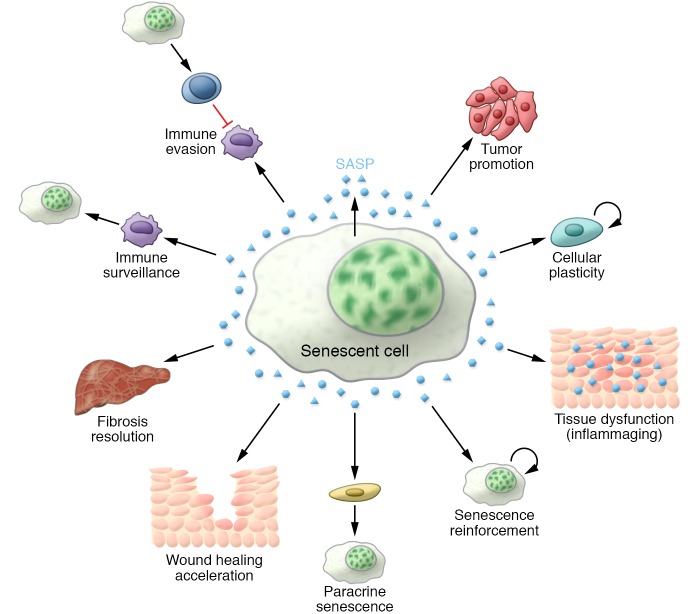
The SASP can have beneficial or detrimental effects (Figure 3). It is important to recognize, however, that the SASP’s effects in specific contexts are pleiotropic. The SASP reinforces the senescence growth arrest in vitro by implementing an autocrine positive-feedback loop. Indeed, knockdown of IL-6R, insulin-like growth factor–binding protein 7 (IGFBP7), or CXCR2, a receptor for IL-8 and related chemokines, prevents senescence (64–66). This autocrine loop contributes to the tumor-suppressive function of senescence. Interestingly, the SASP can also induce nonmalignant proliferating neighbor cells to undergo senescence (termed paracrine senescence) (61, 67, 68). This suggests that senescent cells could also amplify the antitumoral response by limiting the proliferation of nearby cells exposed to similar stressors.
Figure 3. The SASP is an important mediator of the pathophysiological functions of senescent cells.
Scheme summarizing some of the functions associated with the SASP.
Conversely, the senescent secretome can also promote tumorigenesis. In fact, the SASP has an important proinflammatory nature, and inflammatory mediators are powerful drivers of tumor progression. Early work showed that the SASP of senescent fibroblasts can promote proliferation and metastatic features in premalignant epithelial cells or increase tumor vascularization in xenograft transplants (69, 70). More recently, the protumorigenic role of the SASP has been examined using models that better resemble human cancer. For example, the SASP of senescent hepatic stellate cells (HSCs) promotes the proliferation and malignancy of the surrounding hepatocytes in obese mice treated with chemical carcinogens (71). Interestingly, the senescent secretome also mediates the harmful effects of senescent cells that accumulate upon chemotherapy treatment in vivo. In fact, elimination of senescent cells in this context prevents tumor relapse (17).
The interplay between the SASP and the immune response is also complex. On one hand, it is believed that the SASP might have initially evolved as a way to recruit the immune system to eliminate senescent cells. Indeed, during cancer initiation, SASP-dependent recruitment of Th1 cells, NK cells, and macrophages is essential to clear incipient preneoplastic cells and prevent the progression of hepatocellular carcinoma (HCC) (21, 72). On the other hand, the SASP can have immunosuppressive properties (73–75). For instance, Eggert and colleagues (75) showed that when premalignant senescent hepatocytes coexist with liver cancer cells, the SASP-dependent recruitment of immature myeloid cells may promote HCC progression by impairing the function of NK cells, suggesting a multifaceted interaction between the SASP, immune cells, and cancer.
The SASP has also been strongly linked to aging and age-related diseases. Low-level chronic inflammation (also referred to as “sterile inflammation” or “inflammaging”; ref. 76) underlies many age-related pathologies. It seems that the SASP could explain, at least in part, this local inflammation within tissues. Indeed, the elimination of senescent cells reduces levels of proinflammatory cytokines such as IL-6, IL-1α, and TNF-α in fat, kidneys, and skeletal muscle of aged mice (14, 15, 18). However, the relevance of this decreased inflammation in the improvement of age-related diseases after senolysis is not well understood. Further hinting at the SASP’s relevance as a mediator of the detrimental effects of senescent cells in aging is the observation that often only a small percentage of cells in aged tissues are senescent (77). Given the positive impact of eliminating these senescent cells, it is tempting to speculate that SASP suppression may underlie many of the beneficial effects of senolysis.
Altogether, recent discoveries seem to indicate that the deleterious effects of the SASP may outweigh its beneficial properties. Still, the SASP explains important physiological effects exerted by senescent cells. For instance, senescent fibroblasts are induced in response to a cutaneous wound and accelerate wound closure, in part thanks to secreting factors such as the extracellular matrix–associated signaling protein CCN1 and platelet-derived growth factor AA (PDGF-AA), reinforcing the idea that acute, nonpersistent senescence can be beneficial (8, 9). Senescence also limits fibrotic responses (78). For example, senescence acts as a break in the progression of liver fibrosis that is induced by acute liver damage. Dying hepatocytes trigger the proliferation of HSCs, which secrete fibrogenic compounds to form a fibrotic scar. Eventually, these HSCs enter into senescence, and the SASP contributes to fibrotic scar degradation, clearance of the senescent cells, and restoration of tissue homeostasis (79). Finally, in response to tissue damage, factors secreted by senescent cells can also promote stemness (80). The processes that determine the balance between senescence, regeneration, and repair in response to damage remain to be further established.
Irrespective of their functional relevance, components of the SASP have limited utility as biomarkers, as they are not specific to senescence. Only reliable costaining or single-cell profiling will allow us to pinpoint which SASP components are driving the functions of the senescent secretome in specific contexts in vivo.
Regulation of the SASP.
The SASP is regulated at multiple levels, including transcription, translation, mRNA stability, and secretion. The SASP also depends on autocrine and paracrine positive-feedback loops that enhance robust signal amplification. Multiple signaling pathways, including the DDR (81, 82), p38 MAP kinase (33), and cGAS/STING (83–85), have been linked with the regulation of the SASP. Most of these cascades seem to converge on the activation of NF-κB and CCAAT/enhancer-binding protein-β (C/EBPβ).
NF-κB and C/EBPβ are activated and enriched in the chromatin fraction in senescent cells (64, 65, 86) and regulate SASP components by directly controlling transcription of key regulators of the inflammatory SASP such as IL-8 or IL-6. In turn, IL-6 and IL-8 act in an autocrine feed-forward loop to enhance the activity of C/EBPβ and NF-κB and amplify SASP signaling (64, 65). IL-1α also behaves as a master regulator of the SASP (87). Indeed, IL-1α signaling is sufficient to partially recapitulate the inflammatory SASP (68). Moreover, inhibition of the NLRP3 inflammasome, necessary to process and activate IL-1β, can blunt the SASP (68).
The SASP can also be regulated at an epigenetic level. For example, MLL1 inhibition blunts the SASP via a decrease in γ-H2Ax at SASP genes (88), and the histone variant macroH2A1 is a critical component of the positive-feedback loop that maintains SASP expression (89). Perhaps more relevant, SASP induction requires the recruitment of BRD4 to senescence-activated superenhancers that lie adjacent to SASP genes (90).
The mTOR pathway is an important node in SASP regulation. It has been described that mTOR-mediated phosphorylation of the translation repressor protein 4EBP regulates the translation of IL-1α (91) and MAP kinase–activated protein kinase 2 (MAPKAPK2) (92) to control the SASP. MAPKAPK2 inhibits ZFP36L1, an mRNA-binding protein that specifically targets proinflammatory SASP components for mRNA degradation. Thus, mTOR can control the senescent secretome by indirectly regulating the stability of SASP mRNAs (92). Moreover, cells undergoing OIS boost the production of SASP components by coordinating protein synthesis and autophagy in the TOR-autophagy spatial coupling compartment, often abbreviated as TASCC (93). However, the regulation of the SASP by autophagy is complex, as GATA4 controls the SASP by inhibiting autophagy (94).
Most of the regulatory mechanisms reviewed here have a global effect on SASP regulation or preferentially control a proinflammatory SASP subset. In fact, this particular proinflammatory arm is highly conserved among the most studied forms of senescence, including replicative, irradiation-induced, and oncogene-induced senescence. The heterogeneous and variable composition of the SASP, however, suggests the existence of different SASP subsets. For example, a distinct senescent secretome has been described in the context of mitochondrial dysfunction (95), and NOTCH signaling enables a switch between a TGF-β and an inflammatory secretome (60).
SA β-galactosidase activity
The most widely used senescence marker is senescence-associated β-galactosidase (SA β-gal) activity. This enzymatic activity, which is found in many normal cells under physiological conditions (pH 4.0–4.5), is significantly amplified in senescent cells as a result of increased lysosomal content (96, 97). Because of this, histochemical detection of β‑gal activity at pH 6.0 (suboptimal for normal cells) allows specific identification of senescent cells (98). Since SA β-gal activity is detected in most senescent settings, both in vitro and in vivo, it is considered a de facto hallmark of senescence. However, cells deficient in GLB1 (the gene encoding for lysosomal β-gal) do not exhibit impairments in the functional aspects of senescence (97). Another lysosomal enzyme, α-fucosidase, has been proposed as an alternative senescence biomarker (99). The increased lysosomal content of senescent cells, which might reflect increased autophagy (100), is also a feature of cell types such as active macrophages, Kupffer cells, and osteoclasts (101, 102). This highlights the need to use additional markers in combination with SA β-gal activity to characterize senescence. Recently, lipofuscin, an aggregate of oxidized proteins, lipids, and metals that are known to accumulate in aged tissues, was reported to colocalize with SA β-gal activity in senescent cells (103). Although the specificity of lipofuscin accumulation during senescence has not been broadly studied, its potential as a biomarker is appealing, especially since, unlike SA β-gal activity, lipofuscin staining can be performed in paraffin-embedded archival material.
Chromatin reorganization
Senescence is associated with large-scale chromatin rearrangements (104). Besides the already described DDR (26) and the formation of PML bodies (a type of matrix-associated nuclear domain) (105), the most striking chromatin change observed in senescent cells is the formation of senescence-associated heterochromatic foci (SAHFs), which are more prominent in human cells undergoing OIS (106). These foci can be identified by DAPI staining and are characterized by enrichment of repressive marks such trimethylated H3K9 and heterochromatic protein 1 (HP1), accumulation of high-mobility group HMGA proteins, and loss of linker histone H1 (4). Therefore, it was hypothesized that SAHFs may represent a senescence-specific heterochromatic compartment. The SAHFs are the result of the spatial repositioning of preexisting repressive marks rather than being caused by global changes in histone methylation (107). Interestingly, genomic regions contained in the SAHFs are found in lamina-associated domains (LADs) in proliferating cells (108). Upon induction of senescence and loss of lamin B1 (LMNB1), these LADs detach from the nuclear periphery and cluster within the nuclei. Strikingly, Hi-C analysis (a high-throughput variant of chromosome conformation capture) suggests that there is a loss of internal structure and possible decompaction of the DNA enriched in the SAHF core (109), thus calling into question the concept of SAHFs as silencing compartments. Another consequence of the autophagy-mediated degradation of LMNB1 observed in senescence (110) is compromised integrity of the nuclear envelope, which may lead to the observed blebbing of cytoplasmic chromatin fragments (CCFs) (111). These CCFs, via activation of the cGAS/STING pathway, are crucial to initiate a proinflammatory response in senescence and cancer. Indeed, mice deficient in STING show impaired immunosurveillance and decreased tissue inflammation due to reduced SASP (83–85).
Although the diverse global chromatin changes occurring in senescence have clear functional consequences, these changes are not always observed macroscopically or conserved among different cell types or senescence inducers (112). Therefore, it is not straightforward to rely on these global chromatin alterations to identify senescent cells in vivo.
Metabolism, autophagy, and mitochondrial function in senescent cells
Senescent cells display metabolic changes such as increases in glycolysis, mitochondrial metabolism, and autophagy. A clear illustration of this came when Dörr and colleagues showed that the high production of components of the SASP relies on enhanced ATP production mediated by mitochondrial metabolism (i.e., the TCA cycle) and glycolysis (113). The authors suggested that the increased SASP production and secretion lead to a proteotoxic stress that can be attenuated by activation of autophagy. Previous evidence highlighting the role of autophagy in the sustainability of the SASP had reported that the amino acid supply required to maintain the rapid protein turnover that the SASP demands relies on coupling autophagy (autolysosomes) with protein synthesis (93).
The role of mitochondrial metabolism and autophagy in senescence, however, remains controversial. Some studies have shown that inhibition of autophagy facilitates senescence (94, 114). Also, senescent cells display decreased levels of mitophagy (mitochondrial autophagy), which results in a defective mitochondrial network that may contribute to metabolic dysfunction in aging (115). In agreement with this, Garcia-Prat et al. showed that mitophagy contributes to the maintenance of muscle function during aging by preventing senescence in satellite cells (116). Defective mitophagy leads to accumulation of dysfunctional mitochondria and ROS-induced senescence. Similar results were observed in a cell model of parkin-mediated mitophagy (117). Interestingly, depletion of mitochondria impaired senescence by disrupting a positive-feedback loop involving ROS production and a DDR rather than as a result of insufficient energy levels. Indeed, senescent cells lacking mitochondria displayed higher ATP levels due to increased glycolysis. Therefore, it seems that at least in some contexts, the execution of the senescence program is compromised not by insufficient energy levels, but rather by the status of mitochondrial oxidative metabolism. In work related to this concept, Kaplon and colleagues (118) argued that senescent cells must reprogram metabolism in order to support their metabolic demands. They showed that an increase of mitochondrial oxidative respiration through activation of pyruvate dehydrogenase was required for the execution of OIS.
Morphological changes associated with senescence
In cell culture, senescence is normally accompanied by significant morphological changes. Senescent cells become flat, enlarged, and vacuolized, and sometimes appear with multiple or enlarged nuclei. Changes in shape rely on the status of the scaffolding protein caveolin 1 and the Rho GTPases Rac1 and CDC42 (119), and vacuolation has been associated with ER stress caused by the unfolded protein response (120). Senescent cells also form cytoplasmic bridges that allow them to signal to neighboring cells via direct intercellular protein transfer (62). Beyond these examples, the functional significance of most morphological changes associated with senescence is unclear. In vivo, senescent cells appear to preserve the morphology dictated by the architecture of the tissue. However, recent studies have discovered that SA β-gal+ cells in aged mice increase in size (77).
Resistance to apoptosis
Senescence and apoptosis are alternative cell fates that often can be triggered by the same stressors. While we do not have a full understanding of what makes the cell decide between one and another program, mechanisms must be in place to lock those decisions. In this regard, senescent cells are resistant to extrinsic and intrinsic apoptosis (121, 122). Recent studies have suggested that this is a result of the upregulation of BCL-2 family proteins such as BCL-W and BCL-XL (123). This is of extraordinary practical relevance since inhibiting BCL-2 family proteins induces apoptosis on senescent cells (19, 123, 124).
Concluding remarks
Senescence is emerging as a therapeutic target relevant for a wide range of pathologies. Pro-senescent and anti-senescent (senolytic) therapies are showing promising results in preclinical mouse models, and human clinical trials are in progress. One of the most widely used senolytics are BCL-2 family inhibitors that target apoptotic resistance of senescent cells (19, 123, 124). Conversely, drugs that induce senescence (such as palbociclib and other CDK4/6 inhibitors) have shown significant benefits as anticancer agents (125). In agreement with the observations that inhibition of CDK4/6 can induce senescence in certain tumor cell lines in vitro, clinical trials have shown that combining palbociclib with current breast cancer therapies significantly increases median progression-free survival. Interestingly, in order to avoid the possible side effects mediated by the resulting senescent tumoral cells, the pro-senescence therapy with CDK4/6 inhibitors could be combined with senolytics, especially if the immune system is compromised and cannot effectively clear senescent cells.
Overall, it is becoming apparent that it is essential to characterize the mechanisms and functions of senescence cells in each context to design specific, optimal, nontoxic senescence therapies. This will require better characterization of the various subtypes of senescence in vivo and establishing which senescence features can be targeted or enhanced without negatively affecting normal cells. Although emphasis is currently shifting to practical aspects and viability of such therapies, a better understanding of the molecular mechanisms of senescence is essential. The accurate identification of senescent cells in vivo is also important. As indicated in this Review, all in vivo biomarkers for senescence, including SA β-gal activity or p16INK4a expression, may be unreliable depending on the context or owing to technical limitations. Because it seems unlikely that a new universal biomarker of senescence will appear, especially considering the heterogeneity of senescent phenotypes in vivo, single-cell transcriptome and proteome profiling of tissues will be key to understand the nature of these senescent cells, investigate their specific liabilities, and eventually implement therapeutic approaches.
Acknowledgments
Core support from the Medical Research Council (MRC; grants MC-A652-5PZ00 and MC_U120085810) funded the research in JG’s laboratory.
Version 1. 04/02/2018
Print issue publication
Footnotes
Conflict of interest: JG is a consultant to Unity Biotechnology, which also funds research in his laboratory. JG and NH are named inventors in filed patents related to senolytic therapies (GB1708458.3 and GB1708456.7 to JG and NH).
Reference information: J Clin Invest. 2018;128(4):1238–1246.https://doi.org/10.1172/JCI95148.
References
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28(2):99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15(7):482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 6.Storer M, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155(5):1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 7.Muñoz-Espín D, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155(5):1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol. 2010;12(7):676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10(1):51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21(12):1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114(9):1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Childs BG, Baker DJ, Wijshake T, Conover CA, Campisi J, van Deursen JM. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354(6311):472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demaria M, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7(2):165–176. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baar MP, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169(1):132–147.e16. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15(7):397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 21.Kang TW, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479(7374):547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 22.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 23.Jurk D, et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat Commun. 2014;2:4172. doi: 10.1038/ncomms5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helman A, et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22(4):412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamino T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009;15(9):1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 26.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8(7):512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 27.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 28.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 29.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 30.Fumagalli M, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol. 2012;14(4):355–365. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suram A, et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012;31(13):2839–2851. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alimonti A, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120(3):681–693. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30(8):1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 35.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beauséjour CM, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22(16):4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196(1):33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 38.Stewart SA, Ben-Porath I, Carey VJ, O’Connor BF, Hahn WC, Weinberg RA. Erosion of the telomeric single-strand overhang at replicative senescence. Nat Genet. 2003;33(4):492–496. doi: 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- 39.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 41.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7(9):667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 42.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576(1–2):22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 43.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 45.Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6(1):67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 46.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23(10):1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribeiro JD, et al. ZRF1 controls oncogene-induced senescence through the INK4-ARF locus. Oncogene. 2012;32(17):2161–2168. doi: 10.1038/onc.2012.241. [DOI] [PubMed] [Google Scholar]
- 50.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotake Y, Zeng Y, Xiong Y. DDB1-CUL4 and MLL1 mediate oncogene-induced p16INK4a activation. Cancer Res. 2009;69(5):1809–1814. doi: 10.1158/0008-5472.CAN-08-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin N, et al. Interplay between Homeobox proteins and Polycomb repressive complexes in p16INK(4)a regulation. EMBO J. 2013;32(7):982–995. doi: 10.1038/emboj.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith LL, et al. Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell. 2011;8(6):649–662. doi: 10.1016/j.stem.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Cakouros D, et al. Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol Cell Biol. 2012;32(8):1433–1441. doi: 10.1128/MCB.06315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamakoshi K, et al. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J Cell Biol. 2009;186(3):393–407. doi: 10.1083/jcb.200904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burd CE, et al. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013;152(1-2):340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9(2):81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 59.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoare M, et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat Cell Biol. 2016;18(9):979–992. doi: 10.1038/ncb3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson G, et al. A senescent cell bystander effect: senescence-induced senescence. Aging Cell. 2012;11(2):345–349. doi: 10.1111/j.1474-9726.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biran A, et al. Senescent cells communicate via intercellular protein transfer. Genes Dev. 2015;29(8):791–802. doi: 10.1101/gad.259341.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehmann BD, et al. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008;68(19):7864–7871. doi: 10.1158/0008-5472.CAN-07-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Acosta JC, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133(6):1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 65.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 66.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132(3):363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hubackova S, Krejcikova K, Bartek J, Hodny Z. IL1- and TGFbeta-Nox4 signaling, oxidative stress and DNA damage response are shared features of replicative, oncogene-induced, and drug-induced paracrine ‘bystander senescence’. Aging (Albany NY) 2012;4(12):932–951. doi: 10.18632/aging.100520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15(8):978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98(21):12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coppé JP, Kauser K, Campisi J, Beauséjour CM. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem. 2006;281(40):29568–29574. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- 71.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 72.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Mitri D, et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515(7525):134–137. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]
- 74.Toso A, et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 2014;9(1):75–89. doi: 10.1016/j.celrep.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 75.Eggert T, et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30(4):533–547. doi: 10.1016/j.ccell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 77.Biran A, et al. Quantitative identification of senescent cells in aging and disease. Aging Cell. 2017;16(4):661–671. doi: 10.1111/acel.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lujambio A, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153(2):449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ritschka B, et al. The senescence-associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes Dev. 2017;31(2):172–183. doi: 10.1101/gad.290635.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodier F, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glück S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol. 2017;19(9):1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dou Z, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550(7676):402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A. 2017;114(23):E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chien Y, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25(20):2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1α is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci U S A. 2009;106(40):17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Capell BC, et al. MLL1 is essential for the senescence-associated secretory phenotype. Genes Dev. 2016;30(3):321–336. doi: 10.1101/gad.271882.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen H, Ruiz PD, McKimpson WM, Novikov L, Kitsis RN, Gamble MJ. MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence-associated secretory phenotype. Mol Cell. 2015;59(5):719–731. doi: 10.1016/j.molcel.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tasdemir N, et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6(6):612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laberge RM, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17(8):1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herranz N, et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17(9):1205–1217. doi: 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Narita M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332(6032):966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kang C, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349(6255):aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wiley CD, et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016;23(2):303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated β-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113(pt 20):3613–3622. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 97.Lee BY, et al. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging cell. 2006;5(2):187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 98.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hildebrand DG, Lehle S, Borst A, Haferkamp S, Essmann F, Schulze-Osthoff K. α-Fucosidase as a novel convenient biomarker for cellular senescence. Cell Cycle. 2013;12(12):1922–1927. doi: 10.4161/cc.24944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23(7):798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bursuker I, Rhodes JM, Goldman R. β-Galactosidase — an indicator of the maturational stage of mouse and human mononuclear phagocytes. J Cell Physiol. 1982;112(3):385–390. doi: 10.1002/jcp.1041120312. [DOI] [PubMed] [Google Scholar]
- 102.Kopp HG, Hooper AT, Shmelkov SV, Rafii S. β-Galactosidase staining on bone marrow. The osteoclast pitfall. Histol Histopathol. 2007;22(9):971–976. doi: 10.14670/HH-22.971. [DOI] [PubMed] [Google Scholar]
- 103.Georgakopoulou EA, et al. Specific lipofuscin staining as a novel biomarker to detect replicative and stress-induced senescence. A method applicable in cryo-preserved and archival tissues. Aging (Albany NY) 2013;5(1):37–50. doi: 10.18632/aging.100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Adams PD. Remodeling chromatin for senescence. Aging Cell. 2007;6(4):425–427. doi: 10.1111/j.1474-9726.2007.00313.x. [DOI] [PubMed] [Google Scholar]
- 105.Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14(16):2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 106.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 107.Chandra T, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell. 2012;47(2):203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sadaie M, et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 2013;27(16):1800–1808. doi: 10.1101/gad.217281.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chandra T, et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10(4):471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dou Z, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527(7576):105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ivanov A, et al. Lysosome-mediated processing of chromatin in senescence. J Cell Biol. 2013;202(1):129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Micco R, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13(3):292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dörr JR, et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501(7467):421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- 114.Wang Y, et al. Autophagic activity dictates the cellular response to oncogenic RAS. Proc Natl Acad Sci U S A. 2012;109(33):13325–13330. doi: 10.1073/pnas.1120193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61(5):654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.García-Prat L, et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;529(7584):37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 117.Correia-Melo C, et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016;35(7):724–742. doi: 10.15252/embj.201592862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498(7452):109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 119.Cho KA, et al. Morphological adjustment of senescent cells by modulating caveolin-1 status. J Biol Chem. 2004;279(40):42270–42278. doi: 10.1074/jbc.M402352200. [DOI] [PubMed] [Google Scholar]
- 120.Denoyelle C, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8(10):1053–1063. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 121.Wang E. Senescent human fibroblasts resist programmed cell death, and failure to suppress Bcl-2 is involved. Cancer Res. 1995;55(11):2284–2292. [PubMed] [Google Scholar]
- 122.Sasaki M, Kumazaki T, Takano H, Nishiyama M, Mitsui Y. Senescent cells are resistant to death despite low Bcl-2 level. Mech Ageing Dev. 2001;122(15):1695–1706. doi: 10.1016/S0047-6374(01)00281-0. [DOI] [PubMed] [Google Scholar]
- 123.Yosef R, et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat Commun. 2016;7:11190. doi: 10.1038/ncomms11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu Y, et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016;15(3):428–435. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6(4):353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McHugh D, Gil J. Senescence and aging: Causes, consequences, and therapeutic avenues. J Cell Biol. 2018;217(1):65–77. doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]