Abstract
A metagenome wide association (MGWA) study of bacterial host association determinants in Drosophila predicted that LPS biosynthesis genes are significantly associated with host colonization. We were unable to create site-directed mutants for each of the predicted genes in Acetobacter, so we created an arrayed transposon insertion library using Acetobacter fabarum DsW_054 isolated from Drosophila. Creation of the A. fabarum DsW_054 gene knock-out library was performed by combinatorial mapping and Illumina sequencing of random transposon insertion mutants. Transposon insertion locations for 6,418 mutants were successfully mapped, including hits within 63% of annotated genes in the A. fabarum DsW_054 genome. For 45/45 members of the library, insertion sites were verified by arbitrary PCR and Sanger sequencing. Mutants with insertions in four different LPS biosynthesis genes were selected from the library to validate the MGWA predictions. Insertion mutations in two genes biosynthetically upstream of Lipid-A formation, lpxC and lpxB, show significant differences in host association, whereas mutations in two genes encoding LPS biosynthesis functions downstream of Lipid-A biosynthesis had no effect. These results suggest an impact of bacterial cell surface molecules on the bacterial capacity for host association. Also, the transposon insertion mutant library will be a useful resource for ongoing research on the genetic basis for Acetobacter traits.
Keywords: Acetobacter, Drosophila, lipopolysaccharide, Lipid A, mutant library, Mutant Screen Report
Animal associated microbes (‘microbiota’) influence numerous phenotypes of their hosts, including metabolic function, mental health and various diseases (De Palma et al. 2015; Ussar et al. 2015; Cammarota et al. 2015; Verdu et al. 2015; Bienenstock et al. 2015; Petra et al. 2015). The taxonomic and functional complexity of the microbiota challenges our ability to define how these microbes associate with and influence traits of their hosts. The Drosophila melanogaster microbiota is relatively simple, mainly comprised of yeasts, acetic acid bacteria (AABs) and lactic acid bacteria (LABs), together with less abundant but highly prevalent Enterobacteriaceae (Wong et al. 2011; Broderick and Lemaitre 2012; Erkosar et al. 2013; Ma et al. 2015; Cox and Gilmore 2007; Bakula 1969; Brummel et al. 2004; Corby-Harris et al. 2007), and varies in composition and abundance between and within individual flies, including with age (Blum et al. 2013; Broderick et al. 2014; Wong et al. 2013; Ren et al. 2007). This apparent inconstancy is determined by numerous factors, including host genetic selection, environmental sampling, and dietary effects (Blum et al. 2013; Chaston et al. 2015; Wong et al. 2015). For example, the fly microbiota is replenished through the diet, and bacterial loads can be reduced by frequent transfer of flies to sterile diets (Blum et al. 2013; Broderick et al. 2014). Host genetic influences include reciprocal interactions between the host and the diet since fly diets inoculated with a defined fly microbiota vs. the same microbiota and flies, have a different ending microbiota composition (Wong et al. 2015). Also, host genotype can influence microbial composition since different fly genotypes reared from birth with the same starting microbiota can have different ending compositions of microbes (Chaston et al. 2015; Wong et al. 2015). However, under laboratory conditions there is no evidence that the host retains specific bacterial taxa or functions within or across generations, or for a host phylogenetic signal on the identity of associated microbes (Wong et al. 2013). The variability in bacterial abundance and composition is an important factor in animal phenotype studies (Clark et al. 2015; Vandeputte et al. 2017).
Microbial genetic determinants also influence the abundance of bacteria associated with D. melanogaster. For example, the typical laboratory fly associated microbiota do not produce sufficient uracil for the Drosophila gut to elicit an immune response, allowing the insect to respond differently to normal and pathogenic microbes (Lee et al. 2013; Ha et al. 2009). Additionally, D-alanylated teichoic acids on the cell walls of L. plantarum were sensed by the Drosophila enterocytes, enhancing host digestion and promoting growth and maturation of the host (Matos et al. 2017). Acetobacter species that degrade uric acid can outcompete those that do not when associated with Drosophila (Winans et al. 2017). Together, these studies have identified some bacterial genes that influence how bacteria associate with D. melanogaster; our goal was to identify new genes with similar influence.
Because there were no successful approaches for site-directed mutagenesis in fruit fly isolates of Acetobacter, and our preliminary efforts to develop such approaches failed, we created an arrayed transposon insertion library of Acetobacter mutants. Arrayed mutant libraries allow for high-throughput identification of thousands of knock-out mutants at once. One method was described by Goodman et al. (Goodman et al. 2011; Goodman et al. 2009), where transposon insertion mutants were arrayed in 96-well plates, combined into sequencing pools, and sequenced in one sequencing run. The mutants were used to determine bacterial functions necessary for survival in the mouse gut (Goodman et al. 2009). Others have screened arrayed transposon insertion libraries of Klebsiella pneumoniae for antibiotic sensitivity (Ramage et al. 2017); and of L. plantarum for bacterial genes that promote host growth (Matos et al. 2017). In this study we created an arrayed library of 6,418 A. fabarum DsW_054 transposon insertion mutants and report its first use by testing five lipopolysaccharide (LPS) biosynthesis mutants for their ability to associate with D. melanogaster.
Materials and Methods
Bacterial and fly growth media and conditions
D. melanogaster Canton-S flies were grown at 25° on a yeast glucose diet containing 100 g/ liter brewer’s yeast (inactive) (MP Biomedicals), 100 g/ liter glucose (Sigma), 12 g/ liter agar (Apex), and preservatives (0.04% phosphoric acid and 0.42% propionic acid (Sigma)) on a 12-h-light/12-h-dark cycle.
Bacterial strains used in the study are included in Table 1. Media used included lysogeny broth (LB)/agar (Sigma), modified MRS (mMRS) broth/agar and potato dextrose broth/ agar (Sigma). The plasmid bearing E. coli strain was cultured with 50 μg/ml kanamycin and Acetobacter gene knock-out mutants were cultured with 30 μg/ml chloramphenicol and 50 μg/ml kanamycin. All bacterial strains were cultured at 30°. A. fabarum DsW_054 was cultured in potato dextrose broth prior to matings.
Table 1. Bacterial strains used in this study.
| Strain Name | Abbreviation | Preferred Medium | Citation |
|---|---|---|---|
| A. fabarum DsW_054 | mMRS | Winans et al. (2017) | |
| Escherichia coli S17 pJG714 | LB-kan | This study (see Figure S1 in File S1) | |
| A. fabarum DsW_054 Tn5::lpxC | lpxC | mMRS | This study |
| A. fabarum DsW_054 Tn5::lpxB | lpxB | mMRS | This study |
| A. fabarum DsW_054 Tn5::lpxK | lpxK | mMRS | This study |
| A. fabarum DsW_054 Tn5::gmhD_1 | gmhD_1 | mMRS | This study |
| A. fabarum DsW_054 Tn5::gmhD_2 | gmhD_2 | mMRS | This study |
Creation of an arrayed mutant library
An arrayed transposon insertion library was created in two steps: by conjugally transferring the Tn5 transposon vector pJG714 into A. fabarum DsW_054; and by arraying the transposants into 96-well plates. A. fabarum DsW_054 cells were cultured in potato medium for 24 -36 hr at 30° to OD600 = 0.6 and mixed with 18-24 h cultures of E. coli S17-pir (OD600 = 1.0) containing the plasmid pJG714, cultured in LB-kanamycin. Prior to mixing, the cells were washed three times in potato medium, 6 ml A. fabarum DsW_054 were condensed to 100 μl, and 3 ml E. coli were re-suspended in 500 μl. The two strains were mixed in a 1:1 volumetric ratio and transferred to potato medium plates in 50 μl spots. After 4 hr at 30° the cells were collected from the plate in 1ml potato medium, dilution plated (1:50) onto 2X-YPG plates containing kanamycin and chloramphenicol, incubated at 30° for four days, and stored at 4° for no more than 1 week before arraying in 96-well plates. Colonies were individually picked into mMRS broth in 96-well plates to array the transposon insertion library. Each plate was then sealed with Parafilm and incubated with gentle shaking for 48 hr at 30° until most cell densities were between OD600 0.5 and 1.0, and frozen in mMRS-25% glycerol.
Combinatorial mapping and sequencing
To identify the insertion site of each mutant we employed a combinatorial mapping approach using an Eppendorf EpMotion 5075 TMX pipetting robot. A 24 bit binary barcode was assigned to each well of each 96 well plate, corresponding to the presence (1) or absence (0) of the bacteria from each well in the corresponding pooled vials (Goodman et al. 2011). Intermediate sets of 24 pools were created from five 96-well plates at a time in the attached thermocycler feature held at 4° by pipetting 10 μl to each intermediate pool for which a ‘1’ was assigned to that sample of the 96-well plate. The different pools were stored at -20° for a maximum of 2 months. Once all the intermediate pools had been created, a final set of 24 pools was created by mixing all intermediate pools from the same barcode positions (e.g., all ‘Pool 1’ tubes) in equal volumetric ratios. DNA was extracted separately from each of the pools using the DNeasy PowerLyzer Microbial Kit (Qiagen cat# 12255-50).
For Illumina sequencing, each of the 24 final pools was assigned a unique 6 bp indexing barcode that was introduced by PCR (for library preparation details and primer sequences, see File S2). Briefly, DNA was fragmented using a DNA fragmentase, the fragments were C-tailed, and Illumina indexing and sequencing primers (Table S1 in File S1) were added via two rounds of PCR.
Data analysis
The Illumina sequencing data were mapped to the A. fabarum DsW_054 genome using a previously-published TnSeq pipeline (Arnold et al. 2017). Afterward, a 24-bit barcode was assigned to each mapped insertion site in the genome using a threshold of 50 reads per sequencing pool. For example, if there were more than 50 reads for a particular insertion site in sequencing pool 1, a 1 was assigned at the first position in the barcode; if there were less than 50 reads in the second sequencing pool, a 0 was assigned at the second barcode position, etc. We used 50 reads as a cutoff point, but most sites had more than 200 reads for each positive indexing primer. Insertion site barcodes were then matched to the barcodes from the original combinatorial mapping assignments for the well location in the 96-well plate library. The complete mapping to the library is summarized in File S3.
Library validation
Arbitrary PCR was used to validate a subset of the mapped insertion sites for the arrayed library. A transposon specific primer was paired with arbitrary primers for the first round and the second round used a primer specific to the transposon and a primer specific to the tail of the initial arbitrary primers (Table S1 in File S1). A single colony that was suspended in 10 μl H2O and boiled for 10 min at 99° served as the template. For first round arbitrary PCR we mixed 14 μl H2O, 2.5 μl 10x (NH4)2SO4 buffer, 1 μl DMSO, 2 μl MgCl2 (25 mM), 1 ul dNTP (10 μM), 1 μl Arb1 primer (20 μM), 1 μl Arb6 primer (20 μM), 0.5 μl 133 primer (20 μM), 1 μl Taq polymerase 1U/μl (Thermo Scientific), and 1 μl template (boiled cell preparation). We ran the reaction using the following program: 94° for 3:00 followed by six cycles of 94° for 0:20, 30° for 0:20 and 72° for 1:00, followed by 30 cycles of 94° for 0:20, 45° for 0:20, 72° for 1:00 and finished at 72° for 5:00.
This was followed by a second round of arbitrary PCR. For each sample we mixed 31.25 μl H2O, 5 μl 10x (NH4)2SO4 buffer, 2 μl DMSO, 4 μl MgCl2 (25mM), 2 μl dNTP (10 μM), 2 μl Arb2 primer (20 μM), 2 μl 134 primer (20 μM), 1 μl Taq polymerase 1U/μl (Thermo Scientific, 1 μl Template (Product from first round). We ran the solution at 94° for 1:00 followed by 35 cycles of 94° for 0:15, 52° for 0:20, 72° for 1:00, and finish at 72° for 5:00. The 150-700 bp product was visualized on a 1% agarose gel to verify amplification before sequencing via Sanger sequencing. The specific insertion site was manually mapped to the A. fabarum DsW_054 genome using the BLAST feature in RAST (Wattam et al. 2017).
Metagenome-wide-association study
A metagenome wide association study (MGWA) was performed to predict bacterial genes necessary to associate with D. melanogaster using previously published phenotype and genotype data (Chaston et al. 2014). Briefly, in the previous study CFU abundances were collected from pooled homogenates of five whole female flies that had been individually reared with one of 41 different, genome-sequenced bacterial strains. Separately, orthologous groups (OGs) in the 41 bacterial strains were clustered using OrthoMCL, yielding 12,354 OGs. Statistically significant associations between the CFU abundances in D. melanogaster and the presence-absence patterns of the OGs were identified using the R package MAGNAMWAR (Sexton et al. 2018). Data were processed using a linear mixed model with log-transformed CFU abundances as the response variable, and experiment and bacterial treatment as independent random effects. Statistically significant outputs were Bonferroni-corrected.
KEGG pathway analysis
We performed a KEGG pathway analysis on the MGWA results to identify bacterial genetic pathways that were enriched among the significant MGWA results. We assigned KEGG pathway numbers to all OGs in the dataset using ‘BLASTKOALA’ and compared the number of OGs in different KEGG pathways between a reference dataset (all OGs) and a significant dataset (the top 324 significant OGs from the MGWA, based on a p-value less than 0.001) using the ‘KEGG Mapper – Search Pathway’ online tool (Kanehisa et al. 2017; Kanehisa and Goto 2000; Kanehisa et al. 2016). Significant enrichment of KEGG pathways in the top 324 significant OGs was determined by chi-square analysis, with false-discovery-rate (fdr) p-value correction in R. Pathways were only used in the analysis if they had 4 or greater counts in both the top and the reference set of OGs.
Mutant analysis
To verify predictions of the MGWA, mutants from the arrayed library that bore lesions in LPS biosynthesis genes were reared individually with D. melanogaster and their load in adult flies was measured. Monoassociated D. melanogaster were reared as described previously (Koyle et al. 2016). Briefly, adult flies were reared on grape juice agar plates for 16-18 hr to allow for egg collection, eggs were collected and sterilized in two 2.5 min washes of 0.6% hypochlorite solution, rinsed three times in sterile water, and transferred to sterile yeast-glucose diet lacking preservative at a density of 30-80 eggs per vial. Selected transposon insertion mutants (see Table 1) were individually added by inoculating to the vials 50 μl of OD600 = 0.1 normalized bacterial culture. Three separate experiments containing all experimental treatments were performed, each in triplicate. At 5-7 days of age, pools of 5 female flies were lightly anesthetized on CO2 and homogenized in microcentrifuge tubes containing 125 μl MRS and 125 μl Lysing Matrix D ceramic beads (MP Biomedicals 11654034) on a GenoGrinder 2010 homogenizer for 2 min at 1250 rpm. The homogenate was dilution plated on mMRS plates and colony forming units (CFUs) were manually counted. Any flies bearing bacteria other than Acetobacter (determined by visual inspection of colony morphology) were removed from the experiment.
Data and reagent availability
Strains are available upon request. File S1 contains all supplemental figures and tables together with detailed descriptions of other supplemental files. Table S1 in File S1 contains primer sequences used. Table S2 in File S1 contains data from representative conjugation experiments. Tables S3–S6 in File S1 contain pathway essentialiaty predictions for Acetobacter fabarum DsW_054 (Table S3 in File S1), Rhodobacter sphaeroides (Table S4 in File S1), Rhizobium leguminosarum (Table S5 in File S1), and Caulobacter crescentus (Table S6 in File S1). Figure S1 in File S1 is a map of pJG714. File S2 is a detailed protocol for library preparation. File S3 contains the annotated insertion mutant library of A. fabarum DsW_054. File S4 is a script for running the MGWA analysis. File S5 contains the raw phenotype data, the means of which were originally published in (Chaston et al. 2014). File S6 contains the OrthoMCL gene clustering results from work initially published in (Chaston et al. 2014). File S7 contains the MGWA results for host colonization. File S8 describes the Acetobacter conjugation trials. Sequence data are available in the SRA under accession number PRJNA422683.
Results
Creation of an arrayed and mapped transposon insertion library in A. fabarum DsW_054
We constructed a mapped, arrayed transposon insertion library in a strain of Acetobacter isolated from wild Drosophila to enable us to study the effects of gene knockouts in an Acetobacter strain that was otherwise recalcitrant to genetic manipulation. We were initially unable to obtain transposon-insertion-bearing exconjugants (‘tranposants’) from matings that used as recipients two different strains of Acetobacter isolated from laboratory flies, A. pomorum DmCS_004 and A. tropicalis DmCS_005 (Newell and Douglas 2014). As a follow-up, we screened 17 strains of Acetobacter for amenability to genetic modification. Of these, A. fabarum DsW_054, a strain that was isolated from wild-caught Drosophila suzukii (Winans et al. 2017), yielded the greatest number of transposants with a kanamycin-marked Tn5 transposon (Table S2 in File S1; File S8). Pairwise nucleotide alignments of the A. fabarum DsW_054 genome with those of two A. fabarum strains (KR and OG2) recently added to the NCBI WGS database, indicated >98% ANI with both, allowing us to provisionally assign this isolate to the species fabarum (data not shown).
A. fabarum DsW_054 was subjected to further optimization of a conjugation protocol to transfer a plasmid-based mini-Tn5 transposon from donor E. coli cells. The final protocol described in the methods was obtained after varying co-incubation ratios and times with an E. coli donor bearing pJG714, and selecting parameters that maximized transposant recovery. Proof-of-concept arbitrary PCR mapping confirmed that in eight randomly selected colonies, each mutant contained a single, unique transposon insertion site (data not shown).
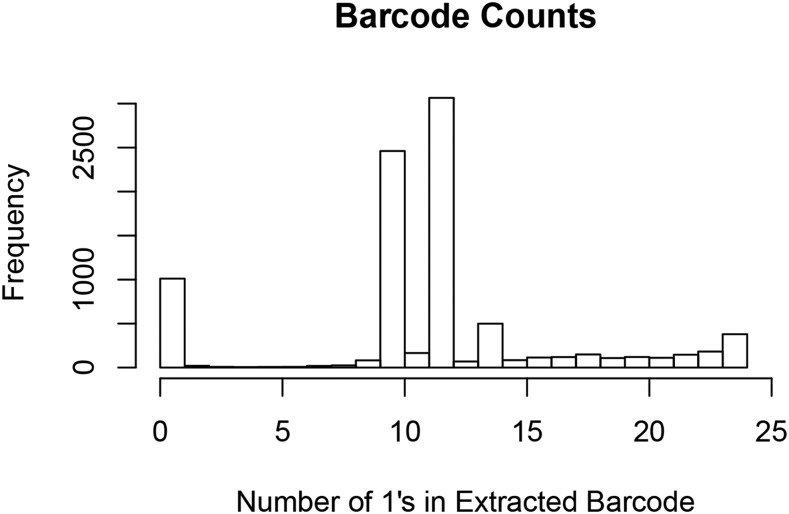
We created a mapped and arrayed transposon insertion library in A. fabarum DsW_054, following the approach of Goodman, et al. (2011). 8,550 mutants were created in groups no larger than 480 mutants at a time, individually transferred to 96-well plates, and the transposon insertion sites in each arrayed mutant were defined in a single Illumina sequencing run by a combinatorial mapping approach. The insertion sites for 6,418 mutants were precisely mapped, with unmapped mutants resulting from insufficient read coverage across all sequencing pools or to duplicate insertion sites in the library (possibly from sister clones). For example, a subset of transposon insertion sites were present in more than 14 sequencing pools, most likely representing sister clones with identical insertion sites that were present in separate wells within the library; whereas the insertion sites present in fewer than 10 sequencing pools were mostly likely not abundant enough to detect in some pools (Figure 1). The relatively large number of insertion sites that were present in just 1 or 2 pools likely result from sequencing errors. To confirm the validity of the mappings, the insertion site was validated manually by arbitrary PCR and Sanger sequencing in 41 of 41 mutants selected from the library, suggesting a high level of accuracy in the mapping of the mutants to their 96-well plate arrayed location. This high validation rate suggests that most of the mutants in the library are mapped to the appropriate location.
Figure 1.
Histogram of sequencing pools in which an insertion sequence was present in the Illumina sequencing run. The combinatorial mapping barcodes assigned each mutant to 10, 12, or 14 sequencing pools, evident as the major peaks in the histogram. Any sequence that was present in anything other than 10, 12 or 14 of the 24 possible sequencing pools could not be mapped to the library and was discarded from further analysis.
Prediction of essential genes in A. fabarum DsW_054
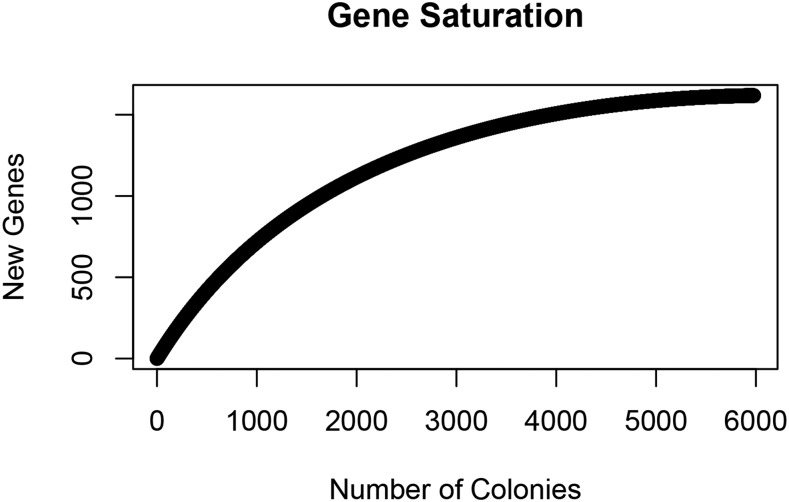
We determined near saturation of the insertion library through analysis of hits within predicted genes. 5,559 mutants mapped within an open reading frame (ORF) called by RAST, representing insertions in 63% of the 2,579 annotated genes in the A. fabarum DsW_054 genome (File S3). To assess the degree of gene saturation represented by the 5,559 ORF-mapped transposon insertion mutants, we performed a rarefaction analysis (Figure 2). The plateau of the curve suggested that the mutant library was well-represented by non-essential A. fabarum DsW_054 genes and that mapping insertion sites in more mutants was unlikely to substantially increase the gene coverage in the collection.
Figure 2.
A rarefaction curve showing the number of new genes with insertions as new colonies were added to the library. The plateau suggests that the addition of mutant colonies would not add a significant number of novel genes to the mutant library.
Based on the gene-level saturation of the mutant library we predicted that the 37% of A. fabarum DsW_054 genes that did not bear any transposon insertions are essential- or are located near (e.g., polar onto) essential-genes for growth in mMRS. A chi-square analysis of the KEGG pathways that were under- or over-represented in the genes bearing at least 1 transposon insertion site in our library suggested that genes encoding ribosomal components or involved in aminoacyl-tRNA biosynthesis, cell cycle, and protein export were essential for growth in mMRS; whereas the ABC transporters, starch and sucrose metabolism and two-component system pathways were enriched for non-essential genes (Table 2). Together, this analysis points to key A. fabarum DsW_054 genes for growth and survival in laboratory culture, the first such analysis of which we are aware for any Acetobacteraceae strain.
Table 2. Prediction of essential pathways in Acetobacter fabarum DsW_054.
| KO | Pathway | No insertions | All genes | p-value | fdr corrected p-value | Predicted essentiality |
|---|---|---|---|---|---|---|
| ko02010 | ABC transporters | 11 | 236 | 0.0005 | 0.01 | nonessential |
| ko03010 | Ribosome | 46 | 52 | 0.0005 | 0.01 | essential |
| ko00970 | Aminoacyl-tRNA biosynthesis | 22 | 24 | 0.0005 | 0.01 | essential |
| ko03060 | Protein export | 13 | 19 | 0.0005 | 0.01 | essential |
| ko04112 | Cell cycle - Caulobacter | 12 | 14 | 0.0005 | 0.01 | essential |
| ko00500 | Starch and sucrose metabolism | 1 | 75 | 0.0015 | 0.03 | nonessential |
| ko02020 | Two-component system | 15 | 170 | 0.0025 | 0.04 | nonessential |
| ko00195 | Photosynthesis | 7 | 7 | 0.0035 | 0.05 | essential |
| ko01120 | Microbial metabolism in diverse environments | 47 | 376 | 0.0040 | 0.05 | nonessential |
| ko00330 | Arginine and proline metabolism | 1 | 53 | 0.0055 | 0.06 | nonessential |
| ko00010 | Glycolysis / Gluconeogenesis | 3 | 66 | 0.0085 | 0.08 | nonessential |
| ko00240 | Pyrimidine metabolism | 28 | 78 | 0.0110 | 0.09 | essential |
| ko02040 | Flagellar assembly | 1 | 42 | 0.0110 | 0.09 | nonessential |
| ko00780 | Biotin metabolism | 11 | 22 | 0.0115 | 0.09 | essential |
| ko01502 | Vancomycin resistance | 5 | 5 | 0.0180 | 0.13 | essential |
| ko01110 | Biosynthesis of secondary metabolites | 107 | 419 | 0.0200 | 0.13 | essential |
| ko00052 | Galactose metabolism | 1 | 41 | 0.0210 | 0.130 | nonessential |
| ko00620 | Pyruvate metabolism | 4 | 69 | 0.0235 | 0.14 | nonessential |
| ko00550 | Peptidoglycan biosynthesis | 11 | 25 | 0.0360 | 0.20 | essential |
| ko01230 | Biosynthesis of amino acids | 49 | 176 | 0.0380 | 0.20 | essential |
| ko00250 | Alanine, aspartate and glutamate metabolism | 13 | 34 | 0.0425 | 0.20 | essential |
| ko00561 | Glycerolipid metabolism | 1 | 31 | 0.0425 | 0.20 | nonessential |
| ko00740 | Riboflavin metabolism | 6 | 10 | 0.0435 | 0.20 | essential |
| ko02024 | Quorum sensing | 12 | 112 | 0.0480 | 0.21 | nonessential |
| ko00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 13 | 33 | 0.0500 | 0.21 | essential |
Genes with no insertions were grouped into functional pathways using KEGG pathway mapper. The ‘no insertions’ column shows the number of genes that have no insertions within our library for each pathway. This was compared to the number of genes within that pathway that are present in A. fabarum (‘All genes’ column) using a chi-square test and the associated p-value and fdr corrected p-value are listed in addition to the predicted essentiality status for each pathway.
Prediction of bacterial pathways that influence bacterial load in D. melanogaster
To predict bacterial genes that are necessary to associate with D. melanogaster we performed a MGWA analysis (File S4). The analysis was performed using previously published data from a survey of CFU abundances in D. melanogaster that were individually associated with each of 41 genome-sequenced bacterial strains (Chaston et al. 2014). An MGWA that associated bacterial CFU abundance data (File S5) with OG presence-absence patterns in the 41 strains (File S6) predicted 324 bacterial genes that influence host association using a p-value cutoff of 0.001 (File S7). Because our previous MGWA analysis had successfully identified genes by looking for enriched functions among top hits, we performed a KEGG enrichment analysis to identify pathways that were enriched in the top 324 hits from the MGWA. After correction for multiple tests, genes from just one KEGG pathway, lipopolysaccharide biosynthesis, were significantly enriched in the top MGWA hits (Table 3). Therefore, MGWA predicted a key role for bacterial LPS biosynthesis in its ability to associate with D. melanogaster.
Table 3. A KEGG pathway analysis of genes predicted to affect host colonization revealed that the LPS synthesis pathway is significant in determining bacterial abundance in the fly gut.
| KO | Pathway | Top Hits | All | p-value | fdr |
|---|---|---|---|---|---|
| ko00540 | Lipopolysaccharide biosynthesis | 9 | 24 | 0.0005 | 0.05 |
| ko00270 | Cysteine and methionine metabolism | 7 | 44 | 0.0350 | 0.89 |
| ko00480 | Glutathione metabolism | 4 | 16 | 0.0355 | 0.89 |
| ko01501 | beta-Lactam resistance | 4 | 21 | 0.0600 | 0.89 |
| ko03020 | RNA polymerase | 2 | 5 | 0.0645 | 0.89 |
| ko00240 | Pyrimidine metabolism | 8 | 61 | 0.0725 | 0.89 |
| ko00450 | Selenocompound metabolism | 3 | 14 | 0.0780 | 0.89 |
The top hits from the MGWA were grouped into functional pathways and compared by chi-square test to the number of all genes in that pathway from the study. The returned p-value and fdr corrected p-value are listed.
Validation of MGWA predictions by mutant analysis
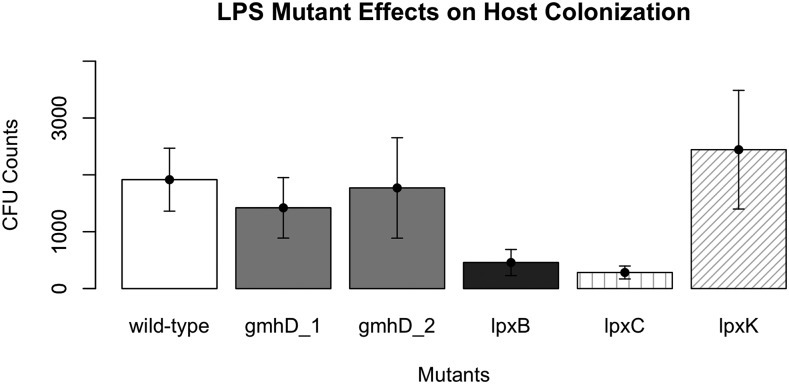
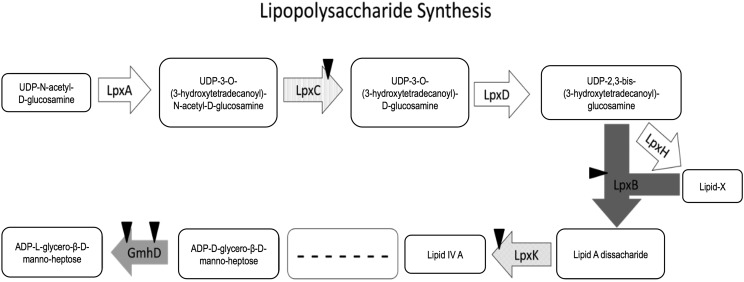
Based on the MGWA predictions, we hypothesized that A. fabarum DsW_054 bearing lesions in LPS biosynthesis genes would have a reduced ability to associate with D. melanogaster. To test this hypothesis, we searched the A. fabarum DsW_054 transposon insertion library for all mutants corresponding to one of the KEGG LPS biosynthesis pathway genes. Five mutants including disruptions in four genes were selected for analysis, and were individually reared with sterile D. melanogaster eggs. CFU load was measured in 5-7 days old adults, revealing that, of the tested genes, lpxB and lpxC mutants that preceded synthesis of Lipid A disaccharide were significantly impaired in their ability to associate with the host relative to wild type A. fabarum DsW_054 (Figure 3). Disruption of two genes downstream of Lipid A disaccharide biosynthesis, gmhD and lpxK, including 2 distinct gmhD lesions, did not significantly alter the ability of A. fabarum DsW_054 to associate with its animal host (Figure 4). There were no differences in growth of the mutants in mMRS broth, suggesting the host association effect did not result from ‘sick’ cells (data not shown). Taken together, these results suggest an important role for A. fabarum DsW_054 Lipid A in D. melanogaster association.
Figure 3.
Mutant analysis of MGWA predictions. Five transposon insertion mutants for LPS biosynthesis genes were individually reared in triplicate in each of three separate experiments with D. melanogaster, and bacterial load was determined in whole fly homogenates when the adult flies were 5-7 days old. Significant differences between treatments were determined by a linear mixed effects model with experimental start and end date included as random effects. Insertion mutants in lpxB and lpxC were significantly less abundant in the flies than the wild-type control, but gmhD and lpxK mutations did not have a significant effect. Means and s.e.m. were derived from all replicate data points (N = 8-9 per treatment).
Figure 4.
LPS biosynthesis pathway. An analysis of the LPS pathway shows that genes biosynthetically upstream, but not downstream, of Lipid A influence bacterial association with D. melanogaster.
Discussion
In this study we report on an MGWA analysis to predict bacterial genes that influence the association with the fruit fly D. melanogaster, the creation of a mapped and arrayed transposon insertion library to identity gene-specific insertions in A. fabarum DsW_054, and the use of the mutant library to test the MGWA-predictions. The library includes 6,418 mutants that were mapped to 1,625 genes and 859 intergenic insertions. A 100% validation rate suggested a high accuracy rate of the mapping. Also, near-saturating coverage of non-essential genes allowed us to make inferences about which genes are essential for A. fabarum DsW_054 growth on mMRS medium. Finally, the host-association tests confirmed the prediction that bacterial LPS biosynthesis genes influence bacterial load in the fruit fly, and identified Lipid A biosynthesis genes as key players for these effects. Follow-up experiments that utilize the rich resources available for interrogating host-microbe interactions in Drosophila are necessary to characterize the molecular basis for these genetic interactions. The mutant library we report in this manuscript adds to those resources.
In this study we identified two Lipid A biosynthesis genes as important for maintaining abundant A. fabarum CFU loads in D. melanogaster, suggesting Lipid A is important for Acetobacter to normally associated with D. melanogaster. However, we note several cautions to this interpretation. First, Lipid A is generally considered to be essential for viability of most gram-negative bacteria, and viable Lipid A mutants are rare in other Proteobacteria (e.g., E. coli). We have not performed any biochemical characterization of our LPS mutants and have no evidence to confirm their role on Lipid A levels in Acetobacter. Thus, it may be possible that these genes exert their influence independent of effects on Lipid A (e.g., polarity). Alternatively, some viable LPS pathway mutants have been isolated in bacteria, such as E. coli (Cabeen et al. 2010; Emptage et al. 2014; Fuhrer et al. 2006; Karsten et al. 2009; Kuo et al. 2016; Langklotz et al. 2011; Powell et al. 2016; Vorachek-Warren et al. 2002; Vuorio and Vaara 1992, 1995; Wang et al. 2014), including complete deletion of gmhD (rfaD) (Wang et al. 2014), temperature-sensitive lpxA and lpxD mutations (Vuorio and Vaara 1992, 1995), and an lpxC gene truncation (Fuhrer et al. 2006; Langklotz et al. 2011). Mutations of other genes involved in Lipid A modification include lpxM, lpxP, lpxL and many genes involved in the formation of O antigen (Cabeen et al. 2010; Emiola et al. 2014; Karsten et al. 2009; Powell et al. 2016; Vorachek-Warren et al. 2002).
Despite these caveats associated with a possible LPS effect, there are strong established relationships between bacterial LPS and animal colonization. Consistent with our current findings, the structure of Lipid A in Francisella tularensis is important in bacterial resistance to fruit fly anti-microbial peptides, and a Lipid A core mutant persisted in lower CFU loads during infection than a wild-type control strain (Vonkavaara et al. 2013). Although the Toll and IMD immune pathways in Drosophila do not respond to LPS, Drosophila can sense LPS through neuron stimulation, allowing for pathogen protection and infection avoidance. It is hypothesized that this occurs through taste as flies eat, allowing them to avoid unwanted bacteria (Soldano et al. 2016), and LPS causes an increase in serotonin production in insects that increases phagocytosis by hemocytes and improves the insect’s ability to fight infection (Qi et al. 2016). LPS is also important for bacterial persistence in other animals. For example, a TnSeq experiment of Snodgrassella alvi colonization in honey bees also revealed that LPS is an important factor in colonization (Powell et al. 2016). In non-insect hosts, E. coli LPS mutants are hyper-susceptible to host immunity in C. elegans ((Kuo et al. 2016)), and LPS is important for E. coli colonization and persistence in sheep (Cornick et al. 2017). Vibrio cholerae LPS mutants had a 30 fold reduction in colonization of the mouse gut (Nesper et al. 2002). Inactivation of PA0011 (involved in Lipid A biosynthesis) in Pseudomonas aeruginosa caused decreased virulence and increased susceptibility to antibiotics (Wang et al. 2016a). Thus, our findings are consistent with a broad base of literature that has established LPS biosynthesis is important for host association across the animal kingdom, even though the mechanisms may vary. The postulated explanation in other animals – that LPS is recognized by the host innate immune system to recognize and defend against potential pathogens prior to infection (Charroux et al. 2009; Kurata 2014) – may not apply in fruit flies since LPS does not appear to stimulate immune activity in Drosophila (Kaneko et al. 2004; Leulier et al. 2003). We propose at least three possible explanations for how mutations in LPS biosynthesis could influence bacterial load in the flies. First, the mutations may reduce bacterial growth when the fly is present, even though there was no defect in bacterial growth in 2xYPG. Second, the mutations may lead to weaker cell membranes that are more sensitive to digestion (Vorachek-Warren et al. 2002; Vuorio and Vaara 1992). Third, the bacteria may influence fly feeding or other behavior preferences (Kim et al. 2017; Fischer et al. 2017) that consequently alter bacterial load. Future experiments are necessary to definitively test these ideas.
An analysis of essential genes in an Acetobacteraceae strain has not been determined previously. A method that samples more deeply than our study is necessary for a comprehensive reporting of essential A. fabarum DsW_054 genes (e.g., by TnSeq), but the saturation of non-essential genes as estimated by a rarefaction curve (Figure 2) suggests some preliminary insights can be gleaned from our work, including that ribosome, aminoacyl tRNA biosynthesis, protein export and cell cycle functions are likely essential for A. fabarum DsW_054 growth in mMRS. As a secondary confirmation for this prediction, we detected congruence between these predictions and TnSeq analyses of the closest A. fabarum DsW_054 relatives for which a study had been performed: Alphaproteobacteria Caulobacter crescentus (Christen et al. 2011), Rhizobium leguminosarum (Perry et al. 2016) and Rhodobacter sphaeroides (Burger et al. 2017). Chi-square tests on the KEGG pathways enriched in predicted essential genes from these three studies confirmed essential roles for ribosome and Aminoacyl-tRNA biosynthesis in all three organisms, along with nonsignificant but trending enrichment for cell cycle and metabolic pathways (see Tables S3–S6 in File S1). One limitation of our analysis A. fabarum DsW_054 is that any gene with at least one insertion is binned as non-essential; this categorization does not take into account genes that are only partially disabled by transposon insertion (e.g., into the 3′ end of a gene) that could be assigned as essential. Regardless, consistency between the essential genes detected in our study and in other conphyletic taxa lend support to these cautious interpretations.
The arrayed and mapped A. fabarum DsW_054 transposon insertion library we created in this study will be a resource for use in any field that studies Acetobacter genetics. Acetobacter species are studied in a diversity of research fields, including as members of insect microbiomes and in commercial production facilities (e.g., fermentation, cellulose production). We focused on Acetobacter as representative members of the Drosophila gut microbiota, and previous studies have identified species-specific influence of Acetobacter strains on numerous traits, including triacylglyeride and glucose content, metabolic rate, development time, fecundity, dietary choices, and egg laying preferences (Fischer et al. 2017; Chaston et al. 2014; Ridley et al. 2012; Morimoto et al. 2017; Leitão-Gonçalves et al. 2017). Acetobacter are also commonly associated with numerous other insect species, including honey bees (Apis mellifera), Anopheles and Aedes mosquitos, leafhoppers (Scaphoideus titanus), and mealybugs (Saccharicoccus sacchari) (Crotti et al. 2010). Acetobacter species are also important in commercial production of acetic acid or fermentation products, such as Kefir, a fermented beverage (Cleenwerck et al. 2008; Garofalo et al. 2015; Gulitz et al. 2011; Moens et al. 2014; Viana et al. 2017; Wang et al. 2016b). Acetobacter species have also been used in other applications in biotechnology, including microbial fuel cell technology to produce both electricity and acetic acid (Tanino et al. 2013), in the production of bacterial cellulose scaffolds to grow cartilage and skin tissue (Yin et al. 2015, Keskin et al. 2017) and is a hopeful candidate for dermal medical applications (Taokaew et al. 2015). As such, this mutant library has the potential to serve as a resource for numerous areas of research into the genetic basis for any of these Acetobacter applications, and we welcome requests for specific strains of interest (see File S3 for full list).
Supplementary Material
Supplemental Material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300530/-/DC1.
Acknowledgments
We thank Andrew Goodman and Mark Mandel for technical advice, and Amber Wise and Carrie Evans for technical assistance. This work was supported by startup funds to J.M.C. from Brigham Young University and by research grants (USDA-AFRI 2015-67013-22915 and NSF IOS-1054980) to J.S.G.
Footnotes
Communicating Editor: D. Schneider
Literature Cited
- Arnold M. F. F., Shabab M., Penterman J., Boehme K. L., Griffitts J. S., et al. , 2017. Genome-Wide Sensitivity Analysis of the Microsymbiont Sinorhizobium meliloti to Symbiotically Important, Defensin-Like Host Peptides. MBio 8(4): e01060–17 10.1128/mBio.01060-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakula M., 1969. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J. Invertebr. Pathol. 14(3): 365–374. 10.1016/0022-2011(69)90163-3 [DOI] [PubMed] [Google Scholar]
- Bienenstock J., Kunze W., Forsythe P., 2015. Microbiota and the gut-brain axis. Nutr. Rev. 73(Suppl 1): 28–31. 10.1093/nutrit/nuv019 [DOI] [PubMed] [Google Scholar]
- Blum J. E., Fischer C. N., Miles J., Handelsman J., 2013. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. MBio 4(6): e00860–13 10.1128/mBio.00860-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick N. A., Buchon N., Lemaitre B., 2014. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio 5(3): e01117–14 10.1128/mBio.01117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick N. A., Lemaitre B., 2012. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3(4): 307–321. 10.4161/gmic.19896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T., Ching A., Seroude L., Simon A. F., Benzer S., 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc. Natl. Acad. Sci. USA 101(35): 12974–12979. 10.1073/pnas.0405207101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger B. T., Imam S., Scarborough M. J., Noguera D. R., Donohue T. J., 2017. Combining Genome-Scale Experimental and Computational Methods To Identify Essential Genes in Rhodobacter sphaeroides. mSystems 2(3): e00015–17 10.1128/mSystems.00015-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen M. T., Murolo M. A., Briegel A., Bui N. K., Vollmer W., et al. , 2010. Mutations in the Lipopolysaccharide biosynthesis pathway interfere with crescentin-mediated cell curvature in Caulobacter crescentus. J. Bacteriol. 192(13): 3368–3378. 10.1128/JB.01371-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G., Ianiro G., Cianci R., Bibbo S., Gasbarrini A., et al. , 2015. The involvement of gut microbiota in inflammatory bowel disease pathogenesis: potential for therapy. Pharmacol. Ther. 149: 191–212. 10.1016/j.pharmthera.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Charroux B., Rival T., Narbonne-Reveau K., Royet J., 2009. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 11(6–7): 631–636. 10.1016/j.micinf.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Chaston J. M., Dobson A. J., Newell P. D., Douglas A. E., 2015. Host Genetic Control of the Microbiota Mediates the Drosophila Nutritional Phenotype. Appl. Environ. Microbiol. 82(2): 671–679. 10.1128/AEM.03301-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston J. M., Newell P. D., Douglas A. E., 2014. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. MBio 5(5): e01631–14 10.1128/mBio.01631-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B., Abeliuk E., Collier J. M., Kalogeraki V. S., Passarelli B., et al. , 2011. The essential genome of a bacterium. Mol. Syst. Biol. 7(1): 528 10.1038/msb.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. I., Salazar A., Yamada R., Fitz-Gibbon S., Morselli M., et al. , 2015. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell Reports 12(10): 1656–1667. 10.1016/j.celrep.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleenwerck I., Gonzalez A., Camu N., Engelbeen K., De Vos P., et al. , 2008. Acetobacter fabarum sp. nov., an acetic acid bacterium from a Ghanaian cocoa bean heap fermentation. Int. J. Syst. Evol. Microbiol. 58(Pt 9): 2180–2185. 10.1099/ijs.0.65778-0 [DOI] [PubMed] [Google Scholar]
- Corby-Harris V., Pontaroli A. C., Shimkets L. J., Bennetzen J. L., Habel K. E., et al. , 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73(11): 3470–3479. 10.1128/AEM.02120-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick N. A., Pitzer J., Helgerson A. F., Madsen M. L., Kurth K. T., et al. , 2017. Use of signature-tagged mutagenesis to identify genes associated with colonization of sheep by E. coli O157:H7. Vet. Microbiol. 201: 177–182. 10.1016/j.vetmic.2017.01.031 [DOI] [PubMed] [Google Scholar]
- Cox C. R., Gilmore M. S., 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75(4): 1565–1576. 10.1128/IAI.01496-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti E., Rizzi A., Chouaia B., Ricci I., Favia G., et al. , 2010. Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microbiol. 76(21): 6963–6970. 10.1128/AEM.01336-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G., Blennerhassett P., Lu J., Deng Y., Park A. J., et al. , 2015. Microbiota and host determinants of behavioural phenotype in maternally separated mice. Nat. Commun. 6(1): 7735 10.1038/ncomms8735 [DOI] [PubMed] [Google Scholar]
- Emiola A., George J., Andrews S. S., 2014. A Complete Pathway Model for Lipid A Biosynthesis in Escherichia coli. PLoS One 10(4): e0121216 10.1371/journal.pone.0121216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emptage R. P., Tonthat N. K., York J. D., Schumacher M. A., Zhou P., 2014. Structural basis of lipid binding for the membrane-embedded tetraacyldisaccharide-1-phosphate 4′-kinase LpxK. J. Biol. Chem. 289(35): 24059–24068. 10.1074/jbc.M114.589986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkosar B., Storelli G., Defaye A., Leulier F., 2013. Host-intestinal microbiota mutualism: “learning on the fly. Cell Host Microbe 13(1): 8–14. 10.1016/j.chom.2012.12.004 [DOI] [PubMed] [Google Scholar]
- Fischer C. N., Trautman E. P., Crawford J. M., Stabb E. V., Handelsman J., et al. , 2017. Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. eLife 6: e18855 10.7554/eLife.18855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer F., Langklotz S., Narberhaus F., 2006. The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol. 59(3): 1025–1036. 10.1111/j.1365-2958.2005.04994.x [DOI] [PubMed] [Google Scholar]
- Garofalo C., Osimani A., Milanovic V., Aquilanti L., De Filippis F., et al. , 2015. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 49: 123–133. 10.1016/j.fm.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Goodman A. L., Wu M., Gordon J. I., 2011. Identifying microbial fitness determinants by Insertion Sequencing (INSeq) using genome-wide transposon mutant libraries. Nat. Protoc. 6(12): 1969–1980. 10.1038/nprot.2011.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A. L., McNulty N. P., Zhao Y., Leip D., Mitra R. D., et al. , 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6(3): 279–289. 10.1016/j.chom.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulitz A., Stadie J., Wenning M., Ehrmann M. A., Vogel R. F., 2011. The microbial diversity of water kefir. Int. J. Food Microbiol. 151(3): 284–288. 10.1016/j.ijfoodmicro.2011.09.016 [DOI] [PubMed] [Google Scholar]
- Ha E. M., Lee K. A., Seo Y. Y., Kim S. H., Lim J. H., et al. , 2009. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in Drosophila gut. Nat. Immunol. 10(9): 949–957. 10.1038/ni.1765 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1): D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1): 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44(D1): D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Goldman W. E., Mellroth P., Steiner H., Fukase K., et al. , 2004. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20(5): 637–649. 10.1016/S1074-7613(04)00104-9 [DOI] [PubMed] [Google Scholar]
- Karsten V., Murray S. R., Pike J., Troy K., Ittensohn M., et al. , 2009. msbB deletion confers acute sensitivity to CO2 in Salmonella enterica serovar Typhimurium that can be suppressed by a loss-of-function mutation in zwf. BMC Microbiol. 9(1): 170 10.1186/1471-2180-9-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin Z., Sendemir Urkmez A., Hames E. E., 2017. Novel keratin modified bacterial cellulose nanocomposite production and characterization for skin tissue engineering. Mater. Sci. Eng. C 75: 1144–1153. 10.1016/j.msec.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Kim G., Huang J. H., McMullen J. G., 2nd, Newell P. D., Douglas A. E., 2017. Physiological responses of insects to microbial fermentation products: Insights from the interactions between Drosophila and acetic acid. J. Insect Physiol. 10.1016/j.jinsphys.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyle M. L., Veloz M., Judd A. M., Wong A. C., Newell P. D., et al. , 2016. Rearing the Fruit Fly Drosophila melanogaster Under Axenic and Gnotobiotic Conditions. J. Vis. Exp. (113): e54219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. J., Chen J. W., Chiu H. C., Teng C. H., Hsu T. I., et al. , 2016. Mutation of the Enterohemorrhagic Escherichia coli Core LPS Biosynthesis Enzyme RfaD Confers Hypersusceptibility to Host Intestinal Innate Immunity In vivo. Front. Cell. Infect. Microbiol. 6: 82 10.3389/fcimb.2016.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata S., 2014. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 42(1): 36–41. 10.1016/j.dci.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langklotz S., Schakermann M., Narberhaus F., 2011. Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all gram-negative bacteria. J. Bacteriol. 193(5): 1090–1097. 10.1128/JB.01043-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. A., Kim S. H., Kim E. K., Ha E. M., You H., et al. , 2013. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153(4): 797–811. 10.1016/j.cell.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Leitão-Gonçalves R., Carvalho-Santos Z., Francisco A. P., Fioreze G. T., Anjos M., et al. , 2017. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 15(4): e2000862 10.1371/journal.pbio.2000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F., Parquet C., Pili-Floury S., Ryu J. H., Caroff M., et al. , 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4(5): 478–484. 10.1038/ni922 [DOI] [PubMed] [Google Scholar]
- Ma D., Storelli G., Mitchell M., Leulier F., 2015. Studying host-microbiota mutualism in Drosophila: Harnessing the power of gnotobiotic flies. Biomed. J. 38(4): 285–293. 10.4103/2319-4170.158620 [DOI] [PubMed] [Google Scholar]
- Matos R. C., Schwarzer M., Gervais H., Courtin P., Joncour P., et al. , 2017. D-Alanylation of teichoic acids contributes to Lactobacillus plantarum-mediated Drosophila growth during chronic undernutrition. Nat. Microbiol. 2(12): 1635–1647. 10.1038/s41564-017-0038-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens F., Lefeber T., De Vuyst L., 2014. Oxidation of metabolites highlights the microbial interactions and role of Acetobacter pasteurianus during cocoa bean fermentation. Appl. Environ. Microbiol. 80(6): 1848–1857. 10.1128/AEM.03344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto J., Simpson S. J., Ponton F., 2017. Direct and trans-generational effects of male and female gut microbiota in Drosophila melanogaster. Biol. Lett. 13(7): 20160966 10.1098/rsbl.2016.0966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesper J., Schild S., Lauriano C. M., Kraiss A., Klose K. E., et al. , 2002. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect. Immun. 70(11): 5990–5996. 10.1128/IAI.70.11.5990-5996.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P. D., Douglas A. E., 2014. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 80(2): 788–796. 10.1128/AEM.02742-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry B. J., Akter M. S., Yost C. K., 2016. The Use of Transposon Insertion Sequencing to Interrogate the Core Functional Genome of the Legume Symbiont Rhizobium leguminosarum. Front. Microbiol. 7: 1873 10.3389/fmicb.2016.01873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petra A. I., Panagiotidou S., Hatziagelaki E., Stewart J. M., Conti P., et al. , 2015. Gut-Microbiota-Brain Axis and Its Effect on Neuropsychiatric Disorders With Suspected Immune Dysregulation. Clin. Ther. 37(5): 984–995. 10.1016/j.clinthera.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. E., Leonard S. P., Kwong W. K., Engel P., Moran N. A., 2016. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. USA 113(48): 13887–13892. 10.1073/pnas.1610856113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. X., Huang J., Li M. Q., Wu Y. S., Xia R. Y., et al. , 2016. Serotonin modulates insect hemocyte phagocytosis via two different serotonin receptors. eLife 5: e12241 10.7554/eLife.12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage B., Erolin R., Held K., Gasper J., Weiss E., et al. , 2017. Comprehensive arrayed transposon mutant library of Klebsiella pneumoniae outbreak strain KPNIH1. J. Bacteriol. 199: e00352-17. 10.1128/JB.00352-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Webster P., Finkel S. E., Tower J., 2007. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6(2): 144–152. 10.1016/j.cmet.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Ridley E. V., Wong A. C., Westmiller S., Douglas A. E., 2012. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One 7(5): e36765 10.1371/journal.pone.0036765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton C. E., Smith H. Z., Newell P. D., Douglas A. E., Chaston J. M., 2018. MAGNAMWAR: An R package for genome-wide association studies of bacterial orthologs. Bioinformatics. 10.1093/bioinformatics/bty001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldano A., Alpizar Y. A., Boonen B., Franco L., Lopez-Requena A., et al. , 2016. Gustatory-mediated avoidance of bacterial lipopolysaccharides via TRPA1 activation in Drosophila. eLife 5: e13133 10.7554/eLife.13133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanino T., Nara Y., Tsujiguchi T., Ohshima T., 2013. Coproduction of acetic acid and electricity by application of microbial fuel cell technology to vinegar fermentation. J. Biosci. Bioeng. 116(2): 219–223. 10.1016/j.jbiosc.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Taokaew S., Phisalaphong M., Newby B. Z., 2015. Modification of Bacterial Cellulose with Organosilanes to Improve Attachment and Spreading of Human Fibroblasts. Cellulose (Lond) 22(4): 2311–2324. 10.1007/s10570-015-0651-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S., Griffin N. W., Bezy O., Fujisaka S., Vienberg S., et al. , 2015. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab. 22(3): 516–530. 10.1016/j.cmet.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte D., Kathagen G., D’Hoe K., Vieira-Silva S., Valles-Colomer M., et al. , 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551(7681): 507–511. 10.1038/nature24460 [DOI] [PubMed] [Google Scholar]
- Verdu E. F., Galipeau H. J., Jabri B., 2015. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 12(9): 497–506. 10.1038/nrgastro.2015.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R. O., Magalhaes-Guedes K. T., Braga R. A., Jr, Dias D. R., Schwan R. F., 2017. Fermentation process for production of apple-based kefir vinegar: microbiological, chemical and sensory analysis. Braz. J. Microbiol. 48(3): 592–601. 10.1016/j.bjm.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonkavaara M., Pavel S. T., Holzl K., Nordfelth R., Sjostedt A., et al. , 2013. Francisella is sensitive to insect antimicrobial peptides. J. Innate Immun. 5(1): 50–59. 10.1159/000342468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorachek-Warren M. K., Ramirez S., Cotter R. J., Raetz C. R., 2002. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 277(16): 14194–14205. 10.1074/jbc.M200409200 [DOI] [PubMed] [Google Scholar]
- Vuorio R., Vaara M., 1992. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 36(4): 826–829. 10.1128/AAC.36.4.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorio R., Vaara M., 1995. Comparison of the phenotypes of the lpxA and lpxD mutants of Escherichia coli. FEMS Microbiol. Lett. 134(2–3): 227–232. 10.1111/j.1574-6968.1995.tb07942.x [DOI] [PubMed] [Google Scholar]
- Wang B., Li B., Liang Y., Li J., Gao L., et al. , 2016a Pleiotropic effects of temperature-regulated 2-OH-lauroytransferase (PA0011) on Pseudomonas aeruginosa antibiotic resistance, virulence and type III secretion system. Microb. Pathog. 91: 5–17. 10.1016/j.micpath.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Wang J., Ma W., Wang Z., Li Y., Wang X., 2014. Construction and characterization of an Escherichia coli mutant producing Kdo(2)-lipid A. Mar. Drugs 12(3): 1495–1511. 10.3390/md12031495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhong H., Liu K., Guo A., Qi X., et al. , 2016b The evaluation of kefir pure culture starter: Liquid-core capsule entrapping microorganisms isolated from kefir grains. Food Sci. Technol. Int. 22(7): 598–608. 10.1177/1082013216628311 [DOI] [PubMed] [Google Scholar]
- Wattam A. R., Davis J. J., Assaf R., Boisvert S., Brettin T., et al. , 2017. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 45(D1): D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans N. J., Walter A., Chouaia B., Chaston J. M., Douglas A. E., et al. , 2017. A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Mol. Ecol. 26(17): 4536–4550. 10.1111/mec.14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C., Chaston J. M., Douglas A. E., 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 7(10): 1922–1932. 10.1038/ismej.2013.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C., Luo Y., Jing X., Franzenburg S., Bost A., et al. , 2015. The Host as the Driver of the Microbiota in the Gut and External Environment of Drosophila melanogaster. Appl. Environ. Microbiol. 81(18): 6232–6240. 10.1128/AEM.01442-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. N., Ng P., Douglas A. E., 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13(7): 1889–1900. 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin N., Stilwell M. D., Santos T. M., Wang H., Weibel D. B., 2015. Agarose particle-templated porous bacterial cellulose and its application in cartilage growth in vitro. Acta Biomater. 12: 129–138. 10.1016/j.actbio.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.