Abstract
Amoebic gill disease (AGD) is one of the largest threats to salmon aquaculture, causing serious economic and animal welfare burden. Treatments can be expensive and environmentally damaging, hence the need for alternative strategies. Breeding for disease resistance can contribute to prevention and control of AGD, providing long-term cumulative benefits in selected stocks. The use of genomic selection can expedite selection for disease resistance due to improved accuracy compared to pedigree-based approaches. The aim of this work was to quantify and characterize genetic variation in AGD resistance in salmon, the genetic architecture of the trait, and the potential of genomic selection to contribute to disease control. An AGD challenge was performed in ∼1,500 Atlantic salmon, using gill damage and amoebic load as indicator traits for host resistance. Both traits are heritable (h2 ∼0.25-0.30) and show high positive correlation, indicating they may be good measurements of host resistance to AGD. While the genetic architecture of resistance appeared to be largely polygenic in nature, two regions on chromosome 18 showed suggestive association with both AGD resistance traits. Using a cross-validation approach, genomic prediction accuracy was up to 18% higher than that obtained using pedigree, and a reduction in marker density to ∼2,000 SNPs was sufficient to obtain accuracies similar to those obtained using the whole dataset. This study indicates that resistance to AGD is a suitable trait for genomic selection, and the addition of this trait to Atlantic salmon breeding programs can lead to more resistant stocks.
Keywords: aquaculture, amoebic gill disease, Salmo salar, disease resistance, animal breeding, fish, GenPred, Shared Data Resources, Genomic Selection
Salmonids are a high-value group of fish species, comprising 16.6% of global fish trade in 2013 (FAO 2016). Demand has grown steadily and is expanding geographically, and Atlantic salmon (Salmo salar) has the highest production volume and value of all the salmonid species (FAO 2016). However, in recent years, Atlantic salmon supply has fluctuated, partly as a result of infectious disease outbreaks in all major salmon producing countries (FAO 2017). These outbreaks are a major threat to sustainable production and future expansion of salmon aquaculture. While solutions to several bacterial and viral diseases (e.g., vaccines) have been widely and routinely applied (Brudeseth et al. 2013), parasitic diseases are currently presenting a substantially greater problem to the industry. In addition to the major economic concern, these parasitic diseases and current treatment strategies can pose serious animal welfare and environmental concerns.
Amoebic gill disease (AGD), primarily caused by Neoparamoeba perurans, has been a perennial problem for salmon aquaculture in Australia, and outbreaks have become increasingly frequent in European salmon farms. It also affects other commercially important salmonids such as rainbow trout (Oncorhynchus mykiss) and chinook salmon (Oncorhynchus tshawytscha), and certain non-salmonid aquaculture species such as turbot (Scophthalmus maximus; Young et al. 2008). While gill disease symptoms are complex, AGD typically presents as multifocal white patches on the gill surface, lesions and epithelial hyperplasia leading to impaired gas exchange, poor growth and ultimately severe morbidity and mortality if untreated (Zilberg and Munday 2000; Adams and Nowak 2003). Current treatment strategies are crude, laborious, stressful to fish, and potentially environmentally damaging; for example involving hydrogen peroxide application or fresh water bathing of affected fish. This results in a large economic burden associated with the costs of treatment and productivity losses due to the disease. Therefore, alternative approaches that help control the impact of AGD are highly desirable.
One such method is improving the resistance of farmed salmon stocks to this disease via selective breeding, the benefits of which can be cumulative and permanent. Several studies have found significant estimates of heritability for disease resistance in aquaculture species (e.g., Silverstein et al. 2009; Gjerde et al. 2011; Yáñez et al. 2014a; Palaiokostas et al. 2016, Tsai et al. 2016b). Harnessing this heritability for genetic improvement in selective breeding programs is a current goal. The high fecundity of aquaculture species, and resulting large full sibling family sizes, facilitates disease challenge testing of close relatives (i.e., full siblings) to enable breeding value estimation in selection candidates. Selection is often more accurate when the relationship between individuals is obtained from genomic data (genomic selection) rather than the pedigree (traditional selection), but it depends on the architecture of the trait as well as other technical variables such as marker density (Daetwyler et al. 2010). For instance, genomic selection has been found to outperform traditional selection in resistance to sea lice in Atlantic salmon (Ødegard et al. 2014; Tsai et al. 2016; Correa et al. 2017b) and in resistance to pasteurellosis in sea bream (Sparus aurata; Palaiokostas et al. 2016). Further, while an initial study found no difference between genomic selection and pedigree-based approaches for resistance to bacterial cold water disease in rainbow trout (Vallejo et al. 2016), a later study with larger sample sizes resulted in doubling of accuracy with the genomic selection approach (Vallejo et al. 2017). One advantage of genomic selection over pedigree-based selection is that it more accurately captures the Mendelian sampling term between closely related individuals in the population—particularly relevant in aquaculture species with large families.
One of the main limitations of genomic selection is the cost; genotyping a large number of animals with a high density SNP panel could be prohibitive for all but the largest aquaculture breeding companies. Several strategies have been proposed to reduce the cost of genotyping for genomic selection in aquaculture via low density SNP panels, including within-family genomic selection (Lillehammer et al. 2013) and the use of genotyping strategies including imputation from low to high density SNPs (Kijas et al. 2016; Tsai et al. 2017). Genotype-by-sequencing technologies are also likely to help reduce costs in the near future given the continuously decreasing costs of sequencing and the advent of new sequencing technologies suitable for low to medium scale SNP genotyping, such as RAD-seq or GT-seq (Robledo et al. 2017). Reducing the cost of genomic selection will be critical to implement genomic selection in most aquaculture breeding programs, and in this sense improving the cost-effectiveness of genomic selection will likely be an important area of research in the coming years (Lillehammer et al. 2013).
Previous studies on host resistance to AGD in salmon have found estimates of heritability ranging from 0.16 to 0.48 (Taylor et al. 2007, 2009). The objectives of this study were a) estimate genetic variance of amoebic gill disease resistance in experimentally challenged Atlantic salmon, b) investigate the architecture of the trait using a single-SNP genome-wide association study (single-SNP GWAS) and regional heritability mapping, c) explore genomic selection using SNP markers and/or pedigree, and d) explore different marker densities with a view to future improvement of cost-effectiveness of genomic selection within commercial breeding programs.
Materials And Methods
Challenge experiments
An AGD challenge experiment using 1,481 Atlantic post-smolt salmon (∼18 months, mean weight ∼700 g) originating from a commercial breeding program (Landcatch, UK) was conducted by distributing the fish equally into 2 × 4 m seawater tanks in the experimental facilities of Machrihanish (Scotland, United Kingdom). Seeder fish with a uniform level of AGD infection were produced by cohabitation with infected fish from an in vivo culture. The challenge was then performed by cohabitation of infected seeder fish at a ratio of 15% seeder to naïve fish, allowing three separate cycles of infection with a treatment and recovery period after the first two (Taylor et al. 2009). For the first two challenges, fresh water treatment was performed 21 days after challenge, followed by a week of recovery. The disease was allowed to progress until the terminal sampling point in the third challenge. Fish were sampled and phenotypes were recorded during three consecutive days. A subjective gill lesion score of the order of severity ranging from 0 to 5 was recorded for both gills (Table 1; Taylor et al. 2016). These gill lesion scores were recorded by a single operator, who referred to pictures to guide classification. Some fish were scored by additional operators, and the scores never differed by >0.5. Further, one of the gills was stored in ethanol for qPCR analysis of amoebic load using Neoparamoeba perurans specific primers. Amoebic load has previously been used as a suitable indicator trait for resistance to AGD in salmon (Taylor et al. 2009). The challenged fish belonged to 312 different families with 1 to 37 fish per family. All fish were phenotyped for mean gill score (mean of the left gill and right gill scores) and amoebic load (qPCR values using Neoparamoeba perurans specific primers, amplified from one of the gills). All phenotypic information is available in File S1.
Table 1. Gill score description.
| Gill score | Level of infection | Description |
|---|---|---|
| 0 | Clear | Healthy red gills, no gross sign of infection. |
| 1 | Very light | One white spot, light scarring or undefined necrotic streaking |
| 2 | Light | 2-3 spots/small mucus patch |
| 3 | Moderate | Established thickened mucus patches or spot groupings up to 20% of the total gill area |
| 4 | Advanced | Established lesions covering up to 50% of gill area |
| 5 | Heavy | Extensive lesions covering most of the gill surface |
All animals were reared in accordance with relevant national and EU legislation concerning health and welfare. The challenge experiment was performed by the Marine Environmental Research Laboratory (Machrihanish, UK) under approval of the ethics review committee of the University of Stirling (Stirling, UK) and according to Home Office license requirements. Landcatch are accredited participants in the RSPCA Freedom Foods standard, the Scottish Salmon Producers Organization Code of Good Practice, and the EU Code-EFABAR Code of Good Practice for Farm Animal Breeding and Reproduction Organizations.
Estimation of Amoebic load
Sampled whole gills were weighed and combined with an equal amount (wt/vol) 10 mM Tris, 1 mM EDTA, pH 8.0. Samples were then homogenized using a Qiagen TissueLyser II (Qiagen, Manchester, UK) following the manufacturers’ recommendations. Total DNA was extracted from 50 µl homogenate using Questgene 9600 DNA extraction kits (Questgene, York, UK) following manufacturers’ protocols. Amoebic load was determined via duplex qPCR reactions using primer/probe combinations targetting a 139 bp N. perurans specific 18S sequence (Fringuelli et al. 2012), and a 66 bp fragment of the Atlantic salmon Elongation Factor α 1 gene (Bruno et al. 2007). DNA was normalized to 50 ng/µl, and 5 µl was combined into 50 µl QPCR duplex reactions comprising: 1X Taqman QPCR reaction mix (Questgene, York, UK), 300 nM N. perurans specific primers, 150 nM N. perurans specific probe, 150 nM ELFα primers, and 75 nm ELFα probe (Table S1). Ampifications were performed using a Biorad iCycler iQ QPCR Detection System. The thermal profile consisted of 95° for 10 min and 45 cycles of 15 s denaturation at 95°/30 s annealing/extension at 56°. Fluorescence in both FAM and HEX channels was acquired during the annealing/extension stage. Ct (threshold cycle) values were recorded and the level of N. perurans load was normalized against the ELF internal control by computing the ratio Equivalent Target Amount (ETA) N. perurans : ETA ELFα.
Genotyping
DNA was extracted from fin tissue samples using the DNeasy 96 tissue DNA extraction kit (Qiagen, UK) and samples were genotyped using an Illumina combined species Atlantic salmon and rainbow trout SNP array (∼17K SNPs, File S2), designed from a subset of SNPs from a higher density array (Houston et al. 2014). Genotypes (File S3) were filtered according to the following criteria: SNP call-rate < 0.9, individual call-rate < 0.9, FDR rate for high individual heterozygosity < 0.05, identity-by-state > 0.95 (both individuals removed), Hardy-Weinberg equilibrium FDR p-value < 0.05, minor allele frequency < 0.05. After this filtering, a total of 1,430 fish and 7,168 SNPs remained for further analysis. The large number of SNPs removed by filtering is due to the lack of informativeness of the rainbow trout SNPs in these Atlantic salmon samples.
Estimation of genetic parameters
Gill score and gill qPCR data were analyzed using linear mixed models, fitting collection date (3 levels) and tank (2 levels) as fixed effects and animal as a random effect. The additive effect was estimated using both the genomic kinship matrix (G-matrix) and the pedigree (A-matrix). Heritabilities were estimated by ASReml 3.0 (Gilmour et al. 2014) fitting the following linear mixed model:
where y is a vector of observed phenotypes, μ is the overall mean of phenotype records, b is the vector of fixed effects of collection date and tank, a is a vector of additive genetic effects distributed as ∼N(0,Gσ2a) or N(0,Aσ2a) where σ2a is the additive (genetic) variance, G and A are the genomic and pedigree relationship matrices, respectively. X and Z are the corresponding incidence matrices for fixed and additive effects, respectively, and e is a vector of residuals. The genomic relationship matrix was constructed by the GenABEL R package (Aulchenko et al. 2007) using the method of VanRaden (VanRaden 2008) and then inverted by applying a standard R function. Phenotypic correlations between traits and genetic correlations were estimated using bivariate analyses implemented in ASReml 3.0 (Gilmour et al. 2014) fitting the linear mixed model described above.
Single-SNP genome-wide association study
The single-SNP GWAS was performed using the GenABEL R package (Aulchenko et al. 2007) by applying the mmscore function (Chen and Abecasis 2007), which accounts for the relatedness between individuals applied through the genomic kinship matrix. Significance thresholds were calculated using a Bonferroni correction where genome-wide significance was defined as 0.05 divided by number of independently segregating SNPs (Duggal et al. 2008) and suggestive as one false positive per genome scan (1/number of independently segregating SNPs). The number of independently segregating SNPs was calculated using Plink v.1.9 (Chang et al. 2015) accounting for linkage disequilibrium among the consecutive SNPs. SNPs showing r2 values > 0.9 were considered linked.
Regional heritability mapping
A regional heritability mapping (RHM) analysis (Nagamine et al. 2012; Uemoto et al. 2013) was performed where the genome was divided into overlapping regions consisting of 20 sequential SNPs and overlapping by 10 SNPs using Dissect v.1.12.0 (Canela-Xandri et al. 2015). The significance of the regional heritability for each window was evaluated using a log likelihood ratio test statistic (LRT) comparing the global model fitting all markers with the model only fitting SNPs in a specific genomic region (File S4). These windows overlap and therefore the significance threshold was determined using a Bonferroni correction using half the number of tested windows.
Genomic prediction
The accuracy of genomic selection was estimated by five replicates of fivefold cross-validation analysis (training set 80%, validation set 20%). The phenotypes recorded in the validation population were masked and breeding values were estimated using ASReml 3.0 using the linear mixed model described above. Prediction accuracy was calculated as the correlation between the predicted EBVs of the validation set and the actual phenotypes divided by the square root of the heritability estimated in the validation population [∼ r(y1 , y2) / 2√h2 ]. Genomic best linear unbiased prediction (GBLUP) was applied to predict the masked phenotypes of the validation sets and the resulting prediction accuracy was compared to that of pedigree-based BLUP (PBLUP). The bias of the EBVs was estimated as the regression coefficient of the phenotypes on the predicted EBVs. Since medium-density SNP array genotyping can be expensive, we also evalutated the impact of reduced SNP density on prediction accuracy by using subsets of the SNP data for the GBLUP. To choose the SNPs for the (pseudo) low density panels we tried two different strategies: 1) we progressively increased the minimum allele frequency threshold in increments of 0.05 (maf, 0.05, 0.10, 0.15, …) resulting in genotype datasets with progresively lower SNP density and progressively higher MAF; and 2) we iteratively removed the SNP showing the lowest mean distance to the previous and the next SNP on the genome, resulting in datasets of evenly spaced genotypes.
Data availability
Primers and probes to perform amoebic load estimation by qPCR are provided in Table S1. Phenotypic data of the fish used in this study is available in File S1. Note that gill scores correspond to an experimental challenge, gill scores higher than 0.5-1 are rarely encountered in Landcatch commercial facilities. Markers included in the SNP array and their position in the Atlantic salmon genome can be found in File S2. Genotypes of the fish used in this study are available in File S3. The regional heritability mapping model is detailed in File S4.
Results And Discussion
The means and standard deviations for AGD resistance traits were 2.79 ± 0.85 and 31.36 ± 3.24 for the gill score and qPCR amoebic load, respectively. Moderate heritability estimates were observed for both phenotypes, which ranged between 0.25 and 0.36 (Table 2), and both the phenotypic and genetic correlations between the two traits were high and positive (0.81 and ∼1 respectively).
Table 2. Heritability estimates for the AGD resistance traits.
| Pedigree | gMatrix | |
|---|---|---|
| Mean gill score | 0.25 ± 0.06 | 0.24 ± 0.04 |
| Amoebic load | 0.36 ± 0.07 | 0.25 ± 0.04 |
A previous study on AGD disease resistance within the Tasmanian Atlantic salmon population found similar heritability estimates, ranging from 0.16 for gross gill score (similar to mean gill score here) to 0.35 for digital image gill score (Taylor et al. 2007). Higher heritability estimates were obtained in the study of Taylor et al. (2009), which varied from 0.23 to 0.48 for mean gill score depending on the number of rounds of re-infection. The highest heritability, 0.48, corresponded to the third challenge trial after two rounds of infection and subsequent freshwater treatment, as in our study. This challenge model is based on results from Taylor et al. (2009) which showed that the gill scores from the third challenge is the most accurate predictor of ultimate survival, potentially implying genetic variation in the adaptive immune response.
Similar heritability estimates were obtained for host resistance to sea lice; ∼0.2 to 0.3 for the North Atlantic sea louse (Lepeophtheirus salmonis; Kolstad et al. 2005; Gjerde et al. 2011; Gharbi et al. 2015; Tsai et al. 2016), and 0.1-0.3 for the Pacific sea louse (Caligus rogercresseyi; Lhorente et al. 2012; Yáñez et al. 2014a; Correa et al. 2017a). Similarly, the heritability of resistance to Gyrodactylus salaris, another ectoparasite mainly affecting wild Atlantic salmon, was estimated to be 0.32 (Salte et al. 2010). These heritabilities are comparable to estimates for host resistance to bacterial and viral infections (Ødegård et al. 2011; Yáñez et al. 2014b), and imply that selective breeding for improved resistance to parasites in salmon is a plausible goal.
Single-SNP genome-wide association analysis and regional heritability mapping
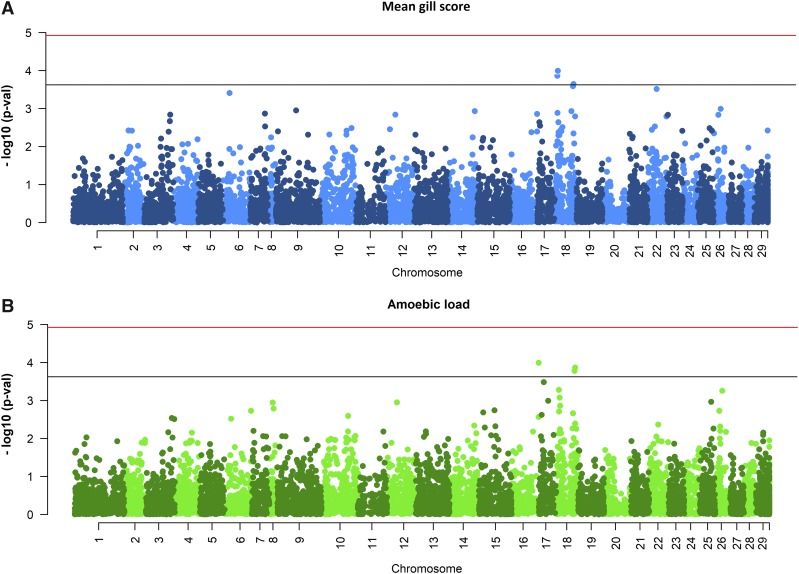
The single-SNP GWAS revealed no major QTL regions that reached the genome-wide significance threshold for gill score or amoebic load (Figure 1). However, there were two suggestive QTL identified for both traits on chromosome 18, seemingly located in two non-overlapping regions around 9-12 Mb and 54-61 Mb respectively, each explaining ∼4% of the additive genetic variance (Table 3). The most significant SNP for amoebic load was observed at the distal end of chromosome 16. There were other genomic regions that either reach suggestive significance but only for one of the traits (i.e., distal end of chromosome 16) or are close (chromosomes 6, 17 or 22), and these could also be QTL of moderate effect (∼3–4% of the additive genetic variance, Table 3) that might have been significant with a larger sample size.
Figure 1.
GWAS for resistance to AGD. Single-SNP GWAS results for A) mean gill score and B) amoebic load are shown. Horizontal bars represent Bonferroni corrected significance (red) and nominal signifcance (black).
Table 3. Top single-SNP GWAS markers for AGD resistance.
| Mean gill score | Amoebic load | ||||||
|---|---|---|---|---|---|---|---|
| Chr. | Position | Explained gen. var. (%) | p-val | Chr. | Position | Explained gen. var. (%) | p-val |
| 18 | 9,010,507 | 4.81 | 1.37E-04 | 16 | 87,305,577 | 4.40 | 1.03E-04 |
| 18 | 61,003,989 | 4.05 | 2.28E-04 | 18 | 61,003,989 | 4.17 | 1.37E-04 |
| 18 | 59,141,833 | 4.13 | 2.59E-04 | 18 | 59,141,833 | 4.21 | 1.67E-04 |
| 22 | 29,458,040 | 3.89 | 3.07E-04 | 17 | 17,603,968 | 3.98 | 3.30E-04 |
| 6 | 20,420,312 | 3.76 | 3.93E-04 | 18 | 9,010,507 | 3.82 | 5.30E-04 |
| 26 | 22,182,178 | 3.21 | 1.03E-03 | 26 | 22,182,178 | 3.41 | 5.60E-04 |
| 9 | 65,305,177 | 3.16 | 1.13E-03 | 18 | 11,619,560 | 3.20 | 8.43E-04 |
| 18 | 54,225,069 | 3.14 | 1.17E-03 | 17 | 31,447,688 | 3.19 | 1.03E-03 |
| 14 | 85,642,477 | 3.14 | 1.18E-03 | 25 | 37,782,067 | 3.05 | 1.10E-03 |
| 18 | 9,896,346 | 3.08 | 1.30E-03 | 12 | 31,597,392 | 3.06 | 1.13E-03 |
| 7 | 46,569,758 | 3.32 | 1.37E-03 | 8 | 13,396,576 | 3.17 | 1.14E-03 |
| 16 | 87,305,577 | 3.10 | 1.40E-03 | 18 | 13,403,715 | 2.99 | 1.39E-03 |
| 12 | 31,597,392 | 3.04 | 1.45E-03 | 8 | 49,527,638 | 2.85 | 1.63E-03 |
| 3 | 82,689,281 | 3.04 | 1.46E-03 | 15 | 49,527,638 | 2.79 | 1.82E-03 |
| 26 | 14,842,966 | 3.08 | 1.47E-03 | 6 | 83815992 | 2.77 | 1.88E-03 |
Chr.: chromosome; gen. var.: genetic variance.
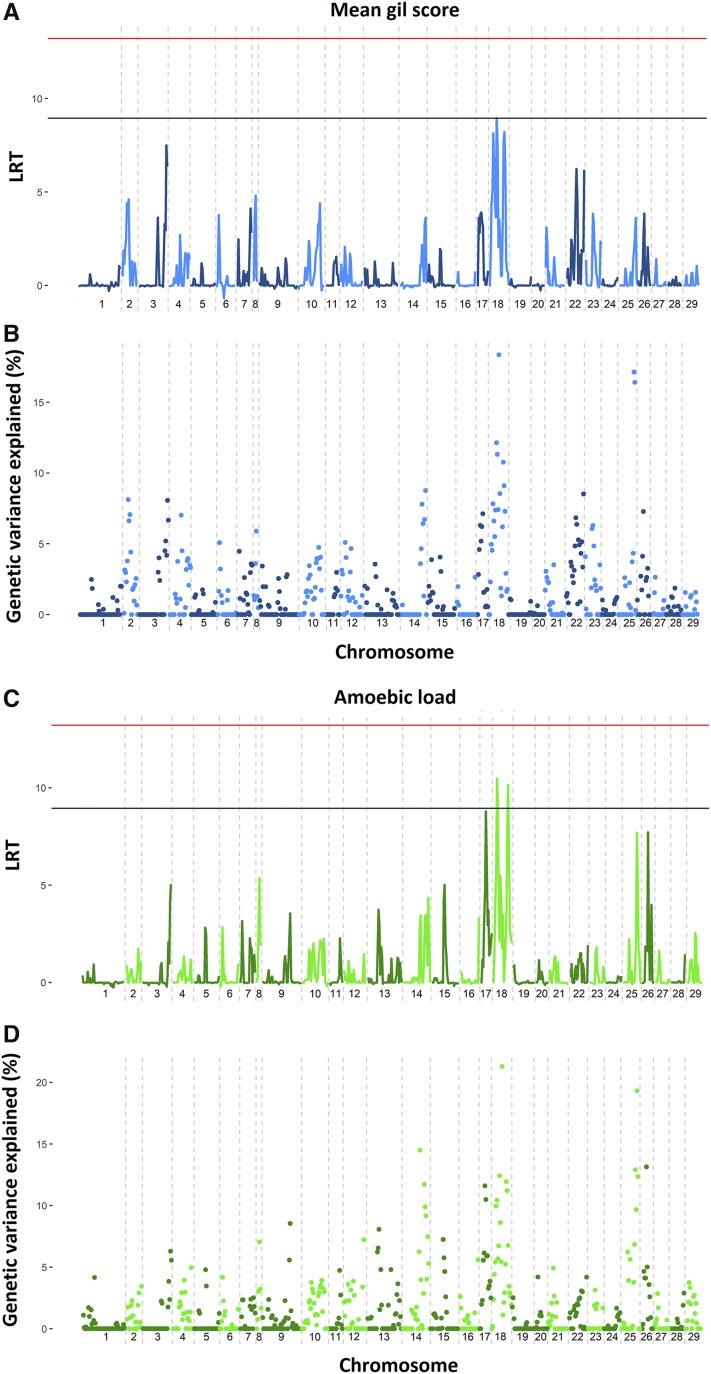
The QTL identified by regional heritability mapping (RHM) were consistent with the results of the single-SNP GWAS, with two regions in chromosome 18 showing the highest significance for both mean gill score and amoebic load (Figure 2). These regions explained between 9.5 and 11.6% of the genetic variance respectively, and contained the most significant SNPs detected by the single-SNP GWAS. Another region in chromosome 18 between 25 and 42 Mb explained ∼20% of the heritability, but its significance was lower. The SNPs in this large region between the two putative QTL may be picking up on effects arising from either or both of the flanking regions due to linkage disequilibrium. Further, regions in chromosomes 17, 25 and 26 almost reached nominal significance for amoebic load, explaining >10% of the genetic variance. The most important discrepancy is in the distal region of chromosome 16, which shows no significant association in RHM but held the most significant marker in the amoebic load single-SNP GWAS. This difference might be explained by the high recombination rates found in the extremes of the chromosomes in Atlantic salmon (e.g., Tsai et al. 2016a); the significant SNP was the penultimate marker in chromosome 16. RHM uses information from several consecutive markers, and has been shown to have an advantage over single-SNP GWAS to explain part of the typical missing heritability of single-SNP association studies and to detect QTL of small effects which otherwise would not be detected using information from single SNPs (Nagamine et al. 2012, Uemoto et al. 2013, Riggio and Pong-Wong 2014; Shirali et al. 2016).
Figure 2.
Regional heritability mapping for AGD resistance. Regional heritability mapping results for mean gill score and amoebic load are shown. A) and C) represent the log-ratio test values for each tested region (20 consecutive SNPs) for mean gill score and amoebic load respectively, horizontal bars represent Bonferroni corrected significance (red) and nominal significance (black). B) and D) represent the percentage of additive genetic variance explained by each region for mean gill score and amoebic load repectively.
Our results point toward a polygenic architecture of resistance to AGD, but potentially including a few QTL explaining moderate levels of the genetic variation. Genotyping additional AGD-challenged and phenotyped samples would help provide evidence in support or against the existance of these QTL. Further, a higher SNP density could possibly identify additional QTL not in linkage disequilibrium with the SNPs in this study, help to fine map the ones reported here, and possibly increase the estimates of genetic variation explained by the QTL.
While a few major disease resistance loci have been described, such as for viral infectious pancreatic necrosis in Atlantic salmon (Houston et al. 2008, Moen et al. 2009), the majority of disease resistance traits for aquaculture species are polygenic in nature (Houston 2017). Polygenic architecture has been observed for host resistance to sea lice (Tsai et al. 2016) and Piscirickettsia salmonis (Correa et al. 2015) in Atlantic salmon, pasteurellosis in gilthead sea bream (Palaiokostas et al. 2016) and Gyridactylus salaris in salmon (Gilbey et al. 2006). Other examples of putative major QTL include whirling disease in rainbow trout, caused by the myxosporean parasite Myxobolus cerebralis, which explains up to 86% of phenotypic variance depending on the family (Baerwald et al. 2011), bacterial cold water disease in trout where 27–61% of the genetic variation is explained by major QTL depending on the line (Vallejo et al. 2017), and Pancreas Disease in Atlantic salmon where approximately 20% of the genetic variation is explained by the largest QTL (Gonen et al. 2015). While resistance to parasitic disease does tend to show a polygenic architecture, and AGD is no exception, the putative QTL region(s) of moderate effect identified merit validation tests in independent populations, and functional genomic and resequencing studies to identify putative underlying genes and mechanisms.
Genomic selection accuracy
Using a fivefold cross-validation analysis, the prediction accuracy with the genomic relationship (G) matrix was ∼18% higher than with the pedigree (A) matrix for both mean gill score and amoebic load, and GBLUP predictions showed practically no bias (Table 4). Prediction accuracies obtained for amoebic load measured by qPCR were ∼20% higher than those of mean gill score, which may be due to the wider range of the amoebic load trait. Taylor et al. (2007) found that gill damage scores obtained using image analysis or histopathology showed high positive genetic correlation, but correlation between these traits and gill score was lower. The prediction accuracy results from the current study suggest genomic selection will significantly outperform pedigree-based selection for AGD resistance, and that both gill score and qPCR measures of amoebic load are useful traits for selection for AGD resistance.
Table 4. Accuracy and bias of genomic selection.
| Pedigree | gMatrix | |||
|---|---|---|---|---|
| Accuracy | Bias | Accuracy | Bias | |
| Mean gill score | 0.51 | 0.90 | 0.62 | 1.00 |
| Amoebic load | 0.60 | 0.88 | 0.70 | 0.99 |
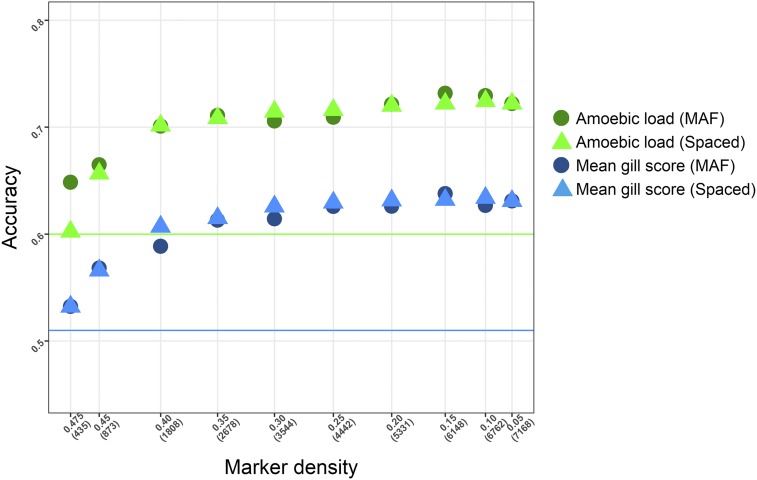
Since genotyping with medium or high-density SNP arrays is relatively expensive, and aquaculture species tend to have closely related animals in training and validation populations (e.g., in ‘sib testing’ schemes), well designed low density genotyping panels may be useful in genomic selection. When SNP density was reduced either via progressive increase in MAF thresholds or selecting evenly-spaced sets of markers, accuracy remained relatively stable until 1,808 SNPs where a gradual drop off in accuracy was observed (Figure 3). However, even at very low SNP density of 435 SNPs the accuracy of prediction was higher using GBLUP than PBLUP, except for Amoebic load estimated using the evenly spaced SNPs which resulted in an accuracy similar to that of PBLUP.
Figure 3.
Prediction accuracy for different SNP densities. Accuracy of genomic prediction (GBLUP) for mean gill score and amoebic load with different SNP densities, selected based on their minimum allele frequencies (MAF) or their position in the genome so the markers are evenly spaced (Spaced). Horizontal lines indicate the accuracy of pedigree selection. X-axis figures represent MAF values and number of SNPs (in parenthesis).
The results for genomic prediction of breeding values are generally consistent with published observations for aquaculture species to date. For host ressitance to sea lice, marked gains in accuracy were observed, from 10 to 52% depending on the population studied (Ødegard et al. 2014; Tsai et al. 2016); and recently different genomic selection models (ssGBLUP, wssGBLUP, BayesB) have been shown to almost double the prediction accuracy for bacterial cold water disease in rainbow trout compared to pedigree-based estimates (Vallejo et al. 2017). Interestingly, both studies on host resistance to sea lice in salmon showed practically no improvement in prediction accuracy when SNP density was increased above 5K (Ødegard et al. 2014; Tsai et al. 2016). As shown in the current study, genomic prediction accuracy is higher compared to pedigree-based prediction even when we use very low density genotyping (a few hundred SNPs). This is somewhat surprising given the size of the salmon genome (∼3 Gb, Lien et al. 2016), but probably reflects the close relationship between the training set and the reference set in the cross validation design – i.e., full and half siblings will occur in both sets. The high accuracy with low marker density may also reflect aspects of the salmon population history, for example relatively low effective population size and past admixture may be expected to result in long-range LD and this may increase the predictive ability of a sparse SNP marker set.
Genotyping costs can be an important hurdle for the application of genomic selection, especially for small companies and breeding programs. For example, in mass spawning species that require genotyping to ascertain the pedigree, genomic selection could potentially be applied without a major genotyping cost increase. Further, this can be combined with genotyping strategies and imputation to improve cost-effectiveness, e.g., Tsai et al. (2017) showed that imputation from 250 SNPs to ∼25K led to an improvement in prediction accuracy of 21% compared to pedigree prediction. Such strategies may increase cost-effectiveness and therefore uptake of genomic selection in aquaculture breeding, with beneficial impact on disease resistance and control.
Conclusions
Host resistance to AGD in Atlantic salmon is moderately heritable (h2 ∼0.25 - 0.30) and can be measured using indicator traits such as gill score or amoebic load measured by qPCR. The genetic architecture of AGD resistance appears to be polygenic, but with two suggestive QTL explaining up to 11% of the genetic variance on chromosome 18, and other non-significant regions in other chromosomes accounting for a similar amount of variance. These possible QTL should be tested in independent populations, and may form the basis for identification of underlying causative genes. Genomic prediction accuracy was substantially higher (∼18%) when using genomic relationships rather than pedigree-based relationships with a ∼7K SNP panel, and remained so even when marker density substantially reduced. Since AGD is a large threat for salmon aquaculture in most major salmon production countries, genomic selection is likely to be an important component of breeding programs to help tackle this disease via genetic improvement of host resistance.
Supplementary Material
Supplemental Material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.118.200075/-/DC1
Acknowledgments
The authors gratefully acknowledge funding from Innovate UK and BBSRC (BB/M028321/1), in addition to BBSRC Institute Strategic Program Grants (BB/P013759/1 and BB/P013740/1). DR is supported by a Newton International Fellowship from The Royal Society (NF160037).
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Adams M. B., Nowak B. F., 2003. Amoebic gill disease: sequential pathology in cultured Atlantic salmon, Salmo salar L. J. Fish Dis. 26(10): 601–614. 10.1046/j.1365-2761.2003.00496.x [DOI] [PubMed] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M., 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23(10): 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- Baerwald M. R., Petersen J. L., Hedrick R. P., Schisler G. J., May B., 2011. A major effect quantitative trait locus for whirling disease resistance identified in rainbow trout (Oncorhynchus mykiss). Heredity (Edinb.) 106(6): 920–926. 10.1038/hdy.2010.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudeseth B. E., Wiulsrød R., Fredriksen B. N., Lindmo K., Løkling K. E., et al. , 2013. Status and future perspectives of vaccines for industrialised fin-fish farming. Fish Shellfish Immunol. 35(6): 1759–1768. 10.1016/j.fsi.2013.05.029 [DOI] [PubMed] [Google Scholar]
- Bruno D., Collet B., Turnbull A., Kilburn R., Walker A. , et al. , 2007. Evaluation and development of diagnostic methods for Renibacterium salmoninarum causing bacterial kidney disease (BKD) in the UK. Aquaculture 269:114–122. [Google Scholar]
- Canela-Xandri O., Law A., Gray A., Woolliams J. A., Tenesa A., 2015. A new tool called DISSECT for analysing large genomic data sets using a Big Data approach. Nat. Commun. 6(1): 10162 10.1038/ncomms10162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C. A. M., Vattikuti S., Purcell S. M., et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. M., Abecasis G. R., 2007. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 81(5): 913–926. 10.1086/521580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa K., Lhorente J. P., López M. E., Bassini L., Naswa S., et al. , 2015. Genome-wide association analysis reveals loci associated with resistance against Piscirickettsia salmonis in two Atlantic salmon (Salmo salar L.) chromosomes. BMC Genomics 16(1): 854 10.1186/s12864-015-2038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa K., Lhorente J. P., Bassini L., López M. E., Di Génova A., et al. , 2017a Genome wide association study for resistance to Caligus rogercresseyi in Atlantic salmon (Salmo salar L.) using a 50 K SNP genotyping array. Aquaculture 472: 61–65. 10.1016/j.aquaculture.2016.04.008 [DOI] [Google Scholar]
- Correa K., Bangera R., Figueroa R., Lhorente J. P., Yáñez J. M., 2017b The use of genomic information increases the accuracy of breeding value predictions for sea louse (Caligus rogercresseyi) resistance in Atlantic salmon (Salmo salar). Genet. Sel. Evol. 49(1): 15 10.1186/s12711-017-0291-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185(3): 1021–1031. 10.1534/genetics.110.116855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal P., Gillanders E. M., Holmes T. N., Bailey-Wilson J. E., 2008. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 9(1): 516 10.1186/1471-2164-9-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO , 2016. The State of World Fisheries and Aquaculture 2016. Contributing to food security and nutrition for all. Food and Agriculture Organization, Rome. [Google Scholar]
- FAO, 2017 GLOBEFISH Highlights – Special Issue. Food and Agriculture Organization, Rome. Available at: http://www.fao.org/3/a-i7095e.pdf. [Google Scholar]
- Fringuelli E., Gordon A. W., Rodger H., Welsh M. D., Graham D. A., 2012. Detection of Neoparamoeba perurans by duplex quantitatitve Taqman real-time PCR in formalin-fixed, paraffin-embedded Atlantic salmonid gill tissues. J. Fish Dis. 35: 711–724. [DOI] [PubMed] [Google Scholar]
- Gharbi K., Matthews L., Bron J., Roberts R., Tinch A., et al. , 2015. The control of sea lice in Atlantic salmon by selective breeding. J. R. Soc. Interface. 12: 0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbey J., Verspoor E., Mo T. A., Sterud E., Olstad K., et al. , 2006. Identification of genetic markers associated with Gyrodactylus salaris resistance in Atlantic salmon Salmo salar. Dis. Aquat. Organ. 71: 119–129. 10.3354/dao071119 [DOI] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Thompson R., 2014. ASReml user guide, Ed. 4th VSN International Ltd., Hemel Hempstead. [Google Scholar]
- Gjerde B., Ødegård J., Thorland I., 2011. Estimates of genetic variation in the susceptibility of Atlantic salmon (Salmo salar) to the salmon louse Lepeophtheirus salmonis. Aquaculture 314(1-4): 66–72. 10.1016/j.aquaculture.2011.01.026 [DOI] [Google Scholar]
- Gonen S., Baranski M., Thorland I., Norris A., Grove H., et al. , 2015. Mapping and validation of a major QTL affecting resistance to pancreas disease (salmonid alphavirus) in Atlantic salmon (Salmo salar). Heredity (Edinb) 115(5): 405–414. 10.1038/hdy.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., Haley C. S., Hamilton A., Guy D. R., Tinch A. E., et al. , 2008. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 178(2): 1109–1115. 10.1534/genetics.107.082974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., Taggart J. B., Cézard T., Bekaert M., Lowe N. R., et al. , 2014. Development and validation of a high density SNP genotyping array for Atlantic salmon (Salmo salar). BMC Genomics 15(1): 90 10.1186/1471-2164-15-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., 2017. Future directions in breeding for disease resistance in aquaculture species. Rev. Bras. Zootec. 46(6): 545–551. 10.1590/s1806-92902017000600010 [DOI] [Google Scholar]
- Kijas J., Elliot N., Kube P., Evans B., Botwright N., et al. , 2016. Diversity and linkage disequilibrium in farmed Tasmanian Atlantic salmon. Anim. Genet. 48(2): 237–241. 10.1111/age.12513 [DOI] [PubMed] [Google Scholar]
- Kolstad K., Heuch P. A., Gjerde B., Gjedrem T., Salte R., 2005. Genetic variation in resistance of Atlantic salmon (Salmo salar) to the salmon louse Lepeophtheirus salmonis. Aquaculture 247(1-4): 145–151. 10.1016/j.aquaculture.2005.02.009 [DOI] [Google Scholar]
- Lhorente J. P., Gallardo J. A., Villanueva B., Araya A. M., Torrealba D. A., et al. , 2012. Quantitative genetic basis for resistance to Caligus rogercresseyi sea lice in a breeding population of Atlantic salmon (Salmo salar). Aquaculture 324–325: 55–59. 10.1016/j.aquaculture.2011.10.046 [DOI] [Google Scholar]
- Lien S., Koop B. F., Sandve S. R., Miller J. R., Kent M. P., et al. , 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533(7602): 200–205. 10.1038/nature17164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehammer M., Meuwissen T. H. E., Sonesson A. K., 2013. A low-marker density implementation of genomic selection in aquaculture using within-family genomic breeding values. Genet. Sel. Evol. 45(1): 39 10.1186/1297-9686-45-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T., Baranski M., Sonesson A. K., Kjøglum S., 2009. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10(1): 368 10.1186/1471-2164-10-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y., Pong-Wong R., Navarro P., Vitart V., Hayward C., et al. , 2012. Localising loci underlying complex trait variation using regional genomic relationship mapping. PLoS One 7(10): e46501 10.1371/journal.pone.0046501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaiokostas C., Ferraresso S., Franch R., Houston R. D., Bargelloni L., 2016. Genomic prediction of resistance to pasteurellosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing. G3 (Bethesda) 6: 3693–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ødegård J., Baranski M., Gjerde B., Gjedrem T., 2011. Methodology for genetic evaluation of disease resistance in aquaculture species: challenges and future prospects. Aquacult. Res. 42: 103–114. 10.1111/j.1365-2109.2010.02669.x [DOI] [Google Scholar]
- Ødegard J., Moen T., Santi N., Korsvoll S. A., Kjøglum S., et al. , 2014. Genomic prediction in an admixed population of Atlantic salmon (Salmo salar). Front. Genet. 5: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggio V., Pong-Wong R., 2014. Regional Heritability Mapping to identify loci underlying genetic variation of complex traits. BMC Proc. 8(Suppl 5): S3 10.1186/1753-6561-8-S5-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo D., Palaiokostas C., Bargelloni L., Martínez P., Houston R. D., 2017. Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquacult. 0: 1–13. 10.1111/raq.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salte R., Bentsen H. B., Moen T., Tripathy S., Bakke T. A., et al. , 2010. Prospects for a genetic management strategy to control Gyrodactylus salaries infection in wild Atlantic salmon (Salmo salar) stocks. Can. J. Fish. Aquat. Sci. 67(1): 121–129. 10.1139/F09-168 [DOI] [Google Scholar]
- Shirali M., Pong-Wong R., Navarro P., Knott S., Hayward C., et al. , 2016. Regional heritability mapping method helps explain missing heritability of blood lipid traits in isolated populations. Heredity 116(3): 333–338. 10.1038/hdy.2015.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein J. T., Vallejo R. L., Palti Y., Leeds T. D., Rexroad C. E., III, et al. 2009. Rainbow trout resistance to bacterial cold-water disease is moderately heritable and is not adversely correlated with growth. J. Anim. Sci. 87(3): 860–867. 10.2527/jas.2008-1157 [DOI] [PubMed] [Google Scholar]
- Taylor R.S., Wynne J. W., Kube P. D., Elliott N. G., 2007. Genetic variation of resistance to amoebic gill disease in Atlantic salmon (Salmo salar) assessed in a challenge system. Aquaculture 272S1: S94–S99. [Google Scholar]
- Taylor R. S., Kube P. D., Muller W. J., Elliott N. G., 2009. Genetic variation of gross gill pathology and survival of Atlantic salmon (Salmo salar L.) during natural amoebic gill disease challenge. Aquaculture 294(3-4): 172–179. 10.1016/j.aquaculture.2009.06.007 [DOI] [Google Scholar]
- Taylor R. S., Huynh C., Cameron D., Evans B., Cook M. T., et al. , 2016. Gill score guide – amoebic gill disease (AGD) management training document, edited by Evans B. Tassal Pty. Ltd., Hobart, Tasmania. [Google Scholar]
- Tsai H. Y., Robledo D., Lowe N. R., Bekaert M., Taggart J. B., et al. , 2016a Construction and annotation of a high density SNP linkage map of the Atlantic salmon (Salmo salar) genome. G3 (Bethesda) 6(7): 2173–2179. 10.1534/g3.116.029009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. Y., Hamilton A., Tinch A. E., Guy D. R., Bron J. E., et al. , 2016b Genomic prediction of host resistance to sea lice in farmed Atlantic salmon populations. Genet. Sel. Evol. 48(1): 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. Y., Matika O., Edwards S. M., Antolín-Sánchez R., Hamilton A., et al. , 2017. Genotype imputation to improve the cost-efficiency of genomic selection in farmed Atlantic salmon. G3 (Bethesda) 7(4): 1377–1383. 10.1534/g3.117.040717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemoto Y., Pong-Wong R., Navarro P., Vitart V., Hayward C., et al. , 2013. The power of regional heritability analysis for rare and common variant detection: simulations and application to eye biometrical traits. Front. Genet. 4: 232 10.3389/fgene.2013.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo R. L., Leeds T. D., Fragomeni B. O., Gao G., Hernandez A. G., et al. , 2016. Evaluation of genome-enabled selection for bacterial cold water disease using progeny performance data in rainbow trout: insights on genotyping methods and genomic prediction models. Front. Genet. 7: 96 10.3389/fgene.2016.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo R. L., Leeds T. D., Gao G., Parsons J. E., Martin K. E., et al. , 2017. Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet. Sel. Evol. 49(1): 17 10.1186/s12711-017-0293-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91(11): 4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Yáñez J. M., Lhorente J. P., Bassini L. N., Oyarzún M., Neira R., et al. , 2014a Genetic co-variation between resistance against both Caligus rogercresseyi and Piscirickettsia salmonis, and body weight in Atlantic salmon (Salmo salar). Aquaculture 433: 295–298. 10.1016/j.aquaculture.2014.06.026 [DOI] [Google Scholar]
- Yáñez J. M., Houston R. D., Newman S., 2014b Genetics and genomics of disease resistance in salmonid species. Front. Genet. 5: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Dyková I., Snekvik K., Nowak B. F., Morrison R. N., 2008. Neoparamoeba perurans is a cosmopolitan aetiological agent of amoebic gill disease. Dis. Aquat. Organ. 78: 217–223. 10.3354/dao01869 [DOI] [PubMed] [Google Scholar]
- Zilberg D., Munday B. L., 2000. Pathology of experimental amoebic gill disease in Atlantic salmon, Salmo salar L., and the effect of pre-maintenance of fish in sea water on the infection. J. Fish Dis. 23(6): 401–407. 10.1046/j.1365-2761.2000.00252.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primers and probes to perform amoebic load estimation by qPCR are provided in Table S1. Phenotypic data of the fish used in this study is available in File S1. Note that gill scores correspond to an experimental challenge, gill scores higher than 0.5-1 are rarely encountered in Landcatch commercial facilities. Markers included in the SNP array and their position in the Atlantic salmon genome can be found in File S2. Genotypes of the fish used in this study are available in File S3. The regional heritability mapping model is detailed in File S4.