Abstract
Ostreid herpesvirus (OsHV) can cause mass mortality events in Pacific oyster aquaculture. While various factors impact on the severity of outbreaks, it is clear that genetic resistance of the host is an important determinant of mortality levels. This raises the possibility of selective breeding strategies to improve the genetic resistance of farmed oyster stocks, thereby contributing to disease control. Traditional selective breeding can be augmented by use of genetic markers, either via marker-assisted or genomic selection. The aim of the current study was to investigate the genetic architecture of resistance to OsHV in Pacific oyster, to identify genomic regions containing putative resistance genes, and to inform the use of genomics to enhance efforts to breed for resistance. To achieve this, a population of ∼1,000 juvenile oysters were experimentally challenged with a virulent form of OsHV, with samples taken from mortalities and survivors for genotyping and qPCR measurement of viral load. The samples were genotyped using a recently-developed SNP array, and the genotype data were used to reconstruct the pedigree. Using these pedigree and genotype data, the first high density linkage map was constructed for Pacific oyster, containing 20,353 SNPs mapped to the ten pairs of chromosomes. Genetic parameters for resistance to OsHV were estimated, indicating a significant but low heritability for the binary trait of survival and also for viral load measures (h2 0.12 – 0.25). A genome-wide association study highlighted a region of linkage group 6 containing a significant QTL affecting host resistance. These results are an important step toward identification of genes underlying resistance to OsHV in oyster, and a step toward applying genomic data to enhance selective breeding for disease resistance in oyster aquaculture.
Keywords: GWAS, OsHV-1, SNP array, linkage map, oysters
A specific variant of the ostreid herpesvirus (OsHV-1-μvar) has been suggested to be the main cause of periodic mass mortality events in farmed Pacific oysters (Crassostrea gigas) worldwide (Segarra et al. 2010), with other contributing factors potentially including Vibrio bacterial infection and elevated temperature (Petton et al. 2015; Malham et al. 2009). Given that Pacific oysters account for 98% of global oyster production, which was estimated at ∼0.6 M tons in 2015, this pathogen is a significant problem for global aquaculture. Due to the current lack of effective options to prevent or control disease outbreaks (e.g., no option for vaccination and limited evidence of effective biosecurity) improving host resistance to OsHV-1 via selective breeding has become a major target.
A significant additive genetic component has been described for survival during OsHV-1 infection, with estimated heritability values ranging from 0.21 to 0.63 (Azéma et al. 2017; Camara et al. 2017; Dégremont et al. 2015a). Substantial efforts are being made to establish selective breeding programs for C. gigas with OsHV-1 resistance as a primary target trait. An encouraging response to selection for resistance has been observed in C. gigas spat after four generations of mass selection (Dégremont et al. 2015b). Modern selective breeding programs for aquaculture species can facilitate the simultaneous selection of multiple traits, including those not possible to measure directly on selection candidates. Genomic tools can facilitate this process, allowing for increase in selection accuracy and rate of genetic gain for target traits, with improved control of inbreeding (Houston 2017). Further, these tools allow investigation of the genetic architecture of key production traits, opening up possibilities for downstream functional studies to discover genes contributing directly to genetic variation. Putative QTL affecting host resistance to OsHV-1 have been identified using a linkage mapping approach (Sauvage et al. 2010), but genome-wide association approaches have not previously been performed in oysters and offer a substantially higher marker density and mapping resolution.
SNP arrays are enabling tools for genetic analysis and improvement of complex traits in farmed animal species. In the past few years, many genomic resources have been developed for C. gigas and include a reference genome assembly (Zhang et al. 2012), and a moderate number of genetic markers, such as microsatellites (Li et al. 2003; Sekino et al. 2003; Sauvage et al. 2009) and SNPs (Fleury et al. 2009; Sauvage et al. 2007; Wang et al. 2015). Importantly, the recent development of medium and high density SNP arrays for oysters (Gutierrez et al. 2017; Qi et al. 2017) raises the possibility of rapidly collecting genotype data for many thousands of SNP markers dispersed throughout the genome. However, only low to medium density linkage maps have been developed, containing a limited number of both microsatellites and SNPs (Li and Guo 2004; Sauvage et al. 2010; Hedgecock et al. 2015; Hubert and Hedgecock 2004). Linkage maps are important tools for the mapping of quantitative-trait loci (QTL), positional cloning, and also for reference genome assembly. Therefore, the development of a high-density linkage map is an important goal to improve resolution of mapping studies. The availability of genomic tools to score tens of thousands of SNP markers in oysters (i.e., SNP arrays) facilitate development of high density linkage maps and high resolution genome-wide association studies into the genetic architecture of traits of economic interest. In addition, these arrays enable testing of genomic selection approaches which are increasingly common in aquaculture breeding, with encouraging empirical data supporting the advantage over pedigree-based approaches (Tsai et al. 2015; Tsai et al. 2016; Vallejo et al. 2017; Dou et al. 2016; Correa et al. 2017).
The aim of this study was to investigate the genetic architecture of resistance to OsHV-1 infection in C. gigas using a large immersion challenge experiment followed by a genome wide association study (GWAS) to identify loci associated with the trait, and the relative contribution of these loci to the heritability of the trait.
Methods
Source of oysters and disease challenge
Oysters used in this study were obtained from multiple crosses of parents provided by Guernsey Sea Farms (UK) and reared at Cefas facilities. The oysters comprised three pair crosses that were created at Cefas (from 3 sires and 2 dams) and each reared separately, while the rest of the crosses (from 14 sires and 14 dams) were obtained as spat from Guernsey Sea Farms and combined into a mixed culture tank at Cefas. Oysters were held at 20 +/− 2 C during post-settlement and fed with a combination of Isocrysis, Tetraselmis, Chaetoceros, Pavlova sp., and ‘Shellfish Diet 1800’ until they reached an appropriate size (>5 mm) at approximately eight months of age. A subsample of approximately 1,000 oysters were then transferred to the challenge tank at 20 +/− 2° for two days for acclimation.
OsHV-1 µVar was isolated from an individual oyster sampled during a 2015 disease outbreak in Poole harbor (UK). Briefly, the gill and mantle were isolated from the diseased oyster, homogenized in filtered sea water, and filtered sequentially through 1.2, 0.8, 0.45 and 0.2 µm filters. Purified virus was then passaged twice by injection through susceptible oysters for proliferation as per previous protocols (Schikorski et al. 2011). Purified virus was pooled from 10 injected oysters and cryopreserved at -80° in 10% glycerol and 10% fetal calf serum with a freezing rate of 1°/min. An aliquot of the oyster herpes virus OsHV-1 μVar was added to the water tank at an end concentration of 2.49x107 copies /ml (empirically assessed by qPCR) with continuous flow. Mortalities were checked every three hours by visual and manual inspection, morbid or dead animals were removed, snap frozen, and stored at -80°. The challenge lasted for 21 days, by which time mortality rate had returned to baseline levels, and all remaining mortalities and survivors were snap-frozen and stored for DNA extraction.
Phenotypic measurements
Survival was coded as a binary trait i.e., 0 (mortality) or 1 (survival). The viral count of all samples was determined by qPCR according to (Martenot et al. 2010), with the additional use of a plasmid-based standard curve cloned for absolute quantification. The estimated copy number was then divided by the weight of the animal (mg) to obtain a measure of the viral load. Viral load values were then normalized by transformation to the logarithmic scale for further analyses.
SNP array genotyping
Genomic DNA was extracted from the whole oyster (minus the shell) using the RealPure genomic DNA extraction kit (Valencia, Spain), quantified on Qubit and the DNA integrity was checked on a 1% agarose gel.. After considering available DNA quality and quantity, only 897 samples were retained for genotyping (33 parents + 864 challenged offspring). Genotyping was carried out at Edinburgh Genomics using the recently developed Affymetrix SNP array for oysters (Gutierrez et al. 2017). After quality control (QC) using the Axiom Analysis Suite v2.0.0.35, 854 samples were retained following the “best practices workflow” with a sample and SNP call threshold of 90% resulted in 23,388 SNPs classified as good quality (‘PolyHighResolution’ and ‘NoMinorHom’ categories), from ∼40 K putative available for C.gigas on the array, and retained for downstream analyses.
Linkage mapping
Linkage maps were constructed using Lep-map 3 (Rastas 2017). Families used for the generation of this map were assigned using Cervus (Kalinowski et al. 2007) as described by Gutierrez et al. (2017), and further confirmed through the IBD module in Lep-map3. Putative erroneous or missing parental genotypes were re-called using the “ParentCall2” module. Linkage groups were identified using the “SeparateChromosomes2” module using a LodLimit = 60 and distortionLod = 1. Data were then filtered to remove markers from families showing deviations expected Mendelian segregation ratios (“dataTolerance=0.001”) and used with the “OrderMarkers2” module to order the markers in the linkage groups. Individuals showing excessive recombination were also removed from the data as this indicated a potential problem with genotyping or family assignment for this individual. The estimated genome coverage of the map was calculated as c = 1 − e−2dn/L, where d is the average spacing of markers, n is the number of markers, and L is the length of the linkage map (Bishop et al. 1983). Only full sibling families were used for the construction of the linkage maps.
Model and heritability estimation
Genetic parameters for the resistance traits were estimated using a linear mixed model approach fitting animal as a random effect using ASReml 4 (Gilmour et al. 2015) with the following model:
where y is the observed trait, u is the vector of additive genetic effects, e is the residual error, and X and Z the corresponding incidence matrices for fixed effects and additive effects, respectively. The (co)variance structure for the genetic effect was calculated either using pedigree (A) or genomic (G) matrices (i.e., u ∼ N(0, Aσa 2) or N(0, Gσa 2)), where G is the genomic kinship matrix and σ2 is the genetic variance. Hence, the narrow sense heritability was estimated by the additive genetic variance and total phenotypic variance, as follows:
where σ2 a is the additive genetic variance and σ2 p is the total phenotypic variance which is a sum of σ 2a + σ 2e . Heritability on the observed binary scale obtained for survival was converted to the underlying liability scale according to Dempster and Lerner (1950). The genomic relationship matrix required for the analysis was obtained according to (VanRaden 2008) using the GenABEL package (Aulchenko et al. 2007) and inverted using a standard ‘R’ function.
Genome-wide association studies
The GWAS was performed using the GenABEL package (Aulchenko et al. 2007) in R. The genotype data were filtered as part of quality control by using the check.markers module to retain SNPs with a MAF > 0.01, call rate >0.90 and allow a deviation from Hardy-Weinberg Equilibrium < 1 × 10−5, leaving 16,223 filtered SNPs for downstream analyses. It is worth noting that approximately 6 K markers failed the QC filter due to the HWE threshold, but inclusion of these SNPs did not significantly influence the results of the GWAS. Association analyses were run using the family-based score test for association (FASTA) using the mmscore function (Chen and Abecasis 2007) with the mixed linear model (MLM) approach used to avoid potential false positive associations derived from population structure. Genotype data were used to calculate the genomic kinship matrix which was fitted in the model alongside the top four principal components as covariates to account for population structure. Additionally, the GWAS was run using the Efficient Mixed-Model Association eXpedited (EMMAX) software (Kang et al. 2010) to perform a form of validation test for SNPs identified as significant in the GenABEL analysis. The genome-wide significance threshold was set to 3.08x10−6 as determined by Bonferroni correction (0.05 / N), where N represents the number of QC-filtered SNPs across the genome, while the suggestive threshold was set as 3.08 × 10−5 (0.5 / N), i.e., allowing 0.5 false positive per genome scan.
Identification of candidate genes
To identify candidate genes potentially underlying the identified QTL for further study, the location of the most significant SNPs on individual contigs and scaffolds was recorded on the C. gigas genome v9 assembly (GCA_000297895.1) (Zhang et al. 2012). The sequences of these scaffolds / contigs were then aligned (using a custom-built blastn database) with the C. gigas gene annotation database. Contig and scaffold sequences for significant SNPs were also aligned using blastn and blastx (using non-redundant protein sequences) from the NCBI database.
Data availability
Linkage map including all mapped markers and their position is given in File S1. Genotype data corresponding to all informative markers for all the individuals involved in this study is given in File S5.
Results
Challenge outcome and trait heritability
At the end of the 21 day disease challenge, 749 oysters had survived while 251 had died during the experiment. From the latter, 71 oysters had no body tissue at the moment of their removal, leaving 181 mortalities suitable for downstream analyses. Therefore, overall mortality was approximately 25%, but in the subset of oysters available for genotyping the mortality was ∼18%. Viral load calculations (copies/mg) showed that mortalities had a higher average viral load (mean 1.1x105, s.e 2.2 x104), than survivors (mean 1.5 x101, s.e 3.1).
A total of 23 full sibling families were identified using the family assignment software. The largest comprised 231 individuals, and the smallest only two individuals. The vast majority of offspring were assigned to a unique parent pair, but a total of seven individuals were assigned to only one parent (five only to a dam and two only to a sire). Making use of the pedigree information, the estimated heritability on observed scale was 0.13 (0.06), corresponding to a value of 0.25 on the underlying liability scale (Table 1). These estimates were slightly lower when using the genomic kinship matrix, with 0.08 ± 0.03 and 0.17 for the observed and liability scale respectively. For viral load, heritability based on pedigree was estimated at 0.19 ± 0.08 and 0.13 ± 0.05 for genomic matrix. (Table 1).
Table 1. Estimated heritabilities for survival and viral load in challenged populations.
| Trait | Method | Heritability (s.e) |
|---|---|---|
| Survival | Observed binary scale (G) | 0.078 (0.037) |
| Underlying liability scale (G) | 0.168 | |
| Observed binary scale (P) | 0.13 (0.058) | |
| Underlying liability scale (P) | 0.25 | |
| Viral load | Log transformed viral load(G) | 0.127 (0.05) |
| Log transformed viral load (P) | 0.19 (0.08) |
Linkage map
The linkage mapping was performed using the 23 full sibling families comprising 809 progenies and 31 parents. On average, 10% of the markers showed evidence of segregation distortion (P < 0.001) in at least one family with at least ten progenies, leaving 21,087 maternally informative markers and 20,528 paternally informative markers for map construction (Table S1).
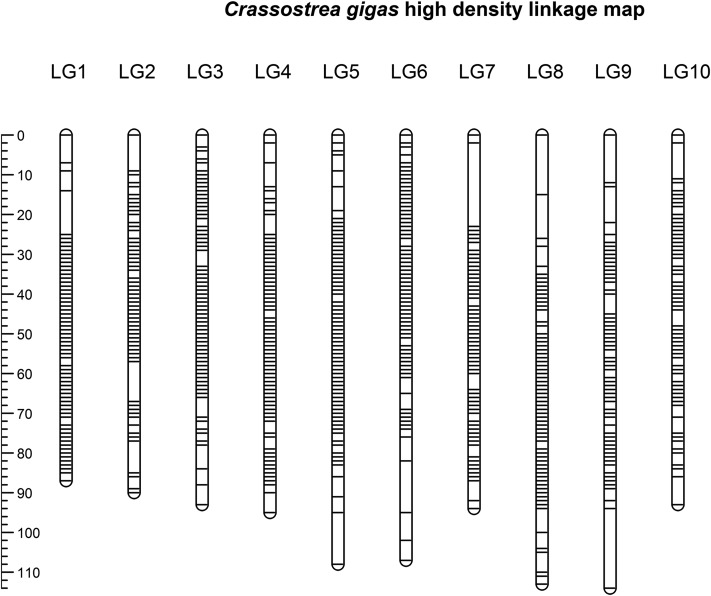
The linkage map contains 20,353 SNPs distributed on 10 LGs (in accordance with the C. gigas karyotype) as shown in Figure 1, with a length of 951 cM for the male map and 994 cM for the female map. The ∼20 K mapped SNPs correspond to 1,921 scaffolds and 149 contigs, according to the latest oyster genome assembly (GCA_000297895.1, Zhang et al. 2012, File S1). These scaffolds and contigs containing mapped SNPs covered approximately 87% of the reference genome length.
Figure 1.
Distribution of SNP markers on the linkage map.
Linkage groups were labeled according to Hedgecock et al. (2015) to keep consistency across C.gigas linkage maps. Our medium density oyster array contains 464 of the SNPs mapped by Hedgecock et al. (2015). From these, 307 were mapped in the current study and their new linkage group assignment fully agrees with their previous assignment (File S2). Likewise, we observed that approximately 38% (734 out 1,921) of the scaffolds with informative markers show evidence of errors in the assembly, due to assignment to at least two distinct LGs in our map (File S3). As expected, the number of LGs associated with scaffolds was positively correlated with scaffold length (Figure S1).
Association analyses
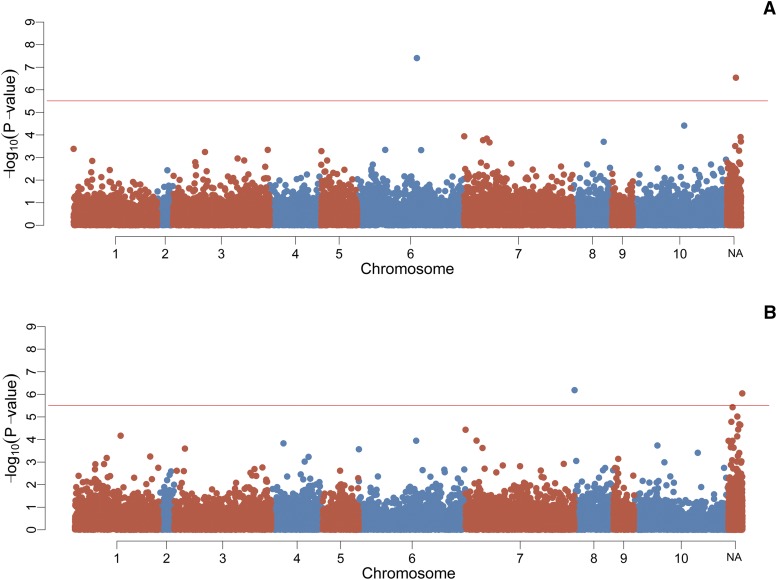
The GWAS for the binary survival trait using the FASTA approach identified two markers showing a genome-wide significant association with the trait (both also significant using EMMAX, with an additional two SNPs significant using EMMAX only), as shown in Table 2, Figure 2A and File S4. Of the ten markers showing the most significant association in the two approaches, four markers are linked to LG 6 but they do not map to the same scaffold, nor are they close together on the linkage map. The proportion of phenotypic variation explained by the top ten markers ranged between 0.019 and 0.047, which implies a polygenic architecture to host resistance, albeit the LG 6 QTL potentially explains a large proportion of the genetic variance given the low heritability estimates.
Table 2. The top ten markers associated with survival.
| SNP ID | LG (position cM) | LG nearest marker (position cM) | Scaffold (position bp) | A1 | A2 | GenABEL | EMMAX | PVE | Nearest Gene |
|---|---|---|---|---|---|---|---|---|---|
| AX-169184215 | LG 6 (42.46) | — | scaffold241 (824,662) | T | G | 3.94E-08* | 4.74E-10* | 0.0473 | CORO1B |
| AX-169192574 | Unassigned | LG 6 (54.61) | scaffold1827 (350,776) | A | G | 2.91E-07* | 7.79E-08* | 0.0411 | MYO10 |
| AX-169208860 | Unassigned | LG 1 (54.37) | scaffold714 (58,763) | G | A | 0.000124 | 5.72E-07* | 0.0224 | CYP1A1 |
| AX-169209993 | LG 7 (9.48) | — | scaffold1599 (493,016) | T | C | 0.000115 | 1.56E-06* | 0.0231 | D2R |
| AX-169207075 | LG 5 (47.54) | — | scaffold57 (142,065) | C | T | 0.004125 | 1.09E-05 | 0.0122 | IFT88 |
| AX-169210119 | Unassigned | LG 6 (29.41) | scaffold198 (583,825) | T | C | 0.000194 | 2.25E-05 | 0.0206 | RANBPM |
| AX-165319118 | LG 5 (25.77) | — | scaffold43494 (138,038) | G | A | 0.000519 | 4.82E-05 | 0.019 | KPNA1 |
| AX-169158711 | LG 6 (42.59) | — | scaffold109 (558,765) | G | A | 0.000468 | 6.48E-05 | 0.0183 | CASP |
| AX-169199571 | LG 10 (42.24) | — | scaffold186 (320,367) | C | T | 3.85E-05 | 7.02E-05 | 0.0247 | AP1AR |
| AX-169168346 | LG 3 (43.83) | — | scaffold1785 (251356) | G | T | 0.000568 | 8.37E-05 | 0.018 | KIF6 |
Genome-wide significant (P < 0.05) markers. A1 & A2, major and minor allele. PVE, phenotypic variation explained by the SNP. The physical position of the SNPs on the Scaffolds are given according to the Pacific oyster reference assembly (Genbank ID GCA_000297895.1).
Figure 2.
Manhattan plots for the GWAS for A) survival and B) Viral load. The position of the SNPs on the X axis is calculated according to the linkage map, Y axis represents the significance value shown as log10 of the p-value. “NA” represent a chromosome that contains markers not assigned to any linkage group. Horizontal red line indicates the genome-wide significance threshold.
The GWAS for the trait of viral load detected two markers showing significant genome-wide association with both FASTA and EMMAX, with an addition eight SNPs identified as significant using EMMAX only (Table 3, Figure 2B and File S4). The SNP showing the most significant association is located in LG 8, however, no other markers are located in the same LG. While most of the markers significantly associated with the trait were not mapped, the nearest mapped SNPs according to their position on the genome scaffolds suggests that three SNPs are located on LG 6. Therefore, it is plausible that there is at least one QTL on LG 6, and this QTL may affect both viral load and the binary trait of survival. The proportion of phenotypic variation in viral load explained by the top ten markers ranged between 0.0209 and 0.037.
Table 3. Top ten markers associated with viral load.
| SNP ID | LG (position cM) | LG nearest marker (position cM) | Scaffold (position bp) | A1 | A2 | GenABEL | EMMAX | PVE | Gene |
|---|---|---|---|---|---|---|---|---|---|
| AX-169203956 | LG 8 (0) | — | scaffold501 (742,989) | C | T | 6.54E-07* | 3.47E-09* | 0.037 | FBN2 |
| AX-169210119 | Unassigned | LG 6 (29.41) | scaffold198 (583,825) | T | C | 9.09E-07* | 9.33E-08* | 0.0349 | RANBPM |
| AX-169172429 | Unassigned | LG 4 (57.01) | scaffold713 (269,794) | T | G | 3.72E-06 | 9.36E-07* | 0.0314 | B3GALNT2 |
| AX-169192982 | Unassigned | LG 6 (43.46) | scaffold1093 (208,087) | C | T | 9.61E-06 | 1.54E-07* | 0.0284 | SKI |
| AX-169167580 | Unassigned | LG 6 (36.95) | scaffold1763 (82,048) | A | G | 1.65E-05 | 8.91E-07* | 0.0277 | CLEC16A |
| AX-169199878 | Unassigned | LG 8 (71.91) | scaffold536 (135,453) | T | C | 2.11E-05 | 1.42E-06* | 0.0269 | TNR |
| AX-169203386 | Unassigned | LG 10 (52.04) | scaffold1301 (188,626) | C | T | 2.27E-05 | 1.13E-06* | 0.0266 | RAPGEF2 |
| AX-169196070 | Unassigned | LG 1 (57.32) | scaffold433 (1,082,890) | G | A | 3.62E-05 | 1.69E-06* | 0.0261 | CARS |
| AX-169199276 | LG 7 (6.68) | — | scaffold128 (550,765) | G | T | 3.70E-05 | 8.44E-07* | 0.0243 | SMARCA5 |
| AX-169194053 | LG 4 (19.39) | — | scaffold728 (174,857) | G | T | 0.000148 | 2.22E-06* | 0.0209 | TNIK |
| AX-169193982 | LG 1 (47.02) | — | scaffold41452 (35,018) | A | G | 6.79E-05 | 6.03E-06 | 0.0243 | U/Pa |
| AX-169192459 | Unassigned | LG 1 (22.66) | scaffold447 (373,487) | G | A | 7.28E-05 | 9.00E-06 | 0.0232 | SCARF2 |
Genome-wide significant (P < 0.05) markers. A1 & A2, major and minor allele. PVE, phenotypic variation explained by the SNP. a U/P indicates uncharacterized protein. The physical position of the SNPs on the Scaffolds are given according to the Pacific oyster reference assembly (Genbank ID GCA_000297895.1).
Discussion
Heritability of OsHV-1 resistance
Estimates of heritability observed for survival to OsHV-1 challenge in the current study were low to moderate (0.078 - -0.25) in comparison to other recent studies that have analyzed resistance to OsHV-1, where estimates have ranged from 0.21 to 0.63 (Dégremont et al. 2015a; Azéma et al. 2017; Camara et al. 2017). Mortality resulting from OsHV-1 exposure in our challenge was relatively low, reaching ∼25% in the overall challenge. The mortality level in the genotyped samples was lower (∼18%), although it is not clear if the dead oysters found with no tissue were affected by the virus or were abnormal at the time of the exposure. It is possible that the population studied may have high level of innate resistance to OsHV-1, considering the low mortality level in ∼8 month old oysters compared to the mortalities typically observed due to OsHV-1 exposure in spat and juvenile oysters (Azéma et al. 2017). Oysters from these families also showed lower mortality levels compared to other batches of oyster spat when using a more established single animal bath OsHV-1 challenges (data not shown), which would support the possibility of a relatively resistant sample of animals. The viral loading data demonstrated that oysters which suffered mortality had higher levels of virus than survivors, and that levels of viral loading in infected animals were in the same range as observed in previous studies (a high of 1.71x106 copies/ mg in this study compared to a high of 2.02x106 previously reported in Sauvage et al. 2010).
Linkage map
The linkage map construction resulted in 10 linkage groups that correspond to the number of chromosomes of C. gigas, successfully mapping ∼20K SNPs. The highest density linkage map for C. gigas to date was described by Hedgecock et al. (2015) and contains ∼1.1K SNPs and microsatellites. Therefore, the linkage map presented in the current study is an improvement to existing resources offering an advance for oyster genomics with potential in assisting future mapping studies, particularly those using the medium density SNP array.
Family assignments were rigorously tested to avoid pedigree errors in the construction of the linkage maps. Distortions from the expected Mendelian segregation were found in ∼10% of the SNPs in the larger families (P < 0.001) (Table S1). Moderate levels of segregation distortion have been commonly observed in oysters (Jones et al. 2013; Hedgecock et al. 2015; Guo et al. 2012) and bivalves in general (Saavedra and Bachère 2006). In the current study, distorted markers were included for the linkage group assignment, but were filtered out for the determination of the order in the LG. It has been argued that distorted markers can affect marker ordering, albeit the effect on map construction has been shown to be minor (Hackett and Broadfoot 2003; Guo et al. 2012).
A measure of the quality of the linkage map was given by overlap with a previous linkage map described by Hedgecock et al. (2015). Several hundred SNPs were successfully re-mapped to the same LG, indicating correct LG definition. Accordingly, reference genome assembly errors observed by Hedgecock et al. (2015) were also observed in our high-density linkage map, where almost ∼40% of the mapped scaffolds were assigned to more than one LG (File S1). This linkage map should be able to provide a good base for the identification of assembly errors and the potential re-assembly of the genome, which seems like a requirement to maximize its utility for future genomics research in this species.
GWAS and associated genes
The association analyses for OsHV-1 survival and viral load suggest that both traits are likely to be impacted by multiple genomic regions, albeit the putative QTL on LG 6 potentially explains a large proportion of the genetic variation. Accordingly, GWAS for survival found SNPs surpassing the genome-wide threshold on LG 6, and SNPs surpassing the suggestive threshold on LG 1, LG 5, & LG 7 (Figure 2, Table 2 and File S4). For the trait of viral load, markers showing a genome-wide significant association were located in LG 8, LG 6LG 10 & LG 4, and suggestive association found in LG 1 & LG 7(Table 3 and File S4). The only previously published study describing genomic regions associated to summer mortality resistance found significant QTL in LG V, VI, VII & IX (which correspond to LG 6, LG 7, LG 8 & LG 10 in our map) in different families (Sauvage et al. 2010). It is noteworthy that LG 6 contains genome-wide significant SNPs for both survival and viral load (and was identified as containing a QTL by Sauvage et al. 2010). In addition, a single SNP (AX-169210119) reached genome-wide significant level for viral load, and the suggestive level for survival. While this SNP was not mapped directly, the nearest mapped SNP was linked to LG 6.
Numerous genes were identified from the genomic regions flanking the most significant SNPs impacting the resistance traits. While the limits defined for screening flanking regions of significant SNPs were defined practically (i.e., the contig / scaffold to which the SNP maps), these genes may represent candidates for future validation, resequencing and functional testing. The SNP showing an association with both survival and viral load (AX-169210119) was located in the RAN Binding Protein 9-like gene which has recently been linked to the interferon gamma signaling pathway (Zhang et al. 2017), and also in viral adhesion and its replication in host cells (Yang et al. 2015). Another gene located near a significant SNP (AX-169184215) is a Coronin gene (CORO1B), from a family of genes that have multi-faceted roles in immune response (Tokarz-Deptuła et al. 2017). Finally, the actin motor protein Myo10 gene is located near AX-169192574, and this gene encodes a protein which is essential for release of Marburgvirus particles from host cells (Kolesnikova et al. 2007). These and other genes may form the basis for downstream functional studies to assess their function in host response to virus in oysters. Further, from a practical breeding perspective, these SNPs may have potential for marker-assisted or genomic selection to improve host resistance in farmed oyster populations. Given the data in the current study do suggest a polygenic or oligogenic nature of resistance to OsHV-1, utilizing all markers to calculate genomic breeding values for resistance may be the most effective approach.
Nevertheless, validation studies are required in independent populations to assess the robustness of the observed association between the significant SNPs and OsHV-1 resistance in oysters, particularly given the unusually low mortality observed in the challenged population. A higher mortality level could potentially provide a higher power of detection for the association analyses that could help confirm any putative QTL.
Conclusion
This is the first GWAS using the a high density SNP panel Pacific oysters, and was enabled by the recent development of a SNP array (Gutierrez et al. 2017). Heritability of resistance to OsHV-1 in oysters was significant, but low to moderate in magnitude. The fact that this heritability was detected using both the pedigree and genomic relationship matrix implies that selective breeding and genomic selection for resistance could be effective. Using the genotype data, a high-density linkage map was constructed for C. gigas, and the GWAS identified numerous markers showing a genome-wide significant association with the traits. The most encouraging QTL was located on LG 6, reaching genome-wide significance for the binary trait of survival, with some evidence of a significant association with viral load. Future analyses will test candidate genes identified by the GWAS, verify trait-associated SNPs in independent populations, and test genomic selection as a tool to enhance host resistance to this problematic pathogen for oyster aquaculture.
Supplementary Material
Supplemental Material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.118.200113/-/DC1.
Acknowledgments
The authors gratefully acknowledge funding from BBSRC and NERC under the UK Aquaculture Initiative (BB/M026140/1, NE/P010695/1) in addition to BBSRC Institute Strategic Program Grants (BB/P013759/1 and BB/P013740/1). SNP array genotyping was carried out by Edinburgh Genomics, The University of Edinburgh. Edinburgh Genomics is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1).
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M., 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23(10): 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- Azéma P., Lamy J.-B., Boudry P., Renault T., Travers M.-A., et al. , 2017. Genetic parameters of resistance to Vibrio aestuarianus, and OsHV-1 infections in the Pacific oyster, Crassostrea gigas, at three different life stages. Genet. Sel. Evol. 49(1): 23 10.1186/s12711-017-0297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. T., Cannings C., Skolnick M., Williamson J. A., Weir B. S., 1983. The number of polymorphic DNA clones required to map the human genome. Statistical Analysis of DNA Sequence Data, B.S. Weir, New York. [Google Scholar]
- Camara M. D., Yen S., Kaspar H. F., Kesarcodi-Watson A., King N., et al. , 2017. Assessment of heat shock and laboratory virus challenges to selectively breed for ostreid herpesvirus 1 (OsHV-1) resistance in the Pacific oyster, Crassostrea gigas. Aquaculture 469: 50–58. 10.1016/j.aquaculture.2016.11.031 [DOI] [Google Scholar]
- Chen W.-M., Abecasis G. R., 2007. Family-Based Association Tests for Genomewide Association Scans. Am. J. Hum. Genet. 81(5): 913–926. 10.1086/521580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa K., Bangera R., Figueroa R., Lhorente J. P., Yáñez J. M., 2017. The use of genomic information increases the accuracy of breeding value predictions for sea louse (Caligus rogercresseyi) resistance in Atlantic salmon (Salmo salar). Genet. Sel. Evol. 49(1): 15 10.1186/s12711-017-0291-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dégremont L., Lamy J.-B., Pépin J.-F., Travers M.-A., Renault T., 2015a New Insight for the Genetic Evaluation of Resistance to Ostreid Herpesvirus Infection, a Worldwide Disease, in Crassostrea gigas. PLoS One 10(6): e0127917 10.1371/journal.pone.0127917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dégremont L., Nourry M., Maurouard E., 2015b Mass selection for survival and resistance to OsHV-1 infection in Crassostrea gigas spat in field conditions: response to selection after four generations. Aquaculture 446: 111–121. 10.1016/j.aquaculture.2015.04.029 [DOI] [Google Scholar]
- Dempster E. R., Lerner I. M., 1950. Heritability of threshold characters. Genetics 35(2): 212–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J., Li X., Fu Q., Jiao W., Li Y., et al. , 2016. Evaluation of the 2b-RAD method for genomic selection in scallop breeding. Sci. Rep. 6(1): 19244 10.1038/srep19244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury E., Huvet A., Lelong C., de Lorgeril J., Boulo V., et al. , 2009. Generation and analysis of a 29,745 unique Expressed Sequence Tags from the Pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: the GigasDatabase. BMC Genomics 10(1): 341 10.1186/1471-2164-10-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A., Gogel B., Cullis B., Welham S., Thompson R., 2015. ASReml user guide release 4.1 structural specification. Hemel hempstead: VSN international ltd [Google Scholar]
- Guo X., Li Q., Wang Q. Z., Kong L. F., 2012. Genetic Mapping and QTL Analysis of Growth-Related Traits in the Pacific Oyster. Mar. Biotechnol. (NY) 14(2): 218–226. 10.1007/s10126-011-9405-4 [DOI] [PubMed] [Google Scholar]
- Gutierrez A. P., Turner F., Gharbi K., Talbot R., Lowe N. R., et al. , 2017. Development of a medium density combined-species SNP array for Pacific and European oysters (Crassostrea gigas and Ostrea edulis). G3 7(7): 2209–2218. 10.1534/g3.117.041780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett C. A., Broadfoot L. B., 2003. Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90(1): 33–38. 10.1038/sj.hdy.6800173 [DOI] [PubMed] [Google Scholar]
- Hedgecock D., Shin G., Gracey A. Y., Den Berg D. V., Samanta M. P., 2015. Second-generation linkage maps for the Pacific oyster Crassostrea gigas reveal errors in assembly of genome scaffolds. G3 5(10): 2007–2019. 10.1534/g3.115.019570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., 2017. Future directions in breeding for disease resistance in aquaculture species. Rev. Bras. Zootec. 46(6): 545–551. 10.1590/s1806-92902017000600010 [DOI] [Google Scholar]
- Hubert S., Hedgecock D., 2004. Linkage Maps of Microsatellite DNA Markers for the Pacific Oyster Crassostrea gigas. Genetics 168(1): 351–362. 10.1534/genetics.104.027342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. B., Jerry D. R., Khatkar M. S., Raadsma H. W., Zenger K. R., 2013. A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genomics 14(1): 810 10.1186/1471-2164-14-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski S. T., Taper M. L., Marshall T. C., 2007. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16(5): 1099–1106. 10.1111/j.1365-294X.2007.03089.x [DOI] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42(4): 348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova L., Bohil A. B., Cheney R. E., Becker S., 2007. Budding of Marburgvirus is associated with filopodia. Cell. Microbiol. 9(4): 939–951. 10.1111/j.1462-5822.2006.00842.x [DOI] [PubMed] [Google Scholar]
- Li G., Hubert S., Bucklin K., Ribes V., Hedgecock D., 2003. Characterization of 79 microsatellite DNA markers in the Pacific oyster Crassostrea gigas. Mol. Ecol. Notes 3(2): 228–232. 10.1046/j.1471-8286.2003.00406.x [DOI] [Google Scholar]
- Li L., Guo X., 2004. AFLP-based genetic linkage maps of the pacific oyster Crassostrea gigas Thunberg. Mar. Biotechnol. (NY) 6: 26–36. [DOI] [PubMed] [Google Scholar]
- Malham S. K., Cotter E., O’Keeffe S., Lynch S., Culloty S. C., et al. , 2009. Summer mortality of the Pacific oyster, Crassostrea gigas, in the Irish sea: the influence of temperature and nutrients on health and survival. Aquaculture 287(1–2): 128–138. 10.1016/j.aquaculture.2008.10.006 [DOI] [Google Scholar]
- Martenot C., Oden E., Travaillé E., Malas J. P., Houssin M., 2010. Comparison of two real-time PCR methods for detection of ostreid herpesvirus 1 in the Pacific oyster Crassostrea gigas. J. Virol. Methods 170(1–2): 86–89. 10.1016/j.jviromet.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Petton B., Bruto M., James A., Labreuche Y., Alunno-Bruscia M., et al. , 2015. Crassostrea gigas mortality in France: the usual suspect, a herpes virus, may not be the killer in this polymicrobial opportunistic disease. Front. Microbiol. 6: 686 10.3389/fmicb.2015.00686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Song K., Li C., Wang W., Li B., et al. , 2017. Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS One 12(3): e0174007 10.1371/journal.pone.0174007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics. 10.1093/bioinformatics/btx494 [DOI] [PubMed] [Google Scholar]
- Saavedra C., Bachère E., 2006. Bivalve genomics. Aquaculture 256(1–4): 1–14. 10.1016/j.aquaculture.2006.02.023 [DOI] [Google Scholar]
- Sauvage C., Bierne N., Lapègue S., Boudry P., 2007. Single Nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster Crassostrea gigas. Gene 406(1–2): 13–22. 10.1016/j.gene.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Sauvage C., Boudry P., De Koning D. J., Haley C. S., Heurtebise S., et al. , 2010. QTL for resistance to summer mortality and OsHV-1 load in the Pacific oyster (Crassostrea gigas). Anim. Genet. 41(4): 390–399. 10.1111/j.1365-2052.2009.02018.x [DOI] [PubMed] [Google Scholar]
- Sauvage C., Boudry P., Lapegue S., 2009. Identification and characterization of 18 novel polymorphic microsatellite makers derived from expressed sequence tags in the Pacific oyster Crassostrea gigas. Mol. Ecol. Resour. 9: 853–855. 10.1111/j.1755-0998.2009.02525.x [DOI] [PubMed] [Google Scholar]
- Schikorski D., Renault T., Saulnier D., Faury N., Moreau P., et al. , 2011. Experimental infection of Pacific oyster Crassostrea gigas spat by ostreid herpesvirus 1: demonstration of oyster spat susceptibility. Vet. Res. (Faisalabad) 42(1): 27 10.1186/1297-9716-42-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra A., Pépin J. F., Arzul I., Morga B., Faury N., et al. , 2010. Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 153(1): 92–99. 10.1016/j.virusres.2010.07.011 [DOI] [PubMed] [Google Scholar]
- Sekino M., Hamaguchi M., Aranishi F., Okoshi K., 2003. Development of Novel Microsatellite DNA Markers from the Pacific Oyster Crassostrea gigas. Mar. Biotechnol. (NY) 5(3): 227–233. 10.1007/s10126-002-0104-z [DOI] [PubMed] [Google Scholar]
- Tokarz-Deptuła B., Malinowska M., Adamiak M., Deptuła W., 2017. Coronins and their role in immunological phenomena. Cent. Eur. J. Immunol. 41(4): 435–441. 10.5114/ceji.2016.65143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.-Y., Hamilton A., Tinch A. E., Guy D. R., Bron J. E., et al. , 2016. Genomic prediction of host resistance to sea lice in farmed Atlantic salmon populations. Genet. Sel. Evol. 48(1): 47 10.1186/s12711-016-0226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.-Y., Hamilton A., Tinch A. E., Guy D. R., Gharbi K., et al. , 2015. Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics 16(1): 969 10.1186/s12864-015-2117-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo R. L., Leeds T. D., Gao G., Parsons J. E., Martin K. E., et al. , 2017. Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet. Sel. Evol. 49(1): 17 10.1186/s12711-017-0293-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 91(11): 4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Wang J., Qi H., Li L., Que H., Wang D., et al. , 2015. Discovery and validation of genic single nucleotide polymorphisms in the Pacific oyster Crassostrea gigas. Mol. Ecol. Resour. 15(1): 123–135. 10.1111/1755-0998.12278 [DOI] [PubMed] [Google Scholar]
- Yang Y.-C., Feng T.-H., Chen T.-Y., Huang H.-H., Hung C.-C., et al. , 2015. RanBPM regulates Zta-mediated transcriptional activity in Epstein–Barr virus. J. Gen. Virol. 96(8): 2336–2348. 10.1099/vir.0.000157 [DOI] [PubMed] [Google Scholar]
- Zhang G., Fang X., Guo X., Li L., Luo R., et al. , 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418): 49–54. 10.1038/nature11413 [DOI] [PubMed] [Google Scholar]
- Zhang J., Cong X., Zhaoqiao J., Yang X., Li M., et al. , 2017. Ran binding protein 9 (RanBPM) binds IFN-λR1 in the IFN-λ signaling pathway. Sci. China Life Sci. 60(9): 1030–1039. 10.1007/s11427-017-9028-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Linkage map including all mapped markers and their position is given in File S1. Genotype data corresponding to all informative markers for all the individuals involved in this study is given in File S5.