Abstract
Splicing of pre-mRNA involves two consecutive trans-esterification steps that take place in the spliceosome, a large dynamic ribonucleoprotein complex situated in the nucleus. In addition to core spliceosomal proteins, each catalytic step requires step-specific factors. Although the Arabidopsis thaliana genome encodes around 430 predicted splicing factors, functional information about these proteins is limited. In a forward genetic screen based on an alternatively-spliced GFP reporter gene in Arabidopsis thaliana, we identified a mutant impaired in putative step II factor PRP18a, which has not yet been investigated for its role in pre-mRNA splicing in plants. Step II entails cleavage at the 3′ splice site accompanied by ligation of the 5′ and 3′ exons and intron removal. In the prp18 mutant, splicing of a U2-type intron with non-canonical AT-AC splice sites in GFP pre-mRNA is reduced while splicing of a canonical GT-AG intron is enhanced, resulting in decreased levels of translatable GFP mRNA and GFP protein. These findings suggest that wild-type PRP18a may in some cases promote splicing at weak, non-canonical splice sites. Analysis of genome-wide changes in alternative splicing in the prp18a mutant identified numerous cases of intron retention and a preponderance of altered 3′ splice sites, suggesting an influence of PRP18a on 3′ splice site selection. The prp18a mutant featured short roots on synthetic medium and small siliques, illustrating that wild-type PRP18a function is needed for a normal phenotype. Our study expands knowledge of plant splicing factors and provides foundational information and resources for further functional studies of PRP18 proteins in plants.
Keywords: alternative splicing, Arabidopsis thaliana, PRP18, second step factor
Messenger RNAs in eukaryotes are derived from primary transcripts (pre-mRNAs) that are subject to extensive co- and post-transcriptional processing involving 5′ cap formation, excision of introns by splicing, and 3′ cleavage and polyadenylation (Moore and Proudfoot 2009). Pre-mRNA splicing occurs in two sequential trans-esterification steps that take place in the spliceosome, a large dynamic ribonucleoprotein (RNP) complex present in the nucleus. The first catalytic step (step I) yields a lariat-3′ exon and a cleaved 5′ exon. The second catalytic step (step II) produces the final mRNA with ligated exons and the spliced out lariat intron (Horowitz 2012; Bertram et al. 2017). In constitutive splicing, the same splice sites are always used for a given intron. By contrast, alternative splicing entails variable splice site usage, which increases transcriptome and proteome diversity. Major modes of alternative splicing include intron retention (IR), exon skipping (ES) and alternative 5‘(donor) and 3′ (acceptor) splice site selection. Alternative splicing is rare in budding yeast but common in plants and animals, in which the majority of multi-intron genes undergo alternative splicing. ES is the major mode of alternative splicing in animal cells whereas IR predominates in plants (Marquez et al. 2012). Alternative splicing in plants is important for development and responses to the environment (Reddy et al. 2013; Staiger and Brown 2013; Filichkin et al. 2015; Deng and Cao 2017).
The spliceosomal cycle involves the step-wise assembly of at least six biochemically and/or structurally defined splicesomal complexes that vary in the composition of proteins and small nuclear (sn) RNAs (Matera and Wang 2014; Yan et al. 2017; Bertram et al. 2017). Core spliceosomal proteins include six Sm/Lsm proteins - B/B’, D1, D2, D3, E, F and G – which form a heptameric ring encircling one of five snRNAs (U1, U2, U4, U5, or U6) to create five distinct snRNPs. In addition to snRNP proteins, numerous non-snRNP proteins are dynamically associated with the spliceosome as the splicesomal cycle proceeds. As an initial step in the splicing process, the U1 and U2 snRNPs recognize the 5′ and 3′ splice sites and conserved branch point of an intron and interact to produce pre-spliceosomal complex A. The addition of a preformed U4/U6.U5 tri-snRNP to complex A yields pre-catalytic complex B. After dissociation of U1 and U4 snRNPs as well as other conformational and compositional changes to the spliceosome, the U2, U5 and U6 snRNPs act at the active sites of two sequentially-formed catalytic complexes, B* and C*, to execute the consecutive step I and II reactions, respectively (Yan et al. 2017; Haselbach et al. 2018). Catalysis is accomplished by two Mg+2 ions that are coordinated by conserved nucleotides in U6 snRNA (Fica et al., 2013). The spliceosome can thus be described as a protein-directed metalloribozyme (Fica et al. 2013; Hang et al. 2015; Yan et al. 2017). The principle protein at the catalytic center is Prp8 (pre-mRNA processing factor8), a U5 snRNP component that provides a scaffold for the pre-mRNA and the U2, U5 and U6 snRNAs to ensure they are properly positioned for the splicing reactions (Hodges et al. 1995; Grainger and Beggs 2005; Yan et al., 2017). Following the two splicing steps, the spliced mRNA and lariat intron are released from the post-spliceosomal complex P (Liu et al. 2017; Wilkinson et al. 2017). The spliceosome then dissociates, liberating individual components to assemble anew at the next intron.
Most information about the protein composition of different spliceosomal complexes has been obtained from proteomic, RNA cross-linking and structural studies performed using Saccharomyces cerevisiae (budding yeast) and metazoan cells (Fabrizio et al. 2009; Wahl et al. 2009; Wahl and Lührmann 2015; Will and Lührmann 2011). Results from similar proteomic and structural analyses are lacking in plants because functional spliceosomal complexes have not yet been isolated from plant cells (Reddy et al. 2012, 2013). Therefore, the predicted protein composition of plant spliceosomes has been largely deduced from comparative sequence analyses. These comparisons have demonstrated that the Arabidopsis thaliana (Arabidopsis) genome encodes approximately 430 spliceosomal proteins and splicing-related factors, the majority of which are conserved in budding yeast and metazoans (Koncz et al. 2012). Only a fraction of these splicing-related proteins have been studied for their contributions to splicing and plant physiology. A number of the splicing factors are encoded by duplicated genes, which can potentially diverge functionally over evolutionary time (Koncz et al. 2012; Reddy et al. 2012, 2013).
Non-snRNP proteins that associate with the spliceosome at a specific step have been identified in budding yeast and humans. Step I factors include Cwc25 and Yju2 (CWC15 and CCDC130, respectively, in humans), which both act to stabilize Prp8 in the catalytic cavity of the B* complex (Wan et al. 2016; Yan et al. 2017; Bertram et al. 2017). In budding yeast, step II requires four proteins not present in the spliceosome at completion of the first trans-esterification step. These include Prp16, a DExD/H –box RNA helicase involved in remodeling the spliceosome after step I to create catalytically-active complex C; Prp22, a DExH-box RNA helicase essential for the second trans-esterification step and release of the mature mRNA from the spliceosome; Slu7, a Zn finger protein; and Prp18, a protein with a unique structure unrelated to any other protein (Horowitz 2012). Slu7 and Prp18 interact with each other and contact Prp8 in complex C* to assist in docking the 3′ splice site-3′ exon into the active site (Zhang and Schwer 1997; Horowitz 2012; Ohrt et al. 2013; Yan et al. 2017; Bertram et al. 2017).
Arabidopsis encodes orthologs of these step I- and step II-specific factors (Koncz et al., 2012), several of which have been studied previously. Of the step I proteins, a Yju2 ortholog, termed CWC16a, was identified in a genetic screen and shown to be important for splicing of a subset of pre-mRNAs (Kanno et al. 2017a). From the step II factors, an ortholog of Prp16, CLUMSY VEIN (CUV), was shown to influence auxin-mediated development and splicing of a selected group of pre-mRNAs (Tsugeki et al. 2015). A Slu7 ortholog, termed SWELLMAP1 (SMP1), was found to be involved in timing of cell cycle arrest during leaf development (Clay and Nelson 2005).
PRP18 is a step II factor that has not yet been studied for its role in pre-mRNA splicing or development in plants. We report here the identification of a prp18a mutant in a forward genetic screen based on an alternatively-spliced GFP reporter gene in Arabidopsis. We describe the phenotypic features of the prp18a mutant and report on genome-wide differential gene expression and alternative splicing profile in the mutant.
Materials And Methods
Plant material
The Arabidopsis transgenic T line harboring an alternatively-spliced GFP reporter gene (referred to hereafter as ‘wild-type’) and the prp18a-1 mutant derived from ethyl methane sulfonate (EMS) mutagenesis of the T line are in the Col-0 ecotype (Kanno et al. 2016, 2017a,b). Seeds of a prp18b-1 T-DNA insertion mutant (SALK_024667C) were provided by the Nottingham Arabidopsis Stock Center (NASC). Plants were cultivated under long-day conditions (22-24°, 16 hr light, 8 hr dark).
Nomenclature of plant generations
M1 mutant plants are grown from seeds treated with EMS and hence are heterozygous for EMS-induced mutations. Self-fertilization (selfing) of M1 plants produces the M2 generation, which is the first generation when a recessive mutation can be homozygous. Further selfing of M2 plants leads to generations M3, M4 and so on. Backcrossing an M2 plant with the parental T line produces the BC1 generation, in which heterozygosity of the original mutation is re-established. Selfing of BC1 plants yields the BC1F2 generation, 25% of which are again homozygous for the respective mutation. BC1F2 plants contain fewer EMS-induced mutations than the original M2 plant. Further selfing of BC1F2 plants produces generations BC1F3, BC1F4 and so forth. Crossing two lines that are homozygous for distinct mutations generates the F1 generation, in which the two mutations are heterozygous. Selfing an F1 plant yields the F2 generation, which segregates the two mutations in a Mendelian manner.
Forward genetic screen, complementation, and phenotype analysis
Details of the forward genetic screen based on an alternatively-spliced GFP reporter gene in the wild-type T line have been described in previous publications (Kanno et al. 2016, 2017a,b). Mutagenesis was accomplished using EMS, which produces almost exclusively G/A to C/T transition mutations (Kim et al. 2006). Screening of mutants was carried out in the M2 generation. Mutants modified in splicing of the GFP pre-mRNA display either a GFP-weak (gfw) or Hyper-GFP (hgf) phenotype relative to the wild-type T line, which shows an intermediate level of fluorescence. So far we have reported five hgf and three gfw mutants retrieved from this screen (Table 1). The gfw4-1 mutant described here was identified by the GFP-weak phenotype of M2 seedlings growing under sterile conditions on solid Murashige and Skoog (MS) medium using a Leica M165FC fluorescence stereomicroscope. A mutation in the PRP18a gene (At1g03140) in gfw4-1 was identified by next generation mapping (NGM) (James et al. 2013) after sequencing pooled DNA isolated from at least 50 BC1F2 seedlings exhibiting a GFP-weak phenotype as described previously (Kanno et al. 2017a,b). Phenotypic analysis of the prp18a-1 mutant was performed on plants of the BC1F3 generation.
Table 1. Mutants identified so far in forward genetic screen.
| Hyper-GFP (hgf) mutant | Name | AGI number | Predicted function in splicing | No. of alleles | Effect on GFP pre-mRNA splicing | Effect of mutation on development | Reference |
|---|---|---|---|---|---|---|---|
| hgf1 | coilin; | At1g13030 | a protein marker protein for Cajal bodies, which are facilitate snRNP maturation | 12* | AU-AC↑ | no | Kanno et al. 2016 |
| hgf2 | CWC16a | At1g25682 | step I factor | 2* | AU-AC↑ | no | Kanno et al. 2017a |
| hgf3 | SMU1 | At1g73720 | which may help to recognize spliceosomal targets for ubiquitination | 1 | AU-AC↑ | no | Kanno et al. 2017a |
| hgf4 | SMFa | At4g30220 | a small nuclear ribonucleoprotein present in snRNPs | 1 | No change | no | Kanno et al. 2017a |
| hgf5 | PRP39a | At1g04080 | U1 snRNP component | 5* | AU-AC↑ | no | Kanno et al. 2017b |
| GFP-weak (gfw) mutant | Name | AGI number | Predicted function in splicing | No. of alleles | Effect on GFP pre-mRNA splicing | Effect of mutation on development | Reference |
|---|---|---|---|---|---|---|---|
| gfw1 | AtRTF2 | At5g58020 | may contribute to ubiquitin-based regulation of the spliceosome | 2 | AU-AC↓, unspliced↑ | embryo lethal | Sasaki et al., 2015; Kanno et al. 2017a |
| gfw2 | PRP8a | At1g80070 | a U5 snRNP component that acts at the catalytic core of the spliceosome | 3 | AU-AC↓, unspliced↑ | embryo lethal | Sasaki et al., 2015; Kanno et al. 2017a |
| gfw3 | RBM25 | At1g60200 | U1 snRNP component | 2 | AU-AC↓, unspliced↑ | low seed set | Kanno et al. 2017b |
| gfw4 | PRP18a | At1g03140 | step II factor | 1 | AU-AC↓, GU-AG↑ | short roots, small siliques | this study |
The mutants obtained so far include a core spliceosomal protein (SMFa); components of the U1 (PRP39a, RBM25) and U5 (PRP8) snRNPs; step I and step II factors transiently associated with the spliceosome (CWC16a and PRP18a, respectively); putative splicing regulatory proteins (RTF2 and SMU1); and one structural protein presumed to be important for snRNP maturation (coilin). So far we have only observed developmental phenotypes with the four identified gfw mutations, two of which are embryo-lethal. The biological significance of these results are not yet clear.
Further screening of the M2 population after publication of the first alleles of coilin, PRP39a and CWC16a has identified three new alleles of coilin (R9H; first intron, 3′ splice site; second intron, 5′ splice site), one new allele of PRP39a (R226*), and one new allele of CWC16a (W18*). These new unpublished alleles are included in the number of alleles shown in this table.
Complementation of the prp18a-1 mutation was achieved by transforming the mutant with a construct containing the PRP18a coding sequence under the control of the 35S promoter and terminator sequences (Pietrzak et al. 1986). Mutant plants (BC1F3 generation) were transformed with this construct using the floral dip method (Clough and Bent 1998) and Agrobacterium binary vector BV-MpPATot SalI (Matzke et al. 2010), which confers resistance to phosphinothricin (PPT). T1 transformants were selected on solid MS medium containing 200 µg/ml cefotaxime and 20 µg/ml PPT. The presence of the prp18a-1 mutation in complemented lines was confirmed by Sanger sequencing.
Testing the effects of a prp18b mutation on GFP fluorescence and plant phenotype
To test whether a homozygous mutation in PRP18b (At1g54590), the paralog of PRP18a, would similarly confer a GFP-weak phenotype and phenotypic defects, we crossed the wild-type T line (T/T) with a homozygous prp18b-1 T-DNA insertion mutant (b/b) (SALK_024667C). Self-fertilization of the F1 plants resulting from the cross (genotype T/-; B/b; the dash denotes hemizygosity for the transgenic T locus) generated a segregating F2 population. F2 seeds were germinated on solid MS medium and screened approximately two weeks later under a fluorescence stereomicroscope for GFP expression, which is observed with a genotype of either T/T or T/- (collectively written hereafter as T/(T)). A subset was transferred to soil for genotyping to identify T/T; b/b plants. T/T;b/b plants in soil were examined for phenotypic features during growth and reproduction. Selfed seedlings of T/T;b/b plants were sown on MS medium and viewed under a fluorescence stereomicroscope to assess GFP expression, and the length of seedling roots on MS medium was noted.
To investigate the viability of double homozygous mutant plants (a/a; b/b), we crossed the homozygous prp18-1 mutant (T/T; a/a) to a b/b plant. Self-fertilization of the F1 plants resulting from this cross (genotype T/-; A/a; B/b) produced a segregating F2 population. The F2 seeds were germinated on solid MS medium and pre-screened under a fluorescence stereomicroscope for a GFP-weak phenotype (indicating a genotype of T/(T); a/a). Selected GFP-weak F2 progeny were transferred to soil for genotyping to identify T/(T); a/a; b/b plants. Primers for detecting prp18a-1 and prp18b-1 alleles are listed in Table S5.
Western blotting
Western blotting to detect GFP protein was carried as described previously using total protein isolated from two week-old seedlings (BC1F3 generation) growing on solid MS medium under a 16 hr light/8 hr dark cycle at 24° (Fu et al. 2015; Kanno et al. 2016, 2017a,b). Monoclonal GFP antibodies were obtained from Roche (Cat. No. 11814 460001). Actin monoclonal antibodies were purchased from Thermo Scientific Pierce (Cat. No. MA1-774).
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR to detect GFP splice variants in the prp18a-1 mutants was conducted using total RNA isolated from two week-old seedlings (BC1F3 generation) growing on solid MS medium as described above using a Plant Total RNA Miniprep kit (GeneMark, Taiwan) according to a published procedure (Sasaki et al. 2015; Kanno et al. 2017a,b). Primers for GFP and actin are listed in Table S5.
RNA-sequencing (RNA-seq)
Total RNA was isolated from two-week-old seedlings (cultivated on MS medium as described above) of the original inbred prp18a-1 mutant, the BC1F3 generation of the prp18a-1 mutant, and the wild-type T line. Library preparation and RNA-seq performed out (biological triplicates for each sample) as described previously (Sasaki et al. 2015; Kanno et al. 2016). Whole genome re-sequencing on the prp18a-1 mutant was conducted to detect remaining EMS-induced second-site mutations that alter splice sites, which were then removed from the analysis of alternative splicing.
RNA-seq reads were mapped to the TAIR10 genome using the following two-step approach. Reads were mapped to the TAIR10 transcriptome using Bowtie2 (Langmead and Salzberg 2012), only alignments of read pairs that were mapped to the same transcript with high identity (>95%) and at least 4bp exact matches in both ends were accepted. Rest reads were mapped to the TAIR10 genome using BLAT (Kent 2002), where alignments with blocks shorter than 8bps were removed. Read counts were computed using RackJ (http://rackj.sourceforge.net/) and normalized into log-count-per-million (logCPM) using the TMM method (Robinson and Oshlack 2010) and the voom method (Law et al. 2014). logCPM values were then transformed into RPKM values. Differentially expressed genes were identified using t-tests on aforementioned RPKM values if the p-value was less than 0.01 and the fold-change was greater than or equal to 2.
To detect the preference of retaining an intron, its intron retention ratio was computed as the average read depth of this intron divided by the average read depth of neighboring exons, and its intron retention ratios in mutant replicates were compared to those in wild-type controls using t-test. In this approach, the underlying null hypothesis assumes that the chance of retaining the intron is the same in both samples, and a significant p-value indicates that the chance is not the same, i.e., a preference of intron retention in one sample. In this study, we classified an intron as intron retention (IR) or more efficient splicing (MES) if its t-test p-value was less than 0.01 and the average intron retention ratio in mutant replicates was two times higher or lower than that in wild-type controls, respectively.
To detect the preference of an exon skipping event or an alternative donor/acceptor event, an approach similar to that for introns was applied. The alternative splicing (AS) ratio was computed as the number of supporting reads of the AS event divided by the number of non-supporting reads of the AS event (splice reads involving one skipped exon for exon skipping events, and splice reads spanning the same exon pair but with other splicing junctions for alternative donor/acceptor events), and log-AS ratios in mutant replicates were compared to those in wild-type controls using t-test. For further confirmation of the AS event, its expression ratios were also computed as supporting read counts divided by unique read counts of its gene, and applied log-expression ratios to t-test. Finally, an exon skipping event or an alternative donor/acceptor event was reported if both p-values were less than 0.01, and it was classified as enhanced or reduced if the average expression ratio in mutant replicates was two times higher or lower than that in wild-type controls, respectively.
Reagent and data availability
Seeds of the homozygous T line are available from the Arabidopsis Biological Resource Center, (ABRC) Ohio State University, under the stock number CS69640. Seeds of the prp18a-1 mutant will be deposited at ABRC and are currently available on request from the Matzke lab.RNA-sequencing data for the prp18 mutants reps 1-6 and the wild-type T control samples for the prp18 mutants reps 4-6 (T wild type reps 1-3) as well as whole genome re-sequencing data for the prp18a-1 mutant are available under Sequence Read Archive (SRA) accession number SRP119240. The wild-type T controls for the prp18 mutant reps 1-3 are under SRA accession numbers SRP093582 (ST biological replicates 4-5) and SRP119240 (T wild type reps 4-6).
Results
Identification of a prp18a mutant in forward genetic screen
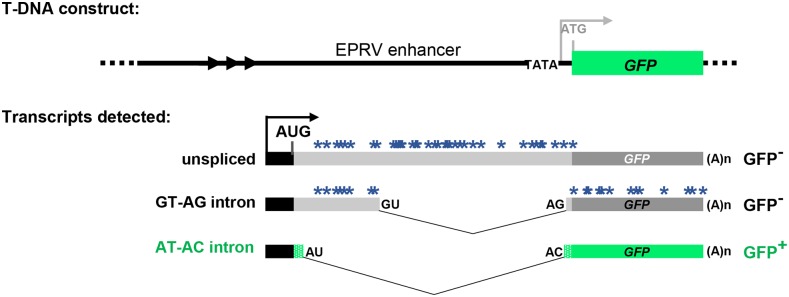
In the wild-type T line used in this study, GFP pre-mRNA is alternatively spliced to yield three main splice variants: a long unspliced transcript, a mid-length transcript arising from splicing a canonical GT-AG intron, and a short transcript resulting from splicing a U2-type intron with non-canonical AT-AC splice sites (Sasaki et al. 2015). Because the unspliced and GU-AG transcripts contain a number of premature termination codons, only the AU-AC transcript represents a bona fide GFP mRNA that can be translated into GFP protein (Figure 1). The three GFP transcripts are present in a balanced ratio in the wild-type T line, which displays an intermediate level of GFP fluorescence (Kanno et al. 2017a,b). Our working hypothesis is that mutations in genes encoding splicing factors will modify the ratio of the three transcripts, leading to either elevated or lowered GFP mRNA levels. These changes will result, respectively, in either a Hyper-GFP (hgf) or GFP-weak (gfw) phenotype compared to the intermediate level of the wild-type T line (Kanno et al. 2016, 2017a,b). Results from the screen so far support this hypothesis: we have identified five hgf mutants and three gfw mutants that vary in the splicing pattern of GFP pre-mRNA (Table 1). Here we report the finding of a fourth mutant, gfw4, in the GFP-weak category.
Figure 1.
Schematic drawing of alternatively-spliced GFP reporter gene. Top: The T-DNA original construct introduced into Arabidopsis contained a GFP reporter gene under the transcriptional control of a minimal promoter (TATA) and upstream viral (EPRV) enhancer element. However, analysis of the wild-type T line revealed that neither the minimal promoter nor the downstream ATG initiation codon (gray letters) is used. Bottom: The T line analysis indicated that transcription of GFP pre-mRNA initiates at an upstream promoter (black bar and arrow) to generate three alternative splice variants that comprise part of the enhancer region (Kanno et al. 2008). These variants include a long unspliced transcript, a middle-length transcript arising from splicing of a canonical GT-AG intron, and a short transcript resulting from splicing a U2-type intron with non-canonical AT-AC splice sites, which are generally considered inefficient splice sites (Crotti et al. 2007). Because the unspliced and GU-AG transcripts contain a number of premature termination codons (blue asterisks), only the AU-AC transcript can be translated into GFP protein. The actual coding sequence of GFP protein (green bars) contains a unique 27 amino acid extension (short stippled green bars) relative to standard GFP (Fu et al. 2015; Kanno et al. 2016). Arrowheads designate a short tandem repeat upstream of the promoter. The black AUG denotes the major translation initiation codon. The distance between the 3′ splice sites for the GT-AG and AT-AC introns is only 3 nt; the non-canonical AC is on the outside (Kanno et al. 2016, 2017a,b).
The gfw4 mutant was identified in an EMS-mutagenized population by the GFP-weak phenotype of M2 seedlings grown on MS medium (Figure 2A). A reduction of GFP protein in the gfw4 mutant was confirmed by Western blotting using an antibody to GFP (Figure 2B). The causal mutation in the gfw4 mutant was determined by next generation mapping (James et al. 2013) to be a G to A transition mutation in the coding region of the gene encoding PRP18a (At1g03140), a putative step II splicing factor that is 420 amino acids in length. The mutation leads to a substitution of an alanine by valine at position 334 in the PRP18a protein (Figure 3A). As the first mutation reported for PRP18a, we designated this allele prp18a-1/gfw4-1.
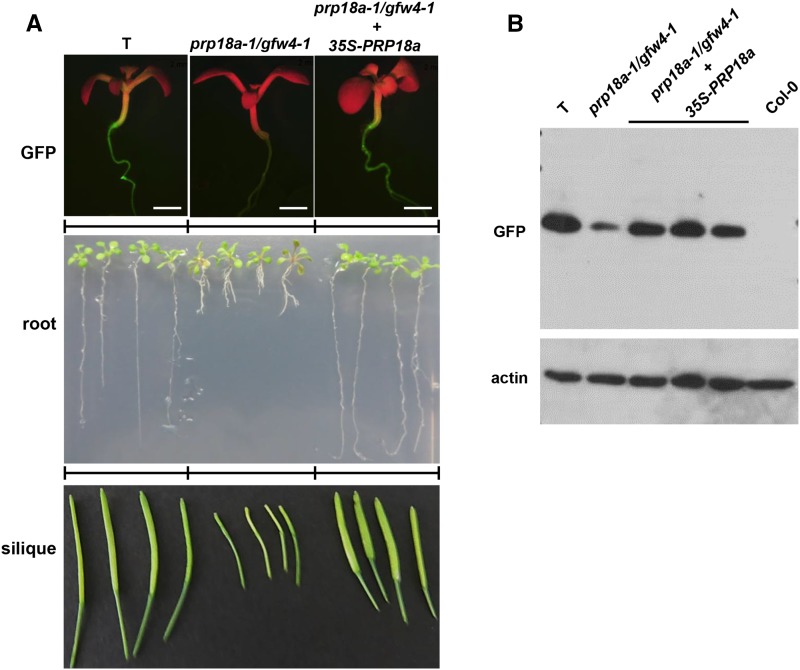
Figure 2.
Phenotypic analysis of the prp18a-1/gfw4-1 mutant and complemented line. A. GFP fluorescence in seedlings (top), root growth of seedlings on solid MS medium (middle) and developing siliques (bottom) of the wild-type T line, the prp18-a-1/gfw4-1 mutant and the prp18a-1/gfw4/1 mutant complemented with a PRP18a transgene under the control of the 35S promoter. B. Western blot analysis of GFP protein in the wild-type T line, prp18a-1/gfw4-1 mutant and three complemented lines. Non-transgenic Col-0 is shown as a negative control. The top panel was probed with an antibody to GFP protein. The bottom panel shows the same blot re-probed with an antibody to actin as a loading control.
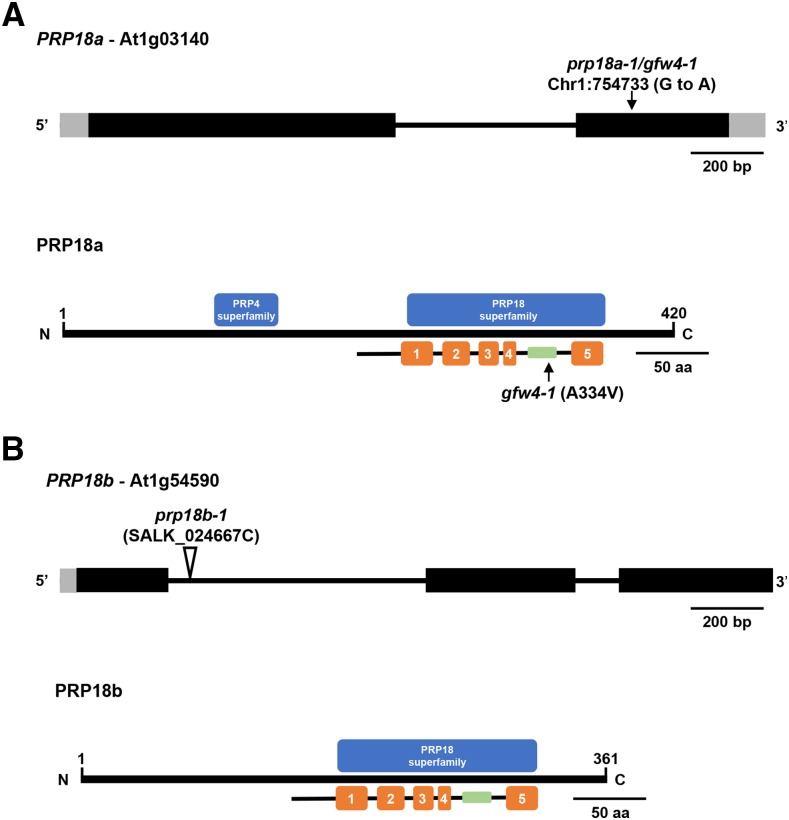
Figure 3.
Gene structures, positions of mutations, and protein domains of PRP18 paralogs in Arabidopsis. (A) The PRP18a gene (At1g03140) contains one intron (thin black bar) and encodes a protein 420 amino acids in length. The G to A transition mutation at position 754733 on chromosome 1 in the prp18a-1/gfw4-1 mutant is indicated (top). The PRP18a protein contains a PRP18 domain, which comprises five α-helices (orange bars). A highly conserved loop (green bar), which is important for PRP18 function, is between the fourth and fifth helices. The prp18a-1/gfw4-1 mutation leads to an alanine to valine substitution at position 334 within this conserved loop. This alanine residue is highly conserved in various plant species (Figure S1). PRP18a also contains a PRP4 domain of unknown function. Intact PRP4 proteins are components of U4/U6 and U4/U6.U5 snRNPs. PRP18 family proteins are conserved in yeasts and metazoans (Figure S2). (B) The PRP18b gene (At1g54590) contains two introns and encodes a protein 361 amino acids in length. Much of the length difference between PRP18a and PRP18b is due to missing N-terminal sequences in PRP18b (Figure S1). The PRP18b protein contains a PRP18 domain but not a recognizable PRP4 domain. The prp18b-1 allele contains a T-DNA insertion in the first intron.
PRP18 is an evolutionarily conserved protein, with orthologs found in other plants, yeasts and metazoans (Figure S1 and Figure S2). Prp18 proteins typically have five alpha-helices separated by four loops, with a highly conserved loop between the fourth and fifth helices (Bacíková and Horowitz 2002; Annamalai 2011). The A334V mutation we identified in the prp18a-1 mutant is in this highly conserved loop region (Figure 3). PRP18a contains two putative nuclear localization signals (Annamalai 2011) and is predicted to be a nuclear protein (http://suba.live/).
Complementation of the prp18a-1 mutant with a construct comprising the wild-type PRP18a coding sequence under the transcriptional control of the 35S promoter and terminator sequences (35Spro-PRP18a-35Ster) restored an intermediate level of GFP fluorescence and increased GFP protein abundance to that observed in wild-type T seedlings (Figure 2A, top and B, respectively). These findings confirm that the prp18a-1 mutation is responsible for the GFP-weak phenotype of the gfw4-1 mutant. The prp18a-1 mutation is recessive, as indicated by the intermediate level of GFP fluorescence in BC1 progeny obtained by backcrossing the mutant to a wild-type T plant.
Phenotypic features of prp18a mutant
The phenotype of the prp18a-1 mutant compared to the wild-type T line was monitored in the BC1F3 generation, which has a reduced number of mutations relative to the original mutant. Mutant prp18a-1 seedlings germinated on solid MS medium at approximately the same time as wild-type seedlings; however, after two weeks of growth, the roots were noticeably shorter in the mutant than in wild-type seedlings (Figure 2A, middle). Despite a tendency to be somewhat bushy and flimsy, adult plants of the prp18a-1 mutant generally resembled wild-type plants in terms of stature and overall appearance (Figure S3). The prp18a-1 mutant flowered around the same time as the wild-type, but the siliques of the mutant developed more slowly and were smaller relative to wild-type (Figure 2A, bottom). The short root and small silique phenotypes of the mutant were both complemented by a 35Spro-PRP18a-35Ster transgene (Figure 2C), demonstrating that these aberrant characteristics were indeed due to the prp18a-1 mutation.
Detection of GFP splicing variants by RT-PCR
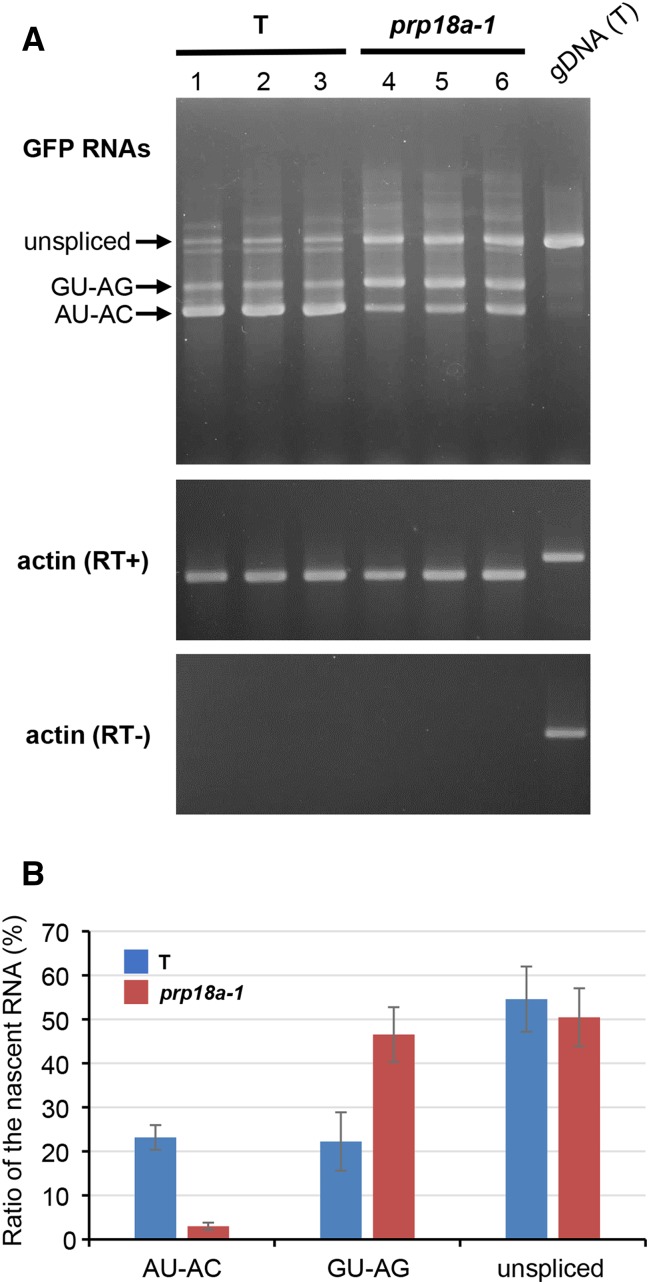
Semi-quantitative RT-PCR was used to examine the splicing pattern of GFP pre-mRNA in the prp18a-1 mutant. The level of the translatable AU-AC transcript was reduced, consistent with the GFP-weak phenotype of the prp18a-1 mutant. However, the level of untranslatable GU-AG transcript appeared to be increased in prp18a-1 relative to the wild-type T line (Figure 4A). This is a unique splicing pattern of GFP pre-mRNA that has not been observed in any other mutant retrieved so far in the screen (Table 1).
Figure 4.
RT-PCR analysis of GFP splice variants in prp18a-1 mutants. (A) Semi-quantitative RT-PCR was used to assess the accumulation of unspliced GFP transcript and two splice variants (resulting from splicing the canonical GT-AG and non-canonical AT-AC introns, respectively) in triplicate samples of the prp18a-1 mutant and the wild-type T line. Actin is shown as a constitutively expressed control. RT- and RT+ panels show experiments with and without reverse transcriptase, respectively. gDNA (T), genomic DNA isolated from T line. (B) The percentages of three major GFP RNA splice variants as determined from an analysis of RNA-seq data (Table S2). The average of five biological replicates is shown. The amount of total GFP transcripts did not change significantly in prp18a-1 mutants.
RNA-sequencing (RNA-seq) analysis
To analyze more comprehensively the effect of a homozygous prp18a-1 mutation on alternative splicing, we carried out RNA-seq using total RNA isolated from two week-old seedlings of the original inbred prp18a-1 mutant (M4 generation), BC1 F3 seedlings of the prp18a-1 mutant, and the wild-type T-line. All samples were run in biological triplicate. Only changes that were statistically significant in both the original mutant and BC1F3 plants are considered here.
The RNA-seq data confirmed the RT-PCR findings on alternative splicing of GFP pre-mRNA: the percentage of translatable AU-AC transcript decreased from 23 to 3% of the total in the prp18a-1 mutant whereas the percentage of the untranslatable GU-AG transcript increased from 22 to 47%. The percentage of unspliced transcript remained roughly the same in the prp18a-1 mutant and wild-type T line (51% and 55%, respectively) (Figure 4B).
A summary of the results of a genome-wide analysis of differentially expressed genes (DEGs) and alternative splicing is shown in Table 2 (Details available in Table S1, Table S2, Table S3 and Table S4). The number of DEGs was relatively modest (174 total), with more up-regulated genes (160) than down-regulated genes (14). No splicing factors were found in either category.
Table 2. DEGs and alternative splicing events in the prp18a-1 mutant.
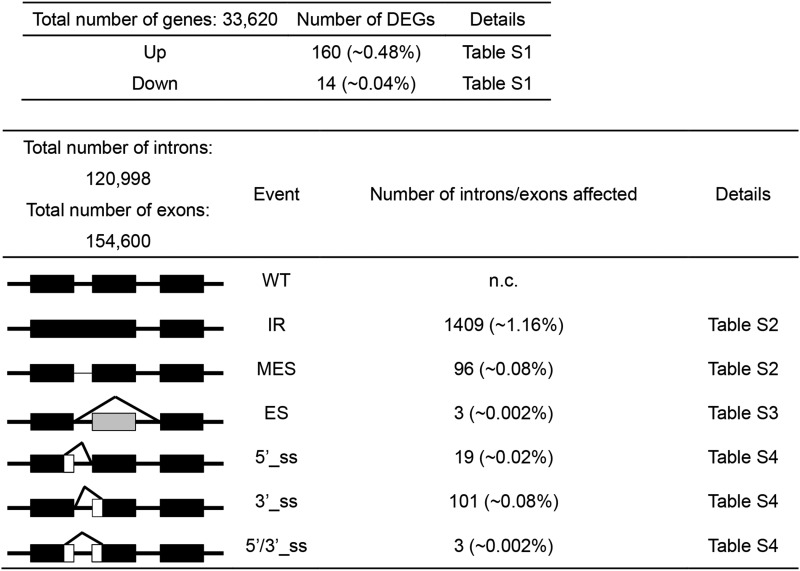 |
The major modes of alternative splicing are shown at the left. Abbreviations: WT, wild-type, IR, intron retention; MES, more efficient splicing; ES, exon skipping; 5′ or 3′_ss, change in 5′ splice site donor or 3′ splice site acceptor; 5′/3′_ss. Change in both 5′ and 3′ splice sites. Numbers in parentheses indicate the percentage of total introns or exons affected. Details of the RNA-seq analysis can be viewed in the indicated supporting tables.
The largest number of alternative splicing events occurred in the category of intron retention (IR), which affected 1409 introns (in 1322 genes). By contrast, only 96 cases (in 89 genes) of more efficient splicing (MES), which involves a decrease in the level of a partially retained intron in wild-type plants, were observed (Table 2). Of the IR events, fifteen involved splicing factors, including PRP39a, PRP40b and BRR2C (Table 3). These three factors were also found to undergo changes in alternative splicing in prp39a mutants, which were identified in the same genetic screen described here (Table 1) (Kanno et al. 2017b). Only three instances of exon skipping were detected. Regarding altered splice site selection, there were approximately five times more cases of altered 3′ splice sites (101) than 5′ splice sites (19) in the prp18a-1 mutant (Table 2)
Table 3. Splicing factors in the intron retention (IR) list in the prp18a-1 mutant.
| Gene | Protein | Predicted function in splicing1 | Reference |
|---|---|---|---|
| At1g04080 | PRP39a | U1 snRNP | Kanno et al. 2017b |
| At1g14650 | SF3a120, SAP114 | 17S U2 snRNP | |
| At1g60900 | U2AF65B | Splice site selection | Park et al. 2017 |
| At1g76860 | Lsm3B | small nuclear ribonucleoprotein | Golisz et al. 2013 |
| At2g16940 | fSAP59, CC1-like splicing factor | recruited prior to B* | |
| At2g29210 | SRm160 | SR-related protein | |
| At2g40650 | PRP38 | U4/U6.U5 tri-snRNP | |
| At2g43370 | U11/U12-35K | U11/U12-specific | Lorkovic et al. 2005 (Table 1) |
| At3g13200 | cwc15 | Putative splicing factor | http://www.arabidopsis.org/index.jsp |
| At3g19670 | PRP40B | U1 snRNP-related | Kang et al., 2009; Kanno et al. 2017b |
| At3g49430 | SR34A | SR protein, putative pre-mRNA splicing factor SF2 | Barta et al. 2008, 2010 |
| At4g02430 | SR34B | SR protein | Barta et al., 2008, 2010 |
| At4g14342 | SF3B10a splicing factor 3B subunit 5 | 17S U2 snRNP | Lorkovic et al. 2005 |
| At5g08290 | Yellow leaf-specific gene 8 (YLS8) | U5 snRNP | |
| At5g61140 | BRR2C | U5 snRNP helicase | Mahrez et al., 2016; Kanno et al. 2017b |
For complete list of IR events, see Table S2.
Except for At3g13200, predicted functions taken from Table S1 of Koncz et al., 2012. References in which a factor is mentioned specifically in a publication are listed (reference information taken from http://www.arabidopsis.org/index.jsp).
Tests of prp18b
In Arabidopsis, PRP18a has an annotated paralog, PRP18b (At1g54590) (http://www.arabidopsis.org/). The PRP18b gene is predicted to encode a protein that is 361 amino acids in length (Figure 3B). However, PRP18b does not appear to be transcribed in two week-old seedlings, which represent the developmental stage analyzed in this study, nor was a transcript detected in an RNA-seq analysis of floral material (Figure S4). Nevertheless we introduced a T-DNA insertion mutation of PRP18b into both the wild-type T line and the prp18a-1 mutant, and examined the prp18b-1 single mutant and the prp18a-1 prp18b-1 double mutant for GFP fluorescence, GFP pre-mRNA splicing and plant phenotype. By these criteria, the prp18b single mutant appeared identical to wild-type plants and the prp18a prp18b double mutant, which was viable, resembled the prp18a-1 single mutant (Figure S4).
Discussion
In a forward genetic screen for mutants showing modified splicing of an alternatively-spliced GFP reporter gene in Arabidopsis, we identified a mutation in the gene encoding PRP18a, an evolutionarily conserved, putative step II splicing factor. Prp18 was first identified in a forward screen in budding yeast for temperature-sensitive splicing mutations (Vijayraghavan et al. 1989) and subsequently shown biochemically to participate in step II of splicing in that organism (Horowitz and Abelson 1993). A recent structural analysis using cryo-electron microscopy (cryo-EM) in budding yeast revealed that Prp18 is enriched in the step II catalytic complex C*, where it has direct interactions with PRP8 and U5 snRNA at the active site (Yan et al. 2017). The mutation we identified leads to an amino acid substitution in a highly conserved loop between helices 4 and 5 of the PRP18a protein. Mutations in this loop region, which is the most evolutionarily conserved part of the PRP18 protein, are known from budding yeast to result in a deficiency in the step II reaction, likely because of disrupted interactions with PRP8 and RNA elements at the catalytic site (Yan et al. 2017). The location of the prp18a-1 mutation, which affects an alanine residue that is conserved in the plant and yeast species examined, is consistent with a loss of function (or partial loss of function) allele. The nature of the prp18a- 1 mutation suggests that Arabidopsis PRP18A acts similarly to budding yeast Prp18 in the step II reaction. However, confirmation of this proposal awaits the development of methods to isolate plant spliceosomal complexes that can be used to analyze splicing reactions in vitro.
PRP18 is not essential in budding yeast but cells lacking this protein grow slowly and are temperature sensitive (Bacíková and Horowitz 2002). Whether PRP18a is essential in Arabidopsis is unclear. The prp18a-1 mutation, which may only lead to partial loss of function, is not lethal but it confers an aberrant phenotype most visible in short roots of seedlings on solid MS medium and small siliques. Although the molecular basis of this phenotype remains to be explored, the findings affirm the necessity of wild-type PRP18a function throughout plant growth and development.
Effect of prp18a-1 mutation on pre-mRNA splicing
There are several differences in how the prp18a-1 mutation affects splicing compared to mutations in other splicing factors identified so far in this screen. First, the splicing pattern of GFP pre-mRNA in prp18a-1, which features decreased splicing of the non-canonical AT-AC intron and increased splicing of the canonical GT-AG intron, is unique. Other gfw mutants generally show reduced splicing efficiency of both the AT-AC and GT-AG introns together with increased levels of unspliced GFP transcript. Conversely, the hgf mutants display increased splicing of the AT-AC intron and decreased splicing of the GT-AG intron accompanied by generally reduced accumulation of the unspliced transcript. One interpretation of these results, which does not imply a specific mechanism, is that wild-type PRP18a protein enhances splicing at non-canonical or inefficient splice sites whereas the HGF factors act in an opposite manner to normally repress splicing at non-canonical and inefficient splice sites. In budding yeast, Prp18 has been reported to suppress splicing at non-canonical sites (Kawashima et al. 2014). Although this finding appears contrary to ours, it nevertheless suggests a role for PRP18 in discriminating between strong and weak splice sites.
A second difference, which was revealed in the genome-wide analysis of splicing, is that the prp18a-1 mutation affects splicing of a higher proportion of splicing factors than other mutants identified in the screen. The significance of this finding is unclear at present but it may suggest that PRP18a is deeply embedded in cross-regulatory networks involving multiple splicing factors, which are thought to coordinate responses of the spliceosome to developmental and environmental cues (Barta et al. 2008).
A third notable difference in splicing in the prp18a-1 mutant compared to other mutants retrieved in the screen is the preponderance of 3′ splice site changes. In prp18a-1, the number of 3′ splice site alterations exceeds 5′ splice site alterations by fivefold. We did not observe such a substantial skew toward either 5′ or 3′ splice site changes in any other mutant, which show at most a twofold difference between changes at the two splice sites (Kanno et al. 2017a,b). The predominance of 3′ splice site alterations in the prp18a-1 mutant might reflect the participation of PRP18a specifically in step II of splicing and potentially 3′ splice site selection (Kawashima et al. 2014).
PRP18 paralogs
The two annotated PRP18 paralogs in Arabidopsis, PRP18a and PRP18b, differ in length and expression level. PRP18a is longer (420 amino acids) and ubiquitously expressed (http://www.arabidopsis.org/index.jsp; eFP Browser) while PRP18b is shorter (361 amino acids) and not expressed in the seedling material examined in this study. Most plant species examined so far have one to two copies of the PRP18 gene (https://phytozome.jgi.doe.gov/pz/portal.html#), which also differ in length in some cases, but determining whether they display different levels of expression requires more detailed analysis.
Arabidopsis PRP18a has additional sequences at the N-terminus and a recognizable PRP4 superfamily domain that are also present in PRP18a orthologs in other plant species, metazoans and Schizosaccharomyces pombe (fission yeast). However, the additional sequences at the N-terminus and the PRP4 domain are largely missing from Arabidopsis PRP18b and budding yeast Prp18. It has previously been suggested that PRP18b may be similar functionally to budding yeast Prp18 whereas PRP18a may have taken on added functions or capabilities endowed by the N-terminal extension (Annamalai 2011). In this context, it is interesting to note that the presence of the additional N-terminal sequences and the PRP4 domain in PRP18a and orthologous proteins is associated with organisms that carry out alternative splicing (fission yeast, plants and metazoans). One conjecture is that these regions help to promote flexibility of splicing patterns and facilitate alternative splicing in those organisms.
General comments on forward screen
Our forward screen, which is still ongoing, has already identified factors specific for each splicing step and for different spliceosomal complexes (Figure 5). PRP18a is the first putative step II-specific factor that we have recovered. It is still not clear why the screen has retrieved these particular splicing factors, several of which (RTF2, CWC16a, SMFa and PRP18a) had not been investigated prior to our studies. The identification of mutants defective at multiple stages of the splicing process, and often only a single member of a paralogous gene pair, hints that the screen may be tapping into a specialized splicing pathway involving a dedicated set of components. We have previously noted links to stress tolerance for coilin and SMFa, both of which were identified in this screen (Kanno et al. 2016, 2017a). These findings may be relevant for the known contribution of alternative splicing to plant responses to stress and environmental signals (Staiger and Brown, 2013: Filichkin et al. 2015). Recent cryo-EM data from budding yeast have demonstrated that the step I factor Yju2 (the ortholog of Arabidopsis CWC16 proteins) and Prp18 interact closely with Prp8 in catalytic core of the B* and C* complexes to facilitate step I and step II reactions, respectively (Galej et al. 2016; Wan et al. 2016; Yan et al. 2017). Our screen has identified mutants defective in all three of these splicing factors, confirming that the alternatively spliced GFP reporter gene system is capable of revealing central, conserved splicing proteins predicted to act directly at the catalytic site. Moreover, our findings validate a key role for these factors in pre-mRNA splicing in plants. Further analysis of these mutants and the additional, uncharacterized mutants emerging from the screen should assist in determining the mechanistic roles and interconnections of a coherent set of splicing factors, and broaden knowledge of alternative splicing in plant growth and development.
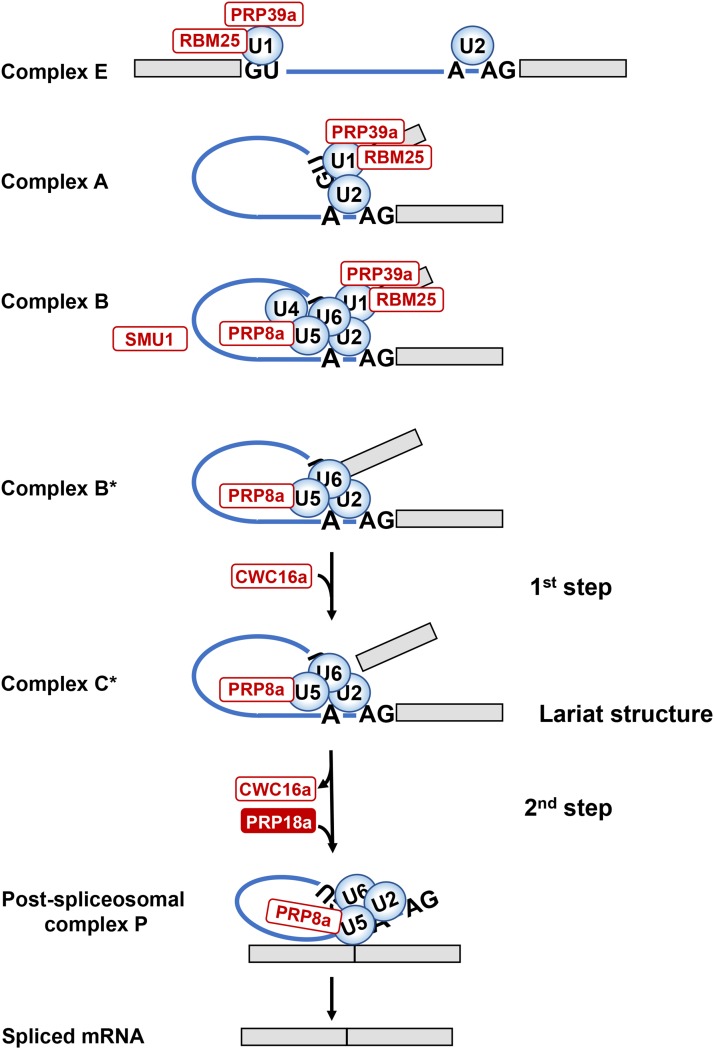
Figure 5.
Schematic depiction of spliceosomal cycle. The main spliceosomal complexes, the two catalytic steps, and the predicted positions of factors identified in the genetic screen (white and red ovals) are shown. In Complex E, the U1/U2 snRNPs recognize 5′ and 3′ splice sites (GU-AG) and adenosine branch point (A) by base-pairing. In pre-spliceosomal Complex A, the U1 and U2 snRNPs interact to bring together 5′ and 3′ splice sites. Complex B (pre-catalytic spliceosome) is created by entry of preformed U4/U6.U5 tri-snRNP. Catalytic Complex B* is formed after dissociation of U1 and U4 snRNPs and other conformational and compositional changes. In complex B*, the U2, U5, and U6 snRNPs are positioned by scaffold protein PRP8a and additional proteins, including step I factor CWC16a, to execute the first step of splicing, which releases the 5′ exon and creates an intron-3′ exon lariat structure. In catalytic Complex C*, the U2, U5 and U6 snRNPs are positioned by PRP8a and additional proteins, including step II factor PRP18a, to carry out the second step of splicing, which excises the intron and joins the two exons. After formation of the Post-spliceosomal complex P, the spliced mRNA is released (Matera and Wang 2014). PRP39a and RBM25 are U1 snRNP components; PRP8a is a constituent of the U5 snRNP; SMU1 is present in the B complex but exits before formation of catalytic complex B*; CWC16a is a step I-specific factor; PRP18a, identified in the present study, is a step II-specific factor. Other factors not shown that were identified in the screen include: SmFa, a core snRNP protein and present in U1, U2, U4 and U5 snRNPs; coilin, which participates in snRNP maturation; and RTF2, which acts at an unknown stage of splicing.
Supplementary Material
Supplemental Material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.118.200022/-/DC1.
Acknowledgments
This work was supported by the Institute of Plant and Microbial Biology (IPMB) at Academia Sinica, additional funds from Academia Sinica, and grants from the Taiwan Ministry of Science and Technology (grant numbers MOST 103-2311-B001-004-MY3, MOST 105-2311-B-001-071 and MOST 106-2311-B-001-023-MY2). We thank Shou-Jen Chou and Ai-Peng Chen in the Genomic Technology core facility at IPMB for preparing libraries for RNA-seq and whole genome re-sequencing, and Jason Fu for technical assistance. The authors declare no conflicts of interest.
Footnotes
Communicating Editor: M. Axtell
Literature Cited
- Annamalai, D., 2011 Expression profiling of second step factors and role of PRP18a, a second step splicing factor in Arabidopsis thaliana. Dissertation, University of Illinois Urbana-Champaign. [Google Scholar]
- Bacíková D., Horowitz D. S., 2002. Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA 8: 1280–1293. 10.1017/S1355838202023099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Lorković Z. J., 2008. Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 326: 83–102. [DOI] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Reddy A. S., 2010. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22(9): 2926–2929. 10.1105/tpc.110.078352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K., Agafonov D. E., Liu W. T., Dybkov O., Will C. L., et al. , 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542(7641): 318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- Clay N. K., Nelson T., 2005. The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell 17(7): 1994–2008. 10.1105/tpc.105.032771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16(6): 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Crotti L. B., Bacíková D., Horowitz D., 2007. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 21(10): 1204–1216. 10.1101/gad.1538207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Cao X., 2017. Roles of pre-mRNA splicing and polyadenylation in plant development. Curr. Opin. Plant Biol. 35: 45–53. 10.1016/j.pbi.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., et al. , 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36(4): 593–608. 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- Fica S. M. N., Tuttle T., Novak N. S., Li J., Lu J., et al. , 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503(7475): 229–234. 10.1038/nature12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S., Priest H. D., Megraw M., Mockler T. C., 2015. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 24: 125–135. 10.1016/j.pbi.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Fu J. L., Kanno T., Liang S. C., Matzke A. J., Matzke M., 2015. GFP loss-of-function mutations in Arabidopsis thaliana. G3. Genes Genomes Genetics 5: 1849–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej W. P., Wilkinson M. E., Fica S. M., Oubridge C., Newman A. J., et al. , 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537(7619): 197–201. 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golisz A., Sikorski P. J., Kruszka K., Kufel J., 2013. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 41(12): 6232–6249. 10.1093/nar/gkt296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger R.J., Beggs J.D., 2005. Prp8 protein: at the heart of the spliceosome. RNA11: 533–57. 10.1261/rna.2220705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J., Wan R., Yan C., Shi Y., 2015. Structural basis of pre-mRNA splicing. Science 349(6253): 1191–1198. 10.1126/science.aac8159 [DOI] [PubMed] [Google Scholar]
- Haselbach D., Komarov I., Agafonov D. E., Hartmuth K., Graf B., et al. , 2018. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172(3): 454–464.e11. 10.1016/j.cell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Hodges P. E., Jackson S. P., Brown J. D., Beggs J. D., 1995. Extraordinary sequence conservation of the PRP8 splicing factor. Yeast 11(4): 337–342. 10.1002/yea.320110406 [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Abelson J., 1993. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 7(2): 320–329. 10.1101/gad.7.2.320 [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., 2012. The mechanism of the second step of pre-mRNA splicing. WIREs RNA 3(3): 331–350. 10.1002/wrna.112 [DOI] [PubMed] [Google Scholar]
- James G. V., Patel V., Nordström K. J., Klasen J. R., Salomé P. A., et al. , 2013. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 14(6): R61 10.1186/gb-2013-14-6-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. H., Feng Y., Vikram M., Jeong I. S., Lee J. R., et al. , 2009. Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch. Biochem. Biophys. 484(1): 30–38. 10.1016/j.abb.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Kanno T., Bucher E., Daxinger L., Huettel B., Böhmdorfer G., et al. , 2008. A structural-maintenance-of-chromosomes hinge domain-containing proteinis required for RNA-directed DNA methylation. Nat. Genet. 40(5): 670–675. 10.1038/ng.119 [DOI] [PubMed] [Google Scholar]
- Kanno T., Lin W. D., Fu J. L., Wu M. T., Yang H. W., et al. , 2016. Identification of coilin mutants in a screen for enhanced expression of an alternatively spliced GFP reporter gene in Arabidopsis thaliana. Genetics 203(4): 1709–1720. 10.1534/genetics.116.190751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Lin W.-D., Fu J. L., Matzke A. J. M., Matzke M., 2017a A genetic screen implicates a CWC16/Yju2/CCDC130 protein and SMU1 in alternative splicing in Arabidopsis thaliana. RNA 23(7): 1068–1079. 10.1261/rna.060517.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Lin W. D., Fu J. L., Chang C. L., Matzke A. J. M., 2017b A genetic screen for pre-mRNA splicing mutants of Arabidopsis thaliana identifies putative U1 snRNP components RBM25 and PRP39a. Genetics 207: 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Douglass S., Gabunilas J., Pellegrini M., Chanfreau G. F., 2014. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet. 10(4): e1004249 10.1371/journal.pgen.1004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12(4): 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Schumaker K. S., Zhu J. K., 2006. EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323: 101–103. [DOI] [PubMed] [Google Scholar]
- Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z., 2012. The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3: 9 10.3389/fpls.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9(4): 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K., 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15(2): R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Li X., Zhang L., Jiang J., Hill R. C., et al. , 2017. Structure of the yeast spliceosomal postcatalytic P complex. Science 358(6368): 1278–1283. 10.1126/science.aar3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic Z. J., Lehner R., Forstner C., Barta A., 2005. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 11(7): 1095–1107. 10.1261/rna.2440305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrez W., Shin J., Muñoz-Viana R., Figueiredo D. D., Trejo-Arellano M. S., et al. , 2016. BRR2a affects flowering time via FLC splicing. PLoS Genet. 12(4): e1005924 10.1371/journal.pgen.1005924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W., Simpson C., Barta A., Kalyna M., 2012. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22(6): 1184–1195. 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Wang Z., 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15(2): 108–121 (erratum: Nat. Rev. Mol. Cell Biol. 15: 294). 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke A. J., Watanabe K., van der Winden J., Naumann U., Matzke M., et al. , 2010. High frequency, cell type-specific visualization of fluorescent-tagged genomic sites in interphase and mitotic cells of living Arabidopsis plants. Plant Methods 6(1): 2 10.1186/1746-4811-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., Proudfoot N. J., 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136(4): 688–700. 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Ohrt T., Odenwälder P., Dannenberg J., Prior M., Warkocki Z., et al. , 2013. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA 19(7): 902–915. 10.1261/rna.039024.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. Y., Lee K. C., Jang Y. H., Kim S. K., Thu M. P., et al. , 2017. The Arabidopsis splicing factors, AtU2AF65, AtU2AF35, and AtSF1 shuttle between nuclei and cytoplasms. Plant Cell Rep. 36(7): 1113–1123. 10.1007/s00299-017-2142-z [DOI] [PubMed] [Google Scholar]
- Pietrzak M., Shillito R. D., Hohn T., Potrykus I., 1986. Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res. 14(14): 5857–5868. 10.1093/nar/14.14.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S., Day I. S., Göhring J., Barta A., 2012. Localization and dynamics of nuclear speckles in plants. Plant Physiol. 158(1): 67–77. 10.1104/pp.111.186700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25(10): 3657–3683. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A., 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11(3): R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Kanno T., Liang S. C., Chen P. Y., Liao W. W., et al. , 2015. An Rtf2 domain-containing protein influences pre-mRNA splicing and is essential for embryonic development in Arabidopsis thaliana. Genetics 200(2): 523–535. 10.1534/genetics.115.176438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J. W., 2013. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25(10): 3640–3656. 10.1105/tpc.113.113803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R., Tanaka-Sato N., Maruyama N., Terada S., Kojima M., et al. , 2015. CLUMSY VEIN, the Arabidopsis DEAH-box Prp16 ortholog, is required for auxin-mediated development. Plant J. 81(2): 183–197. 10.1111/tpj.12721 [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U., Company M., Abelson J., 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 3(8): 1206–1216. 10.1101/gad.3.8.1206 [DOI] [PubMed] [Google Scholar]
- Wahl M. C., Will C. L., Lührmann R., 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136(4): 701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wahl M. C., Lührmann R., 2015. SnapShot: Spliceosome Dynamics I. Cell 161: 1474-e1. [DOI] [PubMed] [Google Scholar]
- Wan R., Yan C., Bai R., Huang G., Shi Y., 2016. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353(6302): 895–904. 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- Wilkinson M. E., Fica S. M., Galej W. P., Norman C. M., Newman A. J., et al. , 2017. Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science 358(6368): 1283–1288. 10.1126/science.aar3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C. L., Lührmann R., 2011. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3(7): a003707 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Wan R., Bai R., Huang G., Shi Y., 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355(6321): 149–155. 10.1126/science.aak9979 [DOI] [PubMed] [Google Scholar]
- Zhang X., Schwer B., 1997. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 25(11): 2146–2152. 10.1093/nar/25.11.2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.