Abstract
Inactivation of the CMP-N-acetylneuraminic acid hydroxylase gene has provided an example of human-specific genomic mutation that results in a widespread biochemical difference between human and nonhuman primates. We have found that, although a region containing a 92-bp exon and an AluSq element in the hydroxylase gene is intact in all nonhuman primates examined, the same region in the human genome is replaced by an AluY element that was disseminated at least one million years ago. We propose a mechanistic model for this Alu-mediated replacement event, which deleted the 92-bp exon and thus inactivated the human hydroxylase gene. It is suggested that Alu elements have played potentially important roles in genotypic and phenotypic evolution in the hominid lineage.
Human-specific traits (upright walking, language ability, etc.) have evolved over time through the emergence and extinction of several hominid species during human evolution (1). The acquisition of such traits accompanies marked changes in morphology and physiology. Humans are most closely related to the African great apes: the chimpanzee (Pan troglodytes), the bonobo (Pan paniscus), and the gorilla (Gorilla gorilla) (2, 3). Research to determine which genes differentiate humans from the great apes has been undertaken (reviewed in ref. 4), and comparative genomic analysis among primates is now underway. Three general classes of genetic differences have been proposed as factors separating humans from the great apes: chromosomal differences, small sequence differences that change gene expression, and biochemical changes resulting from gene inactivation (5). Thus far, three genes are known to have been altered in human-specific manners. One is loss of exon 34 in the tropoelastin gene, which was possibly facilitated by Alu-mediated recombination events (6). However, because exon 35 has already been deleted in catarrhines, loss of an additional exon in the hominid lineage may be of secondary significance. Another is a recent introduction of a premature termination codon into a member of the human type I hair keratin gene cluster (7). Unlike these gene changes, inactivation of the CMP-N-acetylneuraminic acid hydroxylase gene is unique in that it is a single gene and inactivated in the hominid lineage only (8–10).
CMP-N-acetylneuraminic acid (CMP-Neu5Ac) is a nucleotide sugar donor of Neu5Ac, the most common sialic acid in humans. The CMP-Neu5Ac hydroxylase converts CMP-Neu5Ac to the hydroxylated form, CMP-N-glycolylneuraminic acid (CMP-Neu5Gc) (11–14). Sialic acids such as Neu5Ac and Neu5Gc belong to a family of acidic sugars, and they are typically found on the cell surface in all mammals (15–17). These nine-carbon sugars function as ligands in recognition systems mediated by sialic acid-binding lectins, such as CD22, myelin-associated glycoprotein, sialoadhesin, and influenza A virus hemagglutinin (reviewed in ref. 17). Some of these lectins can discriminate between Neu5Ac and Neu5Gc (summarized in table 3 of ref. 18), and expression of the hydroxylase gene thus contributes to regulation of cell–cell interaction mediated by lectins of both endogenous and exogenous origin. It is therefore reasonable to think that the inactivation of the CMP-Neu5Ac hydroxylase gene in humans caused significant changes in several lectin-mediated interactions and possibly contributed to unique features of human evolution (discussed in refs. 8, 10, and 18)
It is known that the human-specific inactivation of the hydroxylase gene resulted from the deletion of a 92-bp exon (8, 9) and a subsequent frameshift in the coding sequence (8). The 92-bp exon encodes a part of the Reiske iron-sulfur-binding region that is essential for the enzyme's activity (8, 9, 19). The truncated hydroxylase therefore cannot convert CMP-Neu5Ac to CMP-Neu5Gc. The same deletion is found in all humans thus far examined, but not in the African apes, so that the 92-bp exon must have been deleted in the early evolution of the hominid lineage (8, 10). To gain insight into the genomic event that produced the human-specific inactivation of the hydroxylase gene, we have performed a comparative genomic analysis of the hydroxylase gene among six hominoids and two cercopithecoids: human, chimpanzee (Pan troglodytes), bonobo (Pan paniscus), gorilla (G. gorilla), orangutan (Pongo pygmaeus), gibbon (Hylobates lar), baboon (Papio anubis), and rhesus monkey (Macaca mulatta). We then propose a model to explain how Alu insertion can delete an exon and thus inactivate a gene.
Materials and Methods
DNA Samples.
Chimpanzee, gorilla, orangutan, and gibbon genomic DNAs were generous gifts from Shintaroh Ueda (University of Tokyo) and Colm O'hUigin (Max Planck Institute, Tübingen, Germany). Human samples were from volunteers in the Varki laboratory or provided by Steven Warren's laboratory (Emory University School of Medicine, Atlanta). Additional great ape samples were from Epstein–Barr virus-transfected lymphoblastoid cell lines obtained from Peter Parham (Stanford University School of Medicine, Stanford, CA). Baboon DNA was kindly provided by Jeffrey Rogers (Southwest Foundation for Biomedical Research Genetics, San Antonio, TX). Rhesus monkey genomic DNA was purchased from CLONTECH.
PCR Products of Chimpanzee Genomic DNA.
By genomic PCR, 10 fragments covering the ≈23-kb region, including the 92-bp exon, were obtained. The PCR primers were designed on the basis of the intron sequence of human CMP-Neu5Ac hydroxylase (GenBank accession no. AB009668). Fragment 1 was generated by using primers CH-18 (5′-TCGCAATAAGAGCACTGGCAAAGAC-3′) and CH-25 (5′-ACAAACCAGAAAGCCCAAGCATGTC-3′). Fragment 2 was generated with primers CH-32 (5′-ACATGCTTGGGCTTTCTGGTTTGTC-3′) and CH-28 (5′-GCTAAGAGGGGAGGACTAATGTGTC-3′). Fragment 3 was generated by using primers CH-9 (5′-TGACACATTAGTCCTCCCCTCTTAG-3′) and CH-13 (5′-CAAATGTTCCCTTCGTGGCAGTGTC-3′). Fragment 4 was generated by using primers CH-8 (5′-CCCTCTTAGCTCTCCTGCCCATGAG-3′) and CH-12 (5′-GAGGGAGGACAGCAACCACCAGAAC-3′). Fragment 5 was generated by using primers CH-34 (5′-TCTGGTGGTTGCTGTCCTCCCTCTC-3′) and CH-36 (5′-AAGCAGGAACCAGACAAGCAGTTTC-3′). Fragment 6 was generated by using primers CH-15 (5′-CTGCTTGTCTGGTTCCTGCTTTTAG-3′) and CH-19 (5′-TAAGTCCCAAGGGTTAGGAGGATTC-3′). Fragment 7 was generated by using primers CH-18 and CH-84 (5′-AGAAGCAAGAGCAGGATGGAGTCAG-3′). Fragment 8 was generated by using primers CH-92 (5′-GCAGAGGGTGCAAGAGAAAGGAGAG-3′) and CH-53 (5′-CTAAAATCCTTGACCCCTAGAATAG-3′). Fragment 9 was generated by using primers CH-10 (5′-TGTGTTGCCAGCATTCTCCCAGTTC-3′) and CH-38 (5′-ACCATATAGCCCAGCAATTCCATTC-3′). Fragment 10 was generated by using primers CH-44 (5′-GTCTATCCTTCTGCCAGTTCCACAC-3′) and CH-106 (5′-AAGAAGGAAACCACATCATCATCTC-3′). The genomic PCR was performed with 20 pmol of each primer and 30 ng of chimpanzee genomic DNA in a total volume of 50 μl containing 200 μM dNTPs and 2.5 units of ExTaq DNA polymerase (TaKaRa) in a TaKaRa ExTaq buffer containing 2 mM MgCl2. A RoboCycler Gradient 96 (Stratagene) was used to produce the following conditions: denaturation at 95°C for 5 min followed by 30 amplification cycles of 95°C for 1 min; 60–62°C for 1 min; 69°C for 7 min; and extension at 69°C for 10 min.
PCR Products of Gorilla Genomic DNA.
The primers CH-54 (5′-CATGGTTCTGCCAATTTTCCCTTTC-3′) and CH-95 (5′-ACACACATGCCCACAACCTGATCTG-3′) were used for genomic PCR. PCR was performed as described in the second section.
PCR Products of Rhesus Monkey Genomic DNA.
The primers MSA-1 (5′-GTCTGTTAGATGCACAAAGCATAAC-3′), MSA-3 (5′-GGTTGATATACTTCATGGTGCTCAC-3′), and CH-96 (5′-AGCTCAGCTCCCTTAACAGGTAATC-3′) were newly designed on the basis of the cDNA and genomic sequences of the human, chimpanzee, and rhesus monkey (GenBank accession nos. AB009668, AF074481, and AB013814). In addition to these primers, CH-9, CH-54, and CH-95 were used for genomic PCR. Twenty picomoles of each primer was used to amplify 100 ng of the rhesus monkey genomic DNA (CLONTECH) in a TaKaRa ExTaq buffer containing 2 mM MgCl2. PCR was performed as described in the section on chimpanzee DNA.
PCR Products of Human, Bonobo, Orangutan, Gibbon, and Baboon Genomic DNAs.
The primers CH-114 (5′-TGGGAAATCATTAGGCATCCACCTG-3′) and CH-148 (5′-TCTTTATTCTGCTGTCTCTGTTCTC-3′) were used for genomic PCR. The PCR conditions were as follows: denaturation at 95°C for 5 min followed by 30 cycles of 95°C for 1 min; 60°C for 1 min; 69°C for 1 min; and extension at 69°C for 10 min.
Sequencing of Genomic PCR Products.
The PCR products were purified by using the QIAquick PCR Purification Kit (Qiagen, Chatsworth, CA) and sequenced directly with an ABI Prism BigDye Terminator Cycle Sequencing FS Ready Reaction Kit (Applied Biosystems). Sequencing primers were produced based on the human intronic sequence (GenBank accession no. AB009668). With use of a GeneAmp PCR system 9600 (Applied Biosystems), cycle sequencing reaction was performed according to the manufacture's instructions. Each reaction sample was analyzed on an ABI Prism 377 fluorescent automated DNA sequencer (Applied Biosystems).
Comparative Genomic Analysis.
DNASIS software (Hitachi, Tokyo) was used for comparative analysis. Repetitive elements on the genomic sequence of each species were detected by using the repeatmasker program at the University of Washington Genomic Center web site (http://www.genome.washington.edu/UWGC/methods.htm).
Analysis of Human AluY Element.
The target human AluY element was picked up by qblastq search with the NR and HTGS databases at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/BLAST/). The primers 0Y-1 (5′-GACGATGCTGAAAAGAGCTGTTTG-3′) and 0Y-2 (5′-CCCTTAGCCCTCAGAAAGATACAC-3′) were designed on the basis of the flanking sequences of selected AluY element (GenBank accession no. AC005692). Twenty picomoles of 0Y-1 and 20 pmol of 0Y-2 were used to amplify each great ape genomic DNA in a 50-μl reaction with 200 μM dNTPs and a TaKaRa ExTaq buffer containing 2 mM MgCl2. The PCR conditions were as follows: denaturation at 95°C for 5 min followed by 30 cycles of 95°C for 1 min; 60°C for 1 min; 69°C for 1 min, and extension at 69°C for 10 min. By using the primers 0Y-1 and 0Y-2 as sequencing primers, sequencing of PCR products was performed as described above.
Results and Discussion
Comparison of Genomic Structure Around the 92-bp Exon.
The 92-bp exon is intact in chimpanzees (six individuals), a bonobo (one individual), gorillas (four individuals), orangutans (three individuals), a gibbon (one individual), a baboon (one individual), and a rhesus monkey (one individual) (Figs. 1 and 2). The chimpanzee samples include representatives of two subspecies, Central and West African chimpanzees. These primates all have an AluSq element ≈350 bp downstream from the 92-bp exon (Figs. 1–3). This AluSq element, subsequently designated as sahAluSq after sialic acid hydroxylase AluSq, belongs to a relatively ancient AluSq subfamily (average age: 44 million years; ref. 22; see Fig. 3). The Alu repetitive family is a primate-specific nonautonomous retroposon and is one of the short interspersed elements (23). The Alu family occupies 10.6% of the human genome (24) and is found on average once every 3 kb (23). Insertion of new Alu elements into the genome seems to occur by means of target-primed reverse transcription of Alu RNA transcript, which is catalyzed by the reverse transcriptase of the L1 non-LTR (long terminal repeat) retroposon (23, 25–27). Such Alu insertions typically accompany insertion-site duplications, so that integrated Alus are flanked by short direct repeats of a duplicated insertion site (23, 26). The 5′-TAAAG-3′ sequence immediately adjacent to both ends of the sahAluSq in the chimpanzee and rhesus monkey (Fig. 1) indicates that this sequence is a direct repeat of the sahAluSq. In the gorilla, the same repeat is found at the 5′ end of the sahAluSq, but the 3′ end repeat has an A → G transition. Comparison of sequences surrounding the sahAluSq element among nonhuman primates reveals that the original target site of sahAluSq insertion was 5′-TAAAGATTGNTTTTT(TTT)AA-3′. The reverse transcriptase encoded by the human L1 non-LTR retroposon is a key enzyme in Alu insertion (23, 26, 27). It contains a domain homologous to the apurinic/apyrimidinic (AP) endonuclease family that can nick DNA by recognizing runs of pyrimidines and purines in a very A+T-rich region (28, 29). This AP endonuclease activity of the reverse transcriptase is essential for target-primed insertion of Alu. The sequence deduced as the target site of the sahAluSq is consistent with these observations. In fact, the sequence contains potential runs that can be recognized by the reverse transcriptase.
Figure 1.
Comparison of genomic nucleotide sequences around the 92-bp exon of various primate CMP-Neu5Ac hydroxylase genes. Hs, Pt, Gg, and Mm refer to the human, chimpanzee, gorilla, and rhesus monkey, respectively. The shaded boxes represent the 92-bp exon deleted in the hominid lineage. The Alu element is represented by the open box. The direct repeats of the sialic acid hydroxylase AluSq (sahAluSq) are underlined. The arrowheads indicate replacement boundaries. The 5′-TAAAGATTAATTTTTATTTTT-3′ sequence, which would have a strong preference to the target-priming by the Alu poly(A) tail, is located in the 5′ region immediately adjacent to the upstream replacement boundary. Dots refer to identical nucleotides in the other primates; dashes indicate gaps used for sequence alignment. In the gap corresponding to the human deletion, the complete sequences of the other primate genes are shown.
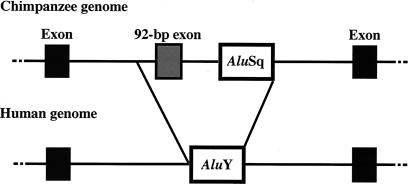
Figure 2.
Schematic comparison of chimpanzee and human CMP-Neu5Ac hydroxylase genomic DNA. In the human genome, the exon to AluSq (E-A) region in the chimpanzee genome is replaced by the sahAluY.
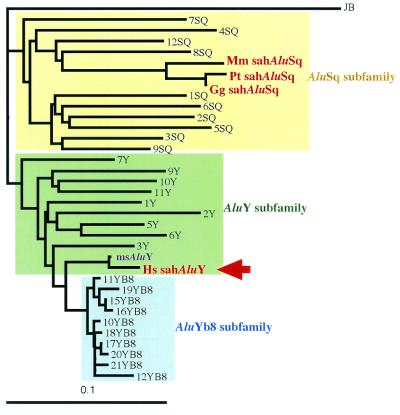
Figure 3.
Phylogenetic tree of Alu subfamilies. Human Alu elements having intact head and tail were randomly selected from both the GenBank database and the on-line database of Alu pairs (http://dir.niehs.nih.gov./ALU/). The tree was made by the neighbor-joining method (20). Distances were calculated with Kimura's two-parameter method (21). The poly(A) tails of sequences were not used in tree-making. The sequence of an AluJb element, which belongs to the old AluJb subfamily, was used as an outgroup. The average age of AluJb, AluSq, AluY, and AluYb8 subfamilies has been estimated at 81, 44, 19, and 3 million years, respectively (ref. 22). The Alu elements shown in Fig. 1 are represented by Hs sahAluY, Pt sahAluSq, Gg sahAluSq, and Mm sahAluSq. msAluY indicates the sequence most similar to the one of sahAluY. Hs, Homo sapiens; Pt, Pan troglodytes; Gg, Gorilla gorilla; Mm, Macaca mulatta.
In humans, we found that an AluY element singly occupies the region of about 800 bp that, in nonhuman primates, still contains the 92-bp exon and the sahAluSq (Figs. 1–3). This AluY element [sialic acid hydroxylase AluY (sahAluY)] belongs to a relatively young Alu subfamily (average age: 19 million years; ref. 22; see Fig. 3). Because the sahAluY occurs at exactly the same chromosomal location for all human samples thus far examined (22 humans from Africa, Europe, and Asia), it is likely that the element has been fixed in the population. However, unlike other genomic regions where the human and the chimpanzee differ by 1–2% (30), the sahAluY in the human and the sahAluSq in the chimpanzee differ from each other by 17% overall. This discrepancy cannot be accounted for by the high nucleotide substitution rate in the Alu family owing largely to the high mutation rate in CpG doublets. It is much more likely that the original human sahAluSq was replaced by a newly disseminated sahAluY. This replacement was human-specific (Fig. 1) and accompanied deletion of a genomic region encompassing the 92-bp exon and the sahAluSq [exon to AluSq region (E-A region)].
The deletion of the 92-bp exon has resulted in fusion of two introns (refs. 8 and 9; Fig. 2). Within the fused region of 22.5 kb, there are six other Alu elements in addition to sahAluY(data not shown): one AluJb, two AluSxs, two AluSqs, and one free left Alu monomer. The Alu density of this region is not especially high and indeed is nearly at the standard level (23). Because all these additional Alu elements are shared by humans and chimpanzees (data not shown), they undoubtedly have not been involved in deleting the E-A region in the human. It seems to be the new sahAluY element that was responsible for the deletion of the E-A region.
AluY Insertion in the Hominid Lineage.
To find the most closely related Alu element to the sahAluY, we performed a blast search at the National Center for Biotechnology Information web site. The closest match is the human AluY, which is located at positions 20358–20638 of Homo sapiens PAC clone RP5–842K16 (GenBank accession no. AC005692; see Fig. 3). The sequence comparison shows that this AluY element [most similar AluY (msAluY)] differs in four non-CpG sites and two CpG sites from the sahAluY. We examined the presence or absence of the orthologous msAluY among the chimpanzee, gorilla, orangutan, and gibbon by genomic PCR and direct sequencing with primers of the 5′ and 3′ flanking sequences of the msAluY. We found that these apes do not possess the ortholog (data not shown). Thus, in addition to sahAluY, there is another AluY specific to humans. Although the detailed chromosomal location is not yet identified, it is possible that insertion of msAluY provides another instance similar to sahAluY. The individual members of different Alu subfamilies are thought to have arisen by amplification of a small subset of “source” genes, which allows subfamilies to evolve in a sequential order (ref. 31; see Fig. 3). One such is sahAluY and another is msAluY, both of which have been inserted uniquely in the hominid lineage. This information would be useful to address the timing of sahAluY dissemination in the hominid genome. This problem will be considered elsewhere, but here we would like to point out that the dissemination could not be very recent. Because there are six nucleotide differences between the two human-specific Alus, it is unlikely that the dissemination occurred within the past 1 million years or so even though the substitution rate in Alus is generally high (30).
Model.
No apparent human-specific sequence feature exists at the boundaries of the E-A region (Fig. 1). It is therefore reasonable to assume that the deletion was triggered by an accidental event that has no sequence preference. A likely possibility is a double-strand break that can be induced by a wide range of factors such as oxidative damage, ionizing radiation, mechanical stress, and action of DNA endonucleases. To model a series of molecular events, we first assume that the deletion was initiated by a double-strand break. We note that a particular sequence of 5′-TAAAGATTAATTTTTATTTTT-3′ is found in the 5′ region immediately adjacent to the upstream deletion boundary in the human, chimpanzee, and gorilla (Fig. 1) and that this sequence is similar to the target site of the sahAluSq and may have a strong preference to the target-priming by the Alu poly(A) tail. The sahAluY in the human can be easily aligned with the sahAluSq in nonhuman primates, although the sequence similarity of the tail region is somewhat lower than that of the head region. Furthermore, both head ends of the sahAluY and sahAluSq elements are identical with the downstream deletion boundary. These findings suggest that free sahAluY RNA transcript interfered in the recombinational repair of a double-strand break because of the target-priming by its poly(A) tail and the annealing to the sahAluSq by its cDNA that resulted from target-primed reverse transcription. On the basis of these considerations, we propose possible molecular mechanisms that caused the Alu-mediated replacement event (Fig. 4).
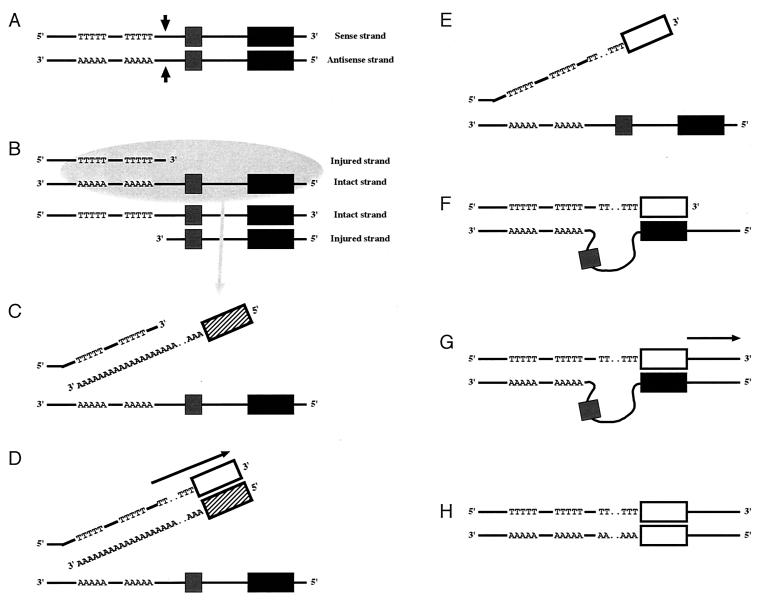
Figure 4.
Model of the Alu-mediated replacement event that occurred in the CMP-Neu5Ac hydroxylase gene in the hominid lineage. (A) Double-strand break indicated by vertical arrows. The solid boxes represent the sahAluSq elements; the shaded box represents the 92-bp exon. The Alu target region containing A/T stretch is located in the 5′ immediately adjacent region of the double-strand break point. (B) Homologous recombination between the injured and intact alleles, after 5′-to-3′ exonucleolytic digestion, which generates a 3′-single-stranded tail. (C) Target-priming to the target site by free sahAluY RNA transcript. Free sahAluY RNA transcript is indicated by both a cross-hatched box and the letter “A,” representing the poly(A) tail. (D) Reverse transcription. An arrow indicates reverse transcription. The open box represents the sahAluY cDNA. (E) Elimination of RNA. (F) Annealing between the sahAluY cDNA and genomic sahAluSq. (G) DNA synthesis. An arrow indicates DNA synthesis. (H) Production of the allele that lacks the E-A region by DNA replication. This allele is derived from the upper strand shown in G.
As a first step, a double-strand break occurs at a position that provides the 5′ deletion boundary (5′ end of the E-A region; see Figs. 1 and 2) (Fig. 4A). Recombinational repair then starts with 5′-to-3′ exonucleolytic digestion of one DNA strand, which leads to the formation of 3′-overhanging single-stranded DNA tails (Fig. 4B). After homologous recombination between the injured and intact alleles, free sahAluY RNA transcript interferes with the repair process through two mechanisms. One mechanism is the target-priming to the 5′-TAAAGATTAATTTTTATTTTT-3′ sequence immediately upstream from the double-strand breakpoint by its poly(A) tail (Fig. 4C). The second mechanism consists of the annealing to the original sahAluSq downstream from the 92-bp exon by its cDNA, which can result from the target-primed reverse transcription (Fig. 4 D–F). The target-priming occurs without enzymatic nicking by L1 reverse transcriptase because the double-strand break provides the nick of DNA (Fig. 4C). The reverse transcription follows the target-priming and produces the sahAluY cDNA from its RNA transcript (Fig. 4D). After elimination of sahAluY RNA transcript (Fig. 4E), the sahAluY cDNA anneals to the genomic sahAluSq (Fig. 4F). The annealing intensity of the Alu tail region is presumed to be somewhat lower than that of the Alu head region because of the regional variation of sequence similarity between the sahAluY and sahAluSq. These actions bring the sahAluSq close to the target site of the sahAluY and block the DNA polymerase extension from the double-strand breakpoint to the head of the sahAluSq (Fig. 4 F and G). DNA synthesis then starts from the head of the sahAluY cDNA (Fig. 4G). Finally, the rearranged allele emerges by DNA replication (Fig. 4H). Thus, Alu-induced incomplete repair of the double-strand break replaces the E-A region by the sahAluY.
The model above (Fig. 4) proposes that a double-strand break repair and Alu target-primed reverse transcription cause an Alu-mediated replacement event. It requires the following structural condition for exon deletion: an exon exists between the tail of a genomic Alu and an Alu target site. The primate genome contains abundant Alu elements (24, 32, 33), and A+T-rich sequences, which can be regarded as potential Alu target sites, frequently occur in the genome. The sequence data from the Human Genome Project confirm that gene-rich regions are Alu-rich (24, 32, 33). Thus, the above-mentioned condition is actually met in the human genome.
The model can also predict Alu conversion (34) when a double-strand break occurs within an Alu element. An inserted Alu is sandwiched in between direct repeats that are derived from their target site (23, 26). In Alu conversion, the poly(A) tail of Alu RNA transcript primes to the adjacent flanking sequence of a genomic Alu element, and therefore Alu cDNA anneals to the genomic Alu element without loop structure (Fig. 4 F and G). This priming introduces a replacement of sequences within an Alu element, leading to Alu conversion. Such an example of Alu conversion has been reported in the low-density lipoprotein receptor gene (34). However, in that case an exon deletion did not follow (34).
Role of Alu Repetitive Family in Primate Evolution.
Because Alu-related events (e.g., Alu insertion and Alu--Alu recombination) can be responsible for diseases caused by abnormal truncations and rearrangements of genes (23, 27), Alus are generally regarded as a disadvantageous or at best neutral agents of organismal evolution. Accordingly, the sahAluY could be a “destructive agent” in primate evolution. However, the situation of inactivation of the hydroxylase gene might be different. As discussed below, inactivation could actually have been favored and fixed in the human population (8, 10).
Genomic Alus are known to be capable of contributing to regulation of gene expression (35–40). Such Alus are referred to as “regulatory Alus” because cis-acting regulatory elements reside within Alus and many Alu classes possess consensus sequences of such regulatory elements (35–39). Hamdi et al. (41) reported that some regulatory Alus are differentially distributed among primates. This finding supports the notion that Alu-related events, such as Alu insertion, Alu–Alu recombination, Alu conversion, and Alu-mediated replacement, might have played significant roles in diversification of primates. In agreement with this idea, the quantitative analyses of the AluYa5 and AluYb8 subfamilies of humans and great apes revealed that the rate of Alu insertion has increased specifically in the hominid lineage (23, 42). The hominid lineage could thus be unique in terms of a high frequency of Alu-related events.
We have provided evidence that Alu insertion caused an exon deletion. It is reported that, although the molecular mechanism has not yet been elucidated, L1 retroposons can transduce surrounding genomic sequences in retrotransposition and induce exon shuffling (43). It is thus possible that L1s have also had an impact on genome evolution as “editing agents.” However, in the human genome, Alus might have played more important roles in rearranging exons than L1s, because, in general, Alus are inserted in gene-rich regions whereas L1s are inserted in gene-poor regions (24, 32, 33).
Pathogen-Mediated Selection of the Alu-Mediated Gene Inactivation.
Many microbial pathogens initiate infection by binding to sialic acids, and some pathogens exert distinct preference for particular types of sialic acids. Influenza viruses show distinct species preference based on Neu5Ac or Neu5Gc expression (44–47), and enterotoxigenic bacteria Escherichia coli K99 adhere specifically to ganglioside GM3(Neu5Gc), but not to GM3(Neu5Ac) in intestinal epithelial cells (48). Furthermore, the amount of human Alu RNA transcripts increases during viral infection (49–51). It is possible that virus infection was somehow related to the hydroxylase inactivation. Thus, a lack of Neu5Gc expression may have conferred protection against infectious pathogens that prefer Neu5Gc. Homo erectus was the first species of Homo whose population expanded widely in mainland Eurasia and Africa (1). The range of H. sapiens expanded further until it included almost every corner of the globe. The genus Homo undoubtedly had to adapt to a wide range of new environments. It is tempting to speculate that the lack of Neu5Gc enabled our ancestors to expand their habitats, first, by evading various animal infectious agents in new environments of H. erectus, and second, by decreasing the infectious risk of H. sapiens from domestication of other vertebrates (some of whose current microbial pathogens are known to prefer Neu5Gc as a binding site) (44–47, 52, 53).
Acknowledgments
We thank Alain Silk for help with cell culture and sequencing, and two anonymous reviewers for their constructive criticisms. This research was supported in part by Japan Society for Promotion of Science Grant 12304046 (to N.T.).
Abbreviations
- Neu5Ac
N-acetylneuraminic acid
- Neu5Gc
N-glycolylneuraminic acid
- sahAluSq
sialic acid hydroxylase AluSq
- sahAluY
sialic acid hydroxylase AluY
- msAluY
most similar AluY
- E-A region
exon to AluSq region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB060157 (human genomic region around the 92-bp exon of CMP-Neu5Ac hydroxylase), AB060158 (chimpanzee genomic region around the 92-bp exon of CMP-Neu5Ac hydroxylase), AB060159 (gorilla genomic region around the 92-bp exon of CMP-Neu5Ac hydroxylase), and AB060160 (rhesus monkey genomic region around the 92-bp exon of CMP-Neu5Ac hydroxylase)].
References
- 1.Klein R G. The Human Career: Human Biological and Cultural Origins. Chicago: Univ. of Chicago Press; 1999. [Google Scholar]
- 2.Satta Y, Klein J, Takahata N. Mol Phylogenet Evol. 2000;14:259–275. doi: 10.1006/mpev.2000.0704. [DOI] [PubMed] [Google Scholar]
- 3.Ruvolo M. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 4.Gagneux P, Varki A. Mol Phylogenet Evol. 2001;18:2–13. doi: 10.1006/mpev.2000.0799. [DOI] [PubMed] [Google Scholar]
- 5.Gibbons A. Science. 1998;281:1432–1434. doi: 10.1126/science.281.5382.1432. [DOI] [PubMed] [Google Scholar]
- 6.Szabó Z, Levi-Minzi S A, Christiano A M, Struminger C, Stoneking M, Batzer M A, Boyd C D. J Mol Evol. 1999;49:664–671. doi: 10.1007/pl00006587. [DOI] [PubMed] [Google Scholar]
- 7.Winter H, Langbein L, Krawczak M, Cooper D N, Jave-Suarez L F, Rogers M A, Praetzel S, Heidt P J, Schweizer J. Hum Genet. 2001;108:37–42. doi: 10.1007/s004390000439. [DOI] [PubMed] [Google Scholar]
- 8.Chou H-H, Takematsu H, Diaz S, Iber J, Nickerson E, Wright K L, Muchmore E A, Nelson D L, Warren S T, Varki A. Proc Natl Acad Sci USA. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. J Biol Chem. 1998;273:15866–15871. doi: 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- 10.Muchmore E A, Diaz S, Varki A. Am J Phys Anthropol. 1998;107:187–198. doi: 10.1002/(SICI)1096-8644(199810)107:2<187::AID-AJPA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 11.Shaw L, Schauer R. Biol Chem Hoppe-Seyler. 1988;369:477–486. doi: 10.1515/bchm3.1988.369.1.477. [DOI] [PubMed] [Google Scholar]
- 12.Muchmore E A, Milewski M, Varki A, Diaz S. J Biol Chem. 1989;264:20216–20223. [PubMed] [Google Scholar]
- 13.Kozutsumi Y, Kawano T, Yamakawa T, Suzuki A. J Biochem (Tokyo) 1990;108:704–706. doi: 10.1093/oxfordjournals.jbchem.a123268. [DOI] [PubMed] [Google Scholar]
- 14.Kawano T, Koyama S, Takematsu H, Kozutsumi Y, Kawasaki H, Kawashima S, Kawasaki T, Suzuki A. J Biol Chem. 1995;270:16458–16463. doi: 10.1074/jbc.270.27.16458. [DOI] [PubMed] [Google Scholar]
- 15.Schauer R. Sialic Acids: Chemistry, Metabolism and Function, Cell Biology Monographs. Vol. 10. New York: Springer; 1982. [Google Scholar]
- 16.Kelm S, Schauer R. Int Rev Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varki A. FASEB J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- 18.Brinkman-Van der Linden E C M, Sjoberg E R, Juneja L R, Crocker P R, Varki N, Varki A. J Biol Chem. 2000;275:8633–8640. doi: 10.1074/jbc.275.12.8633. [DOI] [PubMed] [Google Scholar]
- 19.Schlenzka W, Shaw L, Kelm S, Schmidt C L, Bill E, Trautwein A X, Lottspeich F, Schauer R. FEBS Lett. 1996;385:197–200. doi: 10.1016/0014-5793(96)00384-5. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. Proc Natl Acad Sci USA. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapitonov V, Jurka J. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- 23.Schmid C W. Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 25.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 26.Jurka J. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazazian H H., Jr Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 28.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 29.Martin F, Oliveres M, Lopez M C, Alonso C. Trends Biochem Sci. 1996;21:283–285. [PubMed] [Google Scholar]
- 30.Chen F-C, Li W-H. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid C W, Maraia R. Curr Opin Genet Dev. 1992;2:874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- 32.Dunham I, Hunt A R, Collins J E, Bruskiewich R, Beare D M, Clamp M, Smink L J, Ainscough R, Almeida J P, Babbage A, et al. Nature (London) 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 33.Hattori M, Fujiyama A, Taylor T D, Watanabe H, Yada T, Park H-S, Toyoda A, Ishii K, Totoki Y, Choi D-K, et al. Nature (London) 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 34.Kass D H, Batzer M A, Deininger P L. Mol Cell Biol. 1995;15:19–25. doi: 10.1128/mcb.15.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brini A T, Lee G M, Kinet J-P. J Biol Chem. 1993;268:1355–1361. [PubMed] [Google Scholar]
- 36.Hambor J E, Mennone J, Coon M E, Hanke J H, Kavathas P. Mol Cell Biol. 1993;13:7056–7070. doi: 10.1128/mcb.13.11.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McHaffie G S, Ralston S H. Bone. 1995;17:11–14. doi: 10.1016/8756-3282(95)00131-v. [DOI] [PubMed] [Google Scholar]
- 38.Norris J, Fan D, Aleman C, Marks J R, Futreal P A, Wiseman R W, Iglehart J D, Deininger P L, McDonnell D P. J Biol Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 39.Vansant G, Reynolds W F. Proc Natl Acad Sci USA. 1995;92:8229–8233. doi: 10.1073/pnas.92.18.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fornasari D, Battaglioli E, Flora A, Terzano S, Clementi F. Mol Pharmacol. 1997;51:250–261. doi: 10.1124/mol.51.2.250. [DOI] [PubMed] [Google Scholar]
- 41.Hamdi H K, Nishio H, Tavis J, Zielinski R, Dugaiczyk A. J Mol Biol. 2000;299:931–939. doi: 10.1006/jmbi.2000.3795. [DOI] [PubMed] [Google Scholar]
- 42.Zietkiewicz E, Richer C, Makalowski W, Jurka J, Labuda D. Nucleic Acids Res. 1994;22:5608–5612. doi: 10.1093/nar/22.25.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran J V, DeBerardinis R J, Kazazian H H., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 44.Higa H H, Rogers G N, Paulson J C. Virology. 1985;144:279–282. doi: 10.1016/0042-6822(85)90325-3. [DOI] [PubMed] [Google Scholar]
- 45.Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Virology. 1997;227:493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T, Horiike G, Yamazaki Y, Kawabe K, Masuda H, Miyamoto D, Matsuda M, Nishimura S-I, Yamagata T, Ito T, et al. FEBS Lett. 1997;404:192–196. doi: 10.1016/s0014-5793(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 47.Ito T, Suzuki Y, Suzuki T, Takada A, Horimoto T, Wells K, Kida H, Otsuki K, Kiso M, Ishida H, et al. J Virol. 2000;74:9300–9305. doi: 10.1128/jvi.74.19.9300-9305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smit H, Gaastra W, Kamerling J P, Vliegenthart J F G, de Graaf F K. Infect Immun. 1984;46:578–584. doi: 10.1128/iai.46.2.578-584.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panning B, Smiley J R. Virology. 1994;202:408–417. doi: 10.1006/viro.1994.1357. [DOI] [PubMed] [Google Scholar]
- 50.Panning B, Smiley J R. J Mol Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 51.Russanova V R, Driscoll C T, Howard B H. Mol Cell Biol. 1995;15:4282–4290. doi: 10.1128/mcb.15.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyogashima M, Ginsburg V, Krivan H C. Arch Biochem Biophys. 1989;270:391–397. doi: 10.1016/0003-9861(89)90042-8. [DOI] [PubMed] [Google Scholar]
- 53.Delorme C, Brüssow H, Sidoti J, Roche N, Karlsson K-A, Neeser J-R, Teneberg S. J Virol. 2001;75:2276–2287. doi: 10.1128/JVI.75.5.2276-2287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]