Abstract
The human leukocyte antigen (HLA)-A2-restricted zinc transporter (ZnT)8186–194 and other islet epitopes elicit interferon-γ secretion by CD8+ T cells preferentially in type 1 diabetes (T1D) patients compared with controls. Here, we show that clonal ZnT8186–194-reactive CD8+ T cells express private T-cell receptors and display equivalent functional properties in T1D and healthy subjects. Ex-vivo analyses further revealed that CD8+ T cells reactive to ZnT8186–194 and other islet epitopes circulate at similar frequencies and exhibit a predominantly naïve phenotype in age-matched T1D and healthy donors. Higher frequencies of ZnT8186–194-reactive CD8+ T cells with a more antigen-experienced phenotype were detected in children vs. adults, irrespective of disease status. Moreover, some ZnT8186–194-reactive CD8+ T-cell clonotypes were found to cross-recognize a Bacteroides stercoris mimotope. While ZnT8 was poorly expressed in thymic medullary epithelial cells, variable thymic expressions levels of islet antigens did not modulate the peripheral frequency of their cognate CD8+ T cells. In contrast, ZnT8186–194-reactive cells were enriched in the pancreata of T1D donors vs. non-diabetic and type 2 diabetic controls. Thus, islet-reactive CD8+ T cells circulate in most individuals, but home to the pancreas preferentially in T1D patients. We conclude that the activation of this common islet-reactive T-cell repertoire and progression to T1D likely require defective peripheral immunoregulation and/or a pro-inflammatory islet microenvironment.
Introduction
In the setting of type 1 diabetes (T1D), insulitic lesions are enriched for CD8+ T cells, which are held as the final mediators of islet destruction. Concordantly, preproinsulin (PPI)-reactive CD8+ T-cell clones can lyse β cells in-vitro (1), and β-cell-reactive CD8+ T cells infiltrate the islets of T1D patients (2). Autoimmune CD8+ T cells may therefore provide new biomarkers for disease staging complementary to auto-antibodies (aAbs). While interferon (IFN)-γ-secreting CD8+ T cells detected by enzyme-linked immunospot (ELISpot) distinguish T1D patients from healthy donors (3), the situation is more complex when non-functional human leukocyte antigen (HLA) Class I multimer (MMr) assays are used. Although MMr+CD8+ T cells were often (4), but not invariably (5, 6), found at similar frequencies in both T1D and healthy subjects, they have been reported to exhibit more differentiated effector/memory phenotypes (4, 6) in T1D patients. A rather enigmatic state of ‘benign’ autoimmunity therefore exists in healthy individuals.
We therefore aimed to determine the key features of islet-reactive CD8+ T cells that associate with T1D. We focused our efforts on well-defined HLA-A*02:01 (HLA-A2)-restricted immunodominant epitopes derived from PPI, glutamic acid decarboxylase (GAD), insulinoma-associated protein-2 (IA-2), and islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) (3), and on a highly immunoprevalent zinc transporter (ZnT)8186–194 epitope that we recently described (7). The results indicate that incomplete central tolerance mechanisms allow the survival of an islet-reactive CD8+ T-cell repertoire, which in some individuals may be primed in the presence of defective peripheral immunoregulation and/or a pro-inflammatory islet microenvironment to progress toward T1D.
Results
ZnT8186–194-reactive CD8+ T-cell clones from T1D and healthy donors display equivalent functionality
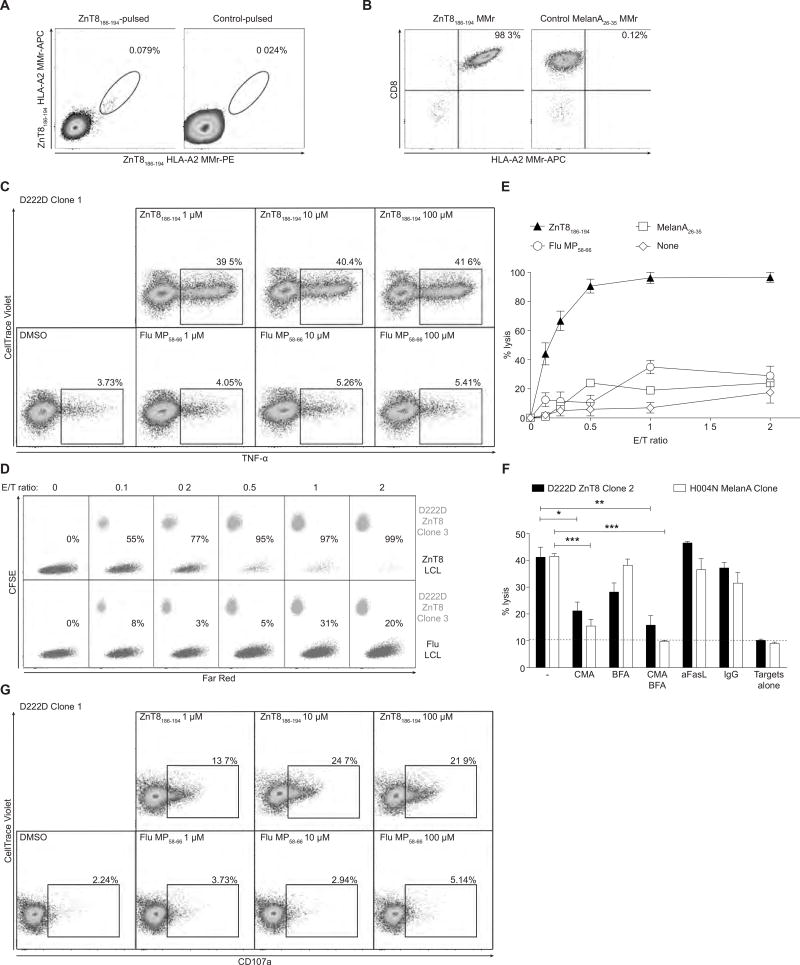
Given that ZnT8186–194-reactive IFN-γ responses are highly prevalent in T1D patients (7), we started by generating ZnT8186–194-reactive CD8+ T-cell clones from a new-onset T1D patient (D222D) (7). Following in-vitro expansion with the ZnT8186–194 peptide (8), HLA-A2 MMr+ cells were labeled with two different fluorochromes (9) and sorted (Fig. 1A). The 3 clones thus generated stained uniformly positive with ZnT8186–194 MMrs (Fig. 1B) and responded to ZnT8186–194 peptide stimulation by secreting tumor necrosis factor (TNF)-α (Fig. 1C), IFN-γ, interleukin (IL)-2 and, to a lesser extent, macrophage inflammatory protein (MIP)-1β in a dose-dependent fashion (Fig. S1A–C). Cytotoxicity was then tested against an HLA-A2+ Epstein-Barr virus (EBV)-transformed B-lymphoblastoid cell line (LCL) pulsed with the cognate ZnT8186–194 peptide. Increasing numbers of clonal CD8+ T cells led to the complete disappearance of ZnT8186–194-pulsed but not control-pulsed targets, with ≥90% lysis at a 1/2 effector/target (E/T) ratio (Fig. 1D–E and S1D–F). This lytic activity was mostly perforin-mediated (Fig. 1F), as it was inhibited significantly by concanamycin A, marginally by brefeldin A (suppressing cytokine secretion) and not at all by a blocking anti-FasL mAb (suppressing Fas-dependent cytotoxicity), and it was associated with CD107a upregulation (Fig. 1G).
Fig. 1. ZnT8186–194-reactive CD8+ T-cell clones from patient D222D.
(A) Frozen-thawed PBMCs were cultured with ZnT8186–194 or no peptide and stained with PE/APC-labeled ZnT8186–194 MMrs. (B) ZnT8186–194 and control MMr stains for one clone obtained from single-sorted ZnT8186–194/ZnT8186–194 double-MMr+ cells. (C) Percent intracellular TNF-α+ D222D clone 1 cells stimulated for 6 h with K562-A2 cells pulsed with ZnT8186–194 or Flu MP58–66 peptide. (D) Percent lysis of FarRed-labeled LCL targets pulsed with ZnT8186–194 (top) or Flu MP58–66 peptide (bottom) and cultured for 24 h with CFSE-labeled D222D clone 3 at increasing E/T ratios. (E) Percent lysis of LCL targets cultured with D222D clones 1, 2 or 3 (mean±SEM; each clone is depicted in Fig. S1D–F). (F) Lysis of cognate peptide-pulsed LCLs cultured for 4 h with D222D clone 2 or a MelanA26–35-reactive clone (E/T 1/1) in the presence of concanamycin A (CMA), brefeldin A (BFA), CMA and BFA, anti-FasL or control IgG1. *p=0.015, **p=0.009, ***p<0.001 by Student’s t test. Results are mean±SEM of triplicate measurements from one of three experiments. (G) Percent surface CD107a+ D222D clone 1 cells stimulated as in (C). For panels A, C, G, gate is on viable CD8+ cells.
Additional ZnT8186–194-reactive CD8+ T-cell clones were generated from other T1D patients and healthy donors. The final panel comprised 16 clones (9 clones from 5 T1D patients and 7 clones from 5 healthy donors), most of which were isolated directly ex-vivo (Table S1). All but one clone (H328C 9B3) stained with HLA-A2 MMrs loaded with ZnT8186–194 (VAANIVLTV), and several of them also with a longer ZnT8185–194 variant (AVAANIVLTV) reported to exhibit similar immunoprevalence (Fig. S2A) (10). Higher staining intensities were observed with ZnT8186–194 MMrs for all clones barring D010R 1D3. Concordantly, ZnT8186–194 bound to recombinant HLA-A2 molecules with higher affinity (KD 15 nM vs. 207 nM) and similar stability (t1/2 1.8 h vs. 2.3 h) relative to the longer ZnT8185–194 peptide (Fig. S2B–C). The ZnT8186–194 epitope was therefore retained for subsequent experiments, except for clone D010R 1D3.
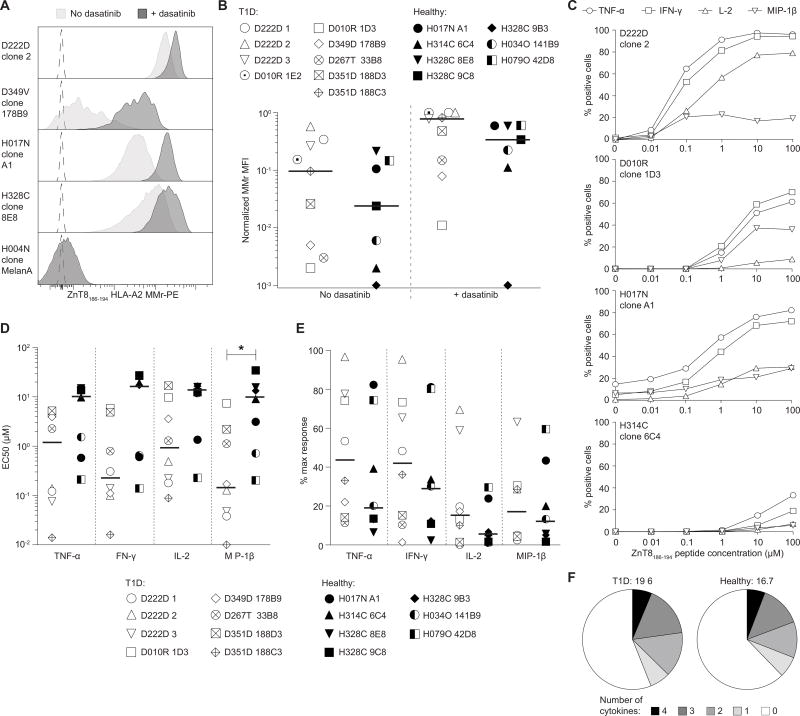
Clones from T1D and healthy donors were first compared for the intensity of ZnT8186–194 MMr staining. The tyrosine kinase inhibitor dasatinib, which stabilizes T-cell receptor (TCR) interactions with peptide-HLA complexes (11), enhanced MMr staining, particularly for MMrlow clones (Fig. 2A). Heterogeneous staining patterns were observed when comparing normalized MMr fluorescence intensities (Fig. 2B). While dasatinib reduced variability, even in its absence no difference was apparent between the T1D and healthy group.
Fig. 2. Ag avidity, Ag sensitivity and polyfunctionality of ZnT8186–194-reactive CD8+ T-cell clones.
(A) ZnT8186–194 MMr staining in the absence (light grey) or presence (dark grey) of dasatinib. The dotted profile indicates the unstained control. (B) ZnT8186–194 MMr median fluorescence intensity (MFI) for the indicated clones in the absence (left) or presence (right) of dasatinib. Bars indicate median values. Results are representative of two separate experiments. (C) The indicated clones were stimulated for 6 h with ZnT8186–194-pulsed K562-A2 cells and percent cytokine+ cells out of viable CD8+ cells calculated. Results are representative of three independent experiments. (D–E) Half maximal effective peptide concentration (EC50; D) and maximal cytokine response (percent cytokine+ cells at optimal peptide concentrations; E) for clones stimulated as above. Bars indicate median values. Results are representative of two to four separate experiments. *p=0.014 by Mann-Whitney test. (F) Polyfunctionality distribution of T1D (left) and healthy clones (right). Percent T cells producing 0 to 4 cytokines among TNF-α, IFN-γ, IL-2 and MIP-1β upon exposure to ZnT8186–194-pulsed K562-A2 cells (100 µM) are shown.
Next, we performed in-vitro recall assays with increasing ZnT8186–194 peptide concentrations. Representative data are shown in Fig. 2C and results summarized in Fig. 2D–E. In line with the MMr staining profiles, the half maximal effective peptide concentration (EC50) required to elicit cytokine responses (TNF-α, IFN-γ, IL-2) and the maximal cytokine responses were again heterogeneous, but not significantly different between T1D and healthy clones (Fig. 2D–E), with one exception noted for the lower MIP-1β EC50 of T1D clones. Clones obtained after in-vitro expansion displayed an equivalent range of antigen (Ag) sensitivities, either high (D222D, H017N) or low (H328C), arguing against a bias compared with clones isolated directly ex-vivo. Moreover, the polyfunctionality index, which reflects the number of cells secreting multiple cytokines (12), was similar for T1D and healthy clones (Fig. 2F). As expected, EC50 values correlated negatively with MMr staining and polyfunctionality indices (Fig. S3).
Collectively, these results show that ZnT8-reactive CD8+ T cells isolated from T1D and healthy subjects exhibit similar Ag avidity, sensitivity and polyfunctionality.
ZnT8186–194-reactive CD8+ T-cell clones from T1D and healthy donors display equivalent β-cell cytotoxicity
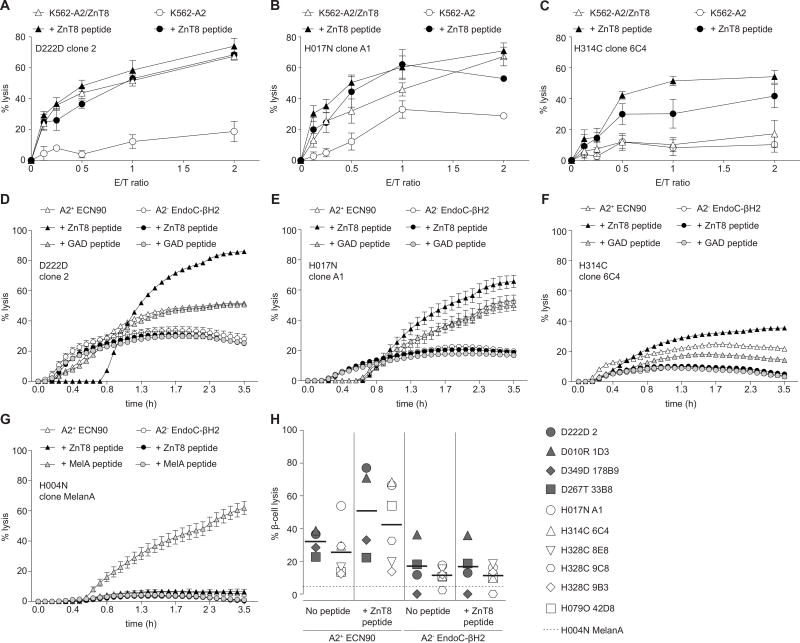
To determine if ZnT8186–194-reactive CD8+ T cells can recognize naturally processed ZnT8 epitopes, we performed cytotoxicity assays using ZnT8-transduced K562-A2 targets (K562-A2/ZnT8). High avidity ZnT8186–194-reactive clones lysed unpulsed K562-A2/ZnT8 targets almost as efficiently as targets pulsed with the ZnT8186–194 peptide, while unpulsed K562-A2 control targets remained largely intact (Fig. 3A–B). In contrast, low avidity ZnT8186–194-reactive clones were unable to lyse K562-A2/ZnT8 targets in the absence of exogenous ZnT8186–194 peptide (Fig. 3C).
Fig. 3. Target cell lysis by ZnT8186–194-reactive CD8+ T-cell clones.
(A–C) Lysis of K562-A2 cells transfected (open triangles) or not (open circles) with a full-length ZnT8 plasmid and cultured for 24 h with clones D222D 2 (A), H017N A1 (B) or H314C 6C4 (C). Filled symbols indicate ZnT8186–194-pulsed target cells (10 µM). Results are presented as mean±SEM of triplicate wells from two separate experiments. (D–G) Real-time cytotoxicity for the indicated clones vs. HLA-A2+ ECN90 (white triangles) or control HLA-A2− EndoC-βH2 β-cell targets (white circles) (E/T 2/1). Black and grey symbols indicate the corresponding targets pulsed with 10 µM ZnT8186–194 or GAD114–122 peptide, respectively. Mean±SEM of triplicate measurements are shown at each time point. Results are representative of at least two separate experiments. (H) Percent maximal HLA-A2+ ECN90 and HLA-A2− EndoC-βH2 β-cell lysis by the indicated clones (T1D, grey symbols; healthy, white symbols; control, horizontal dotted line) in the absence or presence of ZnT8186–194 peptide. Bars indicate median values. Lysis was calculated from the cytotoxicity profiles as in panels D–G.
We then evaluated cytotoxicity against T1D-relevant targets by employing HLA-A2+ ECN90 and control HLA-A2− EndoC-βH2 human β-cell lines. Although the ECN90 but not the EndoC-βH2 line expressed HLA Class I in the unstimulated state, expression levels were similarly upregulated by pretreatment with different cocktails of inflammatory cytokines, without inducing significant β-cell death (Fig. S4A–B). IFN-γ was chosen as the single cytokine that upregulated HLA Class I expression to comparable levels in both lines, and pretreatment was carried out for 18 h prior to a real-time cytotoxicity assay. As observed with K562-A2/ZnT8 targets, high avidity clones lysed unpulsed HLA-A2+ ECN90 cells presenting naturally processed ZnT8-derived epitopes (Fig. 3D–E), while low avidity clones displayed marginal lytic activity (Fig. 3F). Cytotoxicity increased in all cases with the addition of ZnT8186–194 peptide, suggesting more limited natural presentation compared with ZnT8-transduced targets. Lysis of HLA-A2− EndoC-βH2 cells was negligible, and a control clone reactive to the melanocyte self-epitope MelanA26–35 only lysed ECN90 cells in the presence of exogenous MelanA26–35 peptide (Fig. 3G). Microscopic inspection confirmed the lysis measured in real-time cytotoxicity assays (Fig. S4C). β-cell lysis was not different between T1D and healthy clones, either in the absence or presence of exogenous ZnT8186–194 peptide (Fig. 3H). Moreover, IFN-γ-pretreatment of HLA-A2+ ECN90 cells neither upregulated ZnT8 protein expression (Fig. S4D) nor increased the functional activation of co-incubated CD8+ T-cell clones (Fig. S4E–F). Collectively, these results demonstrate that ZnT8186–194-reactive CD8+ T cells display similar cytotoxicity in T1D and healthy subjects and that ZnT8 expression is not modulated by inflammation.
ZnT8186–194-reactive CD8+ T cells display private TCR gene usage but public CDR3β aminoacid sequences
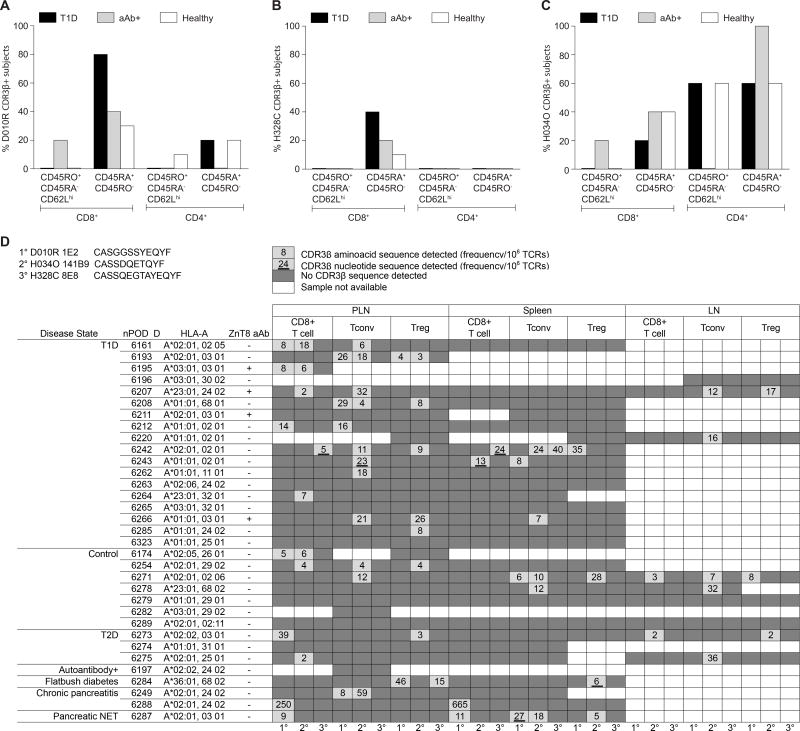
Molecular analysis of expressed TCRα (TRA) and TCRβ (TRB) gene transcripts revealed that no sequences were shared (public) among ZnT8186–194-reactive clones isolated from T1D or healthy subjects (Fig. S5). Of note, the 3 clones from patient D222D harbored an identical TCR. However, this observation did not affect our previous functional comparisons, because the measured parameters were even more similar between T1D and healthy donors when only one D222D clone was considered.
We then interrogated an in-silico database of TRB sequences compiled by high-throughput sequencing of naïve/terminal effector (CD45RA+CD45RO−) or central memory (CD45RO+CD45RA−CD62Lhi) CD4+ and CD8+ T cells obtained from HLA-A2+ recent-onset T1D patients, at-risk aAb+ and healthy subjects (Table S2). The D010R 1E2, H328C 8E8 and H034O 141B9 complementarity-determining region (CDR)3β aminoacid sequences were found among CD8+ and CD4+ T cells isolated from several individuals (Fig. 4A–C), mostly within the CD45RA+CD45RO− pool for CD8+ T cells. Several different TRBV genes were used in conjunction with these identical CDR3β loops to generate ‘mosaic’ sequences. The same CDR3β aminoacid sequences were detected in-silico among the polyclonal TCR repertoires compiled from conventional and regulatory CD4+ and CD8+ T cells isolated from pancreatic lymph node (PLN) and spleen samples by the Network for Pancreatic Organ Donors (nPOD) (Fig. 4D; n=67 identical CDR3β, n=6 identical TCRβ). Most of the CD8+ T-cell hits in PLNs (11/15, 73%) were from HLA-A2+ subjects, but with no obvious association with T1D.
Fig. 4. In-silico search for CDR3β aminoacid sequences from ZnT8186–194-reactive CD8+ T-cell clones.
(A–C) Prevalence of the CDR3β aminoacid sequences from clones D010R 1E2 (A), H328C 8E8 (B) and H034O 141B9 (C) among HLA-A2+ T1D (n=5), aAb+ (n=5) and healthy subjects (n=10), as assessed by in-silico analysis of TCRβ repertoires obtained from the indicated CD8+ and CD4+ T-cell subsets. (D) In-silico search for the same CDR3β aminoacid sequences in the repertoire of CD8+, conventional CD4+ (Tconv; CD127+) and regulatory CD4+ (Treg; CD25+CD127−) T cells obtained from nPOD PLN, spleen and inguinal lymph node (ILN) samples via the online database http://clonesearch.jdrfnpod.org. For each cell type and tissue, the first, second and third columns refer to clones D010R 1E2, H034O 141B9 and H328C 8E8, respectively. Dark and light grey cells indicate negative and positive samples, respectively. Frequencies per 106 TCRs are annotated, and underlining indicates samples with a nucleotide sequence match. White cells indicate unavailable samples. Pancreatic NET, neuro-endocrine tumor.
To extend these findings, we developed D010R 1E2, H328C 8E8 and D222D TRA and TRB clonotype-specific TaqMan real-time quantitative PCR (qPCR) assays (Fig. S6A–B). Applied to CD4+ and CD8+ T-cell cDNA preparations from two independent cohorts of T1D (n=97 and n=53) and healthy subjects (n=97 and n=38), these assays detected the D010R and H328C TRB among CD8+ T cells from a single HLA-A2+ T1D patient in each case (Fig. S6C–D).
Collectively, these results show that ZnT8186–194 recognition is mediated primarily by private clonotypes, some of which share CDR3β aminoacid sequences among individuals to form ‘mosaic’ TCRs.
Circulating islet-reactive CD8+ T cells display similar ex-vivo frequencies and a predominantly naïve phenotype in T1D and healthy subjects
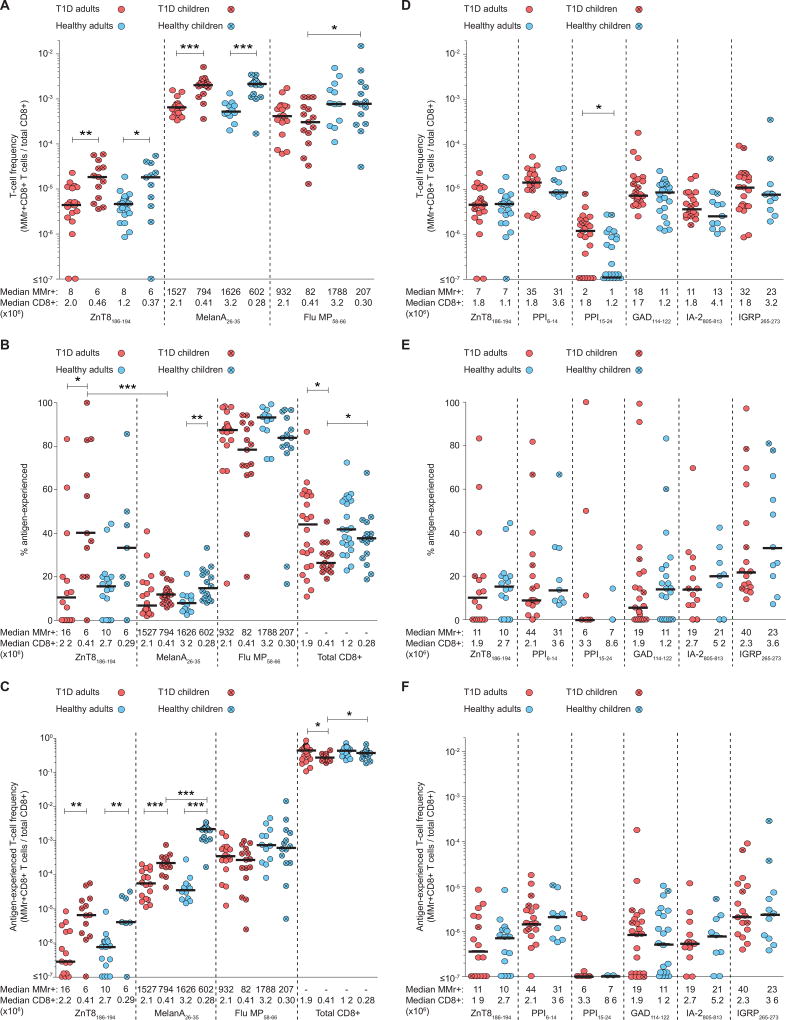
In further experiments, we used ex-vivo combinatorial HLA-A2 MMr assays (9) to analyze ZnT8186–194-reactive CD8+ T cells in 39 HLA-A2+ recent-onset T1D patients (16 children, 23 adults) and 39 age/sex-matched healthy donors (17 children, 22 adults) (Table S3, Fig. S7). Control specificities included MelanA26–35, which is recognized by a large naïve pool in humans (13), and Flu MP58–66, to which most individuals harbor Ag-experienced CD8+ T cells reflecting prior viral exposure. Donors yielding <3×105 total CD8+ T cells or <5 MMr+ cells were excluded from the analysis to avoid undersampling. Parallel ELISpot assays confirmed that ZnT8186–194-reactive IFN-γ responses were more frequent in T1D vs. healthy donors (Fig. S8) (7). In contrast, similar frequencies of ZnT8186–194 and MelanA26–35 MMr+CD8+ T cells were detected in age-stratified T1D and healthy donors (Fig. 5A). Healthy chidren and adults displayed higher frequencies of Flu MP58–66 MMr+CD8+ T cells compared with their age/sex-matched T1D counterparts, while children displayed ~4-fold higher frequencies of ZnT8186–194 and MelanA26–35 MMr+CD8+ T cells compared with adults, irrespective of T1D status.
Fig. 5. Ex-vivo frequencies and Ag-experienced phenotypes of circulating islet-reactive CD8+ T cells.
(A) ZnT8186–194, MelanA26–35 and Flu MP58–66 MMr+CD8+ cells were stained ex-vivo and counted (see Fig. S7). Frequencies out of total CD8+ T cells are depicted for T1D adults (red circles), T1D children (crossed red circles), age/sex-matched healthy adults (blue circles) and children (crossed blue circles). *p≤0.05, **p=0.002, ***p≤0.0003. (B) Percent Ag-experienced cells out of total MMr+ cells. *p≤0.03, **p=0.004, ***p=0.0007. (C) Absolute frequencies of the corresponding Ag-experienced fractions. *p≤0.03, **p≤0.01, ***p≤0.0001. (D) MMr+CD8+ cells reactive to the indicated islet epitopes were stained ex-vivo and counted (see Fig. S10A). Frequencies out of total CD8+ T cells are depicted as in panel A. *p=0.02. (E) Percent Ag-experienced cells out of total MMr+ cells. (F) Absolute frequencies of the corresponding Ag-experienced fractions. Bars display median values. The median number of MMr+ events and total CD8+ T cells analyzed are indicated for each distribution. Significance was determined using the Mann-Whitney test. For panels A and D, data-points with <300,000 CD8+ T cells and <5 MMr+ cells were excluded. For panels B–C and E–F, data-points with <5 MMr+ cells were excluded.
Among ZnT8186–194 MMr+CD8+ cells, the Ag-experienced fraction (CD45RA+CCR7− or CD45RA−CCR7+/−) was similarly limited (generally ≤25% of all ZnT8186–194 MMr+ events) in T1D and healthy adults (Fig. 5B). In contrast, higher proportions of Ag-experienced ZnT8186–194 MMr+ cells were present in T1D children vs. adults, although these values were not different in healthy children. T1D children also harbored significantly higher proportions of Ag-experienced ZnT8186–194 vs. MelanA26–35 MMr+ cells. MelanA26–35-reactive CD8+ T cells were mostly naïve, while Flu MP58–66-reactive CD8+ T cells were mostly Ag-experienced in all groups. Comparable results were obtained using the absolute frequencies of Ag-experienced MMr+CD8+ T cells (Fig. 5C).
Single-cell gene expression analysis of sorted ZnT8186–194 MMr+ cells (Fig. S9A) revealed few differences, with T1D patients displaying higher expression of the Th17-related aryl hydrocarbon receptor (AHR) and the mitotic aurora kinase A (AURKA) and lower expression of the transcriptional activator RAR related orphan receptor A (RORA). In line with our in-vitro data, clonotypic analyses of individual ZnT8186–194 MMr+CD8+ T cells sorted ex-vivo yielded unique CDR3α and CDR3β sequences (Fig. S9B). Of the 21 CDR3β aminoacid sequences obtained, 7 (33%) were found in our in-silico TRB database across all subject groups (Fig. S9C). Among CD8+ T cells, these 7 sequences were again most frequent within the CD45RA+CD45RO− compartment, and predominantly found in T1D vs. healthy subjects. Preferential usage of certain TRBV (mostly TRBV19, 25%) and TRAV (mostly TRAV12-2, 38%) genes was observed (Fig. S9D–E), with TRBV19 also shared among the ZnT8186–194-reactive clones. However, only 8 out of 63 matching sequences (13%) expressed the TRBV gene of the corresponding ZnT8186–194-reactive T cell. Thus, despite biased TRAV and TRBV gene usage, the ex-vivo data confirmed that ZnT8186–194 recognition is mediated primarily by private clonotypes expressing ‘mosaic’ TCRs.
Next, we used an extended combinatorial MMr panel to compare ZnT8186–194-reactive CD8+ T cells with those recognizing other immunodominant β-cell epitopes in adult donors (Fig. S10A). Assay reproducibility across panels was confirmed using separate blood draws from the same individuals (Fig. S10B). As observed for ZnT8186–194, other β-cell-reactive CD8+ T-cell populations displayed similar frequencies (typically 1–50 MMr+ cells/106 CD8+ T cells) in T1D and healthy adults (Fig. 5D). An exception was noted for PPI15–24-reactive CD8+ T cells, whose frequencies were lower than for other β-cell-reactive fractions, and higher in T1D vs. healthy donors. In all instances, the Ag-experienced fraction among MMr+CD8+ T cells was limited in both subject groups (Fig. 5E–F). The few children included in this extended analysis harbored β-cell-reactive CD8+ T cells with a more Ag-experienced phenotype. CD45RA+CCR7+ islet-reactive cells were bona fide naïve, as they were largely CD27+CD28+ and CD95− (Fig. S11).
Collectively, these results show that circulating CD8+ T cells reactive to ZnT8186–194 and other β-cell epitopes occur at similar frequencies and exhibit a predominantly naïve phenotype in T1D and healthy adults, while higher frequencies of total and Ag-experienced ZnT8186–194-reactive CD8+ T cells are present in children, irrespective of T1D status.
Poor promiscuous ZnT8 gene expression in human thymic medullary epithelial cells (mTECs)
The combined observations that CD8+ T cells reactive to ZnT8186–194 and other β-cell epitopes are predominantly naïve and circulate at similar frequencies in T1D and healthy donors are compatible with frequent escape from thymic deletion due to poor islet Ag expression in mTECs (14). We tested this hypothesis by examining ZnT8 (SLC30A8) gene expression in total or immature (AIRE−HLA Class IIlow) vs. mature (AIRE+HLA Class IIhigh) human mTECs from 5 children undergoing heart surgery. As mis-initiated mRNA transcription is described in mTECs (13, 15), forward primers were designed to identify potential alternative start sites via hybridization with exons 5, 6, 7 and 8 of the SLC30A8 gene (Fig. 6A). The ZnT8186–194 region is encoded by exons 7–8, with the exon 8 primer located just downstream. Combinations of these forward primers with reverse primers located either in exon 11 (Fig. 6B) or in the 3’-UTR (Fig. 6C) yielded the expected bands using human islet mRNA. However, mTECs did not express SLC30A8. One sample (#64) displayed a faint band amplified with the exon 8 forward primer, which matched SLC30A8 by sequencing, suggesting low-level mis-initiated transcription starting at exon 8 (i.e. downstream of the ZnT8186–194 region). Collectively, these results demonstrate that ZnT8 is poorly expressed in mTECs, and that SLC30A8 transcription is limited to a mis-initiated mRNA isoform skipping the ZnT8186–194 sequence.
Fig. 6. SLC30A8 and INS gene expression in mTECs and circulating islet-reactive CD8+ T-cell frequencies in HLA-A2+ and HLA-A2− healthy donors.
(A) SLC30A8 RT-PCR strategy. Forward primers spanned exons 5 to 8, reverse primers spanned either exon 11 or the 3’-UTR. The position of the ZnT8186–194-coding region is shown. (B) SLC30A8 expression in mTECs, using the indicated forward primers and the exon 11 reverse primer. (C) SLC30A8 expression in mTECs from donor #64 (previously testing positive with the exon 8 forward primer) and #211 (previously testing negative), using the 3’-UTR reverse primer. (D) INS RT-PCR strategy. Forward primers spanned both or neither of the PPI6–14 and PPI15–24 regions, reverse primers spanned either the 3’-UTR or exon 2 (PCR products 1 and 2, respectively). (E) INS expression in thymuses pooled from 5–8 donors. (F) Ex-vivo MMr+CD8+ cell frequencies in age/sex-matched, EboV- and HCV-seronegative HLA-A2+ and HLA-A2− healthy donors. (G) Percent Ag-experienced MMr+ cells. Bars indicate median values. The median number of MMr+ events is indicated, with a median of 1×106 total CD8+ T cells analyzed. *p≤0.03, **p=0.008, ***p≤0.0004 by Mann-Whitney test. For panels G, data points with <5 MMr+ cells were excluded. NA, not available.
Islet-reactive CD8+ T cells circulate at similar frequencies irrespective of thymic expression of their cognate epitopes
In contrast to ZnT8, other β-cell Ags are expressed by mTECs, including PPI (16, 17), IA-2 (17, 18) and IGRP (19), or by thymic Ag-presenting cells, including GAD65 (20). Moreover, circulating PPI6–14-reactive CD8+ T cells displayed higher frequencies than PPI15–24-reactive CD8+ T cells, despite the fact that the complete INS transcript incorporating both epitopes was detected in the thymus (Fig. 6D–E). In conjunction with the finding that circulating CD8+ T cells reactive to ZnT8186–194 and other islet epitopes also occur at similar frequencies (Fig. 5D), these observations suggest a limited role of thymic Ag gene expression in setting such frequencies.
Potential confounders in this scenario include the efficiency of epitope presentation and the odds of encountering cognate peptide-HLA complexes in the thymic environment. Accordingly, we compared the frequencies of islet-reactive CD8+ T cells in age/sex-matched HLA-A2+ and HLA-A2− healthy adults, reasoning that thymic deletion cannot occur in the absence of the appropriate HLA-A2 restriction (21). To exclude promiscuous presentation by non-HLA-A2 molecules, donors were selected based on HLA-A and HLA-B molecules incapable of binding the selected islet epitopes (Table S4). Moreover, all subjects were seronegative for Ebola (EboV) and hepatitis C virus (HCV). Representative flow cytometry plots are shown in Fig. S12. Except for PPI6–14, HLA-A2-restricted islet-reactive CD8+ T cells occurred at largely equivalent frequencies in HLA-A2+ and HLA-A2− donors (Fig. 6F). Although higher overall relative to most islet specificities, the frequencies of MMr+CD8+ cells recognizing the HLA-A2-restricted foreign epitopes EboV NP202–210 and HCV PP1406–1415 were also similar between HLA-A2+ and HLA-A2− groups. No significant phenotypic differences were observed between groups for any of these MMr+CD8+ T-cell populations (Fig. 6G). As expected, control Flu MP58–66 MMr+CD8+ cells were more prevalent and more Ag-experienced in HLA-A2+ donors.
Collectively, these findings suggest that thymic presentation of these islet epitopes does not trigger significant clonal deletion in HLA-A2+ donors.
ZnT8186–194-reactive CD8+ T cells cross-recognize a Bacteroides stercoris mimotope
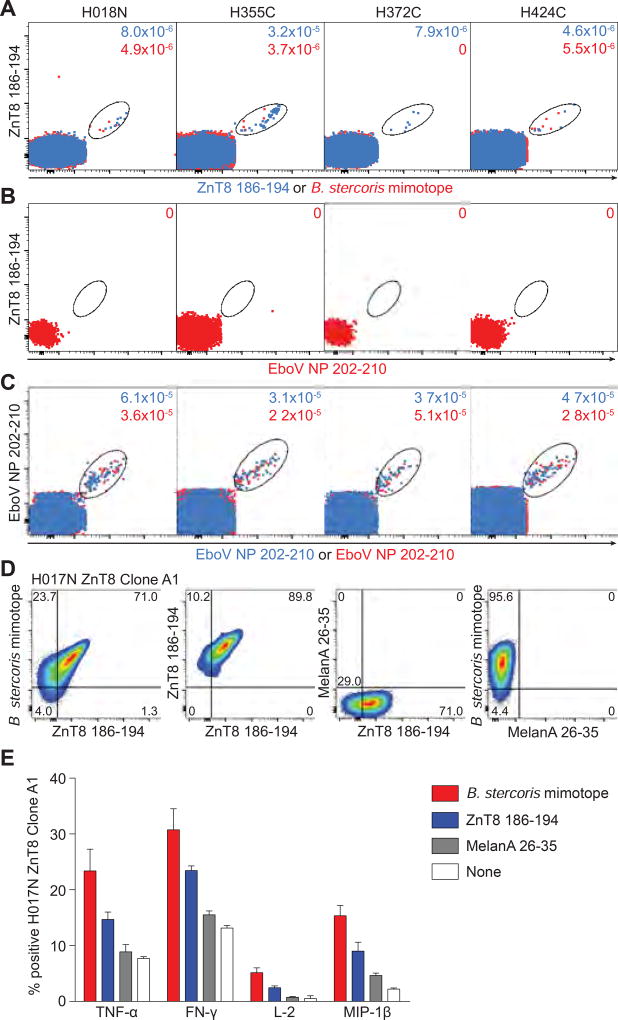
Although islet-reactive CD8+ T cells were predominantly naïve in T1D and healthy adults, substantial Ag-experienced fractions were noted in some individuals, irrespective of disease status (Fig. 5B–5E). Moreover, CD8+ T-cell frequencies correlated with the size of the Ag-experienced fraction for some individual (ZnT8186–194, IGRP265–273) and pooled islet epitopes (Fig. S13A–C), consistent with the patterns observed for Flu MP58–66-reactive CD8+ T cells (Fig. S13D). These observations raise the possibility of cross-priming by unrelated homologous epitopes (mimotopes). Indeed, sequence homology was observed between the ZnT8186–194 epitope (VAANIVLTV) and a peptide (KAANIVLTV) derived from protein WP_060386636.1 of the intestinal commensal Bacteroides stercoris. To assess potential cross-reactivity, we performed ex-vivo MMr assays on duplicate PBMC samples. One sample was stained with pairs of ZnT8186–194 MMrs, while the other was stained with one ZnT8186–194 MMr and one B. stercoris MMr. Similar frequencies were detected for ZnT8186–194-ZnT8186–194 and ZnT8186–194-B. stercoris double-MMr+CD8+ T cells in 3 of 4 donors (Fig. 7A). As a negative control, ZnT8186–194-EboV NP202–210 double-MMr+CD8+ T cells were undetectable (Fig. 7B), while EboV NP202–210-EboV NP202–210 double-MMr+CD8+ T cells were present at similar frequencies in duplicate samples (Fig. 7C). Cross-reactivity with the B. stercoris mimotope was confirmed for 1 of 4 ZnT8186–194-reactive CD8+ T-cell clones via MMr co-staining (Fig. 7D) and in-vitro recall (Fig. 7E), with the B. stercoris mimotope displaying stronger agonist potency than the native ZnT8186–194 peptide. Collectively, these results show that ZnT8186–194-reactive CD8+ T cells can cross-recognize a bacterial mimotope.
Fig. 7. ZnT8186–194-reactive CD8+ T cells cross-recognize a B. stercoris mimotope.
(A–C) Four donors with sizable ZnT8186–194 MMr+CD8+ T-cell fractions were selected. A first PBMC aliquot received PE/BV786-labeled ZnT8186–194 MMrs and PE/BV711-labeled EboV NP202–210 MMrs. For the second aliquot, PE-labeled B. stercoris MMrs replaced the PE-labeled ZnT8186–194 MMrs. (A) Overlay of ZnT8186–194/ZnT8186–194 MMr+ (blue) and ZnT8186–194/B. stercoris MMr+ cells (red). (B) Negative control staining of ZnT8186–194/EboV NP202–210 MMr+ cells. (C) Positive control staining of EboV NP202–210/EboV NP202–210 MMr+ cells from the first and second aliquot. The frequencies of MMr+ out of total CD8+ T cells are indicated. (D) Four ZnT8186–194-reactive CD8+ T-cell clones (D222D 2, D349D 178B9, H017N A1, H328C 9C8) were stained with BV786/PE-labeled ZnT8186–194, PE-labeled B. stercoris and BV650-labeled MelanA26–35 MMrs. The ZnT8186–194/B. stercoris cross-reactive clone H017N is shown, from left to right: ZnT8186–194/B. stercoris MMr+; ZnT8186–194/ZnT8186–194 MMr+; and negative control ZnT8186–194/MelanA26–35 and B. stercoris/MelanA26–35 MMr+ cells. (E) The H017N clone was stimulated with peptide-pulsed LCLs (0.1 µM, 6 h). Percent cytokine+ cells are shown as mean±SEM of two experiments. p=0.008 for Wilcoxon signed-rank comparison of pooled cytokine responses between B. stercoris and ZnT8186–194, MelanA26–35 or no peptide, and between ZnT8186–194 and MelanA26–35 or no peptide.
ZnT8186–194-reactive cells are enriched in the pancreas of T1D donors
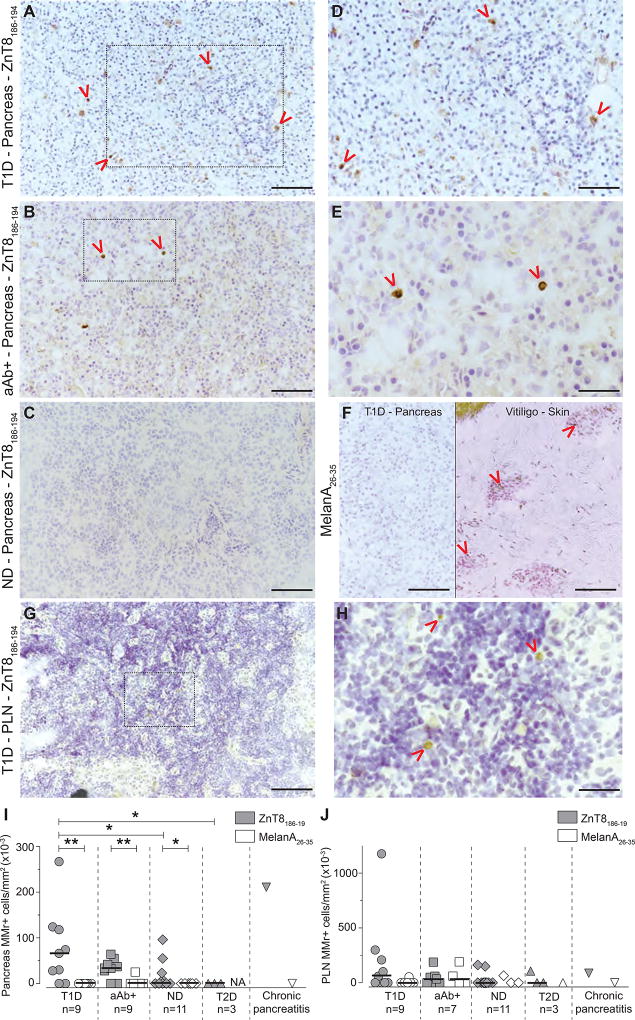
To reconcile the finding that equivalent frequencies of predominantly naïve islet-reactive CD8+ T cells circulate in most individuals, we hypothesized that the T1D-relevant fraction may be sequestered in the pancreas. We therefore performed in-situ ZnT8186–194 MMr staining on frozen pancreatic sections from HLA-A2+ T1D (n=9), aAb+ (n=9), non-diabetic (n=11) and type 2 diabetes (T2D) cases (n=3) from nPOD (Table S5). Representative images are shown in Fig. 8A–E, with scattered ZnT8186–194 MMr+ cells either within islets or the exocrine tissue. Consecutive sections from ZnT8186–194 MMr+ pancreata were probed with MelanA26–35 MMrs, which, in conjunction with positive control vitiligo skin sections, confirmed staining specificity (Fig. 8F). Donor-matched PLN sections were stained in parallel (Fig. 8G–H). While ZnT8186–194 MMr+ cells were significantly more abundant than MelanA26–35 MMr+ cells in T1D, aAb+ and non-diabetic cases, ZnT8186–194 MMr+ cells were enriched in the pancreata of T1D vs. non-diabetic and T2D cases (Fig. 8I). In contrast, ZnT8186–194 MMr+ cell were present at similar densities across all groups in PLN sections (Fig. 8J). Several of these nPOD cases were previously analyzed in-silico for the presence of ZnT8186–194 CDR3β sequences in PLNs (Fig. 4D). These sequences were present in 4/5 cases with ZnT8 MMr+ pancreata and 3/5 cases with ZnT8 MMr+ PLNs (Table S5). A donor with chronic pancreatitis (#6288) and very high ZnT8186–194-reactive CD3Rβ counts among spleen and PLN CD8+ T cells also displayed high densities of ZnT8 MMr+ cells in the pancreas. Collectively, these results show that ZnT8-reactive cells are preferentially enriched in the pancreas of T1D donors.
Fig. 8. In-situ ZnT8186–194 MMr staining of nPOD pancreas and PLN sections.
(A–E) Representative pancreas images (20X magnification; scale bar 100 µm) from cases T1D #6161 (A), aAb+ #6347 (B) and non-diabetic (ND) #6289 (C). Red arrows indicate MMr+ cells and the dotted areas of panels A–B are magnified in panels D (scale bar 80 µm) and E (scale bar 50 µm), respectively. (F) Consecutive sections from ZnT8186–194 MMr+ pancreata were probed with negative control MelanA26–35 MMrs. A representative image from T1D case #6211 is shown on the left, and a positive control staining on skin sections from a vitiligo patient is shown on the right (20X; scale bar 100 µm). (G) Representative PLN image (20X; scale bar 100 µm) from T1D case #6161. (H) Magnification of the dotted area of panel G (scale bar 40 µm). (I–J) Number of ZnT8186–194 and MelanA26–35 MMr+ cells/mm2 section area of pancreas (I) and PLNs (J). Each point represents an individual case (detailed in Table S5). Bars indicate median values. *p≤0.05, **p≤0.009 by Mann-Whitney test. NA, not assessed.
Discussion
In this study, we found that ZnT8186–194-reactive CD8+ T-cell clones exhibited heterogeneous functional profiles, but no consistent differences between T1D and healthy subjects. Most ZnT8186–194-reactive clones originated from naïve precursors and expressed private TCRs. Ex-vivo analyses on larger donor cohorts revealed that the frequency of circulating ZnT8186–194-reactive CD8+ T cells was similar in age-matched T1D vs. healthy donors, but higher in children vs. adults, irrespective of T1D status. A similar pattern was noted for CD8+ T cells recognizing the extra-pancreatic self-epitope MelanA26–35, but the corresponding ZnT8186–194-reactive populations were more Ag-experienced in T1D children. Thus, while most children harbor a larger autoimmune repertoire not restricted to islet Ags, the activation of the islet-reactive fraction occurs preferentially in T1D children, which may reflect a more aggressive islet autoimmunity leading to earlier disease onset. On the other hand, some Ag-experienced β-cell-reactive CD8+ T cells were invariably detected in healthy donors, supporting the possibility that foreign epitopes may prime autoreactive clonotypes expressing promiscuous TCRs (22–24). Indeed, some ZnT8186–194-reactive CD8+ T-cell clonotypes were able to cross-recognize a B. stercoris mimotope. It is noteworthy that B. stercoris belongs to the Bacteroidetes phylum, which is enriched in the gut microbiome of T1D (25) and at-risk aAb+ subjects (26). Circulating CD8+ T cells reactive to other HLA-A2-restricted β-cell epitopes were also detected at equivalent frequencies (1–50 MMr+ cells/106 CD8+ T cells) and with a predominantly naïve phenotype in T1D and healthy adults. This coherent pattern across islet specificities suggests that Ag-driven recruitment involves a limited fraction of naïve precursors, which fits with the paucity of public clonotypes found for ZnT8186–194-reactive CD8+ T cells, as reported for PPI15–24 (4). Interestingly, the lower frequency of Flu-reactive CD8+ T cells observed in T1D vs. healthy donors may reflect impaired anti-viral responses (27).
One strength of our ex-vivo studies is the highly specific and reproducible quantification of MMr+ cells. The observed frequencies of β-cell-reactive CD8+ T cells fall below previous estimates obtained without enrichment (4, 5), but mirror those described using stringent MMr-based magnetic enrichment (22). Although higher frequencies of β-cell-reactive CD8+ T cells have been observed in T1D vs. healthy donors (5), our data align with another study reporting no difference (4). Importantly, this study validates comparisons based on dasatinib-enhanced MMr staining. Although the T1D children in our cohort had a longer disease duration than T1D adults, comparable age-related differences were observed in healthy donors, and similar trends remained when restricting the analysis to more recently diagnosed T1D children. Other studies reporting higher Ag-experienced β-cell-reactive CD8+ T-cell fractions in T1D vs. healthy donors are potentially limited by undersampling and single MMr labeling (4). Moreover, such differences were not observed for all islet epitopes (4), and significant naïve fractions (~40%) were also present in T1D patients (4, 6).
T-cell precursors can escape thymic negative selection due to ‘blindspots’ that result from poor or incomplete expression of certain tissue Ags (13, 17, 28–30), due to alternative splicing and promoter usage and mis-initiated transcription (13–15, 30). These features favor the generation of truncated transcripts lacking certain T-cell epitopes, as observed for ZnT8. However, ZnT8186–194-reactive CD8+ T cells circulated at similar frequencies relative to other islet-reactive populations. Together with the finding that HLA-A2-restricted islet-reactive CD8+ T cells circulated at similar frequencies in HLA-A2+ and HLA-A2− donors, these results show that the thymus does not eliminate all autoreactive CD8+ T cells. Nonetheless, thymic self-Ag expression may ‘prune’ autoreactive clonotypes bearing high-affinity TCRs (22, 31).
Perhaps the most outstanding question raised by our findings relates to the ‘benign’ state of islet autoimmunity ‘licensed’ by incomplete central tolerance. Evidence for such thymic defects in human T1D has been limited to INS polymorphisms, which rank second among T1D susceptibility loci after DQB1 and modulate thymic INS expression (16). Given the ongoing debate surrounding the role of thymic negative selection for autoreactive T cells (22, 32, 33), it is not altogether surprising that even this insulin paradigm does not fully explain T1D development. Indeed, homozygous INS susceptibility alleles are present in ~55% of Caucasians (34), yet very few develop T1D. We propose that incomplete clonal deletion of autoreactive T cells involves several other β-cell Ags besides insulin and is not restricted to T1D patients.
So what distinguishes benign from pathogenic autoimmune T cells? The evidence suggests that neither their blood frequency nor their phenotype are at play. However, naïve T cells circulate perpetually, in contrast to memory T cells. The body load of Ag-experienced autoreactive T cells may therefore be underestimated in the blood of diseased individuals due to tissue sequestration (35). In line with this, the number of ZnT8186–194-reactive cells was higher than for Melan-A26–35-reactive ones in the pancreas, similar between aAb+ and non-diabetic donors, and enriched in the pancreas of T1D donors. Moreover, ZnT8186–194-reactive CD8+ T-cell clones displayed potent lytic activity, much higher than other islet specificities (1). While these findings suggest a pathogenic role for ZnT8-reactive CD8+ T cells, their precise localization and Ag-experienced status within the pancreas and whether their numbers are inversely correlated with age as observed in the blood remain to be verified.
The second possibility is that autoimmune T cells may be differentially regulated in T1D and healthy donors (33, 36), either intrinsically (e.g. anergy or exhaustion) or extrinsically by regulatory T cells. In support of this notion, T1D-specific islet-reactive CD8+ T cells have been repeatedly detected using functional IFN-γ ELISpot readouts (3, 7, 10), which employ unfractionated PBMCs, thus preserving regulatory networks. This observation also argues that peripheral blood can be informative under appropriate assay conditions. ZnT8186–194-reactive CD8+ T cells from T1D patients also expressed higher levels of aurora A kinase, which might hint at increased mitotic activity in the T1D setting. Mirroring this observation, an anergic phenotype has been reported for self-reactive CD8+ T cells in non-autoimmune subjects (22, 36), which may be at least partially imprinted in the thymus (33, 37).
The third possibility is that the central diabetogenic ingredient may be enhanced β-cell vulnerability caused by islet inflammation in the face of similar autoimmune T-cell repertoires across individuals. In this scenario, tolerance to β-cell Ags may depend primarily on T-cell ignorance (32, 37). Three observations are noteworthy in this respect. First, the higher T1D risk at younger age contrasts with the lower risk of other autoimmune diseases, which may indicate greater β-cell vulnerability due to childhood stressors, such as islet-tropic enteroviruses and the metabolic demands imposed by growth. Second, high densities of ZnT8186–194 MMr+ cells were detected in the pancreas of an organ donor with chronic pancreatitis, suggesting that islet-reactive CD8+ T cells can expand under inflammatory conditions that are not autoimmune ab initio. Third, PPI15–24-reactive CD8+ T cells were more frequent in T1D vs. healthy donors, with similar results reported for PPI2–10 (22). These epitopes map to the PPI leader sequence, which may undergo enhanced processing and presentation under metabolic stress (1).
Collectively, the present data pose new challenges toward development of circulating T-cell biomarkers for T1D staging and suggest novel therapeutic strategies based on mimicking ‘benign’ autoimmunity or complementing incomplete central tolerance (38).
Supplementary Material
Acknowledgments
We thank C. Maillard, M. Scotto and S. Rozlan for technical assistance; Univercell Biosolutions for providing the ECN90 β-cell line; K. Kedzierska and her laboratory (University of Melbourne) for help with single-cell TCR sequencing; T. Brusko (University of Florida, Gainesville) for help with the nPOD TCR database; the CyBio platform of the Cochin Institute and DKFZ Flow Cytometry Core Facility for assistance with flow cytometry; T. Loukanov (University of Heidelberg) for providing human thymic tissue; K. Boniface and J. Seneschal (INSERM U1035, Bordeaux) for providing vitiligo skin sections; and S. You for reviewing the manuscript. Members of the ImMaDiab Study Group are listed in the Supplemental Acknowledgments.
Funding: This work was performed within the Département Hospitalo-Universitaire (DHU) AutHorS, supported by the Programme Hospitalier de Recherche Clinique ImMaDiab, Lilly France, the INSERM-Transfert Proof-of-Concept program acDC, the Ile-de-France CORDDIM, and by grants from the JDRF (1-PNF-2014-155-A-V, 2-SRA-2016-164-Q-R), the Aviesan/Astra Zeneca ‘Diabetes and the Vessel Wall Injury’ program, the Société Francophone du Diabète, the Agence Nationale de la Recherche (ANR-2015-CE17-0018-01), and the Fondation pour la Recherche Médicale (Equipe FRM DEQ20140329520), to RM; a JDRF Biomarkers grant (17-2012-598), to KC; an NIH R01 grant (DK052068), to HWD; Lilly and Fondation Bettencourt-Schueller funds, to RS; a European Research Council grant (ERC-2012-AdG), to BK; and funds from JDRF, the Wellcome Trust and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, to the JDRF/Wellcome Trust Diabetes and Inflammation Laboratory (CIMR, University of Cambridge), which provided PBMC samples from the JDRF D-GAP study. Samples from at-risk subjects were obtained through a TrialNet ancillary study to the TN-01 Pathway to Prevention study funded by NIH grants U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085453, U01 DK085461, U01 DK085463, U01 DK085466, U01 DK085499, U01 DK085504, U01 DK085505, U01 DK085509, U01 DK103180, U01-DK103153, U01-DK085476, U01-DK103266 and by JDRF. DAP is a Wellcome Trust Senior Investigator (100326Z/12/Z). This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative T1D research project funded by JDRF. Organ Procurement Organizations partnering with nPOD to provide research resources are listed at www.jdrfnpod.org/our-partners.php. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (INNODIA, no. 115797). This Joint Undertaking receives support from the Union’s Horizon 2020 research and innovation programme and the European Federation of Pharmaceutical Industries and Associations, JDRF, and The Leona M. and Harry B. Helmsley Charitable Trust.
Footnotes
Author contributions: S.C. A.I.L., G.A., K.C., S.P., G.S., K.K., L.N., A.E., T.Ø., J.E.M. and M.K. performed experiments and analyzed data; A.M., K.L., E.L., J.P.B., A.L., V.A., H.W.D., S.B., D.A.P., E.B., M.B., S.C.Z., F.D. and R.S. provided critical reagents, experimental assistance and intellectual input; M.K. and R.M. performed statistical analyses; S.C., A.I.L., G.A., K.C., S.P., D.A.P., B.K. and R.M. designed and interpreted experiments, and wrote the paper; the ImMaDiab Study Group recruited patients and performed clinical phenotyping.
Competing interests: The authors declare that no conflict of interest exists. Some T-cell receptor sequences described herein are covered by Inserm-Transfert patent WO/2017/046335.
Data and materials availability: available materials will be provided with the appropriate material transfer agreement.
References and Notes
- 1.Skowera A, Ellis RJ, Varela-Calvino R, Arif S, Huang GC, Van-Krinks C, Zaremba A, Rackham C, Allen JS, Tree TI, Zhao M, Dayan CM, Sewell AK, Unger W, Drijfhout JW, Ossendorp F, Roep BO, Peakman M. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest. 2008;118:3390–3402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppieters KT, Dotta F, Amirian N, Campbell PD, Kay TW, Atkinson MA, Roep BO, von Herrath MG. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med. 2012;209:51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallone R, Martinuzzi E, Blancou P, Novelli G, Afonso G, Dolz M, Bruno G, Chaillous L, Chatenoud L, Bach JM, van Endert P. CD8+ T-cell responses identify beta-cell autoimmunity in human type 1 diabetes. Diabetes. 2007;56:613–621. doi: 10.2337/db06-1419. [DOI] [PubMed] [Google Scholar]
- 4.Skowera A, Ladell K, McLaren JE, Dolton G, Matthews KK, Gostick E, Kronenberg-Versteeg D, Eichmann M, Knight RR, Heck S, Powrie J, Bingley PJ, Dayan CM, Miles JJ, Sewell AK, Price DA, Peakman M. Beta-cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes. 2015;64:916–925. doi: 10.2337/db14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velthuis JH, Unger WW, Abreu JR, Duinkerken G, Franken K, Peakman M, Bakker AH, Reker-Hadrup S, Keymeulen B, Drijfhout JW, Schumacher TN, Roep BO. Simultaneous Detection of Circulating Autoreactive CD8+ T-Cells Specific for Different Islet Cell-Associated Epitopes Using Combinatorial MHC Multimers. Diabetes. 2010;59:1721–1730. doi: 10.2337/db09-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luce S, Lemonnier F, Briand JP, Coste J, Lahlou N, Muller S, Larger E, Rocha B, Mallone R, Boitard C. Single insulin-specific CD8+ T cells show characteristic gene expression profiles in human type 1 diabetes. Diabetes. 2011;60:3289–3299. doi: 10.2337/db11-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scotto M, Afonso G, Larger E, Raverdy C, Lemonnier FA, Carel JC, Dubois-Laforgue D, Baz B, Levy D, Gautier JF, Launay O, Bruno G, Boitard C, Sechi LA, Hutton JC, Davidson HW, Mallone R. Zinc transporter (ZnT)8(186-194) is an immunodominant CD8+ T cell epitope in HLA-A2+ type 1 diabetic patients. Diabetologia. 2012;55:2026–2031. doi: 10.1007/s00125-012-2543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinuzzi E, Afonso G, Gagnerault MC, Naselli G, Mittag D, Combadiere B, Boitard C, Chaput N, Zitvogel L, Harrison LC, Mallone R. acDCs enhance human antigen-specific T-cell responses. Blood. 2011;118:2128–2137. doi: 10.1182/blood-2010-12-326231. [DOI] [PubMed] [Google Scholar]
- 9.Hadrup SR, Bakker AH, Shu CJ, Andersen RS, van VJ, Hombrink P, Castermans E, Thor SP, Blank C, Haanen JB, Heemskerk MH, Schumacher TN. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods. 2009;6:520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 10.Enee E, Kratzer R, Arnoux JB, Barilleau E, Hamel Y, Marchi C, Beltrand J, Michaud B, Chatenoud L, Robert JJ, van Endert P. ZnT8 Is a Major CD8+ T Cell- Recognized Autoantigen in Pediatric Type 1 Diabetes. Diabetes. 2012;61:1779–1784. doi: 10.2337/db12-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lissina A, Ladell K, Skowera A, Clement M, Edwards E, Seggewiss R, van den Berg HA, Gostick E, Gallagher K, Jones E, Melenhorst JJ, Godkin AJ, Peakman M, Price DA, Sewell AK, Wooldridge L. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J. Immunol. Methods. 2009;340:11–24. doi: 10.1016/j.jim.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen M, Sauce D, Arnaud L, Fastenackels S, Appay V, Gorochov G. Evaluating cellular polyfunctionality with a novel polyfunctionality index. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinto S, Sommermeyer D, Michel C, Wilde S, Schendel D, Uckert W, Blankenstein T, Kyewski B. Misinitiation of intrathymic MART-1 transcription and biased TCR usage explain the high frequency of MART-1-specific T cells. Eur. J. Immunol. 2014;44:2811–2821. doi: 10.1002/eji.201444499. [DOI] [PubMed] [Google Scholar]
- 14.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat. Rev. Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugliese A, Zeller M, Fernandez A, Jr, Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 17.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J. Exp. Med. 2004;199:155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diez J, Park Y, Zeller M, Brown D, Garza D, Ricordi C, Hutton J, Eisenbarth GS, Pugliese A. Differential splicing of the IA-2 mRNA in pancreas and lymphoid organs as a permissive genetic mechanism for autoimmunity against the IA-2 type 1 diabetes autoantigen. Diabetes. 2001;50:895–900. doi: 10.2337/diabetes.50.4.895. [DOI] [PubMed] [Google Scholar]
- 19.Dogra RS, Vaidyanathan P, Prabakar KR, Marshall KE, Hutton JC, Pugliese A. Alternative splicing of G6PC2, the gene coding for the islet-specific glucose-6- phosphatase catalytic subunit-related protein (IGRP), results in differential expression in human thymus and spleen compared with pancreas. Diabetologia. 2006;49:953–957. doi: 10.1007/s00125-006-0185-8. [DOI] [PubMed] [Google Scholar]
- 20.Pugliese A, Brown D, Garza D, Murchison D, Zeller M, Redondo M, Diez J, Eisenbarth GS, Patel DD, Ricordi C. Self-antigen-presenting cells expressing diabetes-associated autoantigens exist in both thymus and peripheral lymphoid organs. J. Clin. Invest. 2001;107:555–564. doi: 10.1172/JCI10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rammensee HG, Bevan MJ. Evidence from in vitro studies that tolerance to self antigens is MHC-restricted. Nature. 1984;308:741–744. doi: 10.1038/308741a0. [DOI] [PubMed] [Google Scholar]
- 22.Yu W, Jiang N, Ebert PJ, Kidd BA, Muller S, Lund PJ, Juang J, Adachi K, Tse T, Birnbaum ME, Newell EW, Wilson DM, Grotenbreg GM, Valitutti S, Quake SR, Davis MM. Clonal deletion prunes but does not eliminate self-specific alphabeta CD8(+) T lymphocytes. Immunity. 2015;42:929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai N, Peng J, Liu F, Gulden E, Hu Y, Zhang X, Chen L, Wong FS, Wen L. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med. 2016;213:2129–2146. doi: 10.1084/jem.20160526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole DK, Bulek AM, Dolton G, Schauenberg AJ, Szomolay B, Rittase W, Trimby A, Jothikumar P, Fuller A, Skowera A, Rossjohn J, Zhu C, Miles JJ, Peakman M, Wooldridge L, Rizkallah PJ, Sewell AK. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J. Clin. Invest. 2016;126:2191–2204. doi: 10.1172/JCI85679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, Hyoty H, Harmsen HJ. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 26.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, Orivuori L, Hakala S, Welling GW, Harmsen HJ, Vaarala O. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diepersloot RJ, Bouter KP, Beyer WE, Hoekstra JB, Masurel N. Humoral immune response and delayed type hypersensitivity to influenza vaccine in patients with diabetes mellitus. Diabetologia. 1987;30:397–401. doi: 10.1007/BF00292541. [DOI] [PubMed] [Google Scholar]
- 28.Giraud M, Taubert R, Vandiedonck C, Ke X, Levi-Strauss M, Pagani F, Baralle FE, Eymard B, Tranchant C, Gajdos P, Vincent A, Willcox N, Beeson D, Kyewski B, Garchon HJ. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–937. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 29.Lv H, Havari E, Pinto S, Gottumukkala RV, Cornivelli L, Raddassi K, Matsui T, Rosenzweig A, Bronson RT, Smith R, Fletcher AL, Turley SJ, Wucherpfennig K, Kyewski B, Lipes MA. Impaired thymic tolerance to alpha-myosin directs autoimmunity to the heart in mice and humans. J. Clin. Invest. 2011;121:1561–1573. doi: 10.1172/JCI44583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein L, Klugmann M, Nave KA, Tuohy VK, Kyewski B. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 2000;6:56–61. doi: 10.1038/71540. [DOI] [PubMed] [Google Scholar]
- 31.Enouz S, Carrie L, Merkler D, Bevan MJ, Zehn D. Autoreactive T cells bypass negative selection and respond to self-antigen stimulation during infection. J. Exp. Med. 2012;209:1769–1779. doi: 10.1084/jem.20120905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legoux FP, Lim JB, Cauley AW, Dikiy S, Ertelt J, Mariani TJ, Sparwasser T, Way SS, Moon JJ. CD4+ T Cell Tolerance to Tissue-Restricted Self Antigens Is Mediated by Antigen-Specific Regulatory T Cells Rather Than Deletion. Immunity. 2015;43:896–908. doi: 10.1016/j.immuni.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis MM. Not-So-Negative Selection. Immunity. 2015;43:833–835. doi: 10.1016/j.immuni.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Bennett ST, Todd JA. Human type 1 diabetes and the insulin gene: principles of mapping polygenes. Annu. Rev. Genet. 1996;30:343–370. doi: 10.1146/annurev.genet.30.1.343. [DOI] [PubMed] [Google Scholar]
- 35.Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, Farber DL. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeda Y, Nishikawa H, Sugiyama D, Ha D, Hamaguchi M, Saito T, Nishioka M, Wing JB, Adeegbe D, Katayama I, Sakaguchi S. Detection of self-reactive CD8(+) T cells with an anergic phenotype in healthy individuals. Science. 2014;346:1536–1540. doi: 10.1126/science.aaa1292. [DOI] [PubMed] [Google Scholar]
- 37.Malhotra D, Linehan JL, Dileepan T, Lee YJ, Purtha WE, Lu JV, Nelson RW, Fife BT, Orr HT, Anderson MS, Hogquist KA, Jenkins MK. Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol. 2016;17:187–195. doi: 10.1038/ni.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culina S, Gupta N, Boisgard R, Afonso G, Gagnerault MC, Dimitrov J, Osterbye T, Justesen S, Luce S, Attias M, Kyewski B, Buus S, Wong FS, Lacroix-Desmazes S, Mallone R. Materno-fetal transfer of preproinsulin through the neonatal Fc receptor protects from autoimmune diabetes. Diabetes. 2015;64:3532–3542. doi: 10.2337/db15-0024. [DOI] [PubMed] [Google Scholar]
- 39.Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, Claesson M, Buus S, Stryhn A. One-pot, mix-and-read peptide-MHC tetramers. PLoS One. 2008;3:e1678. doi: 10.1371/journal.pone.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravassard P, Hazhouz Y, Pechberty S, Bricout-Neveu E, Armanet M, Czernichow P, Scharfmann R. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 2011;121:3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr. Protoc. Immunol. 2011 doi: 10.1002/0471142735.im1033s94. Chapter 10, Unit 10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T cell receptor alphabeta diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003647. 128ra142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonifacio E, Ziegler AG, Klingensmith G, Schober E, Bingley PJ, Rottenkolber M, Theil A, Eugster A, Puff R, Peplow C, Buettner F, Lange K, Hasford J, Achenbach P, Group P-PS. Effects of high-dose oral insulin on immune responses in children at high risk for type 1 diabetes: the Pre-POINT randomized clinical trial. JAMA. 2015;313:1541–1549. doi: 10.1001/jama.2015.2928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.