Abstract
Despite the wide availability of antibiotics, infectious diseases remain a leading cause of death worldwide.1 In the absence of new therapies, mortality rates due to untreatable infections are predicted to rise more than 10-fold by 2050. Natural products (NPs) made by cultured bacteria have been a major source of clinically useful antibiotics. In spite of decades of productivity, the use of bacteria in the search for new antibiotics was largely abandoned due to high rediscovery rates.2, 3 As only a fraction of bacterial diversity is regularly cultivated in the laboratory and just a fraction of the chemistries encoded by cultured bacteria is detected in fermentation experiments, most bacterial NPs remain hidden in the global microbiome. In an effort to access these hidden NPs, we have developed a culture-independent NP discovery platform that involves sequencing, bioinformatic analysis, and heterologous expression of biosynthetic gene clusters (BGCs) captured on DNA extracted from environmental samples (eDNA). Here, we describe the application of this platform to the discovery of the malacidins, a distinctive class of antibiotics that are commonly encoded in soil microbiomes but have never been reported in culture-based NP discovery efforts. The malacidins are active against multidrug-resistant (MDR) pathogens, sterilize MRSA skin infections in an animal wound model, and did not select for resistance under our laboratory conditions.
Keywords: drug discovery, environmental DNA, metagenomics, calcium-dependent antibiotics, natural product
Calcium-dependent antibiotics are a small family of N-acylated cyclic peptides that require calcium for antibacterial activity. Known members of this family contain a conserved Asp-X-Asp-Gly motif that is thought to facilitate calcium binding.4–6 One example, daptomycin, has proved useful clinically in the treatment of multidrug-resistant bacteremia. Calcium-dependent antibiotics were of particular interest to us because individual family members have been shown to have discrete modes of action, targeting either cell wall biosynthesis or cell membrane integrity. Given these differences in modes of action, we hypothesized that the conserved Asp-X-Asp-Gly calcium-binding motif might be indicative of a broader collection of uncharacterized, bacterially encoded antibiotics with diverse mechanisms of action and therapeutic potential.
To test this hypothesis, we undertook a sequence-guided screen of diverse soils for BGCs that encode calcium-binding motifs. Due to the complexity of soil metagenomes, it remains challenging to shotgun sequence deep enough to generate data that is broadly useful for BGC discovery.7 We have developed sequencing strategies that rely on the barcoding of biosynthetic genes using degenerate PCR primers to parse mixtures of BGCs present in environmental samples.7, 8 In this approach, primers targeting conserved NP biosynthetic genes are used to generate PCR amplicon pools containing homologous genes from BGCs present in an eDNA sample (Fig. 1a). Individual next-generation sequencing reads derived from these amplicons (NP sequence tags, NPSTs) are used to predict BGCs present in a sample by comparing them to a database of sequences from characterized BGCs. This analysis is carried out using the bioinformatics platform eSNaPD (environmental Surveyor of Natural Product Diversity: http://esnapd2.rockefeller.edu) that was developed to evaluate metagenome-derived NPSTs.8, 9
Figure 1. Using a culture-independent strategy for the discovery of calcium-dependent antibiotics from the global microbiome.
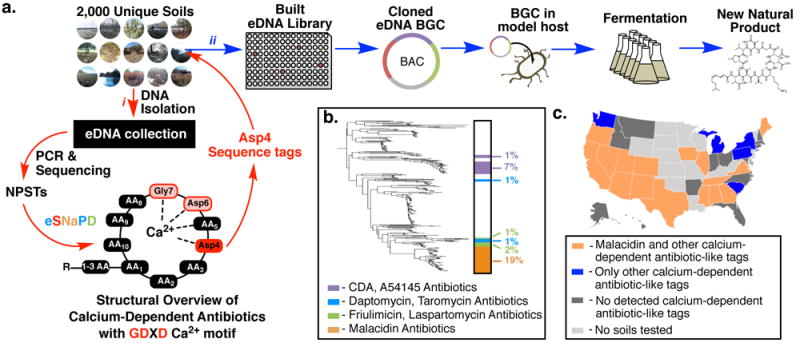
(a) i) Degenerate PCR primers targeting the conserved regions of adenylation domains (AD) found in nonribosomal peptide synthetase genes were used to generate amplicons from an arrayed collection of environmental DNA isolated from 2000 unique soils. The reads from these next-generation sequenced amplicons (natural product sequence tags, NPSTs) were analyzed by eSNaPD (environmental Surveyor of Natural Product Diversity). ii) A desert soil rich in AD-NPSTs from the previously unknown malacidin clade was used to build an arrayed cosmid library. Cosmids harboring all fragments of a targeted biosynthetic gene cluster were assembled and integrated into a heterologous host for production, extraction, and characterization. (b) AD-NPSTs identified by the eSNaPD analysis to be evolutionarily related to the conserved Asp4 ADs of known calcium-dependent antibiotics were used to phylogenetically map the unexplored clades of this larger family across all tested soil microbiomes. The subfamilies of calcium-dependent antibiotics and their relative abundance are illustrated on the phylogenetic tree by color and percent. Across all sampled soil metagenomes, the malacidin antibiotic-clade represents 19% of the NPSTs, and 59% of calcium-dependent antibiotic tags originate from unexplored branches. (c) Geospatial distribution of calcium-dependent antibiotics across sampled US soil metagenomes. US states containing at least one soil with AD-NPSTs from the malacidin clade are indicated in orange. States lacking malacidin tags but still containing calcium-dependent antibiotics NPSTs are indicated in blue. States with at least one sampled soil but no detected calcium-dependent antibiotics NPSTs are highlighted in dark grey.
Known calcium-dependent antibiotics are biosynthesized by nonribosomal peptide synthetases (NRPS). Accordingly, we used primers targeting NRPS adenylation domains (ADs) to track this family of NPs across diverse soil microbiomes. For this study, and as part of our ongoing soil metagenome-driven NP discovery efforts, we expanded our soil collection to over 2,000 soils from ecologically and geographically diverse environments.8 Even using a conservative estimate of 103 unique bacterial species per gram of soil,2 we expect the diversity of bacteria present in this collection to rival that of the largest culture collections. Initially, primers targeting NRPS-ADs were used to screen eDNA isolated from small aliquots of each soil to identify environments predicted to contain gene clusters that encode for unidentified calcium-dependent antibiotics.
Three-quarters of sequenced soils had NPSTs that mapped to at least one adenylation domain from a known calcium-dependent antibiotic biosynthetic gene cluster (Fig. 1b–c, Supplementary Fig. S1). Only 13% of these identified NPSTs cluster at ≥ 95% nucleotide identity to ADs found in characterized calcium-dependent antibiotics and less than 30% of them are found in more than one soil metagenome. Taken together, this indicates that the majority of lipopeptides encoded by the global soil metagenome are likely uncharacterized and that even within our large soil collection, we have only captured a fraction of the biosynthetic diversity that exists within the calcium-dependent antibiotic family.
Phylogenetic analysis of AD sequences from characterized calcium-dependent antibiotics indicated that the domain responsible for incorporating the first aspartic acid (Asp4) in the conserved Asp-X-Asp-Gly motif most closely mapped to functional divergence of BGCs in this family (Supplementary Fig. S1b). We, therefore, focused on eSNaPD data for this domain to track calcium-dependent antibiotic BGCs. A phylogenetic tree derived from tags associated with this domain showed numerous clades not associated with known BGCs, indicating the existence of uncharacterized calcium-dependent antibiotics in soil microbiomes. One distinct, eDNA-specific clade was found in 19% of metagenomes (Fig. 1b), suggesting that the BGCs associated with these tags belong to an abundant and yet uncharacterized family of antibiotics, which we have called the malacidins (metagenomic acidic lipopeptide antibiotic–cidins).
To recover a complete malacidin BGC for use in heterologous expression studies, we retrieved from our soil archive a desert soil (DFD0097) that was rich in NPSTs from the malacidin branch of the AD phylogenetic tree (Fig. 1c). Employing standard soil metagenome cloning methods, a saturating cosmid library was constructed using DNA extracted from DFD0097 soil.10 This 20 million membered library was archived as purified cosmid DNA and E. coli glycerol stocks arrayed in 96-well format, with each library well containing ~20,000 unique clones.10, 11 To expedite the recovery of BGCs, each well of the library was individually screened by PCR using the barcoded AD-targeting primers used to profile soils. Library-derived NPST data were analyzed by eSNaPD to generate a map of BGC information across the arrayed library.
Using this BGC prediction map, overlapping cosmid clones predicted to contain the malacidin BGC were recovered from the library. Sequencing and in silico analysis of these clones suggested that the malacidin BGC spanned 72 kb across three cosmids (DFD0097-644, DFD0097-735, DFD0097-388) (Fig. 2a, Supplementary Table S1, GenBank Accession KY654519). For the purposes of heterologous expression, these three overlapping cosmids were assembled into a contiguous fragment of DNA using transformation-associated recombination in yeast and the E. coli:yeast:Streptomyces shuttle vector, pTARa (Fig. 2b).11 The resulting bacterial artificial chromosome (DFD0097-644:735:388) and the empty pTARa vector were separately conjugated into Streptomyces albus J1074. Extracts from cultures of S. albus harboring DFD0097-644:735:388 were found to exhibit antibacterial activity against Staphylococcus aureus and contain clone specific metabolites (Fig. 2c & 2d). The major clone specific metabolites, malacidin A and B, were isolated from cultures of S. albus DFD0097-644:735:388 and their structures were elucidated using a combination of MS and NMR data. The malacidin structures were supported by a detailed bioinformatic analysis of the BGC (Fig. 2e, Supplementary Fig. S2–S25 and Table S2–S3, Supplemental Discussion).
Figure 2. Maladicin biosynthesis, heterologous expression and structure.
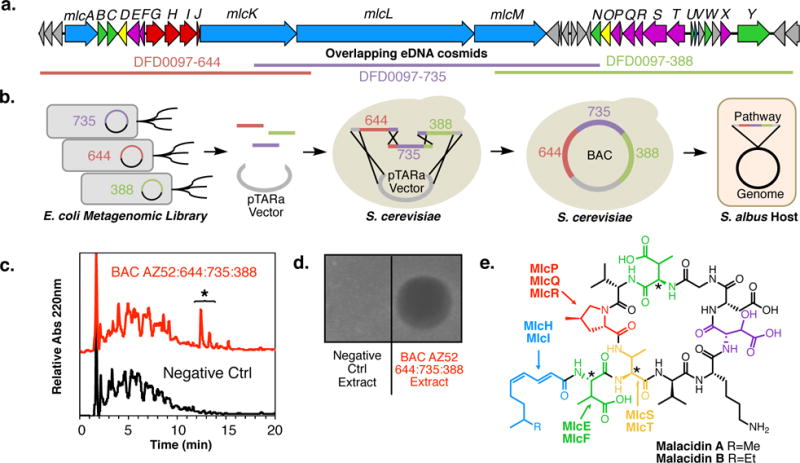
(a) The malacidin biosynthetic gene cluster was recovered on three overlapping cosmid clones and (b) assembled from these three overlapping clones in yeast using transformation-associated recombination (TAR). The resulting bacterial artificial chromosome (BAC) was integrated into S. albus genome for heterologous expression studies. (c) A representative HPLC analysis of crude extracts derived from cultures of S. albus transformed with the malacidin biosynthetic gene cluster (BGC) shows the presence of BGC-specific small molecules. The two primary malacidin peaks are highlighted with an asterisk. (d) Unlike crude extracts of the S. albus host strain alone, only extracts from the S. albus harboring the malacidin BGC showed antibacterial activity when applied to a lawn of S. aureus USA300. Both the (c) HPLC analysis and (d) antibacterial activity are representative of 4 independent fermentations. (e) Malacidin A and B are cyclic lipopeptides containing 8 amino acid macrocycles and polyunsaturated lipids. The malacidins do not contain the conserved DXDG motif seen in all known calcium-dependent antibiotics – incorporating a rare 3-hydroxyl aspartic acid (HyAsp, highlighted in violet) and lacking the spacer residue. Biosynthetic enzymes predicted to be involved in the production of non-proteinogenic amino acid [3-methyl aspartic acid (MeAsp), 4-methyl proline (MePro), and 2,3-diamino 3-methyl propanoic acid (MeDap)] and fatty acid substrates required for the biosynthesis of the maladicins are shown and colored coded according to their activities. Stereocenters in malacidin that were predicted bioinformatically as opposed to through chemical and spectroscopic analysis are denoted with an asterisk.
The malacidins are 10-membered cyclic lipopeptides that only differ by a methylene on the branch at the terminus of their lipid tails. Their peptide cores include 4 nonproteinogenic amino acids (Fig. 2e). Calcium-dependent antibiotics characterized from culture-based discovery programs contain larger, 11 to 13 amino acid rings and completely distinct peptide sequences (Supplementary Fig. S24). The malacidins do not contain the canonical Asp-X-Asp-Gly calcium-binding motif found in known calcium-dependent antibiotics.4 They lack the variable spacer residue found in this canonical motif and contain a ASP-OH, suggesting they either no longer bind calcium or may represent a different calcium-binding motif.4–6 To determine the requirement of calcium for the antibacterial activity of the malacidins, we tested for antibiosis against methicillin-resistant Staphylococcus aureus (MRSA) across a range of calcium concentrations. In these assays, we saw a clear dependence on calcium for antibiosis, indicating that although the malacidins do not contain a canonical Asp-X-Asp-Gly motif, their antibacterial activity remains calcium-dependent (Fig. 3a). Similar experiments using cations other than calcium showed no antibiosis (Supplementary Fig. S26). The malacidins are broadly active against Gram-positive bacteria including MDR pathogens and bacteria resistant to mechanistically diverse, clinically used antibiotics (Table 1, Supplementary Table S4). As the most common form of Staph infections occur on the skin, we elected to test the in vivo efficacy of the malacidins using an animal wound model. Topical administration of malacidin A was successful in sterilizing MRSA infected wounds in a rat model (Fig. 3b). At 24 and 72 hours post infection, malacidin A treatment resulted in no observed bacterial burdens in the wounds. The vehicle treated controls had an average of 5.5 log and 7.0 log of MRSA at 24H and 72H, respectively (Kruskal-Wallis Pvalue <.0001). Likewise, the malacidins showed no significant toxicity or hemolytic activity against mammalian cells at the highest concentrations tested (100–250 μg ml−1, >100 MIC) (Supplementary Fig. S27 and Table S3). Unlike daptomycin, which is unable to treat severe community-acquired pneumonia due to loss of activity in the presence of pulmonary surfactants,12 malacidin A does not share this liability (Fig. 3c). Our experimental efforts to induce resistance to malacidin in the laboratory have so far been unsuccessful. Even after 20 days of exposure to sub-lethal levels of malacidin A, we did not detect any malacidin-resistant S. aureus (Fig. 3d). Whether resistance can arise through horizontal gene transfer from environmental bacteria remains to be seen.
Figure 3. Malacidin, a calcium-dependent antibiotic.
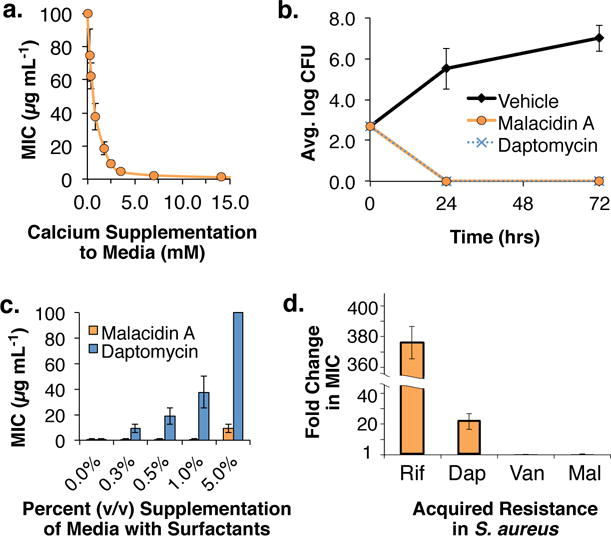
(a) The MIC of malacidin A against MRSA was assessed at various concentrations of calcium and the antibiosis of malacidin A was found to be calcium-dependent. Error bars represent the standard deviation across two replicates over three independent experiments (n = 6). (b) Malacidin A is an effective treatment against MRSA in rat cutaneous wound infections. Error bars represent the standard deviation across replicate wounds (n = 4). (c) Unlike daptomycin, malacidin A activity against S. pneumoniae is largely unaffected by the presence of pulmonary surfactants. Error bars represent the standard deviation across two replicates over three independent experiments (n = 6). (d) After 20 days of repeated exposure to 0.5= MIC of malacidin A (Mal), we did not detect any malacidin-resistant S. aureus. Vancomycin (Van), Daptomycin (Dap), and Rifamycin (Rif) were used as controls in this assay. Error bars represent the standard deviation across three replicates for MIC determination (n = 3).
Table 1. Spectrum of Activity of Malacidin A.
These data in the table are representative of the range of values determined in at least three independent experiments.
| Organism | Acquired Resistance | MIC (μg mL−1) | IC50 (μg mL−1) |
|---|---|---|---|
| S. aureus USA300 | β-lactams (Methicillin, Oxacillin, Penicillin) | 0.2–0.8 | |
| S. aureus USA300 + 10% serum | β-lactams (Methicillin, Oxacillin, Penicillin) | 0.2–0.8 | |
| S. aureus COL | β-lactams | 0.2–0.8 | |
| S. aureus BAA-42 | β-lactams | 0.2–0.8 | |
| S. aureus NRS100 | β-lactams, Tetracycline | 0.2–0.8 | |
| S. aureus NRS108 | β-lactams, Gentamicin, Kanamycin | 0.2–0.8 | |
| S. aureus NRS140 | β-lactams, Erythromycin, Spectinomycin | 0.4–0.8 | |
| S. aureus NRS146 | β-lactams, Vancomycin (VISA) | 0.4–0.8 | |
| E. faecium VRE | Vancomycin (VRE) | 0.8–2.0 | |
| E. faecium Com15 | 0.8–2.0 | ||
| S. pneumoniae | 0.1–0.2 | ||
| S. mutans | 0.1–0.2 | ||
| B. subtilis | 0.2–0.4 | ||
| L. rhamnosus | 0.1–0.2 | ||
| E. coli | >100 | ||
| C. albicans | >100 | ||
| C. neoformans | >100 | ||
| HEK293 | >100 | ||
| MRC5 | >100 |
Characterized calcium-dependent antibiotics function by one of two distinct modes of action (Fig. 4a). Daptomycin displays rapid bactericidal activity by binding cytoplasmic membrane phosopholipids and oligomerizing in the membrane.13, 14 This affects phospholipid synthesis and overall membrane fluidity, ultimately leading to decreased membrane integrity and cell death.15 The potent antibacterial activity of friulimicin and its structural relatives is due to inhibition of bacterial cell wall biosynthesis through binding of the Lipid II precursor, undecaprenyl phosphate (C55-P).16, 17 As the malacidins are structurally distinct from other calcium-dependent antibiotics, we sought to determine whether they function by one of these known mechanisms or a third distinct mode of action. We first assessed the effect of malacidin on membrane integrity (Fig. 4b, Supplementary Fig. S28). No membrane leakage was observed when S. aureus cells pretreated with SYTOX Green or DiBAC4 were exposed to either daptomycin or malacidin in the absence of calcium supplementation. With the addition of calcium, daptomycin-treated S. aureus showed a rapid increase in fluorescence, which is indicative of a loss of membrane integrity. Malacidin, however, did not demonstrate the same effect indicating malacidin and daptomycin have distinct modes of action.
Figure 4. Malacidin mode of action.
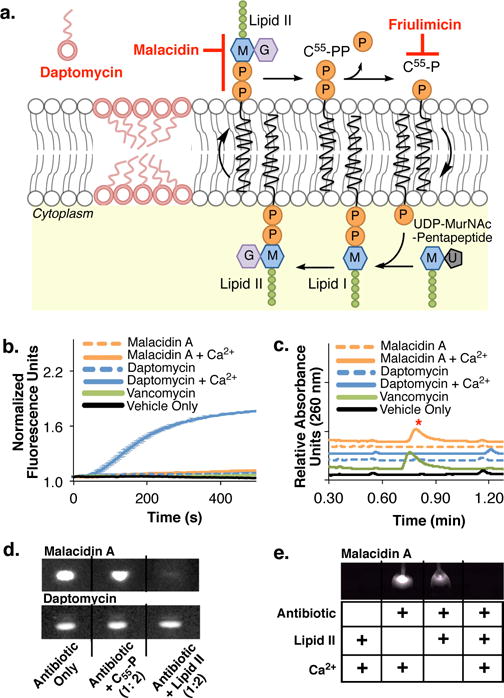
(a) Cartoon showing modes of action of daptomycin, friulimicin and malacidin. (b) In contrast to daptomycin, malacidin A does not cause MRSA membrane leakage in a SYTOX Green fluorescent assay. Error bars represent the standard deviation across three biological replicates (n = 3). (c) As seen with the cell wall biosynthesis inhibitor vancomycin, exposing MRSA to malacidin A results in the accumulation of the cell wall intermediate UDP-MurNAc-pentapeptide. The UDP-MurNAc-pentapeptide peak ([M-H]- = 1148.35) is indicated with a red asterisk on the UPLC- trace. Chromatograms are representative of at least three independent experiments. (d) Interaction of malacidin A and daptomycin with purified cell wall precursors. An interaction is indicated by a reduction of the amount of free antibiotic (visible on the thin-layer chromatography by UV light). (e) The interaction of malacidin A to cell wall precursor, lipid II, is calcium-dependent. Both TLCs in (d) and (e) are representative of at leas two replicate experiments.
As seen with friulimicin and other cell wall intermediate binding antibiotics, S. aureus treated with malacidin accumulates the cell wall precursor undecaprenyl-N-acetylmuramic acid-pentapeptide (UDP-MurNAc-pentapeptide) (Fig. 4c).16, 17 This signaled that the target of malacidin, like that of friulimicin, lies downstream of UDP-MurNAc-pentapeptide formation. Surprisingly, in a TLC mobility shift assay, malacidin did not sequester C55-P, the target of fruilimicin (Fig. 4d).16, 17 Lipid II is the key downstream intermediate of MurNAc-pentapeptide. We therefore tested malacidin for lipid II-binding activity. In this TLC-based mobility shift assay we observed lipid II-dependent disappearance of the malacidin band (Fig. 4d and 4e). Unlike previously characterized calcium-dependent antibiotics, malacidin neither depolarizes the membrane nor binds C55-P but instead appears to interact with lipid II in a calcium-dependent manner. Fortuitously, despite the fact that vancomycin also binds lipid II, the malacidins are active against both vancomycin-intermediate and vancomycin-resistant pathogens.
The malacidins exhibited potent antibacterial activity against Gram-positive pathogens resistant to clinically used antibiotics, including the antibiotic of last resort vancomycin, and did not select for resistance in the laboratory under the conditions of our experiments. The discovery of the malacidins supports our hypothesis that the calcium-dependent antibiotics are a larger than previously thought family of NPs with low susceptibility to resistance and diverse modes of action. Environmental microbes are in a continuous antibiotic arms race that is likely to select for antibiotic variants capable of circumventing existing resistance mechanisms. The sequence-guided metagenomic discovery pipeline outlined here provides a means to interrogate complex environmental metagenomes for these uncharacterized antibiotics by tracking NPSTs that differ from those associated with known antibiotic biosynthetic gene clusters. While metagenome-based antibiotic discovery methods are still in their infancy, the scaling and automation of the pipeline described here should permit the systematic discovery of natural product antibiotics that have until now remained hidden in the global metagenome, providing a potentially powerful approach for combating antibiotic resistance.
Material and Methods
NPST Generation and Sequencing
To add to the diversity of the 185 previously collected soil samples8, 18, an additional 1800 soils were collected for this study from sites throughout the United States. Crude eDNA was extracted from each of these following established protocols.10, 18 Briefly: 25 grams of soil were heated (70 °C) in lysis buffer (100 mM Tris-HCl, 100 mM EDTA, 1.5 M NaCl, 1% (w/v) CTAB, 2% (w/v) SDS, pH 8.0) for 2 hours. Soil particulates were removed from the crude lysate by centrifugation, and eDNA was precipitated from the resulting supernatant with the addition of 0.7 volumes isopropanol. Crude eDNA was collected by centrifugation, washed with 70% ethanol and resuspended in TE. Crude eDNA was then spin column purified (PowerMax soil DNA kit) and employed as template in PCR experiments targeting AD domains as follows: A-domain fragments (~795 bp) were amplified using primers: 5′- GCSTACSYSATSTACACSTCSGG and 5′-SASGTCVCCSGTSCGGTA. These primers are designed to recognize the conserved regions in NRPS A-domains.8, 18 The 5′ ends of the primers were augmented with MiSeq sequencing adapters followed by unique 8 bp barcode sequences identifying the soil metagenome from which they were amplified. PCR conditions: 12 μl reaction, 1× Buffer G (Epicentre), 50 pmol of each primer, 2.5 units Omni Klentaq polymerase (DNA Polymerase Technology) and 100 ng eDNA. Cycle conditions for AD amplification: 95 °C 4 min, (95 °C 30 s, 63.5 °C 30 s, 72 °C 45 s) × 34 cycles, 72 °C 5 min. First-round amplicons contained incomplete Illumina adaptors and therefore required a second round of PCR to append the remainder of the adaptor sequence.19 Amplicons were pooled as collections of 96 samples and cleaned using Agencourt Ampure XP magnetic beads (Beckman Coulter). Cleaned, pooled amplicons were used as template in a second 20-μL PCR using the following reaction conditions: 10 μL of FailSafe Buffer G (Epicenter), 5.8 μL of water, 0.4 μL of each primer (100 μM) (MiSeqForward, CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT; MiSeq Reverse AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT), 0.4 μL of Taq, and 3 μL of cleaned amplicon (50 ng to 100 ng). Amplification proceeded as follows: 95 °C for 5 min, six cycles of 95 °C for 30 s, 70 °C for 30 s, and 72 °C for 45 s, and, finally, 72 °C for 5 min. Prior to sequencing all PCR amplicons were quantified by gel electrophoresis and mixed in an equal molar ratio. The resulting pool was fluorometrically quantified with a HS D1000 ScreenTape (Agilent 2200 TapeStation; Agilent Technologies) and sequenced on an Illumina MiSeq insrument using Reagent Kit v3 (MS-102-3003, Illumina).
Biosynthetic Profiling of NPSTs
Amplicon sequences were analyzed and organized using the eSNaPD (environmental Surveyor of Natural Product Diversity) web based tool as previously described.8, 9, 18 The NRPS-ADs from sequenced and known calcium-dependent antibiotic gene clusters (daptomycin, friulimicin, CDA, laspartomycin, A54145, and taromycin) were added to the eSNaPD reference database of domains from annotated and functionally characterized natural product gene clusters. NPSTs whose closest relatives among all reference ADs were one of these known lipopeptide domains were identified and mapped to soil locations and/or library wells. This analysis, in brief, was completed as previously described19 by debarcoding samples using a paired-end 2×8bp barcode strategy. Debarcoded reads were filtered for quality and 240bp of the forward reads, a single “N” spacer, and 175 bp of the reverse complemented reverse read were concatenated to generate a synthetic amplicon of 416 bp. The reads from each sample were clustered using UCLUST20 to generate the 95% identity centroid sequences (i.e., NPST). Location information was used to map NPSTs back to soil collection locations and/or library wells. The forward read component (the first 240bp) of each NPST were then searched using BlastN21 against a manually curated database of NRPS-AD sequences. NPSTs that returned one of the known calcium-dependent antibiotics as a top match were considered hits. The resulting set of unique hits was used to generate geographic and phylogenetic distribution figures. A multiple sequence alignment of all sequences was generated using MUSCLE22, and the resulting alignment file used to generate a maximum likelihood tree with FastTree.23
Construction and Arraying of Metagenomic Cosmid Libraries
eDNA cosmid libraries were constructed from soil using established protocols.10 Briefly, crude eDNA was isolated from ~0.5 kg of soil as outlined above, and further purified by preparative agarose gel electrophoresis to yield pure high-molecular-weight (HMW) eDNA. HMW eDNA was blunt ended (Epicentre, End-It), ligated into pWEB-TNC (Epicentre), packaged into lambda phage and transfected into E. coli EC100 (Epicentre). Following recovery, transfected cells were inoculated into 8 ml LB with selective antibiotic (12.5 μg mL−1 chloramphenicol) in 24-well plates at a density of ~25,000 clones per well and grown overnight. Matching glycerol stocks and cosmid DNA minipreps were prepared from each well, and arrayed as 768 pools of ~25,000 unique cosmid clones. NPST data was prepared from each library pool by amplifying and sequencing A-domains as described above.
Recovery of Biosynthetic Gene Clusters from eDNA Libraries
Calcium-dependent antibiotic-like NPST sequences identified within metagenomic libraries were automatically assigned to library wells by the barcode parsing functionality of the eSNaPD software package as described above. Specific primers targeting each unique sequence of interest were designed by hand. To recover single clones from library wells, a serial dilution PCR strategy18 was used as follows: Library wells containing targets as one of ∼25,000 unique cosmids were grown overnight to confluence in LB (12.5 μg mL−1 chloramphenicol, 100 μg mL−1 carbenicillin) and diluted to a concentration of 3000 CFU mL−1 as judged by OD600. Then, 384 well plates were inoculated with 100 μL (300 CFU) of the resulting dilution per well, grown to confluence, and screened using real-time PCR, to identify wells containing target clones as 1 of ∼300 clones. Target positive wells were then diluted to a concentration of ∼50 CFU mL−1 and the process repeated to identify wells containing targets as 1 of ∼5 clones. Five clone pools were then plated on solid medium, and target clones identified by colony PCR.
In silico Analysis of Recovered Gene Clusters
Recovered single cosmid clones were pooled and sequenced using ion PGM technology. Reads were assembled into contigs using an assembler program, such as Newbler.24 Overlapping cosmids spanning a single pathway were initially sequenced separately and subsequently assembled into larger contigs. Assembled contigs were then annotated using an in-house pipeline consisting of open reading frame (ORF) predictions with MetaGeneMark25, BLAST search21, and AntiSMASH predictions26 The AntiSMASH predictions employ three prediction algorithms to call the amino acid substrate specificity of an adenylation domain (NRPSPredictor2, Stachelhaus code, and Minowa). These amino acid predictions were used in the initial bioinformatic characterization of clusters to predict chemical structures. Putative functions for new tailoring enzymes in eDNA pathways were assigned based on the predicted function of the closest characterized relative identified by Blast in NCBI.
Assembly of DFD0097-735 pTARa BAC for Heterologous Expression
For assembly of the DFD0097-735 pTARa bacterial artificial chromosome (BAC), transformation-associated recombination (TAR) in yeast was employed.11, 27 Initially, the three overlapping cosmids (DFD0097-644, DFD0097-735, DFD0097-388) containing the full biosynthetic pathway were digested and linearized with DraI. A custom E. coli:yeast:Streptomyces shuttle capture vector, pTARa, containing two 500 bp homology arms to the terminal overlapping cosmid clones was constructed as previously described.11, 27 This vector was subsequently linearized with PmeI and gel purified. The linearized cosmids and capture vector were then cotransformed into S. cerevisiae (BY4727 Δdnl4) using a standard LiAc/ss carrier DNA/PEG yeast transformation protocol.28. Briefly, yeast were grown overnight in 50 mL of YPD media containing G418 (200 μg mL−1) at 30 °C. In the morning, 2 mL of the overnight culture were reinoculated into 50 mL of fresh YPD media containing G418 (200 μg/mL) and grown for ∼4 h (OD600 = 2.0). This culture was harvested by centrifugation (10 min, 3,200 × g), washed twice with sterile 4 °C water, and resuspended in 1 mL of sterile 4 °C water. For each transformation 100 μL of washed cells was transferred to a microfuge tube. The cells were collected by centrifugation (30 s, 18,000 × g) and resuspended in a transformation mix containing 36 μL of 1 M LiAc solution, 50 μL of 2 mg/mL carrier DNA (Salmon sperm DNA) solution, 240 μL of 50% (wt/vol) PEG 3350 solution, and 34 μL of Tris-EDTA containing 2 μg of each cosmid and 1 μg of vector. This transformation mix was incubated at 42 °C for 40 min. Cells were then collected by centrifugation (30 s, 18,000 × g), resuspended in 100 μL of water, and plated on appropriate synthetic composite dropout media agar plates. Agar plates were incubated at 30 °C until colonies appeared. Colonies were checked by PCR. DNA was isolated from PCR-positive yeast clones, transferred into E. coli ET12567/pUZ8002 cells, and then moved into Streptomyces spp. by intergeneric conjugation for heterologous expression.
Heterologous Expression
The assembled BAC, DFD0097-735 pTARa, and an empty pTARa vector control, were separately integrated into the chromosome of Streptomyces albus J1074. Spore suspensions of these recombinant strains were used to seed starter cultures in 50 mL trypticase soy broth (Oxoid). These cultures were grown for 48 h (30 °C/200 rpm) and 0.4 mL of the resulting confluent culture was used to inoculate 50 mL flasks of production media: R5a medium – 100 g L−1 sucrose, 0.25 g L−1 K2SO4, 10.12 g L−1 MgCl2, 10.0 g L−1 D-glucose, 0.1 g L−1 casamino acids, 21 g L−1 MOPS, 2 g L−1 NaOH, 40 μg L−1 ZnCl2 20 μg L−1 FeCl3 6H2O, 10 μg L−1 MnCl2, 10 μg L−1 (NH4)6Mo7O24 4H2O. 50 mL liquid cultures were grown in 125 mL baffled flasks (22°C, 220 rpm) for 14 days.
Isolation of Malacidin A and B
After 14 days, 4 L of cultures were combined, and mycelia were removed by centrifugation at 4,000 × g for 20 minutes. The mycelia-free media supernatant was applied to 150 g of pre-equilibrated Diaion HP-20 resin packed in a column (40 × 220 mm). The HP-20 column was subsequently washed with 2 L of H2O, and then eluted with 2 L of 100 % MeOH. The methanolic elution was concentrated by rotary evaporation, and then combined with 2 g octadecyl-functionalized silica resin (Sigma-Aldrich) per 10 mL concentrate. This resin:concentrate mixture was dried overnight on a Savant Speedvac Concentrator (Thermo-Fisher). The dried-loaded resin was used to dry load a 100 g Gold HP C18 column for medium-pressure reversed-phase chromatography (Teledyne Isco Combiflash Rf150). This chromatography was performed using a linear gradient of 0.1 % acetic acid—acetonitrile from 10–100 % over 20 minutes at 60 mL min−1. To identify column fractions containing active compound, aliquots of 10 mL fractions were analyzed by UPLC-MS. Fractions containing the malacidins were pooled, re-dried on resin, and subjected to a second-round medium-pressure, narrow range reversed-phase chromatography. Using a 100 g Gold HP C18 column, chromatography was performed with a linear gradient of 0.1 % acetic acid-acetonitrile from 30–60 % over 20 minutes at 60 mL min−1. This enabled the initial separation of malacidin A and B containing fractions. Combined fractions containing either malacidin A or B were subsequently cleaned up individually using preparative HPLC (XBridge Prep C18, 10 × 150 mM, 5 μM, Agilent HPLC System) using a linear gradient of 0.1 % trifloroacetic acid-acetonitrile from 30% to 50% over 30 min at a flow rate of 4 mL min−1. Malacidin A and B had a retention time of 12 min and 16 min, respectively. For analysis of purity and detection of fractions by UPLC-MS throughout the isolation process, 5 μL were injected onto a UPLC-MS system (Waters Corporation) and analyzed by a linear gradient of 0.1 % formic acid—acetonitrile from 30–50 % over 3.4 min.
Structural Determination by NMR and ESI-MS/MS
See supplementary discussion for details on structural determination. 1H and 13C NMR spectra were obtained at 600 and 150 MHz, respectively, on a Bruker Avance DMX600 NMR. Spectra were taken at 298 K using either 11.21 mM malacidin A or 7.92 mM malacidin B in 3.59 mM triethylamine in D2O, unless otherwise noted. The chemical shifts were referenced to the methyl group of triethylamine in D2O (δC 8.189, δH 1.292). For ESI-MS/MS, samples in methanol were diluted 1:50 with 50% methanol/0.1% formic acid and infused (5 μL min−1) for analysis by high resolution (60,000@m/z 200)/high mass accuracy MS, MS2 and MS3 (Fusion Lumos, ThermoFischer Scientific). Additionally, each sample was diluted 1:1 with 0.1M ammonium bicarbonate for propionylation of primary amines followed by a 1:25 dilution in methanol/0.1% formic acid and analysis by ESI-MS, MS2 and MS3. Positive ion Electrospray Ionization (ESI) conditions: 3.9 kV, heated capillary set at 300 °C and sheath gas setting of ‘1’. Both ion trap based collision induced dissociation (CID) and beam type fragmentation (HCD) were used. For fragmentation experiments ions were isolated using a window of 2.0 m/z.
Determination of absolute configuration of amino acids of malacidins
Malacidin A and B (0.5 mg) were dissolved in 6N HCl (500 μL) separately and heated at 115 °C for 10 hours. For each antibiotic, four separate reactions were setup. After hydrolysis, the reaction mixtures were cooled in ice-water for 5 min. The reaction solvent was evaporated in vacuo. The dried reaction was resuspended in 500 μL of water and the water was evaporated in vacuo. This process was repeated three times. The hydrolysates, containing free amino acids, were dissolved in 100 μL of 1N NaHCO3. Either 100 μL of L-FDAA (1-fluro-2,4-dinitrophenyl-5-L-alanine amide) or 100 μL of D-FDAA in acetone (10 mg/mL) was added to each of the four vials and they were incubated at 42 °C for 1 hour. To neutralize the reaction, 100 μL of 2N HCl was added to each reaction mixture. Reactions were then diluted with 300 μL of 50% acetonitrile/water. 5 μL of each reaction mixture was analyzed by LC-HRMS with a gradient solvent system (20%–60% acetonitrile/water with 0.1% formic acid over 40 min, Flow rate 0.2 mL/min) on the RP column (Thermo Acclaim 120, C18 2.1 × 150 mm).
Microbial Susceptibility Assays
The malacidins were screened against a panel of assay strains and pathogenic bacteria as indicated in Supplementary Table S3. MIC assays were performed in duplicate in 96-well microtiter plates based on the protocol recommended by the Clinical and Laboratory Standards Institute.29 All presented data are the average of at least three independent assays. Stock solutions of malacidin or daptomycin (2 mg/mL in H2O) were added to the first well in a row and serially diluted (twofold per transfer) across the microtiter plate. CaCl2 was supplemented to media at a final concentration of 15 mM and fetal bovine serum (ATCC) was added to media (1:10) to test the effect of serum. Overnight cultures of bacteria were diluted 5,000-fold, and 50 μL were used as an inoculum in each well. MIC values were determined by visual inspection after 18 h incubation (30 °C, static growth). For the enhanced calcium titration experiments, standard MIC assays were performed with Methicillin-resistant Staphylococcus aureus (MRSA) PFGE strain type USA300 in media supplemented with CaCl2 at: 25.0, 18.8, 14.1, 7.03, 3.52, 2.50, 1.76, 0.880, 0.440, 0.250, 0 mM. To assess the effects of mono- and divalent cations on malacidin activity, standard MIC assays were performed with MRSA USA300 in media supplemented with 15 mM CaCl2, MgCl2, MnCl2, ZnCl2, SrCl2, NaCl, KCl. To evaluate the effects of pulmonary surfactants on activity against a community-acquired pneumonia causing pathogen, standard MIC assays were performed with Streptococcus pneumoniae TCH8431 in media supplemented with both 15 mM CaCl2 and Survanta (beractant, Abbvie) at a final concentration of 5, 1, 0.5, 0.25, or 0 % (v/v).
Mammalian Cytotoxicity Assays
Cytotoxicity against human cell lines was tested using an ATP release assay, CellTitreGlo (Promega), according to the manufacturer’s instructions. HEK293 cells (293FT, Thermo Fisher Scientific, #R700-07) and MRC5 cells (ATCC CCL-171) were grown in complete DMEM media supplemented with 10% FBS, and were inspected visually and tested for mycoplasma contamination using a MycoAlert detection kit (Lonza). Cells were grown to confluence, trypsinized, counted, and plated in 384-well cell culture plates at appropriate density (2500 cells/well for HEK293 and 1000 cells/well for MRC5). The test compound was added 24 hours later in the presence of calcium, and viability was determined after 4.5 h of incubation. Experiments were performed with biological replicates. Hemolytic activity was evaluated by a red blood cell disc diffusion assay. 20 μL stocks of malacidin A and triton-X100 were infused on filter discs, dried completely, and then overlaid on 5% sheep blood agar plates (Hardy Diagnostics). The plates were incubated for 24 hours at 20 °C, then checked for lysis.
Rat Cutaneous Wound Infection Model
Methicillin Resistant Staphylococcus aureus strain MW2 was grown in Mueller Hinton Broth at 37°C with shaking overnight. The culture was centrifuged, supernatant aspirated and the bacteria were gently washed once in sterile saline. The optical density was determined at 600 nm. The bacterial suspension was diluted to provide a challenge inoculum of approximately 500 CFU per wound in a volume of 0.05 mL in sterile 0.9% NaCl. The inoculum count was verified by viable counts on Mannitol Salt Agar plates spread with proper dilutions of the inoculum and incubated at 37°C for 24–48h. For the wound infection model, 8-week-old male (~200 g) Sprague Dawley rats were given two wounds each. Two rats were used at each time point (day 1 and day 3) for each treatment group for a total of 4 rats (8 wounds) per drug. This sample size was statistically calculated based on previous in-house wound burden studies comparing vehicle treated groups with antibiotic controls.30 Rats were randomly selected into the treatment groups. To generate wounds, the rats were anesthetized by intraperitoneal injection of 100 mg kg−1 ketamine +10 mg/kg xylazine and the dorsal side of the rats were shaved with electrical clippers and then depilated with Nair. The exposed skin was wiped with betadine. Two symmetrical wounds on the dorsum of each rat using a 0.8 cm diameter disposable biopsy punch. Sterile polyurethane rings serving as wound chambers were placed over the fresh wounds and attached by surgical adhesive. After the wound creation, rats from each group were infected with 0.05 ml of the bacterial suspension for a final infection dose of 500 CFU per wound. Wounds were covered with Tegaderm visible adhesive dressing, and the rats were rehydrated with physiological saline administered via intraperitoneal injection. The analgesic buprenorphine (0.05 mg kg−1) was administered to minimize pain during surgical recovery. At 30 minutes post infection, rats were given single daily topical treatments of vehicle (25 mM CaCl2 in sterile water), or 0.5 mg Malacidin A or Daptomycin suspended in 25 mM CaCl2 and the wounds were covered in fresh Tegaderm dressing. At 1 day and 3 days post infection, the rats were humanely euthanized and wounds were excised and assessed for bacterial burdens by plating on MSA. Rats were observed twice daily for morbidity and possible signs of acute toxicity. Abnormal clinical signs were recorded if observed. Rats were housed in Public Health Research Institute’s Animal Biosafety Level-2 Research Animal Facility (ICPH RAF), a center of the New Jersey Medical School, Rutgers University (NJMS-Rutgers). The animal facility follows the Public Health Service and National Institute of Health Policy of Humane Care and Use of Laboratory Animals. All experimental protocols were approved by the Rutgers Institutional Animal Care and Use Committee (IACUC).
Selection for Malacidin-resistant mutants
To select for resistant mutants, a single MRSA USA300 colony from a freshly struck plate was inoculated into LB media and grown overnight at 37 °C. The saturated overnight culture was diluted 100-fold, supplemented with a sub-lethal dose (0.5× MIC) of malacidin A, vancomycin, daptomycin, or rifamycin and 15 mM calcium. 200 μL aliquots were then distributed into microtiter plate wells. The next day, 3 μL of from each well was used to inoculate 200 μL of calcium-supplemented media (15 mM calcium) with fresh antibiotic at 0.5× and 4× MIC. This process was repeated for 20 days. In the cases where bacterial growth was observed in the 4× MIC overnight cultures, the resistant culture was plated in successively higher concentrations of antibiotic the following day. This was repeated over the course of the experiment to assess fold change in MIC at Day 0 to Day 20.
Membrane Leakage and Depolarization Assays
The effects of malacidin on membrane integrity was assessed using SYTOX Green. In brief, single colonies of MRSA USA300 were grown in LB media with and without 15 mM CaCl2 to an OD600 of 0.35. 900 μL of cells were mixed with 100 uL 17 μM SYTOX Green dye (Thermo Fisher). The resulting mixture was incubated for 5 minutes at 22 °C, then distributed to a microtiter plate at 50 μL per well. An initial reading of fluorescence pre-antibiotic addition was measured at excitation and emission wavelengths of 488 nm and 523 nm respectively. 50 μL of antibiotics were added to respective wells at a final concentration of 20 μg mL−1. Measurements were immediately collected for 10 minutes. To assess the effects of malacidin on membrane depolarization, similar assays were setup using the membrane potential probe, DiBAC4 (Bis-(1,3-Dibutylbarbituric Acid)Trimethine Oxonol). Single colonies of MRSA USA300 were grown in LB media with and without 15 mM CaCl2 to an OD600 of 0.35. 900 μL of cells were mixed with 100 uL 20 μg mL−1 DiBAC4 dye (Thermo Fisher). The resulting mixture was incubated for 5 minutes at 22 °C, then distributed to a microtiter plate at 50 μL per well. An initial reading of fluorescence pre-antibiotic addition was measured at excitation and emission wavelengths of 492 nm and 515 nm respectively. 50 μL of antibiotics were added to respective wells at a final concentration of 20 μg mL−1. Measurements were immediately collected for 10 minutes. Representative examples from three technical replicates are shown.
UDP-MurNAc-Pentapeptide Accumulation Assay
The intracellular accumulation of the cell wall precursor, UDP-MurNAc-pentapeptide after treatment of MRSA USA300 with malacidin was assessed as previous described.16 In brief, single colonies of MRSA USA300 were grown in LB media with and without 15 mM CaCl2 to an OD600 of 0.6. 1 mL of cells and media were incubated with a final concentration of 130 μg mL−1 chloramphenicol for 15 min at 37 °C. Antibiotics to be assayed were added at 10 μg mL−1 and incubated for 60 min at 37 °C. Vancomycin, known to form a complex with lipid II, was used as positive control. Cells were collected by centrifugation, and resuspended in 30 uL dH2O and incubated in boiling water for 15 minutes. The cell extract was then centrifuged at 14,000 × g. Supernatant was analyzed for UDP-linked cell wall precursors by UPLC-MS system (Waters Corporation). Experiments were performed with biological replicates.
Complex Formation with Cell Wall Precursors
Binding of malacidin to C55-P and lipid II was evaluated by incubating 1 nmol of each purified precursor with 0.5 nmoles of malacidin or daptomycin in 100 mM Tris-HCl, pH 7.5, 0.1 % TX100, 13 mM MgCl2, and with or without 25 mM CaCl2, for 60 min at at 37 °C. Subsequently, the mixture was extracted twice with n-BuOH: 6 M pyridinium acetate buffer, pH 4.2 (3:1, v/v). The butanol fraction was evaporated and the residue was dissolved in CHCl3/MeOH (1:1, v/v). The resuspension was analyzed for the loss of unbound malacidin or daptomycin to a complex by TLC analysis using chloroform/methanol/water/ammonia (88:48:10:1, v/v/v/v) as the solvent and detection by 254/366 nm visualization. Experiments were performed with biological replicates.
Data Availability
The DNA sequence to the malacidin gene cluster has been deposited as GenBank Accession KY654519. The eSNaDP2.0 software is available at http://esnapd2.rockefeller.edu/. The remaining data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Fred Rubino and Dan Kahne for discussion and an aliquot of lipid II. HEK293 cells and MRC5 cells were kindly provided by the High-throughput Screening Resource Center at the Rockefeller University. This work was supported in part by a grant from the Gates Foundation and NIH U19AI109713. B.M.H. was supported by NIH Grant F32 AI124479. Z.C.-P. was supported by NIH Grant F32 AI1100029.
Footnotes
Author Contributions: B.M.H. and S.F.B. designed research; B.M.H., S.H.K., M.K., J.G.O., M.A.T., J.M., A.E., and H.M. performed research; B.M.H., S.H.K., and Z.C.-P. analyzed data; and B.M.H. and S.F.B. wrote the paper.
Competing Interest: The authors declare no competing financial interest.
Supplementary Information: Supplementary figures and tables are provided to accompany the main text and methods.
References
- 1.O’Neill J. The Review on Antimicrobial Resistance. 2016;2016 [Google Scholar]
- 2.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 3.Reddy BV, et al. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Applied and environmental microbiology. 2012;78:3744–3752. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strieker M, Marahiel MA. The structural diversity of acidic lipopeptide antibiotics. Chembiochem : a European journal of chemical biology. 2009;10:607–616. doi: 10.1002/cbic.200800546. [DOI] [PubMed] [Google Scholar]
- 5.Jung D, Rozek A, Okon M, Hancock RE. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chemistry & biology. 2004;11:949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Bunkoczi G, Vertesy L, Sheldrick GM. Structure of the lipopeptide antibiotic tsushimycin. Acta crystallographica Section D, Biological crystallography. 2005;61:1160–1164. doi: 10.1107/S0907444905017270. [DOI] [PubMed] [Google Scholar]
- 7.Katz M, Hover BM, Brady SF. Culture-independent discovery of natural products from soil metagenomes. Journal of industrial microbiology & biotechnology. 2016;43:129–141. doi: 10.1007/s10295-015-1706-6. [DOI] [PubMed] [Google Scholar]
- 8.Owen JG, et al. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11797–11802. doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy BV, Milshteyn A, Charlop-Powers Z, Brady SF. eSNaPD: a versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chemistry & biology. 2014;21:1023–1033. doi: 10.1016/j.chembiol.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady SF. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nature protocols. 2007;2:1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, et al. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers. 2010;93:833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman JA, Mortin LI, Vanpraagh AD, Li T, Alder J. Inhibition of daptomycin by pulmonary surfactant: in vitro modeling and clinical impact. The Journal of infectious diseases. 2005;191:2149–2152. doi: 10.1086/430352. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T, Taylor SD, Palmer M, Duhamel J. Membrane Binding and Oligomerization of the Lipopeptide A54145 Studied by Pyrene Fluorescence. Biophysical journal. 2016;111:1267–1277. doi: 10.1016/j.bpj.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straus SK, Hancock RE. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochimica et biophysica acta. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Muller A, et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider T, et al. The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrobial agents and chemotherapy. 2009;53:1610–1618. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleijn LH, et al. Total Synthesis of Laspartomycin C and Characterization of Its Antibacterial Mechanism of Action. Journal of medicinal chemistry. 2016;59:3569–3574. doi: 10.1021/acs.jmedchem.6b00219. [DOI] [PubMed] [Google Scholar]
- 18.Owen JG, et al. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charlop-Powers Z, et al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14811–14816. doi: 10.1073/pnas.1615581113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular biology and evolution. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T, Luo Y, Chen Y, Li X, Yu J. BIGrat: a repeat resolver for pyrosequencing-based re-sequencing with Newbler. BMC research notes. 2012;5:567. doi: 10.1186/1756-0500-5-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic acids research. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic acids research. 2011;39:W339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallifidas D, Brady SF. Reassembly of functionally intact environmental DNA-derived biosynthetic gene clusters. Methods in enzymology. 2012;517:225–239. doi: 10.1016/B978-0-12-404634-4.00011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature protocols. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 29.Cockerill FR. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria at Grow Aerobically; Approved Standard#x02014;Ninth Edition. Clinical and Laboratory Standards Institute; Wayne, PA: 2012. [Google Scholar]
- 30.Zhao Y, et al. Carbohydrate-derived fulvic acid is a highly promising topical agent to enhance healing of wounds infected with drug-resistant pathogens. J Trauma Acute Care Surg. 2015;79:S121–129. doi: 10.1097/TA.0000000000000737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequence to the malacidin gene cluster has been deposited as GenBank Accession KY654519. The eSNaDP2.0 software is available at http://esnapd2.rockefeller.edu/. The remaining data that support the findings of this study are available from the corresponding author upon request.


