SUMMARY
The ability to recognize self and to recognize partnering cells allows microorganisms to build social networks that perform functions beyond the capabilities of the individual. In bacteria, recognition typically involves genetic determinants that provide cell surface receptors or diffusible signaling chemicals to identify proximal cells at the molecular level that can participate in cooperative processes. Social networks also rely on discriminating mechanisms to exclude competing cells from joining and exploiting their groups. In addition to their appropriate genotypes, cell-cell recognition also requires compatible phenotypes, which vary according to environmental cues or exposures as well as stochastic processes that leads to heterogeneity and potential disharmony in the population. Understanding how bacteria identify their social partners and how they synchronize their behaviors to conduct multicellular functions is an expanding field of research. Here we review recent progress in the field and contrast the various strategies used in recognition and behavioral networking.
Keywords: Self-recognition, myxobacteria, type VI secretion system, cooperation, conflict
Introduction
Living in multifaceted natural communities, many species have developed the ability to identify other related individuals and form social bonds. Social recognition is a common trait across a broad range of species, including mammals (Penn & Frommen, 2010), plants (Chen et al., 2012), insects (Leonhardt et al., 2016) and single-celled organisms (Pathak et al., 2013). Like higher organisms, microbes monitor and respond to their neighbors, including distinguishing between conspecific individuals and distinct microbial species (Strassmann et al., 2011, Stubbendieck & Straight, 2016). Recognition of social partners allows microbes to conduct sophisticated group behaviors that increase their fitness. Notably, some unicellular species have taken steps toward multicellularity by assembling related individuals into tightly bound cooperative groups (Du et al., 2015). A well-known example is the social amoeba Dictyostelium discoideum, a single-celled slime mold that uses an aggregation strategy to build spore-filled multicellular fruiting bodies. D. discoideum uses an allorecognition strategy whereby cells identify clonemates and close relatives through heterotypic interactions between polymorphic adhesion proteins (TgrB1 and TgrC1) (Hirose et al., 2011). Capitalizing on the advantages of such unicellular systems, studies of the molecular and evolutionary basis of microbial social networks have proliferated over the past decade.
Bacteria are found within diverse growth niches in which different species are inevitably in a constant struggle for limited resources. To better compete, bacteria can form social groups in which the abilities of the many exceed those of the individual. Within natural microbial habitats, diversity of taxonomic units is high, and recent findings suggest that cell-cell recognition between microbes strongly influences their social outcomes (Wall, 2016). In this minireview we discuss different recognition strategies developed by social bacteria and their roles in establishing functional community-based networks. Along with genetic-based recognition systems, we also discuss how physiological or phenotypic variation between related individuals influences recognition and social networking. As this review underscores, single-celled bacterial species are experimentally robust systems for studying the molecular basis of recognition and its social consequences.
Genetic Recognition in Bacteria
Genetic recognition relies on the detection of perceptible cues, such as diffusible chemical signals or cell surface receptors. One function of recognition among closely related individuals is the synchronization of group responses, which leads to cooperative social functions (Papenfort & Bassler, 2016). The specificity of cell-cell recognition often limits the benefits of cooperation to kin and potentially avoids adverse interactions with non-kin. In other cases, inter-species recognition leads to cooperation. For example, bacterial colonization and biofilm development typically involve interactions between different species (Burmølle et al., 2014). Taxonomic diversity can lead to benefits such as increased resistance to antibiotics and broader metabolic capabilities as compared with monoculture biofilms (Elias & Banin, 2012). In this section, we discuss examples of intra- and inter-species recognition and their social functions (Fig. 1).
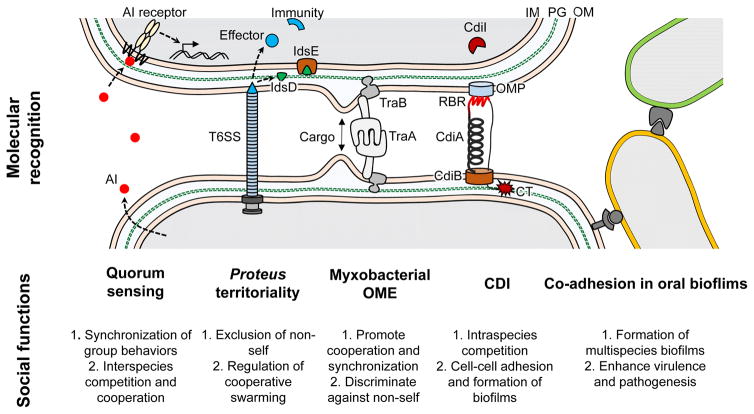
Fig. 1. Social recognition in bacteria.
Schematics of representative recognition systems used by social bacteria. AI, autoinducer; OMP, outer membrane protein; IM, inner membrane; PG, peptidoglycan; OM, outer membrane; CT, C-terminal toxin; RBR, receptor binding region; T6SS, type VI secretion system. See text for details.
1. Self-recognition and outer membrane exchange in myxobacteria
Myxobacteria are a group of soil-dwelling Gram-negative rods that exhibit complex social behaviors. During vegetative growth, cells form multicellular groups referred to as swarms that collectively move and forage for food. The sociality of myxobacteria is exemplified by their ability to aggregate into multicellular fruiting bodies analogous to social slime molds. Within myxobacterial groups, individual cells make frequent contact with each other by gliding motility (Kaiser, 2003). We recently identified a contact-dependent mechanism involving self-recognition that we named outer membrane exchange (OME) (Pathak et al., 2013). During this process, copious amounts of outer membrane (OM) material are transferred between cells when they make physical contact (Nudleman et al., 2005, Wei et al., 2011). Cargo includes lipoproteins, OM phospholipids, lipopolysaccharide (LPS) and toxins (Cao et al., 2015, Vassallo et al., 2017). Because of the diversity of the cargo, OME serves as a platform for coordinating social functions. Participation in OME requires cells to make two key proteins, TraA and TraB, whereas individuals lacking either protein are excluded from OME (Pathak et al., 2012). TraA is a cell surface receptor, and it forms a functional adhesion with TraB (Cao & Wall, 2017) (Fig. 1). Overexpression of TraA/B leads to tight cell-cell binding (Vassallo et al., 2015). Recognition specificity occurs by an N-terminal variable domain within TraA that governs homotypic interactions between receptors (Pathak et al., 2013). TraA from different environmental isolates is highly polymorphic within this domain, whereas other regions of TraA are conserved within conspecific isolates. TraA polymorphisms restrict OME such that it occurs between clonemates and close relatives that produce identical or nearly identical TraA receptors.
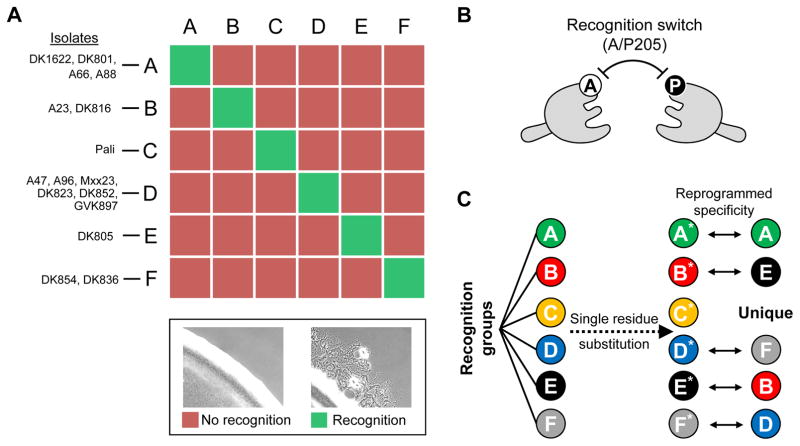
The high level of sequence variation in environmental TraA receptors suggests that there are diverse recognition groups in nature. To date we have experimentally tested TraA receptors from 16 Myxococcus xanthus environmental isolates and found that they belong to six recognition groups (Pathak et al., 2013) (Fig. 2A). Notably, TraA polymorphisms facilitate the formation of distinct social groups (Cao & Wall, 2017). For example, isogenic cells overproducing different TraA receptors form distinct cell clusters (aggregates) during growth in liquid medium. By chimeric allele analysis and site-directed mutagenesis, we discovered the malleable nature of TraA specificity, which provides a molecular explanation for how diversity of recognition arose in natural populations (Cao & Wall, 2017). For example, we found a conserved single residue switch within the TraA variable domain (Fig. 2B), and substitutions at this position, A205P or P205A, can alter the specificity of TraA recognition (Fig. 2C). By creating the corresponding substitutions in receptors that belong to six defined recognition groups, we created a unique allele that recognizes only itself; it does not recognize its parental allele or any other TraA receptor (Fig. 2C). The malleable nature of TraA likely allows it to tolerate many spontaneous sequence changes within its variable domain during evolution, which in turn can lead to changes in specificity and the diversification of myxobacterial social groups.
Fig. 2. TraA recognition in myxobacteria.
A) 16 M. xanthus environmental isolates form six distinct recognition groups. Members of each group are shown. Representative micrographs of experimental assay that determines TraA recognition specificity is shown (bottom). In brief, TraA cell-cell recognition between two strains results in motility (flares) at the edge of a mixed colony(Pathak et al., 2013). B) Single amino acid substitutions in a TraA (A/P205) switch leads to a change in homotypic recognition specificity between receptors (Cao & Wall, 2017). C) TraA homotypic recognition reprogrammed through single residue substitutions (A205P or P205A) from six recognition groups. TraA receptors after A/P substitutions are indicated with asterisks, and their recognition specificities are shown on the right.
TraA serves as the first layer of recognition for determining OME partners. A second layer of recognition further distinguishes true clonemates for cooperation (Dey et al., 2016, Vassallo et al., 2017). This second layer involves the exchange of polymorphic toxins that reside in the OM. Such toxins, encoded within a highly variable prophage-like region of the genome, lead to the death of recipient cells that lack the cognate immunities. This dual recognition strategy ensures high selectivity in the sharing of OM components that can happen only between individuals with compatible TraA receptors and cognate toxin-immunity pairs. This form of kin discrimination also results in visible boundaries between two approaching swarm populations that express compatible TraA receptors but non-cognate toxin-immunity pairs, whereas swarms composed of identical cells freely merge together.
Recognition among clonemates and close siblings leads to beneficial interactions through the sharing of cellular goods. For example, OME can repair damaged cells by diluting defective materials in unhealthy cells and replenishing missing components from healthy cells (Vassallo et al., 2015). Such behaviors increase the fitness of the population as a whole, which requires a minimum number of cooperative healthy cells to carry out multicellular functions (Vassallo & Wall, 2016). In contrast, different environmental isolates that produce compatible TraA receptors are likely to antagonize each other through the delivery of polymorphic toxins. This discrimination strategy likely plays a key role in regulating social interactions and ensuring related individuals form uniform and cohesive groups to conduct sophisticated social functions.
2. Contact-dependent inhibition
Similar to OME, contact-dependent inhibition (CDI) is a bacterial recognition system that involves physical interactions between cells through surface-embedded receptors (Fig. 1). It was initially discovered in Escherichia coli isolate EC93, where it was shown to specifically inhibit the growth of other E. coli strains, i.e., not other species, by a mechanism that involves toxin delivery (Aoki et al., 2005, Jones et al., 2017). In Gram-negative bacteria, the core of a CDI system consists of three components: CdiA, CdiB and CdiI. CdiA/B are two-partner secretion proteins. CdiA is a long filamentous protein that harbors a C-terminal toxin domain (CT), and the transport of CdiA across the OM relies on the OM β-barrel transporter CdiB. Upon binding to receptors on neighboring cells through its receptor-binding region (RBR), CdiA releases its toxin domain, which is subsequently delivered into the cytoplasm of target cells. The presence of a cognate immunity protein, CdiI, which binds and inactivates the toxin, protects producer cells from self-intoxication (Aoki et al., 2010). Growth of related non-self target cells lacking the cognate antitoxin is inhibited, usually through the disruption of the membrane proton-motive force or nucleic acid degradation. Specificity in this system occurs because the CdiA-CT and CdiI proteins are highly polymorphic across different bacterial isolates (Aoki et al., 2010). Cells that contain a CDI system have a competitive advantage over cells lacking immunity by inhibiting their growth. As described below, recent findings have also uncovered cooperative roles for CDI (Jones et al., 2017).
Self-recognition in CDI was revealed by elucidating the binding interactions between CdiA and cell surface receptors (Ruhe et al., 2013). BamA, a key component of the OM β-barrel assembly machine (BAM), was first identified as a receptor for CdiA from E. coli EC93. Although BamA is highly conserved among the Gram-negative bacteria, the sequence variations within its surface-exposed loops restrict CdiAEC93 binding to E. coli strains. CdiA proteins from other E. coli isolates can bind different receptors, but again they bind receptors only from related bacteria. For example, the heterotrimeric complex of osmoporins OmpF and OmpC serves as a receptor for CdiA from E. coli EC536 (Beck et al., 2016). Sequence variation within the surface-exposed residues in OmpC among different isolates prevents the binding and intoxication of distant strains. Therefore, CDI is suggested to serve as a tool for recognizing close relatives, and the subsequent toxin delivery step discriminates social partners.
CDI also promotes cooperative interactions. For example, CDI systems facilitate auto-aggregation and biofilm formation (Ruhe et al., 2015, Anderson et al., 2014). The adhesion properties of CdiA allow social interactions between sibling cells, whereby the specific recognition between CdiA and receptors excludes unrelated cells from engaging in CDI-dependent cell-cell adhesion (Jones et al., 2017). In addition, recent work suggests that the exchange of toxin modules between siblings leads to cell-cell signaling and changes in gene expression that contribute to biofilm formation (Garcia et al., 2016). Thus, as our knowledge about the CDI system has increased, it has become apparent that this mechanism encompasses more than simply inhibitory processes.
3. Territoriality in Proteus mirabilis
Another cell contact-dependent mechanism that involves recognition has been described in Proteus mirabilis, a Gram-negative bacterium that collectively swarms over surfaces and forms a visible boundary (Dienes line) between colonies of different identities. Boundary formation in P. mirabilis represents an example of territoriality behavior, which is also observed in other bacteria such as Bacillus subtilis (Stefanic et al., 2015) and M. xanthus (Vassallo et al., 2017). In P. mirabilis, a functional T6SS is required for Dienes line formation (Alteri et al., 2013). Different models underlying Proteus territoriality have been proposed (Fig. 1). One model invokes the T6SS as a weapon to deliver toxin effectors to non-self P. mirabilis cells that lack cognate immunity (Alteri et al., 2013). Thus, when swarming colonies meet, T6SS-mediated attack leads to cell death and the formation of an inter-strain boundary. In a second model, T6SS delivers polymorphic Ids (Identification of self) proteins between swarming cells, which in turn govern Proteus territorial behaviors (Gibbs et al., 2008, Saak & Gibbs, 2016). For example, when IdsD is delivered into neighboring cells, it specifically interacts with a cognate IdsE protein. Swarming populations expressing matching D–E pairs have the same identity and merge, whereas populations producing divergent D–E proteins are not recognized as self and form a boundary between swarms. The exchange of Ids proteins between cells does not have fatal consequences (Saak & Gibbs, 2016). Instead, the non-cognate D–E pairs result in negative swarm regulation. Therefore, IdsD is suggested to act as a regulatory factor for multicellular swarming, and D–E recognition allows cells to communicate during collective movements and facilitates swarm expansion of kin cells. Importantly, the recognition and antagonism in Proteus territoriality helps to exclude non-siblings from swarms and leads to a dominant strain within a niche(Gibbs & Greenberg, 2011). This model is consistent with clinical findings that a single genotype of P. mirabilis usually dominates during urinary tract infections (Gibbs & Greenberg, 2011).
4. Quorum sensing
Besides cell contact-dependent mechanisms, bacteria are capable of using diffusible factors to sense their surroundings and recognize self. Quorum sensing (QS) is a well-known bacterial cell-cell communication process, during which individuals process extracellular signals to synchronize group behaviors (Miller & Bassler, 2001) (Fig. 1). QS systems are found in diverse bacterial species. The signaling molecules synthesized by cells are termed autoinducers (AIs), and their concentration within an environmental niche reflects the population density of AI-producing cells. Once a population reaches a threshold density (i.e., a quorum), the signaling cells collectively change gene expression in a QS-regulated manner to generate a group response. QS promotes the fitness of bacteria by assembling individual cells into cohesive units that display various group activities, such as bioluminescence, virulence factor secretion, antibiotic production, social motility, sporulation and biofilm formation (Papenfort & Bassler, 2016). QS ensures that individuals do not frivolously undertake an activity when only a limited number of cells are present. For example, bioluminescence production is energetically costly and will not generate a fitness gain when undertaken by only a few cells. Notably, the mechanisms underlying QS systems can differ significantly from one species to another, and AIs synthesized by different organisms are also chemically diverse (Papenfort & Bassler, 2016). The QS receptors can exhibit extraordinary ligand-binding specificity, which allows cells to precisely identify AIs produced by related individuals within a heterogeneous population (Hawver et al., 2016). In addition, QS can occur between divergent bacterial species. For example, AI-2, one of the most common and broadly recognized communication signals, serves as a means for inter-species interactions that facilitates the development of multispecies biofilms (Rickard et al., 2008). QS also plays a role in interbacterial competition. In Vibrio cholera, QS controls the activation of type VI secretion system (T6SS) (Zheng et al., 2010, Shao & Bassler, 2014), a mechanism that allows V. cholera cells to kill other bacterial species and create a favorable niche free of non-kin competitors (MacIntyre et al., 2010).
5. Oral biofilm
Human oral biofilms represent one of the best-studied microbial ecosystems, within which various recognition mechanisms occur among cells either through direct physical contacts or through the exchange of diffusible chemicals. The oral cavity is a complex environment that consist of saliva (liquid), teeth (hard surface) and epithelial tissues (soft surface) and is exposed to a fluctuating amount of nutrients, changes in temperature and mechanical perturbations. Approximately 500 distinct species have been shown to reside within oral environments (Zaura et al., 2009). To colonize and survive in this niche, different microbial species recognize and interact with each other to create multispecies biofilms (Kuramitsu et al., 2007). For example, specific coaggregation between different microbial species plays a key role in the colonization, organization and growth of oral biofilms (Kolenbrander et al., 2010) (Fig. 1). The initial colonizers first adhere to the oral surfaces, which then allows other species (early, middle or late colonizers) to subsequently bind to form multispecies communities. Within oral biofilms, different bacterial species communicate by cell-cell contact between surface receptors as well as by QS molecules (e.g., AI-2), which likely synchronizes gene expression and cell behaviors (Bassler et al., 1997). In addition, the metabolism of different cells may influence other residents living within the same biofilm (Kuramitsu et al., 2007). Such metabolic interactions can be either antagonistic or cooperative, depending on whether the metabolic products of one organism are adverse or suitable for the growth of others. Oral biofilms thus serve as an attractive and relatively well understood model for studying complex inter-species recognition and social interactions.
Physiological Heterogeneity and Recognition in Bacteria
Genetic diversity inherent to microbial life in natural environments is accompanied and complicated by physiological heterogeneity within clonal populations (Lidstrom & Konopka, 2010, Stewart & Franklin, 2008). Such physiological variability is widespread and results in the formation of subpopulations with qualitatively different phenotypes, and as described here it can influence cell-cell recognition and social interactions. We define physiological heterogeneity as cell-to-cell variations in measurable parameters between clonal cells that exhibit differences in, for example, morphology, growth rate, age, cellular damage loads and gene expression patterns (Lidstrom & Konopka, 2010). In general, these variations are caused by differences in microenvironments (e.g., nutrient and oxygen gradients in biofilms) or emerge as a consequence of stochastic differences in gene expression or phase variation under relatively homogenous environments (Ackermann, 2015, Serra et al., 2013, Stewart & Franklin, 2008). Stochastic variations are distributed in populations in the form of ‘noise’, e.g., random fluctuations in gene expression, and can take distinct forms. For instance, bistability or multistability occurs when gene expression patterns in clonal populations segregate into two or more, respectively, stable states or phenotypes (Veening et al., 2008). These distinct phenotypes can be epigenetically inherited through several rounds of cell division (Casadesús & Low, 2013). Cellular differentiation leads to even greater physiological differences, whereby subpopulations of clonal cells acquire distinct morphologies and specialized functions (e.g., heterocysts in cyanobacteria and developmental differentiation in myxobacteria) (Wolk et al., 1994, O’Connor & Zusman, 1991). One example of bimodality stabilized by a positive feedback loop is the development of competent cells in B. subtilis. When B. subtilis enters the phase of late exponential growth, QS initiates stabilization of the ComK transcription factor levels in cells, and in turn ComK activates a regulon responsible for competence and its own synthesis. Because ComK levels stochastically fluctuate between individual cells, only a fraction of cells (~10%) will reach the ComK threshold level needed to create a positive feedback loop, and the population will transiently bifurcate into competent and non-competent cells (Maamar & Dubnau, 2005). In addition to competent cells, B. subtilis forms other distinct morphotypes such as matrix producers, motile and non-motile cells, producers of extracellular proteases and cells that sporulate. Differentiation into these cell types is mutually exclusive, as regulatory mechanisms involved in gene expression control lead to particular phenotypes while simultaneously repressing genes governing other cell types(Lopez et al., 2009).
Physiological Heterogeneity: the Good and the Bad
In fluctuating environments, the presence of cells with distinct physiological states often enables a clonal population to better adapt to sudden environmental changes. This strategy allows populations to spread risk or bet-hedge against changing conditions (Veening et al., 2008). Persister cells, which are in a state of dormancy, are one example of such a mechanism. The advantage of dormancy is that those cells are resistant to certain stressors, such as antibiotics that act on metabolically active cells. After the insult has passed, persisters become metabolically active and re-populate the biofilm (Lewis, 2010, Maisonneuve & Gerdes, 2014). Although this strategy provides direct benefits to persisters under stressful conditions, it can also be viewed as a social trait influenced by kin selection, as suggested by mathematical modeling by Gardner et al. (Gardner et al., 2007). Specifically, by being dormant, persisters decrease the level of competition within a population that faces resource exhaustion. Reduced reproductive output by persisters means reduced individual fitness and, as such, persistency can be viewed as an altruistic trait, with the benefits of it, according to the model, limited to close relatives. This behavior is in accordance with Hamilton’s rule stating that altruistic traits will be favored by selection as long as the reproductive cost to the actor performing the behavior is outweighed by the reproductive benefit of the recipient multiplied by its relatedness to the actor. (Hamilton, 1963, Hamilton, 1964). Another example where physiological heterogeneity impacts antibiotic resistance was recently described in E. coli. Here, asymmetric distribution of the antibiotic efflux pump AcrAB-TolC during cell division leads to pumps clustering at the old pole, which in turn makes progeny cells with old poles more resistant to antibiotics compared to their daughter cells with newer poles (Bergmiller et al., 2017).
Division of labor is another example of how physiological heterogeneity benefits bacterial populations. Here, subpopulations undertake the task of producing goods that are used by the whole community or a subpopulation. The expression of virulence factors in Salmonella typhimurium in a murine colitis model is a case where physiological heterogeneity benefits the population through both division of labor and bet-hedging. During infection, a fraction of the S. typhimurium population will express type 3 secretion system 1 (T3SS-1), a major virulence determinant, and use it to invade host intestinal mucosa evoking an inflammatory response. Products of this response, such as tetrathionate, help S. typhimurium outcompete commensal microbes found in the intestinal tract. These metabolites are used by a S. typhimurium subpopulation residing in the gut lumen but are not available to the T3SS-1–producing cells. Additionally, T3SS-1–expressing cells are slow growing and exhibit higher levels of antibiotic tolerance compared with their siblings that do not express T3SS-1, a trait that can be considered a bet-hedging strategy, similar to persisters.
In contrast, physiological heterogeneity can potentially be detrimental to the fitness of a population. Social bacteria, in particular, rely on their ability to synchronize cellular responses and engage in collective behaviors. As discussed above, QS is one of the means for controlling the physiological state of a population, where a behavioral change depends on population density and signaling In some instances, population fitness is compromised by the failure of cells to synchronize a response, thus blocking the formation of biofilms (Parsek & Greenberg, 2005), secretion of virulence factors (Smith & Iglewski, 2003) or survival during the stationary phase (Goo et al., 2012). This failure to synchronize can result from either genetic (e.g. presence of social mutants) or physiological heterogeneity. Furthermore, the production of public goods—secreted factors shared by a community—is energetically costly, and an inability to develop a synchronized response hinders the fitness of a population. For example, Vibrio fischeri uses ~20% of its metabolic potential to produce luciferase which results in bioluminescence only if produced on a population-wide scale. Bioluminescence produced by V. fischeri is beneficial to its symbiont organism, marine bobtail squid, which in turn provides a niche and food for the bacterium (Ruby, 1996). If V. fischeri cells fail to produce a homogenous or synchronized response, insufficient levels of bioluminescence will be made and their symbiotic relationship and fitness will be compromised. In another example, developmental aggregation of myxobacteria may be inhibited by physiological heterogeneity. This multicellularity-by-aggregation strategy requires that cellular behaviors be synchronized, which is obtained through population density dependent intercellular signaling (Kaiser, 2004, Zhang et al., 2012). Moreover, as indicated above, we suggest that the mixing of cellular components by OME facilitates the development of a homogeneous cell population that is better suited to conducting synchronized functions (Vassallo et al., 2015).
Physiological Recognition as a Function of Social Integration
Plants can carry out self/non-self recognition based on their physiological state rather than genetic identity (Gruntman & Novoplansky, 2004, Falik et al., 2006). Here, genetically identical individuals are perceived as alien after a short period of separation during which their physiological state has changed (Gruntman & Novoplansky, 2004). In general, cells recognize each other through phenotypic properties shaped by their genotype and expression thereof, which is influenced by environmental and stochastic fluctuations. Although not as well studied as genotype recognition, the process of bacterial recognition and discrimination based on physiological states also influences social interactions. We recently described a mechanism that myxobacteria use to address physiological heterogeneity within a mixed population consisting of growing and starving cells. The question we asked was whether physiologically distinct siblings would cooperate, antagonize or maintain neutral interactions. To test this, we created auxotroph strains and mixed them with their parent strain. Notably, on minimal medium, where the auxotrophs were starving for a metabolite, they were antagonized (killed) by their prototroph siblings. In contrast, when strains were mixed and placed on minimal media with the missing auxotroph metabolite or on rich medium, the strains grew equally well and interacted harmoniously. Similarly, when auxotrophs and prototrophs were mixed on starvation agar they harmoniously interacted. These findings show that antagonism only occurs under conditions when strains are physiologically different; otherwise their interaction are cooperative. We further found that antagonism depends on T6SS and gliding motility, and we identified a novel effector-immunity pair, TsxEI, that mediates killing (V. Troselj, A. Treuner-Lange, L. Søgaard-Andersen, D. Wall, submitted). Antagonism is caused by decreased levels of a specific immunity protein (TsxI) in starving cells, which makes them susceptible to intoxication. In contrast, within a homogeneously starving population, T6SS-mediated killing is not detected, indicating that starvation downregulates T6SS function. We hypothesize that the biological purpose of this sibling antagonism is to recognize and eliminate less-fit cells from the population or to delay the onset of development by cannibalizing cells that enter the developmental program prematurely. Likewise, when a population develops a consensus response to starvation, the cells synchronize their behavior and commit to development.
Another example of sibling discrimination based on physiological states is seen in B. subtilis cannibalism. When exposed to nutrient limitation or stress, a subpopulation of B. subtilis accumulates phosphorylated Spo0A (Spo0A~P), the master regulator of sporulation. Spo0A~P controls both sporulation and matrix production, and its accumulation to a threshold level is regulated by multiple input signals. Whereas high levels of Spo0A~P initiate sporulation, lower levels trigger extracellular matrix production in cells and simultaneously initiate a pathway involved in cannibalism that is mediated by two toxins, Skf and Sdp, with co-expression of cognate antitoxins conferring immunity to the producer cells. As Spo0A levels vary stochastically among cells, a subpopulation of cells will not reach the threshold level of Spo0A~P for becoming matrix and toxin producers. These cells, therefore, remain susceptible to the Skf and Sdp toxins and are lysed and cannibalized by their siblings (López et al., 2009, González-Pastor et al., 2003, González-Pastor, 2011). Furthermore, cells that reach Spo0A~P levels high enough to initiate sporulation can also cannibalize non-sporulating cells using the same mechanism. In both cases, cannibalism delays the onset of sporulation for toxin producers by providing nutrients for prolonged vegetative growth.
Monitoring or policing cell populations by identifying and eliminating individuals that pose a threat is a feature of eukaryotic organisms that use surveillance or immunity systems. However, eukaryotes are not alone in this ability, as bacteria have also evolved systems to monitor their populations. One example is how social mutants (cheaters) are dealt with among social bacteria. Cheaters are non-cooperative individuals that utilize public goods without contributing to their production. Consequently, these cooperative behaviors are vulnerable to exploitation, as cheaters do not pay the metabolic cost of producing the public goods and therefore have a fitness advantage over cells that do produce them. This means that cooperative individuals are at risk of being outcompeted unless the population has mechanisms that control and/or eliminate cheaters (Hibbing et al., 2010, Wang et al., 2015, Velicer et al., 2000, Manhes & Velicer, 2011). This policing behavior helps address the problem of genotypic and physiological heterogeneity that impedes social cooperativity. QS is a trait that is vulnerable to exploitation. One example for how policing occurs involves Pseudomonas aeruginosa populations, in which QS cooperators produce cyanide, which inhibits the growth of QS mutants but not of the cooperators (Wang et al., 2015). Similarly, in Burkholderia thailandensis, T6SS expression is induced by QS in cooperators, which renders QS mutants susceptible to T6SS-mediated poisoning (Majerczyk et al., 2016). Whereas these mechanisms keep social mutants from exploiting public goods, they can also be used against siblings that are physiologically different. Namely, cells that are genetically equipped to engage in QS can fail to do so because of gene expression variability or microenvironment differences that renders them blind and/or unresponsive to the QS signal. In these cases, the non-cooperative cells are discriminated against and face the same fate as QS mutants.
Social bacteria may oscillate between homogeneous and heterogeneous populations based on their temporal needs and environmental cues. As discussed above, both states have an adaptive value in fluctuating environments that likely depends on the species’ lifestyle. Thus, social bacteria that engage in collective behaviors benefit from mechanisms that enable cell-cell signaling, recognition and synchronization of behaviors (Fig. 3). In contrast, populations can also successfully adapt by differentiating into different cell types (Fig. 3). Regardless of the final outcome, bacteria discriminate not only between self and non-self but also between their phenotypic or physiological states. Therefore, physiological differences also need to be taken into account to understand how bacteria recognize and cooperate with each other.
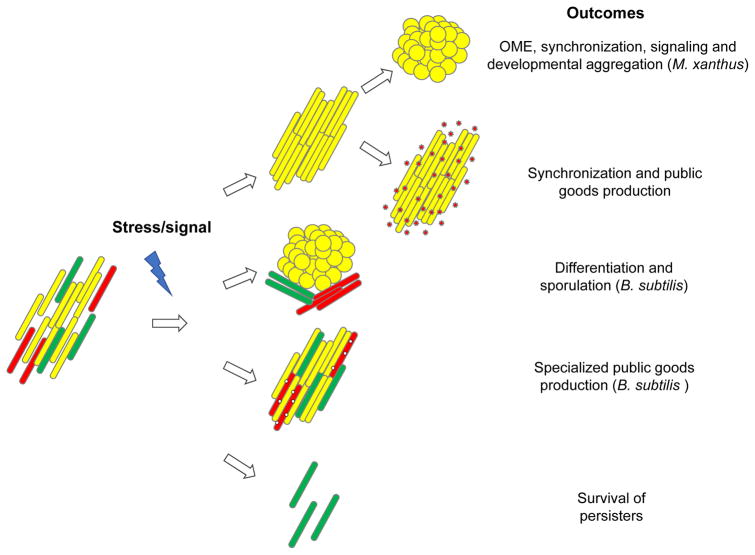
Fig. 3. Physiological heterogeneity and integration of social functions.
An environmental cue (a stressor or a signal) induces a response in a physiologically heterogeneous bacterial population, as represented by the red, yellow and green rods. Depending on the species and the nature of the signal, the social responses vary and can lead to population synchronization or differentiation (top to bottom).
Conclusion
Genetic recognition enables bacteria to communicate and establish homogeneous populations with siblings in which cooperative behaviors are limited to close relatives. In social bacteria, kin recognition facilitates complex social behaviors that depend on the population density of like individuals. Additionally, molecular recognition systems can mediate inter-species relationships in diverse and stratified multispecies communities found in natural environments or in eukaryotic hosts. Bacterial interactions and the ability of bacteria to form complex communities are also shaped by physiological heterogeneity and the ways in which it is recognized and managed within populations. Physiological heterogeneity is a layer of complexity in bacterial social networks that needs to be better addressed in future research to allow a complete understanding of bacterial behavior. This understanding may help us manipulate bacterial social behavior for medical, ecological or industrial purposes, given the application and development of tools that enable precise tracking of cell-to-cell differences in populations.
Originality Significance Statement.
The authors confirm that this minireview is original and describes recent advances in recognition and social interactions in bacteria.
Acknowledgments
This work was supported by National Institutes of Health Grant GM101449 to D.W. and grant 2P20GM103432.
Abbreviations
- T6SS
type VI secretion system
- OME
outer membrane exchange
- CDI
contact dependent inhibition
Footnotes
The authors declare they have no conflicts of interest
References
- Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nature reviews. Microbiology. 2015;13:497. doi: 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Garcia EC, Cotter PA. Kind Discrimination and Competitive Exclusion Mediated by Contact-Dependent Growth Inhibition Systems Shape Biofilm Community Structure. PLOS Pathogens. 2014;10:e1004076. doi: 10.1371/journal.ppat.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CtK, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- Arnoldini M, I, Vizcarra A, Peña-Miller R, Stocker N, Diard M, Vogel V, Beardmore RE, Hardt WD, Ackermann M. Bistable expression of virulence genes in salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS biology. 2014;12:e1001928. doi: 10.1371/journal.pbio.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. Journal of bacteriology. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CM, Willett JLE, Cunningham DA, Kim JJ, Low DA, Hayes CS. CdiA Effectors from Uropathogenic Escherichia coli Use Heterotrimeric Osmoporins as Receptors to Recognize Target Bacteria. PLOS Pathogens. 2016;12:e1005925. doi: 10.1371/journal.ppat.1005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmiller T, Andersson AM, Tomasek K, Balleza E, Kiviet DJ, Hauschild R, Tkačik G, Guet CC. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science. 2017;356:311–315. doi: 10.1126/science.aaf4762. [DOI] [PubMed] [Google Scholar]
- Burmølle M, Ren D, Bjarnsholt T, Sørensen SJ. Interactions in multispecies biofilms: do they actually matter? Trends in Microbiology. 2014;22:84–91. doi: 10.1016/j.tim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Cao P, Dey A, Vassallo CN, Wall D. How myxobacteria cooperate. Journal of Molecular Biology. 2015;427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Wall D. Self-identity reprogrammed by a single residue switch in a cell surface receptor of a social bacterium. Proceedings of the National Academy of Sciences. 2017;114:3732–3737. doi: 10.1073/pnas.1700315114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J, Low DA. Programmed heterogeneity: epigenetic mechanisms in bacteria. Journal of Biological Chemistry. 2013;288:13929–13935. doi: 10.1074/jbc.R113.472274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJW, During HJ, Anten NPR. Detect thy neighbor: Identity recognition at the root level in plants. Plant Science. 2012;195:157–167. doi: 10.1016/j.plantsci.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Dey A, Vassallo CN, Conklin AC, Pathak DT, Troselj V, Wall D. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. Journal of bacteriology. 2016;198:994–1004. doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Kawabe Y, Schilde C, Chen Z-h, Schaap P. The Evolution of Aggregative Multicellularity and Cell–Cell Communication in the Dictyostelia. Journal of Molecular Biology. 2015;427:3722–3733. doi: 10.1016/j.jmb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS microbiology reviews. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- Falik O, de Kroon H, Novoplansky A. Physiologically-mediated self/non-self root discrimination in Trifolium repens has mixed effects on plant performance. Plant signaling & behavior. 2006;1:116–121. doi: 10.4161/psb.1.3.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EC, Perault AI, Marlatt SA, Cotter PA. Interbacterial signaling via Burkholderia contact-dependent growth inhibition system proteins. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:8296–8301. doi: 10.1073/pnas.1606323113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A, West SA, Griffin AS. Is bacterial persistence a social trait? PLoS One. 2007;2:e752. doi: 10.1371/journal.pone.0000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs KA, Greenberg EP. Territoriality in Proteus: advertisement and aggression. Chemical reviews. 2011;111:188–194. doi: 10.1021/cr100051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs KA, Urbanowski ML, Greenberg EP. Genetic Determinants of Self Identity and Social Recognition in Bacteria. Science (New York, NY) 2008;321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS microbiology reviews. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- González-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Goo E, Majerczyk CD, An JH, Chandler JR, Seo YS, Ham H, Lim JY, Kim H, Lee B, Jang MS. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proceedings of the National Academy of Sciences. 2012;109:19775–19780. doi: 10.1073/pnas.1218092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman M, Novoplansky A. Physiologically mediated self/non-self discrimination in roots. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3863–3867. doi: 10.1073/pnas.0306604101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD. The evolution of altruistic behavior. The American Naturalist. 1963;97:354–356. [Google Scholar]
- Hamilton WD. The genetical evolution of social behaviour. II. Journal of theoretical biology. 1964;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- Hawver LA, Jung SA, Ng WL. Specificity and complexity in bacterial quorum-sensing systems. FEMS microbiology reviews. 2016;40:738–752. doi: 10.1093/femsre/fuw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nature reviews Microbiology. 2010;8:15. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S, Benabentos R, Ho HI, Kuspa A, Shaulsky G. Self-recognition in social amoebae is mediated by allelic pairs of Tiger genes. Science. 2011;333:467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Low DA, Hayes CS. Can’t you hear me knocking: contact-dependent competition and cooperation in bacteria. Emerging Topics in Life Sciences. 2017;1:75–83. doi: 10.1042/ETLS20160019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Coupling cell movement to multicellular development in myxobacteria. Nature Reviews Microbiology. 2003;1:45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Signaling in myxobacteria. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Micro. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiology and molecular biology reviews: MMBR. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt Sara D, Menzel F, Nehring V, Schmitt T. Ecology and Evolution of Communication in Social Insects. Cell. 2016;164:1277–1287. doi: 10.1016/j.cell.2016.01.035. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annual review of microbiology. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nature chemical biology. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS microbiology reviews. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Molecular microbiology. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Molecular microbiology. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- Majerczyk C, Schneider E, Greenberg EP. Quorum sensing control of Type VI secretion factors restricts the proliferation of quorum-sensing mutants. Elife. 2016;5:e14712. doi: 10.7554/eLife.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhes P, Velicer GJ. Experimental evolution of selfish policing in social bacteria. Proceedings of the National Academy of Sciences. 2011;108:8357–8362. doi: 10.1073/pnas.1014695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annual Reviews in Microbiology. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- O’Connor KA, Zusman DR. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. Journal of bacteriology. 1991;173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Micro. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Greenberg E. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends in microbiology. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS genetics. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak DT, Wei X, Dey A, Wall D. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS genetics. 2013;9:e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DJ, Frommen JG. Kin recognition: an overview of conceptual issues, mechanisms and evolutionary theory. Animal behaviour: evolution and mechanisms. 2010:55–85. [Google Scholar]
- Rickard AH, Campagna SR, Kolenbrander PE. Autoinducer-2 is produced in saliva-fed flow conditions relevant to natural oral biofilms. Journal of Applied Microbiology. 2008;105:2096–2103. doi: 10.1111/j.1365-2672.2008.03910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri–Euprymna scolopes light organ symbiosis. Annual Reviews in Microbiology. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- Ruhe ZC, Townsley L, Wallace AB, King A, Van der Woude MW, Low DA, Yildiz FH, Hayes CS. CdiA promotes receptor-independent intercellular adhesion. Molecular microbiology. 2015;98:175–192. doi: 10.1111/mmi.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor Polymorphism Restricts Contact-Dependent Growth Inhibition to Members of the Same Species. mBio. 2013:4. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saak CC, Gibbs KA. The Self-Identity Protein IdsD Is Communicated between Cells in Swarming Proteus mirabilis Colonies. Journal of bacteriology. 2016;198:3278–3286. doi: 10.1128/JB.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck G, Mika F, Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio. 2013;4:e00103–00113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Bassler BL. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Molecular microbiology. 2014;92:921–930. doi: 10.1111/mmi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Current opinion in microbiology. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Stefanic P, Kraigher B, Lyons NA, Kolter R, Mandic-Mulec I. Kin discrimination between sympatric Bacillus subtilis isolates. Proceedings of the National Academy of Sciences. 2015;112:14042–14047. doi: 10.1073/pnas.1512671112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nature reviews Microbiology. 2008;6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annual review of microbiology. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- Stubbendieck RM, Straight PD. Multifaceted Interfaces of Bacterial Competition. Journal of Bacteriology. 2016;198:2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS pathogens. 2011;7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, Wall D. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E2939–2946. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo CN, Cao P, Conklin A, Finkelstein H, Hayes CS, Wall D. Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. eLife. 2017:6. doi: 10.7554/eLife.29397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo CN, Wall D. Tissue repair in myxobacteria: A cooperative strategy to heal cellular damage. BioEssays. 2016;38:306–315. doi: 10.1002/bies.201500132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404:598. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- Wall D. Kin recognition in bacteria. Annual review of microbiology. 2016;70:143–160. doi: 10.1146/annurev-micro-102215-095325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proceedings of the National Academy of Sciences. 2015;112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Molecular microbiology. 2011;81:315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- Wolk CP, Ernst A, Elhai J. The molecular biology of cyanobacteria. Springer; 1994. Heterocyst metabolism and development; pp. 769–823. [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy ‘core microbiome’ of oral microbial communities. BMC Microbiology. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ducret A, Shaevitz J, Mignot T. From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS microbiology reviews. 2012;36:149–164. doi: 10.1111/j.1574-6976.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proceedings of the National Academy of Sciences. 2010;107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]