Abstract
Heme-based tryptophan dioxygenases are established immunosuppressive metalloproteins with significant biomedical interest. Here, we synthesized two mechanistic probes to specifically test if the α-amino group of the substrate directly participates in a critical step of the O atom transfer during catalysis in human tryptophan 2,3-dioxygenase (TDO). Substitution of the nitrogen atom of the substrate to a carbon (probe 1) or oxygen (probe 2) slowed the catalytic step following the first O atom transfer such that transferring the second O atom becomes less likely to occur, although the dioxygenated products were observed with both probes. A monooxygenated product was also produced from probe 2 in a significant quantity. Analysis of this new product by HPLC coupled UV–vis spectroscopy, high-resolution mass spectrometry, 1H NMR, 13C NMR, HSQC, HMBC, and infrared (IR) spectroscopies concluded that this monooxygenated product is a furoindoline compound derived from an unstable epoxyindole intermediate. These results prove that small molecules can manipulate the stepwise O atom transfer reaction of TDO and provide a showcase for a tunable mechanism by synthetic compounds. The product analysis results corroborate the presence of a substrate-based epoxyindole intermediate during catalysis and provide the first substantial experimental evidence for the involvement of the substrate α-amino group in the epoxide ring-opening step during catalysis. This combined synthetic, biochemical, and biophysical study establishes the catalytic role of the α-amino group of the substrate during the O atom transfer reactions and thus represents a substantial advance to the mechanistic comprehension of the heme-based tryptophan dioxygenases.
Graphical abstract

Introduction
Tryptophan is an essential but also the least abundant amino acid. The vast majority of tryptophan not reserved for protein synthesis takes a route for degradation through the kynurenine pathway for energy metabolism and also for providing a pool of metabolites including quinolinic acid for de novo nicotinamide adenine dinucleotide (NAD) biosynthesis.1 The first and committed step of the kynurenine pathway is carried out by two hemedioxygenases, tryptophan 2,3-dioxygenase (TDO, EC 1.13.11.11) and the homologue enzyme indoleamine 2,3-dioxygenase (IDO, EC 1.13.11.17) in two isoforms. TDO is a homotetrameric protein with high substrate specificity for l-Trp (Figure 1). This protein is mostly found in the liver and also present at low expression levels in other tissues. IDO is a monomeric enzyme with broader substrate specificity and is located in nonhepatic tissues. Both enzymes utilize a mononuclear b-type heme catalyzing the same tryptophan oxidation reaction by inserting molecular dioxygen to cleave the pyrrole ring, forming N-formylkynurenine (NFK) (Scheme 1).2 Of note, TDO served as one of the first demonstrations of the incorporation of both O atoms of molecular oxygen into an organic substrate by an enzyme.3 Other than mediating tryptophan degradation, IDO and TDO are also the representative metabolic enzymes used in mammals to maintain proper amplitude and duration of immune responses and prevent unrestrained immune activation in healthy cells. However, malignant tumors recruit this mechanism for immune evasion, which makes this enzyme a promising drug target for developing cancer immunotherapies.4
Figure 1.
Ternary complex of human TDO with L-Trp and molecular oxygen bound in the active site based on the crystal structure (PDB entry: 5TI911)
Scheme 1.
Mechanistic Model of Stepwise Oxygen Insertion via an Epoxyindole Intermediate
The mechanism of TDO/IDO has been long debated for either a Criegee-type rearrangement, implicated dioxetane pathway, or a novel epoxyindole-based mechanism (Scheme S1).5–12 In the past decade, mechanistic studies have focused on the differentiation between a stepwise (through an epoxyindole) or a concerted (through a dioxetane) oxygen insertion mechanism. Recently, compelling evidence from multiple groups supports the stepwise oxygen insertion mechanism.8–9, 11, 13–14, 15,16–17
Of note, the model shown in Scheme 1 is a novel mechanism in heme chemistry. It predicts an alkylperoxo radical intermediate followed by an epoxyindole and a histidine-ligated compound II-like species (an oxoferryl heme). Interestingly, the O atom transfer from the compound II-like species to the epoxyindole intermediate predicts the involvement of the α-amino group of the tryptophan substrate. Such a high-valent intermediate has been observed in the catalytic pathway of IDO8, 13 and the enzyme reactivation process of TDO.17 The possible participation of the ammonium group in catalysis was initially suggested from studies of dye-sensitized photooxygenation of tryptophan in model studies.18 This interaction was later echoed by EPR/ENDOR and computational studies leading to a proposal that the ammonium group stabilizes the Fe(III)-superoxide in the initial reaction steps of the catalytic cycle.9, 14 In particular, quantum mechanics/molecular mechanics (QM/MM) simulations have suggested that the ammonium group in tryptophan is crucial for epoxyindole ring opening.9, 15 However, this possibility has not been explored experimentally. In particular, a substrate lacking the α-amino group, that is, indole propionic acid7, can function as an alternate substrate of human IDO,13 which casts into question the role of the α-amino group in catalysis.
We hypothesized that if the proton transfer from the ammonium group of tryptophan is critical for opening the ring of the epoxyindole intermediate, then replacing the ammonium moiety with a substituent that is less able to donate a proton will slow the TDO enzyme reaction. In this study, we probed the TDO mechanism by employing two synthesized mechanistic probes (tryptophan analogueues), 1 and 2 (Figure 2). Here, we report a study of these probes with human TDO. The results indicate the crucial role of the ammonium group in proton transfer assisted epoxyindole ring opening and strongly support the stepwise oxygen insertion mechanism via the epoxyindole intermediate and compound II pathway.
Figure 2.
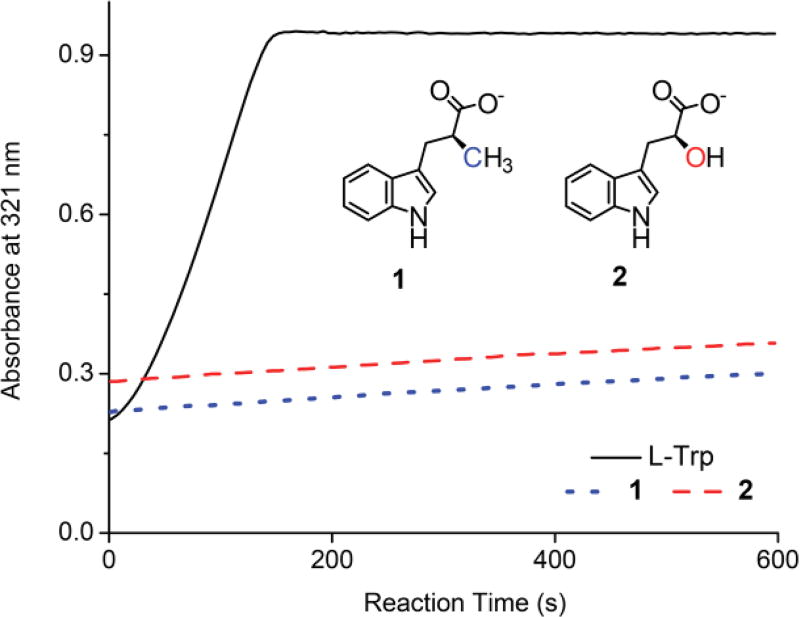
Chemical structure of the synthetic probes and comparison of reaction rates of human TDO with l-Trp and the probes. With 200 µM of l-Trp, the NFK formation by 1.0 µM TDO was complete within 150 s. Conversely, at 200 µM, both probes 1 and 2 reacted ca. 500-fold slower under the same conditions. The inset shows the chemical structures of 1 and 2.
Materials and Methods
Organic Synthesis of Probes 1 and 2
Methyl- and hydroxyl-containing tryptophan analogue probes 1 and probe 2 were prepared by adapting previously reported protocols (see Supporting Information Scheme S2 and the associated text for synthetic routes and characterization details).19–21 Methyl-based analogue probe 1 was synthesized from indole-3-carboxaldehyde using a three-step sequence involving Wittig olefination, reduction, and hydrolysis.19 Hydroxyl-containing analogue probe 2 was generated from indole using a SnCl4-mediated epoxide opening followed by saponification.20 The detailed chemical analysis and spectroscopic characterization of these probes are included in Supporting Information.
Enzymatic Reaction
The cloning, expression, and purification of human TDO has been described in a previous report.22 The enzyme assays were conducted using a modified procedure published previously.16–17 In a typical reaction for product analyses and structural identification, 1 mM of each mechanistic probe and L-tryptophan were reacted with 50 µM of purified human TDO at room temperature for a day. The enzyme reaction mixtures were filtered using 10 kDa molecular weight cut off centrifugal filter unit (Millipore). Filtered samples were analyzed using a Thermo Scientific Ultimate-3000SD HPLC rapid separation system equipped with photodiode array detector. Samples injected were eluted using gradient mixing solvent A, 0.1% FA in 90:10 water:acetonitrile, and solvent B, acetonitrile at the flow rate of 1.2 mL/min. InertSustain C18 column with the particle size of 5 µm and the dimension of 4.6 ID × 100 mm (GL Sciences Inc.) was used for HPLC analysis and sample preparation for further MS and NMR analysis.
Inhibition Mechanism Study
The inhibition assays were conducted in a buffer system containing 50 mM Tris-HCl, 200 mM NaCl, 5% ν/ν glycerol (pH 8.0) at room temperature. The reaction was initiated by adding 1 µM enzyme to the mixture containing 1 mM L-ascorbate and L-Trp (20 – 3200 µM) in the absence or presence of probe 1 or 2 at three different concentrations. Steady-state formation of NFK was monitored at 321 nm (extinction coefficient, 3752 M−1cm−1) using a Perkin Elmer Lambda 25 UV-visible spectrophotometer.
The data were analyzed using OriginPro (OriginLab, Northampton, Massachusetts, USA). Global fittings were conducted across the datasets using the following equation:
in which parameters, kcat, Km, Ki1, and Ki2 are shared.
NMR spectroscopy
All NMR spectra were recorded on a Bruker (Billerica, MA) Avance III HD 500 MHz spectrometer equipped with a 5-mm Prodigy Probe at 303 K running TopSpin 3.5 pl 6. Spectra were recorded in D2O. One dimension 1H (pulse sequence: zg30) spectra were recorded with 1 s relaxation delays, 64 k data points, and multiplied with an exponential function for a line-broadening of 0.3 Hz before Fourier transformation. 13C (pulse sequence: udeft) spectra were recorded with 10 s relaxation delays, 21k data points, and multiplied with an exponential function for a line-broadening for 2 Hz before Fourier transformation. Multiplicity edited HSQC (pulse sequence: hsqcedetgpsisp 2.3) spectra were acquired with spectral widths of 16 ppm for 1H and 220 ppm for 13C, 2k × 128 data points, and with a relaxation delay of 2 s. HMBC (pulse sequence: hmbcetgpl3nd) spectra were acquired with spectral widths of 13 ppm for 1H and 220 ppm for 13C, 4k × 128 data points, and with a relaxation delay of 5 s. 1D gradient selected NOESY (pulse sequence: selnogp) were recorded with a mixing time of 300 ms, a relaxation delay of 2 s, and multiplied with an exponential function for a line-broadening of 1 Hz before Fourier transformation.
Mass Spectrometry
Mass spectra were collected on a maXis plus quadrupole-time of flight mass spectrometer equipped with an electrospray ionization source (Bruker Daltonics) and operated in the negative ionization mode. Samples were introduced via syringe pump at a constant flow rate of 3 µL/min. The critical source parameters are summarized as follows: capillary voltage, 3500 V with a set end plate offset of −500 V; nebulizer gas pressure, 0.4 bar; dry gas flow rate, 4.0 L/min; source temperature, 200 °C. Mass spectra were the average of 1 min of scans collected at a rate of 1 scan per second in the range 50 ≤ m/z ≤ 1500. Compass Data Analysis software version 4.3 (Bruker Daltonics) was used to process all mass spectra.
IR Spectroscopy
Infrared spectra were measured at a Shimadzu FTIR-8400S Fourier transform infrared spectrometer using KBr pellets.
Results
Mechanistic Probes as Alternative Substrates and Inhibitors
At physiological pH, the ammonium group of l-Trp may donate a proton if needed during catalysis. Two mechanistic probes were designed by changing the α-amino group of the native substrate to either a methyl group (1) or a hydroxyl group (2) (Figure 2, inset). The methyl α-group of 1 would be unable to donate a proton, and the α-hydroxyl group of 2 could be a hydrogen bond donor or reluctant proton source. We synthesized the two probes and performed biochemical analyses with isolated human TDO.
The ability for TDO to activate molecular oxygen using 1 and 2 as the substrate in place of l-Trp was assessed by following product formation as we have previously done with 1-methyl-L-tryptophan with a bacterial TDO.16 After incubating with TDO, each probe generated a new peak centered near 321 nm similar to that observed from the reaction with l-Trp, suggesting that both probes are alternate substrates. The catalytic rate using these probes as the substrate in place of l-Trp was compared to that with l-Trp (Figure 2). The data showed that the reaction with the probes proceeded at a much slower rate than that with the native substrate.
Both probes were found to be reversible inhibitors of TDO. The Ki values determined from the global fitting of the double reciprocal plots were 320 ± 43 and 444 ± 32 µM for 1 and 2, respectively (Figure 3), indicating these are relatively weak inhibitors. Although the plots show a mixed inhibition pattern, these probes are predominately competitive inhibitors. The secondary Ki values obtained are ca. 3.6–30-fold higher than the primary Ki values, which are higher than the Km values obtained from the fitting. This inhibition pattern might derive from the fact that TDO is a homotetrameric enzyme which exhibits cooperativity, and its heme centers are inequivalent.21 Additionally, this enzyme has a secondary l-Trp binding site remote from the catalytic heme center for regulating the enzyme stability.11, 22 As such, two distinct binding modes could arise from binding to two inequivalent hemes or the regulating site.
Figure 3.
Lineweaver–Burkplots from the inhibitory assays of (A) 1 and (B) 2.
Product Analysis by HPLC Coupled to UV–Vis Spectroscopy
We analyzed the enzymatic reaction products by HPLC with a diode array detector (Figure 4). The product of the native substrate, NFK eluted 0.5 min earlier than L-Trp. The peak at 7.2 min was identified as a decay product of NFK (Figure S1, Table S1, Scheme S3 and Figure S2). A similar spontaneous deamination product can be formed from NFK.23–24 The peaks corresponding to L-Trp and NFK showed characteristic absorption spectra at 280 and 321 nm, respectively. Figure 4A shows the HPLC separations of three sets of experiments on l-Trp, probes 1 and 2, monitored at 280, 294, and 321 nm, respectively, to maximize the opportunity to observe potential products. The absorption spectrum of each identified eluent was recorded by an online diode-array spectrometer (Figure 4B). The enzyme reaction mixture of probe 1 showed the unreacted probe 1 peak with the retention time of 8.5 min, and a new peak (1a) eluted at 7.5 min. The species showed spectral feature similar to that of NFK, suggesting a dioxygenated product of probe 1 was produced. Likewise, the HPLC separation of the reaction mixture with probe 2 contained an NFK-like peak (2a) with the retention time of 6.3 min and unreacted probe at 7 min. Intriguingly, the reaction mixture also contained an additional new peak (2b) that was not observed in the reaction with either probe 1 or L-Trp. This new peak eluted 0.3 min earlier than the NFK-like peak and showed UV-vis absorption spectra at 294 nm, which was distinct from that of L-Trp, probe 1, probe 2 and their corresponding NFK products (Figure 4B).
Figure 4.
Separation of reaction mixtures by HPLC. (A) Superimposed HPLC chromatograms obtained from separate single-wavelength (black trace 280 nm; red trace, 294 nm; blue trace, 321 nm) detections for (a) l-Trp (control), (b) reaction mixture of L-Trp, (c) reaction mixture of 1, and (d) reaction mixture of 2. (B) Absorption properties of each of the fractions obtained by an online diode array UV–vis spectrometer coupled with the HPLC system.
High-Resolution MS Analysis of the Reaction Products Proving O Atom Transfer to the Probes
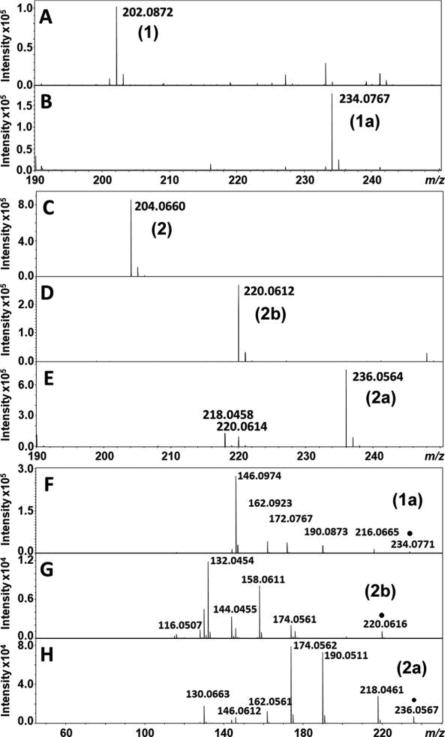
Each eluent from the HPLC separation was further analyzed by high-resolution MS. The fraction containing probe 1 reaction product with the retention time of 7.5 min showed an increase of 32 Da (1.8 ppm mass accuracy) compared to the unreacted compound, indicating 1a, the dioxygenated product of probe 1 (Figure 5 A and B, range 190 ≤ m/z ≤ 250 shown), was produced. Fractions containing the reaction products of probe 2 with the retention time of 6.0 and 6.3 min showed increases of 16 and 32 Da, respectively, suggesting the mono- (2b) and dioxygenated (2a) products of probe 2 (Figure 5, C–E). Theoretical and experimentally measured accurate masses are summarized in Table 1. Tandem MS analyses were also carried out, and the chemical structures and the corresponding masses are shown in Scheme S4. The results were consistent with a dioxygenated product (1a) from probe 1 and two products, a mono- (2b) and a dioxygenated (2a) product from probe 2 (Figure 5, F–H). Together with the HPLC-coupled UV-vis data, we conclude that an NFK-like dioxygenation product was produced on each of the mechanistic probes and a single-oxygen inserted product from 2.
Figure 5.
Negative ion ESI mass spectra of the LC fractions containing (A) 1, (B) 1a, (C) 2, (D) 2b, and (E) 2a. Tandem mass spectra confirming the identity of (F) 1a, (G) 2b, and (H) 2a. Parent ions are indicated with “●”.
Table 1.
Calculated and Experimentally Measured Accurate Masses for Probes 1, 2, and Their Oxygenated Products Detected by ESI Mass Spectrometrya
| molecular formula |
m/z theoretical |
m/z observed |
mass accuracy (ppm) |
|
|---|---|---|---|---|
| 1 | C12H13NO2 | 202.0874 | 202.0872 | −1.0 |
| 1bb | C12H13NO3 | 218.0823 | N/A | N/A |
| 1a | C12H13NO4 | 234.0772 | 234.0767 | −2.1 |
|
| ||||
| 2 | C11H11NO3 | 204.0666 | 204.0660 | −2.9 |
| 2b | C11H11NO4 | 220.0615 | 220.0612 | −1.4 |
| 2a | C11H11NO5 | 236.0564 | 236.0564 | 0.0 |
Ion species are [M-H+]−.
1b is monooxygenated product of probe 1, which was not detected in this study.
Determining the Chemical Structure of the Monooxygenated Product by NMR and IR Spectroscopies
Next, the monooxygenated product 2b from the TDO reaction of probe 2 was analyzed by NMR to determine if it would be an epoxyindole intermediate or a decayed derivative (Figure 6, additional NMR data and analyses are included in Supporting Information Table S2 and Figures S3–S7). 1H-NMR spectra contained a unique signal at 5.46 ppm (labeled “11” in Figure 6A), which could correlate with an epoxyindole product. However, the instability of the initial epoxyindole intermediate might enable rearrangement to two other cyclized forms with the same exact mass and same signal “11” in 1H NMR (Scheme 2). 1D-selective NOESY on proton “2” determined the stereochemistry of the newly formed stereocenter (Figure 6B). The signal of the proton “11” indicates both protons “11” and “2” should be on the same side. 13C NMR assigned or listed all of the 11 peaks of carbons (Figure 6C). Together with 1H NMR, the edited HSQC and HMBC spectra were obtained. The HSQC spectrum showed C–H bond connectivity except for the four quaternary carbons at positions 4 (88.2 ppm), 5 (130.1 ppm), 10 (149.4 ppm), and 1 (178.6 ppm) (Figure 6D). Finally, the HMBC data provide compelling evidence that the as-isolated monooxygenated product, 2b, is a cyclized compound with a five-membered ring (Figure 6E). The unique cross peak between proton “11” and carbon at position 2 (78.6 ppm) proves its connectivity with 3-bond correlation, which rules out the epoxyindole itself. The lack of the cross peak between proton “11” and carbon at position 1 (178.6 ppm) also supports the structure with a new five-membered ring generated on the probe 2 (Scheme 2). A proposed mechanism of PrnB in the pyrrolnitrin biosynthesis pathway also predicts the similar five-membered ring compound.25 Finally, our NMR analysis revealed that the monooxygenated product 2b from probe 2 is (2S,3aS,8aR)-3a-hydroxy-3,3a,8,8a-tetrahy-dro-2H-furo[2,3-b]indole-2-carboxylic acid.
Figure 6.
NMR analysis of 2b. (A) 500 MHz 1H NMR in D2O, (B) 1D-selective NOESY spectrum, (C) 125 MHz 13C NMR, (D) HSQC spectrum, CH and CH3’s red, CH2’s blue, and (E) HMBC spectrum.
Scheme 2.
Summary of Reaction Products of Probe 2
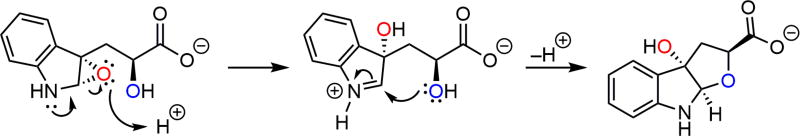
The monooxygenated product 2b from probe 2 was further analyzed by infrared (film) and the spectral features were ob-served at 3352, 2924, 2852, 1611, 1592, 1484, 1430, 1330, 1301, 1257, 1195, 1122, 1108, 1031, 998, 971, 943, 881, 861, 800, 748, 700, and 592 cm−1 (Figure S8). The IR signal at ~1600 is indicative of an acyclic carboxylate, which is consistent with the proposed structure shown in Scheme 2. A proposed pathway leading to the monooxygenated product 2b from the putative epoxide intermediate of probe 2 is shown in Scheme 3.
Scheme 3.
Proposed Mechanism of the Formation of the Observed Monooxygenated Product 2b from Reaction of Probe 2
Discussion
Key NMR Assignments and Evidence for the Ammonium Involvement in Catalysis
TDO is an enzyme discovered some 82 years ago,26 yet it is one of the hottest inhibition targets for drug development.4, 27–31 The mechanistic understanding of its action will potentially help in designing intermediate- or transition state-based analogues to effectively inhibit this enzyme. While IDO has a more relaxed substrate specificity than does TDO, human TDO is more specific to the chemical structure of the substrate. Thus, the test with human TDO for the tryptophan analogue-based mechanistic probes as alternate substrates may have a better chance to reveal the mechanism shared by these heme-dependent dioxygenases. With a dioxygenated product observed from both probes as a result of multiple turnovers, we would expect the TDO reaction to proceed through a pathway that resembles the catalytic cycle with l-Trp.
NMR analysis revealed that monooxygenation of probe 2 generated a furoindoline, which presumably arose from isomerization of unstable epoxyindoline.32 The chemical shifts and splitting patterns in the 1H and 13C NMR spectra were consistent with furoindoline 2b, and assignment was made with the assistance of HSQC NMR data. The identity of the monooxygenated species as furoindoline 2b rather than an epoxyindoline was supported by HSQC NMR data, which revealed a 3-bond correlation between the proton at position 11 and the carbon at position 2. Further, the relative stereochemistry was assigned on the basis of 1D selective NOESY, which showed that protons at positions 2 and 11 resided on the same face of the tetrahydrofuran ring system. The unambiguous detection of a monooxygenated product from probe 2 delivers compelling experimental evidence indicating that the O atom transfer takes place step-by-step, ruling out the dioxetane mechanism.
The formation of an epoxyindole species as an intermediate has been implicated from a resonance Raman study8 and the LC–MS detection of a minor cyclic aminoacetal byproduct.33 Together with the aminoacetal byproduct from L-Trp, the presence of 2b from the monooxygenation of probe 2 through the proposed mechanism suggested in Scheme 3 strongly supports an epoxyindole intermediate predicted in the stepwise oxygen insertion mechanism.
At physiological pH, tryptophan is zwitterionic. Hence, the role of the ammonium group in the mechanism was first proposed by Nakagawa and colleagues in a model study in 1977,18 which, unfortunately, received little attention in the ensuing 30 years. This proposal was reintroduced and further developed by two independent computational studies, a molecular dynamics and hybrid QM/MM methods7, 15 and an ONIOM study,9 in 2010–2012. However, the proposed role of the ammonium group from these computational studies is diverse, and there is so far no experimental evidence to support the proposed function directly. The results described in our synthetic probe study, the ca. 500-fold decrease in reaction rate upon substitution of the amino group and the identification of the detected furoindole compound 2b, provide strong experimental support for the participation of the α-amino group of the substrate in catalysis.
Dioxygneation versus Monooxygenation—The Determining Factor
The substitution of the nitrogen atom of the substrate ammonium group to oxygen sufficiently reduces the efficiency of the O atom transfer from the compound II-like species to the epoxyindole intermediate, such that it reaches the limit for the acid-catalyzed ring-opening of the epoxyindole at the later part of the catalytic pathway. Thus, both monooxygenated and dioxygenated products are produced. A significant amount of the monooxygenated intermediate is released to the solution and its instability leads to the epoxyindole ring-opening product. Although the substituted hydroxyl group is less capable of donating a proton to the epoxyindole, it may stabilize the epoxyindole by H-bonding interactions, making it longer lived at the active site. Of note, a TDO homologues protein MarE in the maremycin biosynthetic pathway in Streptomyces sp. B9173 is reported to generate a monooxygenated product, presumably via an epoxyindole intermediate.34 In contrast, the carbon-substitution as shown in probe 1 cannot stabilize the epoxyindole.
It is, however, intriguing that methyl substitution yields only a dioxygenated product but not a detectable monooxygenated product. The lack of monooxygenation product may arise from the inability of the α-methyl group to facilitate an acid-catalyzed regiospecific ring-opening of the epoxyindole, so that a protein component at the distal pocket is eventually recruited for assisting the catalytic cycle. Likewise, the α-deaminated substrate, IPA, functions as an alternate substrate of human IDO, although this substrate requires a long lag phase (ca. 50 s) to form product and, in that case, also monooxygenated product13. In the active site of TDO, the binding mode of L-Trp is dictated by the interaction between the L-amino acid moiety and surrounding residues (Figure 1). Arg144 forms a salt bridge with the carboxyl group in a bidentate fashion. The main chain amino group of Thr342 also involves H-bonding with the substrate carboxyl group. Thr342 and the carboxyl group of heme interact with the ammonium group of the substrate through H-bonding. The substrate specificity of TDO for L-Trp is accomplished by these interactions. In the primary structure level, Arg144 and Thr342 are also conserved in the IDO system. Recently, the crystal structure of hIDO in complex with L-Trp and cyanide as an oxygen surrogate became available.37 The structure revealed that the binding mode of L-Trp in hIDO is almost identical to that in hTDO. This is quite surprising because broader substrate specificity of IDO has been believed based on a more relaxed binding pocket than hTDO, lacking the tight non-covalent interaction that allows only L-Trp in TDO active site.11, 35–36 An identical substrate binding mode of L-Trp between TDO and IDO suggests similar oxygen insertion mechanisms but different kinetic details. IPA and probe 1, which both lack the α-amino group, would suffer from poor binding to the TDO active site because they do not provide a hydrogen-bonding partner to Thr342 and the heme carboxyl group in TDO. Because the insertion of two oxygen atoms is observed for IPA and probe 1, another proton source other than ammonium group and its substitution to hydroxyl group might be involved in the epoxyindole ring opening. The loss of a functionally important atom could cause a change in the rate-limiting step so that the accumulation of a monooxygenated intermediate is affected.
Although the present work provides evidence to support only a specific step of the catalytic cycle, it helps refine the catalytic pathway of this group of dioxygenases, whose mechanism-based inhibitors are much needed. After several decades of considerable efforts of screening and biochemical studies, only a few compounds have entered preclinical and Phase I trials to target immune escape of cancers via IDO/TDO inhibition. Among these prodrugs, only one is a TDO inhibitor (i.e., indole LM10),28 primarily due to its stringent substrate specificity. A thorough understanding of the catalytic pathway would help design more synthetic compounds that target both IDO and TDO.
The stepwise oxygen insertion model is composed of two parts (Scheme 1). The first half starts with molecular dioxygen becoming a bridging molecule between the heme iron and the organic substrate. This could be accomplished by either an electrophilic addition or an iron-bound superoxide radical attack to the l-Trp, which requires further investigation despite extensive studies.7, 10, 38 The second half involves a histidine-ligated ferryloxo (a compound II-like species)8, 13 and an epoxyindole intermediate. The formation of a substrate-based epoxide intermediate has been found in other systems such as nonheme iron extradiol dioxygenase and Rieske dioxygenase.39–40 The formation of an epoxyindole intermediate accompanied by a compound II-like species defines the key characteristic of heme dioxygenase mechanism. The high-valent Fe species is responsible for oxygen insertion and the pyrrole ring opening to complete the dioxygenation reaction. This part of the reaction starts with the proton transfer-assisted epoxyindole ring opening coupled with a nucleophilic ferryl oxygen addition reaction.
From a mechanistic perspective, the dioxygenation mechanism of TDO and IDO does not share the well-established strategy derived from the studies of the cytochrome P450 enzyme superfamily, which invokes a thiol-ligated ferryl-oxo porphyrin π cation radical (FeIV=O[porphyrin·+]) known as compound I. Two oxidizing equivalents (relative to the initial ferric state of the enzyme) are placed on the heme cofactor in the compound I type of oxidant.41 Instead, a compound II-like species is the key oxidant, and it is responsible for performing the critical O atom transfer to the organic substrate. The compound II-like species is one-electron more reduced than compound I. Therefore, it was once thought that the compound II species might be incapable of oxygen insertion. However, the P450 enzyme reaction starts from the ferric species and obtains electrons from both the organic substrate and the NAD(P)H, whereas in the heme-based dioxygenases the ferrous, not the ferric, form is the catalytic active form. Thus, the compound II-like species in tryptophan dioxygenases indeed carries two oxidizing equivalents (relative to its catalytic active ferrous state) and is apparently competent for performing an O atom transfer. A surprising example of Compound II reactivity is found in the heme-thiolate peroxygenase APO.42 The ferryl hydroxide form of APO-Compound II shows high reactivity toward benzylic C–H bond with the bond dissociation energy up to 85 kcal/mol. In the P450 mechanism, the resting enzyme accepts electrons and protons from NAD(P)H and undergoes an O–O bond cleavage to generate the compound I species. After the intended oxidation to an organic substrate, the P450 enzyme returns to the initial ferric, but not ferrous, state. Similarly, catalases, peroxidases, peroxygenases, and some other heme-dependent enzymes employ an adapted compound I-based strategy. Hence, from a bioinorganic perspective, the mechanism illustrated in Scheme 1 is distinct and thus deserves more attention.
Concluding Remarks
By using the ammonium ion of the substrate as an independent variable in a mechanistic pursuit, we characterized the mono-oxygenated product of an N-to-O atom-substituted probe in the reaction with human TDO. Direct product analysis and structural determination revealed that the product is derived from the proposed epoxyindole intermediate, thereby proving the catalytic function of the substrate α-amino group and corroborating the stepwise oxygen insertion mechanism for heme dioxygenase. In the TDO-catalyzed reaction, the enzyme utilizes not only the heme center and protein residues, but also the substrate itself in the mechanism of oxidation.
Supplementary Material
Acknowledgments
This work was supported in whole or part by the National Institutes of Health (NIH) grants GM108988 (to A.L.) and GM124661 (to. R.A.A.), and the Lutcher Brown Distinguished Chair Endowment fund (to A.L.). The mass spectrometry facility was sponsored by National Institutes of Health Grant G12MD007591. The NMR spectrometer used at UTSA is a shared instrument sponsored by the National Science Foundation (NSF) under award no. 1625963. Support for the NMR instrumentation at KU was provided by NSF Academic Research Infrastructure grant no. S10RR024664 and NSF Major Research Instrumentation grant no. 0320648. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of National Institutes of Health.
Abbreviations
- TDO
tryptophan 2,3-dioxygenase
- IDO
indoleamine 2,3-dioxygenase
- NFK
N-formylkynurenine
- IPA
indole propionic acid
Footnotes
- Scheme S1, providing the mechanistic models of both 507 stepwise and concerted oxygen insertion; Scheme S2 and associated text, providing the details of chemical synthesis and characterization of the molecular probes of 1 and 2; Figure S1, Table S1, Scheme S3, and Figure S2, providing the identification of the peak at 7.2 min in Figure 4Ab; Scheme S4, providing the chemical structures and masses for the tandem mass spectrometry analysis; Table S2, providing the key numbers of NMR shifts; and Figures S3–S8, providing the NMR and IR spectroscopic characterization of the TDO reaction products from the synthetic probes (PDF).
The authors declare no competing financial interest.
References
- 1.Stone TW, Darlington LG. Nat. Rev. Drug. Discov. 2002;1:609–20. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 2.Hayaishi O, Katagiri M, Rothberg S. J. Am. Chem. Soc. 1955:5450–5451. [Google Scholar]
- 3.Hayaishi O, Rothberg S, Mehler AH, Saito Y. J. Biol. Chem. 1957;229:889–896. [PubMed] [Google Scholar]
- 4.Cervenka I, Agudelo LZ, Ruas JL. Science. 2017;357:1–8. doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- 5.Raven EL. J. Biol. Inorg. Chem. 2017;22:175–183. doi: 10.1007/s00775-016-1412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sono M, Roach MP, Coulter ED, Dawson JH. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 7.Capece L, Lewis-Ballester A, Batabyal D, Di Russo N, Yeh S-R, Estrin DA, Marti MA. J. Biol. Inorg. Chem. 2010;15:811–823. doi: 10.1007/s00775-010-0646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis-Ballester A, Batabyal D, Egawa T, Lu C, Lin Y, Marti MA, Capece L, Estrin DA, Yeh SR. Proc. Natl. Acad. Sci.U. S. A. 2009;106:17371–6. doi: 10.1073/pnas.0906655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung LW, Li X, Sugimoto H, Shiro Y, Morokuma K. J. Am. Chem. Soc. 2010;132:11993–2005. doi: 10.1021/ja103530v. [DOI] [PubMed] [Google Scholar]
- 10.Makino R, Obayashi E, Hori H, Iizuka T, Mashima K, Shiro Y, Ishimura Y. Biochemistry. 2015;54:3604–3616. doi: 10.1021/acs.biochem.5b00048. [DOI] [PubMed] [Google Scholar]
- 11.Lewis-Ballester A, Forouhar F, Kim SM, Lew S, Wang Y, Karkashon S, Seetharaman J, Batabyal D, Chiang BY, Hussain M, Correia MA, Yeh SR, Tong L. Sci. Rep. 2016;6:35169. doi: 10.1038/srep35169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng J, Liu A. Arch. Biochem. Biophys. 2014;544:18–26. doi: 10.1016/j.abb.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Booth ES, Basran J, Lee M, Handa S, Raven EL. J. Biol. Chem. 2015;290:30924–30. doi: 10.1074/jbc.M115.695684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davydov RM, Chauhan N, Thackray SJ, Anderson JL, Papadopoulou ND, Mowat CG, Chapman SK, Raven EL, Hoffman BM. J. Am. Chem. Soc. 2010;132:5494–500. doi: 10.1021/ja100518z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capece L, Lewis-Ballester A, Yeh SR, Estrin DA, Marti MA. J. Phys. Chem. B. 2012;116:1401–13. doi: 10.1021/jp2082825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng J, Dornevil K, Liu A. J. Am. Chem. Soc. 2012;134:12209–12218. doi: 10.1021/ja304164b. [DOI] [PubMed] [Google Scholar]
- 17.Fu R, Gupta R, Geng J, Dornevil K, Wang S, Zhang Y, Hendrich MP, Liu A. J. Biol. Chem. 2011;286:26541–26554. doi: 10.1074/jbc.M111.253237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa M, Watanabe H, Kodato S, Okajima H, Hino T, Flippen JL, Witkop B. Proc. Natl. Acad. Sci. U. S. A. 1977;74:4730–3. doi: 10.1073/pnas.74.11.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuda K, Toyoda H, Nishio H, Nishida T, Dohgo M, Bingo M, Matsuda Y, Yoshida S, Harada S, Tanaka H, Komai K, Ouchi S. J. Agr. Food Chem. 1998;46:4416–4419. [Google Scholar]
- 20.Deechongkit S, You SL, Kelly JW. Org. Lett. 2004;6:497–500. doi: 10.1021/ol036102m. [DOI] [PubMed] [Google Scholar]
- 21.Gupta R, Fu R, Liu A, Hendrich MP. J. Am. Chem. Soc. 2010;132:1098–109. doi: 10.1021/ja908851e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira P, Shin I, Sosova I, Dornevil K, Jain S, Dewey D, Liu F, Liu A. Mol. Genet. Metab. 2017;120:317–324. doi: 10.1016/j.ymgme.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bova LM, Wood AM, Jamie JF, Truscott RJ. Invest. Ophthalmol. Vis. Sci. 1999;40:3237–44. [PubMed] [Google Scholar]
- 24.Tomek P, Palmer BD, Kendall JD, Flanagan JU, Ching LM. Biochim. Biophys. Acta. 2015;1850:1772–80. doi: 10.1016/j.bbagen.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhu X, van Pee KH, Naismith JH. J. Biol. Chem. 2010;285:21126–33. doi: 10.1074/jbc.M110.120485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotake Y, Masayama I. Z. Physiol. Chem. 1936;243:237–244. [Google Scholar]
- 27.Schwarcz R, Stone TW. Neuropharmacology. 2017;112:237–247. doi: 10.1016/j.neuropharm.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ananieva E. World J. Biol. Chem. 2015;6:281–289. doi: 10.4331/wjbc.v6.i4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Front. Immunol. 2014;5:673. doi: 10.3389/fimmu.2014.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breda C, Sathyasaikumar KV, Sograte Idrissi S, Notarangelo FM, Estranero JG, Moore GG, Green EW, Kyriacou CP, Schwarcz R, Giorgini F. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5435–40. doi: 10.1073/pnas.1604453113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilotte L, Larrieu P, Stroobant V, Colau D, Dolusic E, Frederick R, De Plaen E, Uyttenhove C, Wouters J, Masereel B, Van den Eynde BJ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2497–502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haynes SW, Ames BD, Gao X, Tang Y, Walsh CT. Biochemistry. 2011;50:5668–5679. doi: 10.1021/bi2004922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basran J, Efimov I, Chauhan N, Thackray SJ, Krupa JL, Eaton G, Griffith GA, Mowat CG, Handa S, Raven EL. J. Am. Chem. Soc. 2011;133:16251–7. doi: 10.1021/ja207066z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zou Y, Brock NL, Huang T, Lan Y, Wang X, Deng Z, Tang Y, Lin S. J. Am. Chem. Soc. 2017;139:11887–11894. doi: 10.1021/jacs.7b05517. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto H, Oda S, Otsuki T, Hino T, Yoshida T, Shiro Y. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2611–6. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forouhar F, Anderson JL, Mowat CG, Vorobiev SM, Hussain A, Abashidze M, Bruckmann C, Thackray SJ, Seetharaman J, Tucker T, Xiao R, Ma LC, Zhao L, Acton TB, Montelione GT, Chapman SK, Tong L. Proc. Natl. Acad. Sci. U. S. A. 2007;104:473–8. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis-Ballester A, Pham KN, Batabyal D, Karkashon S, Bonanno JB, Poulos TL, Yeh SR. Nat. Commun. 2017;8:1693. doi: 10.1038/s41467-017-01725-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basran J, Booth ES, Lee M, Handa S, Raven EL. Biochemistry. 2016;55:6743–6750. doi: 10.1021/acs.biochem.6b01005. [DOI] [PubMed] [Google Scholar]
- 39.Rivard BS, Rogers MS, Marell DJ, Neibergall MB, Chakrabarty S, Cramer CJ, Lipscomb JD. Biochemistry. 2015;54:4652–64. doi: 10.1021/acs.biochem.5b00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Li J, Liu A. J. Biol. Inorg. Chem. 2017;22:395–405. doi: 10.1007/s00775-017-1436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rittle J, Green MT. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Ullrich R, Hofrichter M, Groves JT. Proc. Natl. Acad. Sci. U. S.A. 2015;112:3686–91. doi: 10.1073/pnas.1503340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.