Abstract
Agrobacterium infection and regeneration of the putatively transformed plant from the explant remains arduous for some crop species like peanut. Henceforth, a competent and reproducible in planta genetic transformation protocol is established for peanut cv. CO7 by standardizing various factors such as pre-culture duration, acetosyringone concentration, duration of co-cultivation, sonication and vacuum infiltration. In the present investigation, Agrobacterium tumefaciens strain EHA105 harboring the binary vector pCAMBIA1301–bar was used for transformation. The two-stage selection was carried out using 4 and 250 mg l−1 BASTA® to completely eliminate the chimeric and non-transformed plants. The transgene integration into plant genome was evaluated by GUS histochemical assay, polymerase chain reaction (PCR), and Southern blot hybridization. Among the various combinations and concentrations analyzed, highest transformation efficiency was obtained when the 2-day pre-cultured explants were subjected to sonication for 6 min and vacuum infiltrated for 3 min in Agrobacterium suspension, and co-cultivated on MS medium supplemented with 150 µM acetosyringone for 3 days. The fidelity of the standardized in planta transformation method was assessed in five peanut cultivars and all the cultivars responded positively with a transformation efficiency ranging from minimum 31.3% (with cv. CO6) to maximum 38.6% (with cv. TMV7). The in planta transformation method optimized in this study could be beneficial to develop superior peanut cultivars with desirable genetic traits.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1231-1) contains supplementary material, which is available to authorized users.
Keywords: Acetosyringone, Arachis hypogaea (L.), GUS histochemical assay, In planta genetic transformation, Sonication, Vacuum infiltration
Introduction
Arachis hypogaea (L.), universally known as peanut or groundnut, is an important legume crop of the Fabaceae family. Peanut is widely cultivated for its edible oil in tropical and subtropical regions. China is the largest producer of peanuts (13.65 million tonnes) followed by India with an annual production of 7.09 million tonnes (FAOSTAT 2014). Peanut has high nutritional value and contains 49.24% fat, 25.80% protein, 4.72% soluble sugar, vitamin B, E, and 30 essential nutrients (USDA 2016). It contains antioxidants like resveratrol and p-coumaric acid, and is a source of biologically active flavonoids, isoflavones, and polyphenols (Janila et al. 2013). Hence it has greater potential to reduce hunger and malnutrition across the globe (Enserink 2008; Krishna et al. 2015). Apart from being an important food crop, peanut oil is also used in butter production, preparation of desserts, snack products and soup (Rustom et al. 1996). Due to the above said benefits and ever increasing population, the consumption and usage of groundnut are gradually increasing. However, gradual reduction in cultivable land due to the urbanization, and several abiotic and biotic stress factors are limiting the groundnut production. Hence, there is a requirement to develop peanut cultivars with value-added traits, to achieve higher yield under abiotic and biotic stresses such as drought, salinity, cold and pathogen infections.
Till date, genetic engineering by plant transformation is known to play a major role in developing superior cultivars with the desired genetic traits. Agrobacterium-mediated genetic transformation and biolistic transformation are most widely using methods for the production of genetically modified crops. These methods require well established regeneration protocols for developing transformed plants from suitable explants. Majority peanut transformation protocols utilized tissue culture-based regeneration system to obtain transformed plants from several explants, including, hypocotyl, epicotyl, leaf, embryonic callus, and cotyledonary node (Franklin et al. 1993; Qiusheng et al. 2005; Anuradha et al. 2008; Iqbal et al. 2012; Chu et al. 2013; Hsieh et al. 2017). However, the regeneration process is labor-intensive, time-consuming, requires skilled technicians, and might also lead to somaclonal variations (Mayavan et al. 2015). The problems associated with tissue culture-based transformation could be overcome by following the in planta transformation protocols (Subramanyam et al. 2015).
The in planta transformation is straightforward, quick, does not require proficient technicians and remarkable somaclonal variations do not occur (Manickavasagam et al. 2015). Successful in planta transformation was reported for the first time by Feldmann and Marks (1987) using Arabidopsis, and since then several researchers adopted in planta transformation for several recalcitrant crops, including radish, tomato, brinjal, okra, and snake gourd with some amendments to improve the transformation efficiency (Park et al. 2005; Yasmeen et al. 2009; Subramanyam et al. 2013, 2015; Manickavasagam et al. 2015). These amendments include pre-culture duration, different Agrobacterium strains, acetosyringone concentration, sonication, and vacuum infiltration. Various explants such as seed, epicotyl, shoot apical node, pollen tube, flower and whole fruit were used for the in planta transformation (Jaganath et al. 2014). Among these, seed is considerably the best explant for in planta transformation since, seeds are available throughout the year, easy to handle and screening of putatively transformed plants raised from the seeds is easy and efficient (Manickavasagam et al. 2015). Rohini and Rao (2000) have already reported in planta transformation for peanut using mature seed embryo axis with a transformation efficiency of 3.3%. In their report, the method was not evaluated against different cultivars. The success and the transformation efficiency of Agrobacterium-mediated in planta genetic transformation also depended on the genotype used.
Hence, the present study was aimed to standardize the in planta transformation protocol with higher transformation efficiency using groundnut cv. CO7. We also used the standardized protocol to evaluate the genotype effect on in planta transformation using other popular groundnut cultivars.
Materials and methods
Seed source and surface sterilization
Seeds of peanut cv. CO7 were obtained from Tamil Nadu Agricultural University (TNAU), Coimbatore, Tamil Nadu, India. The seeds were propagated in the greenhouse and used for the experiments.
Freshly harvested, dried and clean seeds were selected for surface sterilization. The seeds were disinfected with 70% (v/v) ethanol for 1 min, followed by soaking them in 0.1% (w/v) mercuric chloride for 6 min. After surface sterilization, seeds were rinsed 5–6 times in sterile distilled water for 1 min each. The sterilized seeds were incubated in sterile distilled water for 24 h under complete darkness on an orbital shaker (100 rpm). A longitudinal cut was made in imbibed seeds, and seed coat was removed. One-half of the seed (cotyledon with full embryo) was used as an explant to evaluate the germination percentage, minimum inhibitory concentration of BASTA® and Agrobacterium-mediated in planta genetic transformation.
Assessment of seed germination
The half seed explants (100 explants) were inoculated onto MS basal medium (Murashige and Skoog 1962) and incubated for 15 days at 25 ± 2 °C under 16 h photo period at a light intensity of 50 µmol m−2 s−1. After 15 days of inoculation, the seedling with prominent root and shoot was considered as germinated seed and the germination percentage was calculated.
Minimum inhibitory concentration (MIC) of BASTA®
The optimum and effective concentration of BASTA® to select the transformed seedlings was determined by inoculating the control explants onto the MS basal medium comprising various concentrations (0, 2, 4, 6, 8 or 10 mg l−1) of BASTA® (Bayer crop science, Pinkenba, Australia). The cultures were incubated for 15 days at 25 ± 2 °C under 16 h photo period at a light intensity of 50 µmol m−2 s−1. The specific BASTA® concentration wherein the explants failed to progress into seedlings with prominent roots and shoot was considered as the MIC for selecting the transformants. In addition, different BASTA® concentrations (0, 50, 100, 150, 200, 250 or 300 mg l−1) were sprayed on 40-day-old control plants grown under the greenhouse conditions. After 7 days of treatment, the plants were visually observed. The minimum concentration of BASTA® that triggered maximum chlorosis, necrosis, and leaf burning was considered as MIC to screen the 40-day-old putatively transformed plants from the non-transformed plants.
Agrobacterium tumefaciens strains and binary vector
A. tumefaciens strain EHA105 (kindly provided by Rafael Perl Treves, Bar-Ilan University, Israel) harboring the binary vector pCAMBIA1301–bar was used to standardize the in planta genetic transformation. EHA 105 is an L, l-succinamopine strain with a C58 chromosomal background harboring pEHA105 virulence helper plasmid (Hood et al. 1986, 1993). The T-DNA region (Supplementary Fig. 1) of the binary vector consists of hptII gene cassette (CaMV35SP: hptII: 35s poly A), gusA gene cassette (CaMV35SP: gusA: nospolyA) and a bar gene cassette (CaMV35SP: bar: nospolyA). The Agrobacterium strain was preserved in AB agar medium (Agrobacterium minimal medium) fortified with 10 mg l−1 rifampicin (SRL, Mumbai, India) and 50 mg l−1 kanamycin (SRL, Mumbai, India).
Preparation of Agrobacterium suspension
A single A. tumefaciens colony was inoculated into an Erlenmeyer flask (250 ml) containing sterile 50 ml LB (Luria–Bertani) liquid medium supplemented with appropriate antibiotics. The flasks were incubated at 28 °C for 24 h in a shaker incubator set at 180 rpm. The bacterial cells were collected from the actively growing Agrobacterium culture by centrifugation at 6000 rpm for 10 min and the bacterial cell pellet was diluted to an OD600 of 0.8 using infiltration medium (MS liquid medium with 5% sucrose; pH 5.7) containing various concentrations of acetosyringone (0, 50, 100, 150 or 200 µM). The Agrobacterium suspensions were then incubated for 1 h on an incubator shaker set at 180 rpm and 28 °C, and then used to standardize the in planta transformation.
In planta transformation
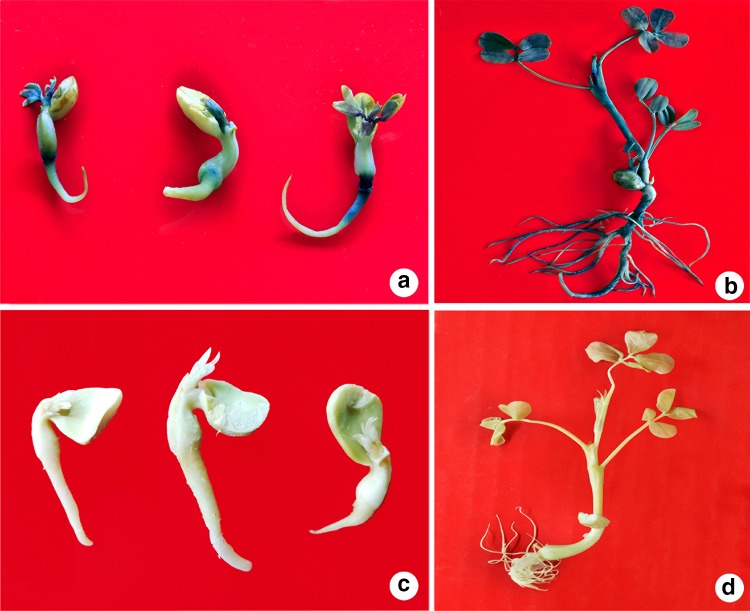
The half seed explants prepared from the peanut cv. CO7 (Fig. 1a) were used to standardize the various parameters affecting in planta transformation which includes, pre-culture duration, acetosyringone concentration, duration of co-cultivation, sonication, and vacuum infiltration. Prior to the Agrobacterium infection, half seed explants (Fig. 1b) were pre-cultured for different time intervals (0, 1, 2, 3 or 4 days) in a 50-ml liquid MS basal medium. During the pre-culture, the cultures were kept in the incubator with 100 rpm at 25 ± 2 °C under complete darkness. The pre-cultured explants were inoculated into Agrobacterium suspension containing various concentrations (0, 50, 100, 150, 200 or 250 µM) of acetosyringone and then subjected them to sonication for 0, 2, 4, 6, 8 or 10 min using a water bath sonicator (1510 Branson, Branson Ultrasonics, Kanagawa, Japan). The sonicated explants were swiftly transferred into the fresh Agrobacterium suspension and exposed to vacuum infiltration at 750 mm of Hg for different time durations (0, 1, 2, 3, 4 or 5 min). After 1 h post vacuum infiltration in bacterial suspension, the explants were separated and dry blotted on sterilized filter paper for 10 min followed by co-cultivation for various time intervals (0, 1, 2, 3, 4 or 5 days) on MS basal medium containing different concentrations (0, 50, 100, 150, 200 or 250 µM) of acetosyringone. Afterwards, the explants were rinsed with sterile liquid MS medium containing 250 mg l−1 cefotaxime (Alken laboratories, Mumbai, India) to wash away the excess of adhering Agrobacterium cells. The explants were then blotted dry on sterile filter paper and inoculated into the MS medium supplemented with 250 mg l−1 cefotaxime and 4 mg l−1 BASTA®. The cultures were incubated for 15 days at 25 ± 2 °C under 16 h photoperiod with a light intensity of 50 µmol m−2 s−1. The whole set of experiment was repeated 3 times and for each treatment 100 explants were used. After 15 days, well-developed plantlets were transplanted into paper cups having sterile planting mixture (3:1:1 v/v/v red soil, coconut shell powder and river sand) and placed in a growth chamber for hardening. After hardening the putatively transformed plants were transplanted into pots having a sterile planting mixture, placed in the greenhouse for acclimatization. The second round of selection was done by spraying 250 mg l−1 BASTA® on 40-day-old plants and those plants surviving the selection process were allowed to grow further and used them for screening the presence of transgenes (bar, hpt II, and gus A genes).
Fig. 1.
In planta Agrobacterium-mediated genetic transformation of peanut cv. CO7 using half seed as an explant. a Mature dry seeds of peanut cv. CO7 used for preparing the half seed explants; b half seed explants made from imbibed seeds (black arrows direct the embryonic area); c co-cultivation of half seed explants infected with A. tumefaciens EHA105 carrying pCAMBIA1301–bar plasmid; d non-transformed seedling on MS medium containing 4 mg l−1 BASTA®. (white arrow indicates the necrosis of the embryonic axis); e, f putatively transformed seedling development after 2 and 10 days of inoculation, respectively, on MS medium comprising 4 mg l−1 BASTA®; g, h putatively transformed plantlet development after 15 and 20 days of inoculation, respectively, on MS medium comprising 4 mg l−1 BASTA®; i hardened putatively transformed peanut plants maintained in the growth chamber; j 40-day-old acclimatized putatively transformed peanut plant after 1 week of BASTA® spray (250 mg l−1) in the greenhouse; k 40-day-old acclimatized non-transformed (control) plant with dried leaves after 1 week of BASTA® spray (250 mg l−1) under the greenhouse conditions; l transformed peanut plant with healthy leaves after 6 weeks of BASTA® spray (250 mg l−1) in the greenhouse
GUS histochemical analysis
The expression of the β-glucuronidase (gus A) gene in the putatively transformed seedlings and plantlets was detected using the GUS histochemical analysis as outlined by Jefferson et al. (1987). The respective tissue/plant samples were incubated for 24 h in GUS assay buffer (Mayavan et al. 2015) at 37 °C. The tissue samples were then subjected to dechlorophylization for 24 h using methanol.
Molecular analysis of putatively transformed plants
The GUS assay positive plants were selected for molecular analysis. The transgene integration into the genome of putatively transformed plants was confirmed by polymerase chain reaction (PCR) and Southern blot hybridization. Genomic DNA from the putatively transformed and control plants were isolated by using the method of Dellaporta et al. (1983) with minor modifications. The leaf tissue was powdered in liquid nitrogen and immediately transferred to microcentrifuge tube containing 1 ml of extraction buffer [100 mM Tris–HCl (pH 8.0), 25 mM EDTA (pH 8.0), 1.5 M NaCl, 2% CTAB, 0.2% β-mercaptoethanol (v/v) and 1% PVP (w/v)] and mixed well to form a slurry and incubated at 65 °C for 60 min. The extract was mixed with equal volume of chloroform:isoamyl alcohol (24:1 v/v) and centrifuged (Remi, Mumbai, India) at 12,000 rpm for 12 min at room temperature. To the supernatant equal volume of isopropanol and half volume of 5 M NaCl were added and incubated at room temperature for 30 min followed by centrifugation at 12,000 rpm for 12 min at room temperature. The pellet was retrieved and washed with 70% ethanol by centrifugation at 12,000 rpm for 12 min at 4 °C. Dried DNA pellet was dissolved in 1X TE buffer, [10 mM Tris–HCl (pH 8.0) and 1.0 mM EDTA] and 3 µl of RNase A (10 mg/ml) was added and incubated for 1 h at 37 °C, followed by extraction twice with chloroform:isoamyl alcohol (24:1) by centrifugation at 12,000 rpm for 12 min at room temperature. To the aqueous phase cold absolute ethanol was added and DNA was collected in the form of a pellet by centrifugation at 12,000 rpm for 10 min followed by washing with 70% ethanol (twice). The pellet was air dried and the DNA was dissolved in 100 µl of 1X TE buffer.
The presence and integration of T-DNA region in the peanut genome was confirmed by PCR with the primers specific to amplify the bar (barFP: 5′-atcgtcaaccactacatcgagac-3′; barRP: 5′-ccagctgccagaaacccacgtc-3′), hptII (hptFP: 5′-atgaaaagcctgaactcaccgcgacg-3′; hptRP: 5′-ctatttcttgccctcggacgagtgct-3′), and gus A (gusFP: 5′-actcgacggcctgtgggcattcagtctg-3′; gusRP: 5′-cactgaccggatgccgacgcgaag-3′) genes. The PCR program consisted of initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 61 °C (bar) or 55 °C (hptII and gus A) for 1 min, extension at 72 °C for 30 s, and the reaction was finally concluded at 72 °C for 10 min. The plasmid pCAMBIA1301–bar served as a positive control, whereas the genomic DNA from control plant served as the negative control. The PCR products were electrophoresed on a 1.0% agarose gel.
Southern blot hybridization was carried out as previously described by Sambrook et al. (1989). Genomic DNA was extracted from PCR-positive putatively transformed and control peanut plants. About 10 µg of genomic DNA and 5 µg of pCAMBIA1301–bar plasmid were subjected to overnight digestion with EcoRI enzyme and size fractioned on a 1.0% agarose gel. The fragmented DNA from the agarose gel was alkali blotted onto a Hybond N+ nylon membrane (GE healthcare, Buckinghamshire, UK). The PCR amplified, and alkaline phosphatase (ALP) labeled (GE healthcare, Buckinghamshire, UK) bar gene was used to prepare a probe for hybridization. The hybridization, stringent washing of the membrane, chemiluminescent development using CDP-Star substrate, and detection were performed according to the manufacturer’s instructions (GE healthcare, Buckingham Shire, UK). The binary vector pCAMBIA1301–bar and genomic DNA from control plant served as positive control and negative control, respectively.
Stable inheritance analysis
For segregation analysis, seeds were harvested from open pollinated Southern blot hybridization-positive plants. The seeds collected from five different transformed lines were labeled as S1, S2, S3, S4, and S5. The seeds were sterilized and inoculated on MS medium supplemented with BASTA® (4 mg l−1). After 15 days, the plantlets with a shoot and well-developed roots were hardened and acclimatized. The transgene inheritance in the progeny plants was proved by GUS histochemical analysis and PCR using primers that specifically amplify the coding region of gus A gene.
Influence of genotype on in planta transformation
Following standardization of the various transformation constraints using cv. CO7, similar transformation settings were adopted to check the effects of genotype on the efficiency of the developed in planta transformation technique with the other 4 popular peanut cultivars (CO6, TMV-7, TMV-2, and VR13).
Statistical analysis
The data collected during all the experiments were subjected to one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test (DMRT), and the statistical analysis was done at the level of P < 0.05 using SPSS 16.0 (SPSS Inc. USA).
Results
Determination of seed germination rate
The germination ratio directly influences the transformation efficiency. The seeds of all the 5 (CO7, CO6, TMV-7, TMV-2, and VR13) cultivars inoculated on MS medium, displayed a good germination percentage which ranged from 82 to 91.5% (Supplementary Fig. 2). Among the five cultivars, CO7 exhibited highest germination percentage (91.5%). Hence, we used cv. CO7 for the standardization of various parameters influencing the transformation of peanut.
Fig. 2.
Screening of putatively transformed seedlings and plantlets by GUS assay. a Putatively transformed seedlings; b putatively transformed plantlet; c non-transformed (control) seedlings; d non-transformed (control) plantlet
MIC of BASTA®
In the present study, supplementing BASTA® in the medium significantly influenced the germination percentage of explants. The germination percentage was gradually reduced with an increase in the concentration of BASTA®. At 4 mg l−1 BASTA® in the medium, all the control explants failed to develop into the seedlings with prominent roots and shoots. In addition, 40-day-old control plants sprinkled with BASTA® (250 mg l−1) showed severe chlorosis, necrosis, stunting growth, and finally withered in a week. Therefore, 4 mg l−1 and 250 mg l−1 of BASTA® were considered as optimal concentrations to select the putatively transformed seedlings and 40-day-old plants, respectively.
Impact of pre-culture on transformation efficiency
In the present investigation, gus A gene expression was also observed in the seedlings and plantlets developed from un pre-cultured explants (Table 1). However, pre-culturing the explants prior to the Agrobacterium infection significantly improved the transformation efficiency (Table 1). The percentage of germinating explants and survival of plants after BASTA® (250 mg l−1) spray, and the gus A gene expressing plants increased steadily with an increase in the pre-culture duration. Among the various durations, 2-day pre-culture of the seed explants prior to the Agrobacterium infection resulted in higher transformation efficiency of 8.0% (Table 1). Beyond 2 days of pre-culture, the transformation rate gradually reduced (Table 1).
Table 1.
Influence of different factors with variables on the transformation efficiency of peanut cv. CO7
| Treatment | No. of seeds infected | Mean no. of germinated seeds on selection mediumA | Mean no. of plants surviving after BASTA® sprayB | Mean no. of plants expressing gus A gene | Transformation efficiency (%) |
|---|---|---|---|---|---|
| Pre-culture duration (days) | |||||
| 0 | 100 | 19.3 ± 0.23c | 8.6 ± 0.13d | 3.3 ± 0.20d | 3.3 ± 0.20d |
| 1 | 100 | 20.6 ± 0.24b | 10.3 ± 0.15b | 5.6 ± 0.18ba | 5.6 ± 0.18ba |
| 2 | 100 | 22.6 ± 0.34a | 12.6 ± 0.28a | 8.0 ± 0.26a | 8.0 ± 0.26a |
| 3 | 100 | 18.0 ± 0.25d | 9.3 ± 0.19c | 6.3 ± 0.25b | 6.3 ± 0.25b |
| 4 | 100 | 16.3 ± 0.16e | 7.0 ± 0.25e | 4.6 ± 0.25c | 4.6 ± 0.25c |
| Duration of co-cultivation (days) | |||||
| 1 | 100 | 22.6 ± 0.34d | 12.6 ± 0.28d | 8.0 ± 0.26d | 8.0 ± 0.26d |
| 2 | 100 | 24.3 ± 0.21c | 14.3 ± 0.22c | 10.3 ± 0.15b | 10.3 ± 0.15b |
| 3 | 100 | 28.0 ± 0.25a | 18.6 ± 0.33a | 14.3 ± 0.22a | 14.3 ± 0.22a |
| 4 | 100 | 25.3 ± 0.10b | 15.3 ± 0.33b | 9.3 ± 0.19c | 9.3 ± 0.19c |
| 5 | 100 | 19.6 ± 0.25e | 8.0 ± 0.25e | 5.6 ± 0.22e | 5.6 ± 0.22e |
| Acetosyringone concentration (µM) | |||||
| 0 | 100 | 28.0 ± 0.25d | 18.6 ± 0.33da | 14.3 ± 0.22ca | 14.3 ± 0.22ca |
| 50 | 100 | 30.6 ± 0.39b | 21.3 ± 0.27c | 16.3 ± 0.16ba | 16.3 ± 0.16ba |
| 100 | 100 | 32.3 ± 0.24ab | 22.6 ± 0.18b | 17.0 ± 0.29b | 17.0 ± 0.29b |
| 150 | 100 | 33.0 ± 0.25a | 24.6 ± 0.22a | 19.6 ± 0.24a | 19.6 ± 0.24a |
| 200 | 100 | 29.3 ± 0.31c | 19.3 ± 0.23d | 15.0 ± 0.27c | 15.0 ± 0.27c |
| 250 | 100 | 27.0 ± 0.25e | 16.0 ± 0.21e | 13.3 ± 0.24d | 13.3 ± 0.24d |
| Duration of sonication (min) | |||||
| 0 | 100 | 33.0 ± 0.25e | 24.6 ± 0.22e | 19.6 ± 0.24e | 19.6 ± 0.24e |
| 2 | 100 | 35.3 ± 0.28d | 26.6 ± 0.25d | 20.3 ± 0.27da | 20.3 ± 0.27da |
| 4 | 100 | 38.6 ± 0.16b | 28.3 ± 0.25c | 22.6 ± 0.25c | 22.6 ± 0.25c |
| 6 | 100 | 40.3 ± 0.29a | 31.6 ± 0.20a | 25.3 ± 0.10a | 25.3 ± 0.10a |
| 8 | 100 | 37.3 ± 0.26c | 29.3 ± 0.26b | 23.3 ± 0.24b | 23.3 ± 0.24b |
| 10 | 100 | 30.6 ± 0.22f | 21.0 ± 0.22f | 20.6 ± 0.24d | 20.6 ± 0.24d |
| Duration of vacuum infiltration (min) | |||||
| 0 | 100 | 40.3 ± 0.29e | 31.6 ± 0.20e | 25.3 ± 0.10e | 25.3 ± 0.10e |
| 1 | 100 | 42.6 ± 0.21d | 34.3 ± 0.16d | 27.0 ± 0.22d | 27.0 ± 0.22d |
| 2 | 100 | 45.3 ± 0.34b | 36.6 ± 0.20c | 29.3 ± 0.26c | 29.3 ± 0.26c |
| 3 | 100 | 47.0 ± 0.21a | 38.3 ± 0.22a | 33.6 ± 0.28a | 33.6 ± 0.28a |
| 4 | 100 | 44.3 ± 0.26c | 37.6 ± 0.29b | 31.3 ± 0.24b | 31.3 ± 0.24b |
| 5 | 100 | 34.6 ± 0.31f | 25.0 ± 0.25f | 19.6 ± 0.25f | 19.6 ± 0.25f |
Prior to the Agrobacterium infection, the explants were pre-cultured and sonicated for various time intervals. The sonicated explants were inoculated into the A. tumefaciens, EHA 105 harboring pCAMBIA1301–bar plasmid, suspension and infected them with the aid of vacuum infiltration for various time intervals. Following infection, infected explants were co-cultivated for various time intervals on MS medium containing different concentrations of acetosyringone
Transformation efficiency = no. of GUS-positive plants/no. of infected seeds × 100. Mean values of three separate trials (±) with standard errors. In each column (for specific factor), numbers with different letters indicate they are considerably different from each other according to Duncan’s multiple range test at a probability level of 5%
AInfected explants were propagated in MS medium supplemented with 4 mg l−1 BASTA®
B40-day-old plants were sprinkled with 250 mg l−1 BASTA®. Visual observations and the data were documented after 1 week
Influence of co-cultivation on transformation efficiency
In the current study, 2-day pre-cultured explants were co-cultivated for 1–5 days with A. tumefaciens strain EHA105. The plantlets developed from the explants co-cultivated with Agrobacterium at all-time intervals showed gus A gene expression. However, higher transformation efficiency was achieved from the explants co-cultivated for 3 days. About 18.6% of co-cultivated explants survived and converted to plants that showed resistance to 250 mg l−1 BASTA®, while 76.8% of them showed gus A gene expression with 14.3% transformation efficiency (Table 1). Beyond 3 days of co-cultivation, the explant germination rate was significantly reduced, which ultimately led to a lower transformation efficiency (Table 1).
Effect of acetosyringone on transformation efficiency
The selection of optimum concentration of acetosyringone plays an important role in obtaining higher transformation efficiency. In the current study, the percentage of seed germination, plant survival after BASTA® spray, number of plants expressing gus A gene, as well as the transformation efficiency steadily improved with increasing acetosyringone concentration, and the highest transformation efficiency of 19.6% was observed when acetosyringone was provided at a concentration of 150 µM (Table 1). Supplementation of acetosyringone in the infection and co-cultivation medium beyond 150 µM resulted in a gradual reduction of the transformation efficiency (Table 1).
Impact of sonication on transformation efficiency
The duration of sonication also strongly influenced the survival and regeneration of the explants. The germination percentage of the Agrobacterium-infected explants, number of plants survived against BASTA® spray, and the plants with gus A gene expression, improved with increasing sonication duration and 6 min sonication was found as an ideal duration to attain 25.3% of transformation efficiency (Table 1). Longer sonication duration (beyond 6 min) showed adverse effects on germination of seeds which finally led to a reduction in the efficiency of transformation (Table 1).
Effect of vacuum infiltration on transformation efficiency
In the current study, the period of vacuum infiltration played an important role in achieving higher transformation percentage. Among the various time durations, 3 min vacuum infiltration of sonicated seed explants (6 min sonication) in Agrobacterium suspension resulted in highest transformation efficiency of 33.6% (Table 1). Above 3 min vacuum infiltration, a gradual reduction in the germination rate was observed, which eventually resulted in a reduction of transformation efficiency (Table 1).
Selection of transformed peanut plants
In the current study, the Agrobacterium cells adhering to the co-cultivated explants (Fig. 1c) were removed by washing them with sterile liquid MS medium containing cefotaxime (250 mg l−1) and then inoculated them onto the MS medium supplemented with 4 mg l−1 BASTA®. The explants which were not transformed failed to develop into the seedlings (Fig. 1d). The putatively transformed explants developed into seedlings with prominent roots and shoot (Fig. 1e, f), and within 15 days they grew into plantlets (Fig. 1g, h). The individual plantlets were transplanted into the paper cups having sterile planting mixture (Fig. 1i) and allowed to harden for 15 days in the growth chamber (Fig. 1i). The second round of selection was applied to 40-day-old plants by spraying BASTA® (250 mg l−1). Following BASTA® spray even after 1 week, the putatively transformed plants were healthy and grew further (Fig. 1j). While, the chimeric and non-transformed plants exhibited chlorosis, necrosis and leaf burning (Fig. 1k), which finally perished within a week after BASTA® spray. The BASTA®-resistant putatively transformed plants were allowed to grow (Fig. 1l) and subjected to further analysis.
GUS histochemical analysis
The putatively transformed seedlings and plantlets displayed a strong blue color (Fig. 2a, b) indicating the integration and expression of gus A gene in the plant genome. The non-transformed (control) seedlings and plantlets did not develop a blue color (Fig. 2c, d).
Molecular confirmation of putatively transformed plants
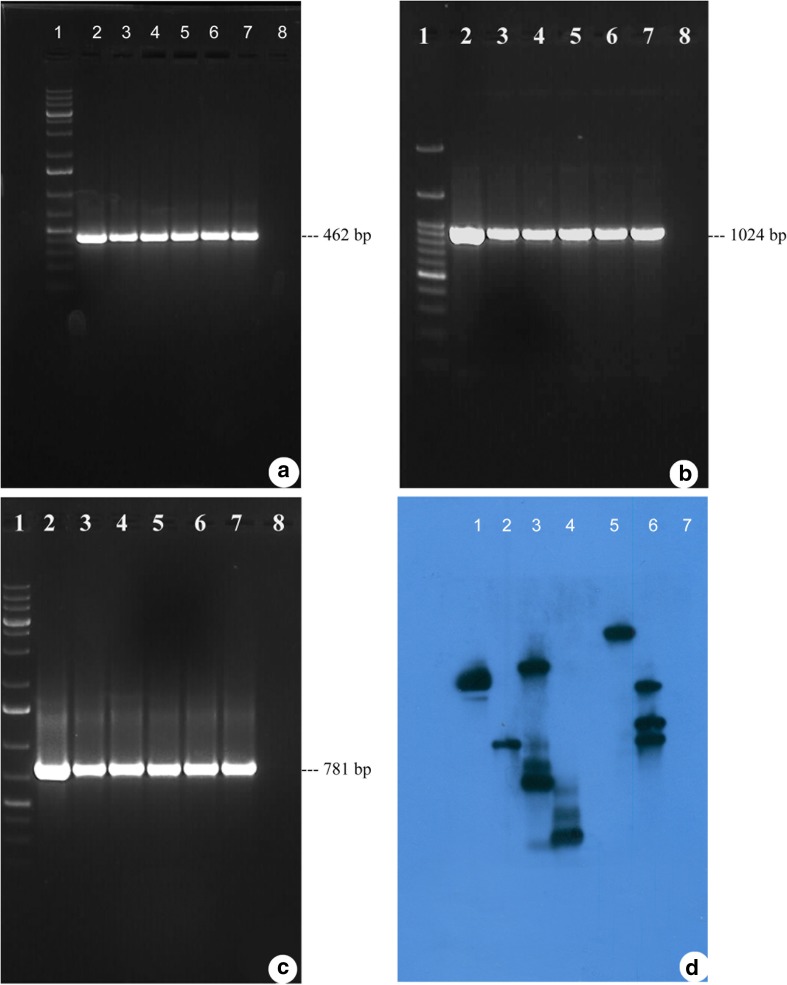
The transgene integration into the genome of the putatively transformed plants was confirmed by PCR and Southern blot hybridization. The presence of amplified fragment of 462 bp (Fig. 3a, lanes 3–7), 1024 bp (Fig. 3b, lanes 3–7), and 781 bp (Fig. 3c, lanes 3–7) in the genomic DNA from putatively transformed plants and binary vector pCAMBIA1301–bar (Fig. 3a–c, lane 2) confirmed the presence and integration of bar, hptII and gus A genes, respectively, into the plant genome. Further, the genomic DNA of the control plant did not show any amplification (Fig. 3a–c, lane 8).
Fig. 3.
Screening of putatively transformed plants by PCR and Southern blot hybridization. a, b, c PCR analysis of putatively transformed peanut plants using bar, hptII and gus A gene-specific primers, respectively. Lane 1, 1 kb plus DNA ladder; lane 2, pCAMBIA1301–bar plasmid as a positive control; lanes 3–7, genomic DNA from putatively transformed peanut plants; lane 8, control (non-transformed) peanut plant genomic DNA as a negative control. d Southern blot hybridization of PCR-positive putatively transformed peanut plants. Lanes 1–4 and 6, genomic DNA from putatively transformed peanut plants; lane 5, pCAMBIA1301–bar plasmid as a positive control; lane 7, control (non-transformed) peanut plant genomic DNA as a negative control
The integration and the copy number of the bar gene in the putatively transformed plant genome was confirmed by Southern blot hybridization. The putatively transformed plants showed a single copy (lane 2), two copies (lane1), three copies (lanes 4 and 6), and five copies (lane 3) of the integrated bar gene in their genome (Fig. 3d). The binary vector pCAMBIA1301–bar plasmid showing a hybridization signal, acted as a positive control (Fig. 3d, lane 5), while genomic DNA isolated from the non-transformed (control) plants did not develop any hybridization signal and acted as a negative control (Fig. 3d, lane 7).
Stable inheritance analysis of transformed plants
Ten seeds (T1) were collected from each of the 5 Southern blot hybridization-positive plants (S1, S2, S3, S4 and S5) of cv. CO7 and germinated on the medium containing 4 mg 1−1 BASTA®. The germinated seedlings with healthy roots and shoot exhibited intense blue color after X-gluc staining. Further, the PCR amplification of gus A gene in the progeny plants (Fig. 4a–e; Table 2) clearly demonstrated the stable transgene inheritance into their respective progenies following Mendelian inheritance ratio (3:1).
Fig. 4.
Analysis of gene segregation in the transformed peanut plants (T1 plants) by PCR using gus A gene-specific primers. a–e Transformed peanut plant lines S1, S2, S3, S4, and S5, respectively
Table 2.
Analysis of gene segregation in the transformed (T1) peanut cv. CO7 plants by PCR
| Transformed line | No. of plants analyzed | No. of PCR-positive plants | No. of PCR-negative plants |
|---|---|---|---|
| S1 | 10 | 7 | 3 |
| S2 | 10 | 8 | 2 |
| S3 | 10 | 7 | 3 |
| S4 | 10 | 8 | 2 |
| S5 | 10 | 8 | 2 |
Influence of genotype on transformation efficiency
The in planta transformation protocol optimized for cv. CO7 (Fig. 5) was used to transform other four popular groundnut cultivars (CO6, TMV2, TMV7 and VR13). All the tested cultivars responded well for in planta transformation with more than 30% transformation efficiency (Table 3). Among the five cultivars evaluated, cv. TMV7 showed higher transformation efficiency of 38.6% (Table 3). The obtained results indicate that optimized in planta transformation method developed in this study is cultivar independent and could be applied to a wide range of peanut cultivars.
Fig. 5.

Flowchart representation showing various steps of in planta genetic transformation of peanut cv. CO7
Table 3.
Competency of in planta transformation methodology on different peanut cultivars
| Cultivars used for transformation | No. of seeds infected | Mean no. of germinated seeds on selection mediuma | Mean no. of plants surviving after BASTA® sprayb | Mean no. of plants expressing gus A gene | Transformation efficiency (%) |
|---|---|---|---|---|---|
| CO7 | 100 | 44.3 ± 0.26d | 35.6 ± 0.32d | 31.3 ± 0.24e | 31.3 ± 0.24e |
| CO6 | 100 | 47.0 ± 0.21b | 38.3 ± 0.22c | 33.6 ± 0.28d | 33.6 ± 0.28d |
| TMV2 | 100 | 45.6 ± 0.25ca | 40.0 ± 0.25b | 36.3 ± 0.21b | 36.3 ± 0.21b |
| TMV7 | 100 | 49.3 ± 0.30a | 44.3 ± 0.26a | 38.6 ± 0.25a | 38.6 ± 0.25a |
| VR13 | 100 | 46.0 ± 0.21c | 39.6 ± 0.38ba | 35.0 ± 0.25c | 35.0 ± 0.25c |
The explants from diverse peanut cultivars were pre-cultured for 2 days, sonicated for 6 min, and then vacuum infiltrated for 3 min in A. tumefaciens EHA105 harboring pCAMBIA1301–bar plasmid suspension. The infected explants were incubated for 3 days on MS medium containing 150 µM acetosyringone
Transformation efficiency = no. of GUS-positive plants/no. of infected seeds × 100. Mean values of three separate trials (±) with standard errors. In each column, numbers with different letters indicate they are considerably different from each other according to Duncan’s multiple range test at a probability level of 5%
aInfected explants were propagated in MS medium supplemented with 4 mg l−1 BASTA®
b40-day-old plants were sprinkled with 250 mg l−1 BASTA®. Visual observations and the data were documented after 1 week
Discussion
Production of transgenic plants within a short period by avoiding tissue culture phase is always advantageous for biotechnological improvement of crop plants. Previous reports from our research group mainly focused on using seed as an explant, and obtained higher transformation efficiency for sugarcane, brinjal, snake gourd, and okra (Mayavan et al. 2013; Subramanyam et al. 2013, 2015; Manickavasagam et al. 2015). Although a seed-based in planta transformation for peanut was reported previously by Rohini and Rao (2000), lower transformation efficiency (3.3%) obtained in their study made us investigate the various parameters to improve the peanut transformation efficiency.
Screening of transformants is the foremost and vital step in the production of transformed plants. Efficient selection agent at an optimum concentration prevents the development of non-transformants, but allows the transformed tissue/plant to develop further. Hence, it is necessary to choose an efficient selection marker and stringent concentration of selection agent for screening the putatively transformed plants during the in planta transformation. The bar gene conferring resistance to BASTA®, is considered as an efficient selection marker for screening the putatively transformed plants during the in planta transformation (Subramanyam et al. 2015). Moreover, based on the previous reports (Mayavan et al. 2013; Subramanyam et al. 2015; Manickavasagam et al. 2015) from our research group, we strongly believe that 2-stage selection completely eliminates the non-transformants. Hence, we determined the MIC of BASTA® initially during the germination of seed and later on 40-day-old plants, which was determined as 4 and 250 mg l−1, respectively. Similar to the present study, Subramanyam et al. (2015) screened the putatively transformed snake gourd seedlings in the medium containing 10 mg l−1 BASTA® and 1-month-aged plants by sprinkling BASTA® (150 mg l−1). Mayavan et al. (2013) reported that the transformed sugarcane seedlings and 2-month-aged plants were selected by using 2 mg l−1 and 2 g l−1 BASTA®, respectively. In the same way, 2-stage selection processes completely eliminated the development of non-transformed plants of Brassica rapa, brinjal, and okra (Qing et al. 2000; Subramanyam et al. 2013; Manickavasagam et al. 2015).
Based on our investigation, it has been proven that pre-culturing explants prior to Agrobacterium infection is not necessarily essential to obtain transformed peanut plants. However, pre-culturing prior Agrobacterium infection significantly improved the transformation efficiency. Pre-culturing the explants results in active cell division and, the actively dividing cells are more prone to T-DNA delivery and integration (Mariashibu et al. 2013). Pre-culture also averts the negative effects of Agrobacterium and selection agent on cell division (Mayavan et al. 2013). We observed that pre-culturing the explants for 2 days prior to infection, drastically enhanced the transformation efficiency by 4.7%. The results of the current study were in agreement with those of Athmaram et al. (2006), Mayavan et al. (2013) and Subramanyam et al. (2013) who observed higher transformation efficiency in pre-cultured peanut somatic embryos, sugarcane, and brinjal seeds, respectively. On the contrary, some researchers reported the negative effect of pre-culture on the transformation efficiency of peanut, citrange, and strawberry (Egnin et al. 1998; Cervera et al. 1998; Haddadi et al. 2015).
During the co-cultivation, T-DNA releases from the Agrobacterium and integrates into the genome of the plant cell within a stipulated time and this duration depends on the plant species and the Agrobacterium strain used for infection (Uranbey et al. 2005). The peanut transformation efficiency enhanced further by 6.3% when co-cultivated for 3 days. However, beyond 3 days of co-cultivation, the overgrown Agrobacterium restricted the explant germination and ultimately led to a reduction in the transformation efficiency. Similar observations were made by Mayavan et al. (2013) in sugarcane where the transformation efficiency gradually increased and higher efficiency (24.2%) was observed after co-cultivating seed explants for 3 days; however, beyond 3 days the transformation efficiency declined owing to the overgrowth of Agrobacterium. Manickavasagam et al. (2015) also reported the importance of optimum co-cultivation duration (3 days) in enhancing the efficiency of in planta transformation for okra. Park et al. (2005) and Chen et al. (2010) too reported that co-cultivation for 3 days improved in planta transformation efficiency in radish and switch grass, respectively.
Another important factor that might influence transformation efficiency is acetosyringone. Plant tissues when wounded release phenolic compounds. The phenolic compounds act as signal molecules to attract the Agrobacterium towards the wounding site and activate the vir genes essential for transfer and integration of the T-DNA into the plant cell (Subramanyam et al. 2011). However, some plant species secrete limited amount of phenolics and hence to accelerate the transformation efficiency it is necessary to provide exogenous phenolic compounds during the Agrobacterium-mediated genetic transformation. Acetosyringone is the most widely used phenolic compound in the Agrobacterium-mediated genetic transformation (Stachel et al. 1985; Fortin et al. 1992), and in our study, we observed that supplementation of acetosyringone (150 µM) enhanced transformation rate by 5.3%. However, further increase in acetosyringone concentration, reduced transformation efficiency probably owing to the increase in the concentration of the alcohol that was used for dissolving acetosyringone (Subramaniam et al. 2009). Similar interpretations were made by Manickavasagam et al. (2015) and Subramanyam et al. (2013, 2015) wherein they observed increased transformation efficiency with an optimum concentration of acetosyringone, and a higher concentration of acetosyringone negatively affected the seed germination and transformation efficiency in okra, brinjal, and snake gourd, respectively.
Apart from using exogenous supplements to enhance transformation, physical treatment of explants might also enhance the transformation rate by rendering the explants more amenable to undergo infection. Sonication is a mechanical process to create the microcavities across the explant, and these microcavities act as a channel for Agrobacterium to reach the meristematic region of the explant (Mayavan et al. 2015). In addition, mechanical wounding by sonication may also release the phenolic compounds necessary for the Agrobacterium to infect the meristematic cells of the explant. Sonication of the explant before the Agrobacterium infection considerably enhanced the transformation percentage of several crops including radish, chick pea, Brassica napus, and brinjal (Park et al. 2005; Pathak and Hamzah 2008; Li et al. 2009; Subramanyam et al. 2013). In this study, sonication for 6 min enhanced transformation efficiency further by 5.7%. Results acquired in this study are in consensus with the earlier reports of Mayavan et al. (2013), Subramanyam et al. (2015) and Manickavasagam et al. (2015) who observed improved transformation efficiency by sonicating the seeds of sugarcane, snake gourd, and okra, respectively, for an optimum duration. Further, they also reported the negative role of longer duration of sonication on seed germination and transformation efficiency, which might be due to the unrecoverable damage caused on the explant surface by excessive sonication.
During the vacuum infiltration, a negative pressure is produced by the vacuum generated, which eliminates air from the microcavities and forms hollow spaces across the explant surface. The Agrobacterium suspension is forced into the empty spaces with an increase in the pressure, which effectively infects the meristematic cells of the explant (Mariashibu et al. 2013). The vacuum infiltration was successfully employed to infect the sonicated seed explants of radish, kidney bean, and citrus (Park et al. 2005; Liu et al. 2005; De Oliveira et al. 2009). In our study, vacuum infiltration following sonication enhanced transformation efficiency by 8.3%, further increase in the vacuum infiltration above optimal duration resulted in reduced transformation efficiency which might be due to the over accumulation of Agrobacterium in tissue crevices. Similar observations were recorded for okra and snake gourd, in which 3 min vacuum infiltration of sonicated seed explants with Agrobacterium suspension significantly improved the transformation efficiency, while longer vacuum infiltration resulted in lower transformation efficiency due to the uncontrolled growth of Agrobacterium (Manickavasagam et al. 2015; Subramanyam et al. 2015).
Following genetic transformation of explants, applying stringent selection pressure with an efficient selection agent at a specific developmental stage is a crucial step to eliminate the chimeric and non-transformed plants. Hence, we followed 2-stage selection process as suggested by Mayavan et al. (2013) and Subramanyam et al. (2015) for sugarcane and snake gourd, respectively. Screening the transformants at seedling stage using 4 mg l−1 BASTA® and at 40-day-old plant stage with 250 mg l−1 BASTA®, we were able to recover transgenic plants with reduced incidence of escapes. Segregation is also an important phenomenon to consider during the production of transgenic plants. Hence, we evaluated the segregation by seed germination assay, GUS histochemical analysis and PCR. The obtained results proved that the transgene was segregated into their progeny plants in 3:1 ratio.
The reliability of the transformation protocol depends on the wide applicability over different cultivars. Hence, the in planta transformation procedure established (Fig. 5) in this study was evaluated against a wide range of peanut cultivars including CO6, TMV7, TMV2 and VR13. All the tested cultivars responded positively to in planta transformation and developed BASTA®-resistant putatively transformed plants. GUS histochemical analysis confirmed the gus A gene expression in the putatively transformed plants which indicates that the in planta genetic transformation protocol developed in the present study could be used to transform wide range of peanut cultivars.
Conclusion
In this study, an efficient Agrobacterium-mediated in planta genetic transformation methodology was developed for peanut. Various parameters such as, pre-culture duration, acetosyringone concentration, duration of co-cultivation, sonication, and vacuum infiltration were standardized to achieve higher transformation efficiency. The segregation study showed that the transgene was positively inherited in the progeny plants. The amicability of the standardized procedure was evaluated against various peanut cultivars, and all the tested peanut cultivars responded well for transformation and produced higher transformation efficiencies. As per our knowledge, this is the first report on evaluating the various elements influencing in planta transformation of peanut. The optimized in planta genetic transformation methodology established in this study could be beneficial to improve various peanut cultivars with desirable genetic traits.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (TIFF 76 kb) Supplementary Fig. 1 Illustration of the T-DNA region of the binary vector pCAMBIA1301–bar
Supplementary material 2 (TIFF 2296 kb) Supplementary Fig. 2 Evaluation of the germination percentage of 5 diverse peanut cultivars. One hundred seeds from each cultivar were used for the germination test. Mean values of three separate trials (±) with standard errors
Acknowledgements
Sivabalan Karthik, is grateful to Jawaharlal Nehru Memorial Fund, New Delhi, India, for the award of Jawaharlal Nehru Scholarship (Ref no: SU-1/88/2016-17/79) for his doctoral research.
Abbreviations
- CaMV 35S
Cauliflower mosaic virus 35S promoter
- gusA
β-Glucuronidase gene
- hptII
Hygromycin phosphotransferase
- MS
Murashige and Skoog medium
- nos Poly A
Nopaline synthase terminator
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1231-1) contains supplementary material, which is available to authorized users.
Sivabalan Karthik and Gadamchetty Pavan have contributed equally to this work.
References
- Anuradha TS, Divya K, Jami SK, Kirti PB. Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens. Plant Cell Rep. 2008;27:1777–1786. doi: 10.1007/s00299-008-0596-8. [DOI] [PubMed] [Google Scholar]
- Athmaram TN, Bali G, Devaiah KM. Integration and expression of Bluetongue VP2 gene in somatic embryos of peanut through particle bombardment method. Vaccine. 2006;24:2994–3000. doi: 10.1016/j.vaccine.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Cervera M, Pina JA, Juarez J, Navarro L, Pena L. Agrobacterium-mediated transformation of citrange: factors affecting transformation and regeneration. Plant Cell Rep. 1998;18:271–278. doi: 10.1007/s002990050570. [DOI] [PubMed] [Google Scholar]
- Chen X, Equi R, Baxter H, Berk K, Han J, Agarwal S, Zale J. A high-throughput transient gene expression system for switchgrass seedlings. Biotechnol Biofuels. 2010;3:9. doi: 10.1186/1754-6834-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Bhattacharya A, Wu C, Knoll JE, Ozias-Akins P. Improvement of peanut (Arachis hypogaea L.) transformation efficiency and determination of transgene copy number by relative quantitative real-time PCR. In Vitro Cell Dev Biol Plant. 2013;49:266–275. doi: 10.1007/s11627-013-9518-8. [DOI] [Google Scholar]
- De Oliveira MLP, Febres VJ, Costa MGC, Moore GA, Otoni WC. High-efficiency Agrobacterium-mediated transformation of citrus via sonication and vacuum infiltration. Plant Cell Rep. 2009;28:387–395. doi: 10.1007/s00299-008-0646-2. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA mini preparation: version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Egnin M, Mora A, Prakash CS. Factors enhancing Agrobacterium tumefaciens-mediated gene transfer in peanut (Arachis hypogaea L.) In Vitro Cell Dev Biol Plant. 1998;34:310–318. doi: 10.1007/BF02822740. [DOI] [PubMed] [Google Scholar]
- Enserink M. The peanut butter debate. Science. 2008;322:36–38. doi: 10.1126/science.322.5898.36. [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2014) Agricultural data. http://www.fao.org/faostat/en/#data/QC/visualize Accessed 16 Nov 2017
- Feldmann KA, Marks MD. Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol Gen Genet. 1987;208:1–9. doi: 10.1007/BF00330414. [DOI] [Google Scholar]
- Fortin C, Nester EW, Dion P. Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J Bacteriol. 1992;174:5676–5685. doi: 10.1128/jb.174.17.5676-5685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin CI, Shorrosh KM, Trieu AN, Cassidy BG, Nelson RS. Stable transformation of peanut callus via Agrobacterium-mediated DNA transfer. Transgenic Res. 1993;2:321–324. doi: 10.1007/BF01976172. [DOI] [PubMed] [Google Scholar]
- Haddadi F, Aziz MA, Abdullah SN, Tan SG, Kamaladini H. An efficient Agrobacterium-mediated transformation of strawberry cv. Camarosa by a dual plasmid system. Molecules. 2015;20:3647–3666. doi: 10.3390/molecules20033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Helmer GC, Fraley RT, Chilton MD. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in the region of pTiBo542 outside the T-DNA. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgen Res. 1993;2:208–218. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- Hsieh YF, Jain M, Jianping Wang J, Gallo M. Direct organogenesis from cotyledonary node explants suitable for Agrobacterium-mediated transformation in peanut (Arachis hypogaea L.) Plant Cell Tissue Org Cult. 2017;128:161–175. doi: 10.1007/s11240-016-1095-1. [DOI] [Google Scholar]
- Iqbal MM, Nazir F, Ali S, Asif MA, Zafar Y, Iqbal J, Ali GM. Over expression of rice chitinase gene in transgenic peanut (Arachis hypogaea L.) improves resistance against leaf spot. Mol Biotechnol. 2012;50:129–136. doi: 10.1007/s12033-011-9426-2. [DOI] [PubMed] [Google Scholar]
- Jaganath B, Subramanyam K, Mayavan S, Karthik S, Elayaraja D, Udayakumar R, Manickavasagam M, Ganapathi A. An efficient in planta transformation of Jatropha curcas (L.) and multiplication of transformed plants through in vivo grafting. Protoplasma. 2014;251:591–601. doi: 10.1007/s00709-013-0558-z. [DOI] [PubMed] [Google Scholar]
- Janila P, Nigam SN, Pandey MK, Nagesh P, Varshney RK. Groundnut improvement: use of genetic and genomic tools. Front Plant Sci. 2013;4:23. doi: 10.3389/fpls.2013.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan NW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna G, Singh BK, Kim EK, Morya VK, Ramteke PW. Progress in genetic engineering of peanut (Arachis hypogaea L.)—a review. Plant Biotechnol J. 2015;13:147–162. doi: 10.1111/pbi.12339. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao DG, Wu YJ, Tian X. A simplified seed transformation method for obtaining transgenic Brassica napus plants. Agric Sci China. 2009;8:658–663. doi: 10.1016/S1671-2927(08)60261-8. [DOI] [Google Scholar]
- Liu Z, Park BJ, Kanno A, Kameya T. The novel use of a combination of sonication and vacuum infiltration in Agrobacterium-mediated transformation of kidney bean (Phaseolus vulgaris L.) with lea gene. Mol Breed. 2005;16:189–197. doi: 10.1007/s11032-005-6616-2. [DOI] [Google Scholar]
- Manickavasagam M, Subramanyam K, Ishwarya R, Elayaraja D, Ganapathi A. Assessment of factors influencing the tissue culture-independent Agrobacterium-mediated in planta genetic transformation of okra [Abelmoschus esculentus (L.) Moench] Plant Cell Tissue Org Cult. 2015;123:309–320. doi: 10.1007/s11240-015-0836-x. [DOI] [Google Scholar]
- Mariashibu TS, Subramanyam K, Arun M, Mayavan S, Rajesh M, Theboral J, Manickavasagam M, Ganapathi A. Vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in Indian soybean cultivars. Acta Physiol Plant. 2013;35:41–54. doi: 10.1007/s11738-012-1046-3. [DOI] [PubMed] [Google Scholar]
- Mayavan S, Subramanyam K, Arun M, Rajesh M, Dev GK, Sivanandhan G, Jaganath B, Manickavasagam M, Selvaraj N, Ganapathi A. Agrobacterium tumefaciens-mediated in planta seed transformation strategy in sugarcane. Plant Cell Rep. 2013;32:1557–1574. doi: 10.1007/s00299-013-1467-5. [DOI] [PubMed] [Google Scholar]
- Mayavan S, Subramanyam K, Jaganath B, Sathish D, Manickavasagam M, Ganapthi A. Agrobacterium-mediated in planta genetic transformation of sugarcane setts. Plant Cell Rep. 2015;34:1835–1848. doi: 10.1007/s00299-015-1831-8. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Park BJ, Liu Z, Kanno A, Kameya T. Transformation of radish (Raphanus sativus L.) via sonication and vacuum infiltration of germinated seeds with Agrobacterium harbouring a group 3 LEA gene from B. napus. Plant Cell Rep. 2005;24:494–500. doi: 10.1007/s00299-005-0973-5. [DOI] [PubMed] [Google Scholar]
- Pathak MR, Hamzah RY. An effective method of sonicated assisted Agrobacterium-mediated transformation of chickpea. Plant Cell Tissue Org Cult. 2008;93:65–67. doi: 10.1007/s11240-008-9344-6. [DOI] [Google Scholar]
- Qing CM, Fan L, Lei Y, Bouchez D, Tourneur C, Yan L, Robaglia C. Transformation of Pakchoi (Brassica rapa L. ssp. chinensis) by Agrobacterium infiltration. Mol Breed. 2000;6:67–72. doi: 10.1023/A:1009658128964. [DOI] [Google Scholar]
- Qiusheng Z, Bao J, Likun L, Xianhua X. Effects of antioxidants on the plant regeneration and gus expressive frequency of peanut (Arachis hypogaea) explants by Agrobacterium tumefaciens. Plant Cell Tissue Org Cult. 2005;81:83–90. doi: 10.1007/s11240-004-3176-9. [DOI] [Google Scholar]
- Rohini VK, Rao KS. Transformation of peanut (Arachis hypogaea L.): a non-tissue culture based approach for generating transgenic plants. Plant Sci. 2000;15:41–49. doi: 10.1016/S0168-9452(99)00160-0. [DOI] [Google Scholar]
- Rustom IYS, Lopez-Leiva MH, Nair BM. Nutritional, sensory and physicochemical properties of peanut beverage sterilized under two different UHT conditions. Food Chem. 1996;56:45–53. doi: 10.1016/0308-8146(95)00153-0. [DOI] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. New York: Cold Spring Harbor Press; 1989. [Google Scholar]
- Stachel SE, Messens E, Van Montagu M, Zambryski PC. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature. 1985;318:624–629. doi: 10.1038/318624a0. [DOI] [Google Scholar]
- Subramaniam S, Samian R, Midrarullah Rathinam X. Preliminary factors influencing transient expression of gus A in Dendrobium savin white protocorm-like bodies (PLBs) using Agrobacterium-mediated transformation system. WASJ. 2009;7:1295–1307. [Google Scholar]
- Subramanyam K, Subramanyam K, Sailaja KV, Srinivasulu M, Lakshmidevi K. Highly efficient Agrobacterium-mediated transformation of banana cv. Rasthali (AAB) via sonication and vacuum infiltration. Plant Cell Rep. 2011;30:425–436. doi: 10.1007/s00299-010-0996-4. [DOI] [PubMed] [Google Scholar]
- Subramanyam K, Rajesh M, Jaganath B, Vasuki A, Theboral J, Elayaraja D, Karthik S, Manickavasagam M, Ganapathi A. Assessment of factors influencing the Agrobacterium-mediated in planta seed transformation of brinjal (Solanum melongena L.) Appl Biochem Biotechnol. 2013;171:45–468. doi: 10.1007/s12010-013-0359-z. [DOI] [PubMed] [Google Scholar]
- Subramanyam K, Arunachalam C, Thaneswari RM, Sulaiman AA, Manickavasagam M, Ganapathi A. Highly efficient Agrobacterium-mediated in planta genetic transformation of snake gourd (Tricosanthes cucumerina L.) Plant Cell Tissue Org Cult. 2015;123:133–142. doi: 10.1007/s11240-015-0821-4. [DOI] [Google Scholar]
- Uranbey S, Sevimay CS, Kaya MD, Ipek A, Sancak C, Basalma D, Er C, Ozcan S. Influence of different co-cultivation temperatures, periods and media on Agrobacterium tumefaciens-mediated gene transfer. Biol Plant. 2005;49:53–57. doi: 10.1007/s10535-005-3057-z. [DOI] [Google Scholar]
- USDA (2016) Food composition database. https://ndb.nal.usda.gov/ndb/search/list?qlookup=16087 Accessed 16 Nov 2017
- Yasmeen A, Mirza B, Inayatullah S, Safdar N, Jamil M, Ali S, Choudhry MF. In planta transformation of tomato. Plant Mol Biol Rep. 2009;27:20–28. doi: 10.1007/s11105-008-0044-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (TIFF 76 kb) Supplementary Fig. 1 Illustration of the T-DNA region of the binary vector pCAMBIA1301–bar
Supplementary material 2 (TIFF 2296 kb) Supplementary Fig. 2 Evaluation of the germination percentage of 5 diverse peanut cultivars. One hundred seeds from each cultivar were used for the germination test. Mean values of three separate trials (±) with standard errors