Abstract
Familial hypercholesterolemia (FH) is the most common inherited form of dyslipidemia and a major cause of premature cardiovascular disease. Management of FH mainly relies on the efficiency of treatments that reduce plasma low-density lipoprotein (LDL) cholesterol (LDL-C) concentrations. MicroRNAs (miRs) have been suggested as emerging regulators of plasma LDL-C concentrations. Notably, there is evidence showing that miRs can regulate the post-transcriptional expression of genes involved in the pathogenesis of FH, including LDLR, APOB, PCSK9, and LDLRAP1. In addition, many miRs are located in genomic loci associated with abnormal levels of circulating lipids and lipoproteins in human plasma. The strong regulatory effects of miRs on the expression of FH-associated genes support of the notion that manipulation of miRs might serve as a potential novel therapeutic approach. The present review describes miRs-targeting FH-associated genes that could be used as potential therapeutic targets in patients with FH or other severe dyslipidemias.
Keywords: Apolipoprotein B, Low-density lipoprotein cholesterol, LDL receptor, LDLRAP1, microRNA, PCSK9
Necessity of New Therapeutic Options for FH Patients
Familial hypercholesterolemia (FH) is a frequent, severe, and mostly autosomal dominant genetic disorder associated with elevated plasma low-density lipoprotein cholesterol (LDL-C) levels that predisposes patients to premature cardiovascular disease (CVD), especially if remain undiagnosed or inadequately treated [1–3].
The disorder most commonly results from loss-of-function mutations in the LDLR gene encoding LDL receptor protein, and genes encoding for proteins that interact with the receptor, including apolipoprotein B (APOB), proprotein convertase subtilisin/kexin type 9 (PCSK9), or LDLR adaptor protein 1 (LDLRAP1) [4, 5]. These pathogenic mutations cause deficient clearance of circulating LDL particles via hepatic LDLR leading to increased plasma LDL-C levels from birth and deposition in the arterial wall, thus accelerating atherosclerosis and the risk of premature CVD [6, 7].
Statins, ezetimibe, bile acid sequestrants, and more recently PCSK9 inhibitors are the main therapeutic drugs for the treatment of heterozygous FH (HeFH); all of which work solely or predominantly via increased LDLR activity and LDL-C clearance. Additional LDL-C-lowering can be achieved in homozygous FH (HoFH) patients by decreasing production of LDL-C, or its precursors, with either the microsomal triglyceride transfer protein (MTP) inhibitor, lomitapide, or the antisense oligonucleotide which reduces apoB synthesis, mipomersen. LDL-C reduction can also be achieved by regular LDL apheresis which mechanically removes LDL. Statins and ezetimibe are the most commonly used cholesterol-lowering drugs and are now generically available at low cost in most countries [8–10]. Nevertheless, despite the 50 to 60% LDL-C reduction achievable by these two generally well-tolerated agents, a large number of FH patients still do not reach the recommended LDL-C goals due to their very high baseline LDL-C levels [2, 3]. In HeFH patients who already have evidence of CVD, less than 10 to 20% achieve a LDL-C level below 70 mg/dL on conventional drug therapy [11]. In HoFH patients, where the typical phenotype has untreated LDL-C > 500 mg/dL and at only minimal to moderate residual LDLR activity, even high doses of atorvastatin or rosuvastatin reduce mean LDL-C by only 22–25% [12, 13], and ezetimibe achieves an additional 20% reduction [14], thus few, if any, HoFH patients can reach anywhere near optimal LDL-C levels.
Therefore, alternative therapeutics either alone or in combination with current approved therapies can be considered to reduce residual disease burden in patients with FH [15]. In this context, modulation of microRNAs (miRs)-dependent gene expression represents a promising approach.
RNA-Based Therapeutic Approaches
In the past decade, RNA-based approaches have shown potential as novel therapies for human disorders. These therapeutic approaches are indebted to the advances in knowledge of the RNA field. As revealed by ENCODE (Encyclopedia of DNA Elements) project, a multi-center study aiming to find a collection of functional elements in the human genome via sequencing RNA from various sources, more than 90% of the human genome contains non-coding RNAs that can affect other coding sequences of the genome [16]. Several new classes of non-coding RNAs associated with the most disparate and critical functions have been identified [17]. Among these, small interfering RNAs (siRNAs) and miRs have attracted considerable interest for drug discovery and development because of their important role in gene regulation [18].
RNA Interference Pathway
miRs and siRNAs are endogenously transcribed non-coding short hairpin RNAs that inhibit gene expression through RNA interference (RNAi) pathways [19–21]. RNAi is a highly conserved cellular mechanism present in the most eukaryotic cells that interferes with post-transcription steps and consequently silences the expression of homologous genes [21, 22]. Either double-stranded miRs or siRNAs, composed of a passenger strand (sense strand) and a guide strand (antisense strand), can interact with and activate the RNA-induced silencing complex (RISC). The passenger strand is cleaved by the endonuclease argonaute 2 (AGO2) of the RISC, while the guide strand remains associated with the RISC. Subsequently, the guide strand directs the active RISC to target mRNA that is cleaved by AGO2 component [22, 23]. miRs inhibit the expression of genes by hybridizing guide strand to partially complementary binding sites typically localized in the 3′ untranslated regions (3′ UTR) of target mRNAs, while siRNA guide strand only binds to mRNA that is fully complementary to it, causing specific gene silencing [24, 25]. In rare cases, mRNAs contain highly complementary miRNA-binding sites and therefore miRs guide the sequence-specific cleavage of the mRNA in a process similar to that mediated by siRNAs [25]. Mechanistically, efficient inhibition is either supplied by interfering with translation or by predisposing mRNAs to degradation that is initiated by deadenylation and decapping of the mRNAs [24].
Therapeutic Potential of RNAi-Based Therapeutics
The therapeutic potential of miRs and siRNAs has been shown in the treatment of many different diseases such as cancers [26–29], infections [29–32], and cardiovascular diseases [33, 34]. The siRNA-based therapeutic approaches involve the delivery of a synthetic siRNA into the target cells aimed at suppressing the expression of a specific mRNA, and provide a gene silencing effect [35]. The siRNA-based cholesterol-lowering agent inclisiran (formerly ALN-PCS) has recently successfully completed a phase II trial in 497 patients with a baseline LDL-C of ~130 mg/dL [36]. Patients receiving a single subcutaneous injection of 300 mg of inclisiran achieved mean LDL-C reductions of 51 and 45% at Days 60 and 90, respectively (p < 0.0001 compared to placebo). Inclisiran was generally well tolerated with treatment emergent adverse events of 54% both in patients randomized to placebo or inclisiran, and with no differences between inclisiran doses. Injection site reactions (ISRs) with inclisiran were seen in 3.2% of patients, and were mild or moderate, and mostly transient. Inclisiran thus appears to provide a durable reduction in PCSK9 and LDL-C levels with small and infrequent subcutaneous injections [36].
In contrast to the siRNA approach, therapeutic procedures based on miRs embrace two different strategies including miR inhibition and miR replacement. The former strategy exploits antisense therapy to suppress the action of the endogenous miRs by using synthetic single-stranded RNAs acting as miR antagonists with sequences complementary to the endogenous miR [37]. The miR antagonists include anti-miRs, locked-nucleic acids (LNA) or antagomiRs carrying chemical modifications that magnify the affinity for the target miR and trap the endogenous miR in a configuration that cannot be incorporated into RISC complex, or cause degradation of the endogenous miR [38].
In the case of replacement strategy, synthetic miRs (miR mimics) are applied to mimic the function of the endogenous miRs causing mRNA inhibition/degradation, and exert a gene silencing effect [38].
Although siRNAs have been frequently used for silencing the target genes, and even a few siRNAs are studied in clinical trials [39], there are several fundamental disadvantages which considerably limit therapeutic use of siRNA methodology. The most important drawbacks include scrambling in the biogenesis of the endogenous miR, evocation of the interferon response, and the off-target effects [40–43]. On the other hand, the unique biogenesis and mechanism of miR action enables it to be freed from aforementioned limitations. Impressive advantages of miR-based tools, such as specificity and safety, and the fascinating feature of multiple-targeting potential nominate miRs as a useful therapeutic approach [44].
miRs as Therapeutic Target and Tool for FH Therapy
Since miRs are known to have a pivotal role in LDL-C metabolism [45] and dysregulated miRs have been frequently found to be involved in the pathogenesis of FH and various CVDs, they have been suggested as potential targets for therapeutic intervention [46–49].
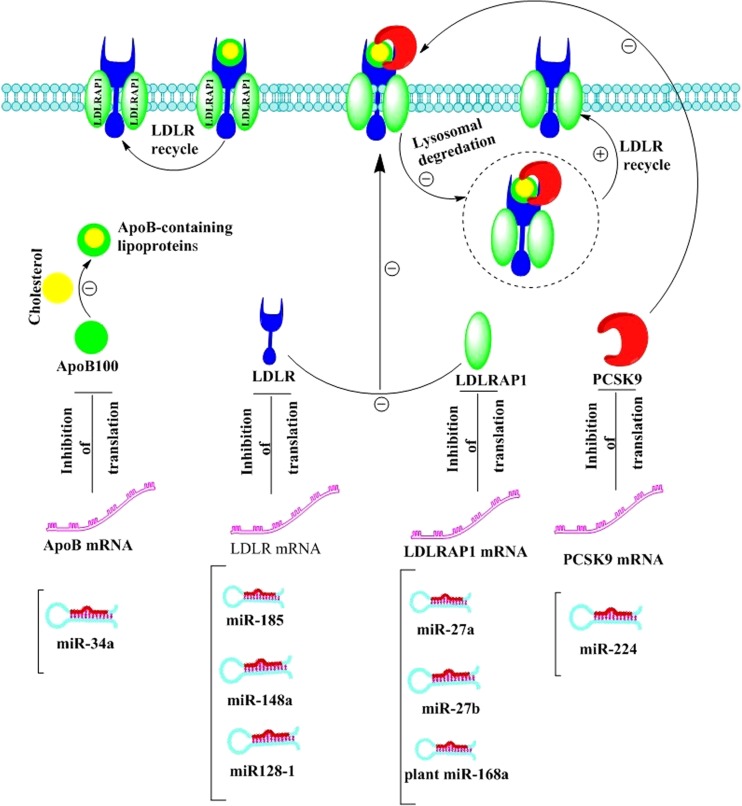
Different lines of evidence support the legitimacy of considering miRs as therapeutic targets for FH teatmenttr. First, miR-based therapy with miR-34- and miR-122-based drugs has reached phase 2 clinical trial development [50–52]. Second, a number of in vitro and in vivo studies have revealed the importance of miRs in controlling plasma LDL-C through modulating LDLR, apoB, and PCSK9 expression. Hence, in attention to critical and well-established role of LDLR, APOB, and PCSK9 genes in LDL-C hemostasis and metabolism, therapeutic manipulation of related miRs, via miR mimics or inhibitors, can be an efficient and attractive approach for lowering elevated LDL-C and reducing risk of CV events in FH patients (Fig. 1).
Fig. 1.
A schematic view of miR-mediated regulation of FH-related genes
LDLR-Targeting miRs
To date, a large number of mutations in the LDLR gene have been found to be associated with FH [5]. LDLR gene mutations may affect function and structure of LDLR protein leading to decreased affinity of the receptor to ligands such as apoB-containing lipoproteins [53] and/or to deficiency of LDLR internalization [54] which consequently result in abnormal accumulation of LDL-C in plasma [38].
As emerged bioinformatically by genome-wide association studies (GWAS), LDLR was predicted as a target gene for miR-130b, miR-301b, miR-148a, and miR-128-1, thereby they may have the potential to regulate circulating LDL-C levels [55]. In vitro studies on human and mouse hepatoma cells have shown that a large number of miRs, including miR-27a, miR-27b, miR-185, miR-148a, miR-128-1, miR-130b, and miR-301b, can directly modulate LDLR expression through post-transcriptional regulation via targeting the 3’ UTR of LDLR [56–60] (Table 1). Among these miRs, as revealed by in vivo studies on genetically modified mice, only miR128-1, miR-148a, and miR-185 could significantly change plasma LDL-C levels. Inhibition of miR128-1 and miR-148a expression markedly reduced circulating levels of plasma LDL-C in ApoE−/− and Ldlr−/+ mice, respectively [58, 59]. Inhibition of miR-185 expression was also associated with significantly less plasma LDL-C levels and slow atherosclerotic plaque progression [60]. In addition to the aforementioned validated miRs, a wide range of miRs, including hsa-miR-130a-3p, bta-miR-17-5p, bta-miR-146a, bta-miR-146b, hsa-miR-27a-3p, hsa-miR-17-5p, and many other miRs have been predicted by Miranda to interact with the 3’ UTR region of LDLR mRNA. Finally, a recent study by He et al. revealed that miR-195 inhibited lipopolysaccharide-mediated intracellular cholesterol accumulation and LDL cell uptake by decreasing LDLR gene expression [62]. These findings illustrate that inhibition of miR128-1, miR-148a, and miR-185 might be an efficient therapy to decrease plasma LDL-C levels in patients with FH.
Table 1.
An overview of putative therapeutic miRs validated to directly target FH-related pathogenic genes
| Regulating miR | Validation method | Condition | Prediction program | Ref. |
|---|---|---|---|---|
| LDLR-regulating miRs | ||||
| miR-27a | Luciferase reporter assay, real-time PCR, Western blot | HepG2 cells | TargetScan, miRanda |
[56] |
| miR-27b | Luciferase reporter assay, real-time PCR, Western blot | Cos-7 cells | TargetScan, miRWalk, miRanda | [57] |
| miR-128-1 | Luciferase reporter assay, real-time PCR | HEK293T cells, C57BL/6 J and Apoe −/− mice |
TargetScan | [58] |
| miR-130b | ||||
| miR-301b | ||||
| miR-148a | Luciferase reporter assay, real-time PCR, Western blot, ribonucleoprotein immunoprecipitation | HepG2 cells, Hepa cells, Huh7 cells |
TargetScan, miRWalk, and miRanda | [59] |
| miR-185 | Luciferase reporter assay, real-time PCR, Western blot | HepG2 cells | miRanda | [60] |
| PCSK9-regulating miRs | ||||
| miR-224 | Luciferase reporter assay, real-time PCR, Western blot | U251 and U87 cells | DIANA microT | [61] |
| mi-27a | Luciferase reporter assay, real-time PCR, Western blot | HepG2 cells | TargetScan, miRanda |
[56] |
| miR-195 | overexpressing or knocking down miR-195, real-time PCR, Western blo | HepG2 and Huh7 | – | [62] |
| miR-132 | Luciferase reporter assay, real-time PCR, Western blot | LNCaP and C4-2B cells | TargetScan | [63] |
| miR-499 | Luciferase reporter assay, real-time PCR, Western blot | HepG2 and Huh7 cells | miRBase | [64] |
| MiR-342 | Luciferase reporter assay, real-time PCR, Western blot | LNCaP and C4-2B cells | TargetScan | [65] |
| miR-185 | Luciferase reporter assay, real-time PCR, Western blot | HepG2 and THLE-2 cells | miRStart | [65, 66] |
| apoB-containing lipoproteins-regulating miRs | ||||
| miR-34a | Luciferase reporter assay | C57BL/6 mice | ND | [67] |
| miR-30c | Luciferase reporter assay | Cos-7 cells | TargetScan | [68] |
| Luciferase reporter assay | Male C57Bl/6 J and L-MTP
-/- mice, Huh7 cells |
TargetScan | [69] | |
| miR-122 | Luciferase reporter assay | Hepa cells Gene knockout mice |
TargetScan | [70] |
| Luciferase reporter assay | AML 12 cells, HeLa cells |
TargetScan | [71] | |
| LDLRAP1- regulating miRs | ||||
| miR-27a | Luciferase reporter assay, real-time PCR, Western blot | HepG2 cells | TargetScan, miRanda |
[56] |
| miR-27b | Luciferase reporter assay, real-time PCR, Western blot | Cos-7 cells | Targetscan, miRWalk, miRanda | [57] |
| miR168a | Luciferase reporter assay, real-time PCR, Western blot | C57BL/6 J mice, HepG2, and Caco-2 cells | ND | [72] |
ND not define
PCSK9-Targeting miRs
PCSK9 is an intriguing protein with a ubiquitous expression and a range of partially unexplored functions [73–75]. PCSK9 is well-known for its homeostatic role in cholesterol metabolism through regulation of LDLR fate via binding to the extracellular EGF-A domain of the hepatic LDLR and promoting its lysosomal degradation. In rare (less than 0.5%) of FH patients, a missense gain-of-function mutation in the PCSK9 gene results in elevated plasma PCSK9 levels, increased LDLR degradation and impaired recycling leading to reduced LDLR-mediated clearance of LDL-C from the bloodstream and increased plasma LDL-C levels [76, 77].
PCSK9 inhibition has emerged as an effective, approved and marketed LDL-C-lowering therapy where monoclonal antibodies (mAbs) to PCSK9 profoundly reduce LDL-C in FH patients [78–82]{Sahebkar, 2013 #48; Sahebkar, 2013 #49}. Evolocumab (Repatha®) and alirocumab (Praluent®) are now approved by regulatory authorities in the USA, Europe, Japan, and many other countries for the treatment of HeFH where LDL-C is inadequately reduced by maximal doses of statins. In addition, evolocumab is approved at a dose of 420 mg monthly for the treatment of HoFH. Preliminary findings from post-hoc or exploratory analysis of phase III trials with evolocumab and alirocumab provided encouraging evidence that the LDL-C reductions resulted in further reductions in CVD when added to existing statin with or without ezetimibe therapy [83, 84]. The CVD benefit has recently been confirmed in the Further Cardiovascular Outcomes Research With PCSK9 Inhibition in Subjects With Elevated Risk (FOURIER) trial with evolocumab with 27,500 patients (15% risk reduction of primary endpoint and 20% significant risk reduction of key secondary outcomes) [85].
PCSK9 mAbs at the currently approved doses (140 mg biweekly or 420 mg monthly) have a relatively short duration of effect, necessitating every 2- or 4-week administration; however, this may not be a concern for the management of patients owing to the satisfactory adherence of patient to these mAbs in phase III trials [86–88]. In addition, the cost-effectiveness of PCSK9 mAbs for the treatment of FH, which could be life-long, is debatable [89]. These issues highlight the need for durable and less expensive PCSK9 inhibitors that could potentially serve as alternatives to PCSK9 mAbs in order to reduce the cost of treatment. miR-based therapeutic approaches can be regarded as such an alternative for PCSK9 inhibition.
Although there are not many verified miRs for PCSK9 regulation, some recent studies have shown that PCSK9 can be regulated directly and indirectly by some miRs. As verified by luciferase reporter method, miR-224 directly targets PCSK9 mRNA and significantly downregulates its expression [61]. Notably, miR-27a was reported to target promoter region of the PCSK9 gene and up-regulate its expression [56]. Also, miR-195 was able to increase PCSK9 gene expression in liver cancer cells stimulated with lipopolysaccharide [62]. Such inducing effect of the miR is supported by evidence showing miRs can enhance expression of target genes via interaction with promoter region instead of 3’ UTR of the mRNAs [90] (Table 1).
Furthermore, PCSK9 expression has been reported to be modulated by miRs regulating SREBP transcription factors. The proximal promoter of PCSK9 gene harbors a functional sterol regulatory element (SRE) that is targeted by sterol-responsive element binding proteins (SREBPs) in response to alterations in intracellular cholesterol levels [76, 77]. miR-132 and miR-499 have been recently identified to indirectly downregulate SREBP-1c [63] that is known to be a PCSK9 transcription factor [64, 76]. It was also found that miR-185 and miR-342 can inhibit the expression of SREBP-1 and -2, and therefore, reduce cholesterol synthesis [65, 66]. Given that SREBPs are the crucial transcription factors upregulating PCSK9 gene, miRs inhibiting these transcription factors can be potentially exploited for FH miR therapy using miR mimetics.
On the other hand, several miRs were recently predicted to bind to the 3′ UTR of PCSK9 mRNA. These include miR-18a-5p, miR-148, miR-323-5P, miR-570, miR-584t, miR-663-b, miR-922, miR-3919, miR-3974, miR-4509, miR-4690-5p, miR-4732-5p, miR-4795-5P, miR-5586-3P, and miR-6134 [91]. There are also several miRs presented by miRTarBase database, which are predicted, by Miranda, to interact with 3′ UTR of PCSK9, including hsa-miR-335-5p, hsa-miR-124-3p, hsa-miR-215-5p, hsa-miR-192-5p, hsa-miR-6864-5p, hsa-miR-6515-5p, hsa-miR-4656, hsa-miR-6797-5p, hsa-miR-1249-5p, hsa-miR-6875-5p, hsa-miR-4721, hsa-miR-6761-5p, hsa-miR-3126-5p, hsa-miR-4802-5p, hsa-miR-4446-3p, hsa-miR-6873-5p, hsa-miR-7845-5p, hsa-miR-6856-5p, hsa-miR-6758-5p, hsa-miR-6882-5p, hsa-miR-1303, hsa-miR-744-3p, hsa-miR-6501-5p, hsa-miR-4423-5p, hsa-miR-367-5p, hsa-miR-6764-5p, hsa-miR-1304-3p, hsa-miR-1915-3p, hsa-miR-1208, and hsa-miR-193a-5p. The aforementioned miRs merit further studies to be experimentally verified as direct regulators of PCSK9.
apoB-Containing Lipoproteins-Targeting miRs
LDL-C-lowering approaches acting via modulation of LDLR activity or expression are ineffective in patients with HoFH with LDLR activity less than 2% [92–94]. In this condition, elevated plasma cholesterol can be modulated via reducing production of atherogenic apoB-containing lipoproteins including LDL-C and lipoprotein(a). This approach has been found to lower plasma cholesterol in HoFH patients by reducing apoB expression using mipomersen, an antisense oligonucleotide (ASO), and by inhibiting the activity of MTP by using lomitapide [95–98]. Side effects of both mipomersen and lomitapide, such as fatty liver and elevation in plasma transaminases [99], call for the necessity of alternative agents—with different mechanisms of action—modulating plasma cholesterol without causing side effects.
Hepatic production of atherogenic apoB-containing lipoproteins has been identified to be regulated by three miRs, including miR-34a [67], miR-30c [68, 69], and miR-122 [70, 71] (Table 1). The miRs have been reported to reduce production of liver apoB-containing lipoproteins through inhibition of HNF4α and MTP mRNA expression. Both HNF4α and MPT have critical roles in the assembly of apoB-containing lipoproteins. HNF4α is a transcription factor that binds to the promoter regions of APOB and MTP genes to enhance their expression [100–102]. It was found that miR-34a reduces HNF4α levels causing reduction of apoB and MTP expression [67, 103]. MTP is an important chaperone that transfers lipids to apoB peptide [104, 105]. miR-30c inhibits MTP expression via binding to the 3′ UTR of MTP mRNA and induces post-transcriptional degradation leading to reduced assembly of apoB-containing lipoproteins [68, 69]. In the case of miR-122, it decreases MTP expression by yet an unknown mechanism [106].
In addition, a number of miRs proposed to interact with 3′ UTR of apoB100 mRNA has been predicted by Miranda and presented in miRTarBase database. These include mmu-miR-122-5p, hsa-miR-885-5p, hsa-miR-7151-5p, hsa-miR-6869-5p, hsa-miR-502-5p, hsa-miR-1911-5p, hsa-miR-499b-5p, hsa-miR-500b-3p, hsa-miR-6862-3p, hsa-miR-6784-3p, and hsa-miR-624-5p.
To sum up, overexpression of miR-122, miR-34a, and miR-30c, as well as other predicted miRs, can be therapeutically useful to reduce production and plasma levels of atherogenic apoB-containing lipoproteins independent of LDLR targeting in FH, particularly in homozygous patients. However, the interactions and effectiveness of these miRs needs to be experimentally verified by using miR mimetics.
LDLRAP-Targeting miRs
Low-density lipoprotein receptor adaptor protein 1 (LDLRAP1) is a cytosolic protein containing an N-terminal phosphotyrosine-binding (PTD) domain that interacts with the cytoplasmic tail of the LDLR and facilitates circulating plasma LDL-C clearance. Through clathrin-mediated endocytosis of hepatic LDLR, PTD domain of LDLRAP1 binds to the internalization sequence of the cytoplasmic tail of LDLR, and acts as an adaptor for LDLR endocytosis in the hepatocytes. A rarely-occurring recessive form of FH, named autosomal recessive hypercholesterolaemia, is caused by loss-of-function mutations in LDLRAP1 gene leading to defection in LDLR internalization and subsequently, the reduced clearance of plasma LDL-C [5, 107].
miR-27a, miR-27b, and plant miR-168a have been found to directly target LDLRAP1 expression, and consequently modulate LDLR activity (Table 1). As validated by luciferase reporter assays, 3′ UTR of LDLRAP1 mRNA is directly and specifically targeted by miR-27a [56] and miR-27b [57]. Although in vitro study on human hepatic Huh7 cells revealed that over-expression of miR-27b can profoundly downregulate mRNA expression of LDLRAP1 gene, in vivo study on wild type mice showed that modulation of miR-27b cannot affect plasma levels of LDL-C and other indices of lipid profile [57]. miR-27a was also found to downregulate mRNA level of LDLRAP1 in HepG2 cells [56], while its in vivo effect on plasma LDL-C is unknown and needs to be studied. Furthermore, miR168a is a plant miR abundant in rice and has been found to be one of the most enriched exogenous plant-based miRs in the sera of Chinese peoples. In vitro and in vivo mechanistical studies revealed that miR168a has a high degree of complementarity with the exon 4 of mammalian LDLRAP1, which can bind to the human or mouse LDLRAP1 mRNA, inhibit LDLRAP1 expression in hepatocytes, and result in reduced clearance of LDL-C from mouse plasma [72]. As predicted by Miranda algorithm, there are several miRs in miRTarBase database which are proposed to interact with 3′ UTR of LDLRAP1 mRNA, including hsa-miR-124-3p, mmu-miR-124-3p, hsa-miR-9-5p, hsa-miR-615-3p, and hsa-miR-92a-3p.
In summary, miR inhibition using antisense approach can be a useful strategy to suppress aforementioned miRs, resulting in enhanced LDLRAP1 activity and improved clearance of plasma circulating LDL-C.
Concluding Remarks
miRs regulating critical genes closed to FH, including LDL, APOB, PCSK9, and LDLRAP1, can be considered as potential therapeutic targets for FH patients. LDLR can be upregulated through inhibition of miRs including miR128-1, miR-148a, and miR-185 and consequently enhances liver clearance of LDL-C. In the case of PCSK9, use of therapeutic miRs mimic miR-224, miR-132 and miR-499, miR-185, and miR-342, and also inhibition of miR-27a can downregulate PCSK9 expression, and therefore, can also lower elevated LDL-C in FH patients. Notably, reduction of atherogenic apoB-containing lipoproteins, as an efficient therapeutic approach in HoFH, can be achieved via miR-122, miR-34a and miR-30c mimics, which are known to reduce atherogenic lipoproteins independent of LDLR. In the case of LDLRAP1, inhibition of miR-27a and miR-27b can be a reliable and efficient approach for upregulation of LDLRAP1 protein and enhancing LDLR activity leading to improved clearance of LDL-C from sera of hypercholesterolemic patients. Overall, among the aforementioned miRs, miR-27a is suggested to be the most putative therapeutic target as it can regulate three of the 4 important photogenic genes in FH patients including LDLR, PCSK9, and LDLRAP1.
Funding
There is no funding source.
Conflict of Interest
Dr. Banach has served on speaker’s bureau of Abbott/Mylan, Abbott Vascular, Actavis, Akcea, Amgen, Biofarm, KRKA, MSD and Sanofi-Aventis, served as a consultant to Abbott Vascular, Amgen, Daichii Sankyo, Esperion, Lilly, MSD and Resverlogix, and received grant from Sanofi and Valeant. Dr. Stein has received consulting fees related to development of PCSK9 inhibitors from Amgen, Regeneron/Sanofi, Genentech/Roche and BMS and other LDL drugs from AstraZeneca, Catabasis, CymaBay, Gemphire. He is also an advisor on lipid altering drugs for CVS/Caremark. Other authors have no competing interests to declare.
Ethical Approval
This is an extensive review and therefore no ethical approval is necessary.
Informed Consent
This is an extensive review and therefore no informed consent is necessary.
Contributor Information
Maciej Banach, Phone: +48 42 639 37 71, Email: maciejbanach77@gmail.com.
Amirhossein Sahebkar, Phone: 985138002288, Email: amirhossein.sahebkar@uwa.edu.au.
References
- 1.Najam O, Ray KK. Familial hypercholesterolemia: a review of the natural history, diagnosis, and management. Cardiology and therapy. 2015;4(1):25–38. doi: 10.1007/s40119-015-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collaboration, E.A.S.F.H.S et al. Pooling and expanding registries of familial hypercholesterolaemia to assess gaps in care and improve disease management and outcomes: rationale and design of the global EAS familial Hypercholesterolaemia studies collaboration. Atheroscler Suppl. 2016;22:1–32. doi: 10.1016/j.atherosclerosissup.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Vallejo-Vaz AJ, et al. Familial hypercholesterolaemia: a global call to arms. Atherosclerosis. 2015;243(1):257–259. doi: 10.1016/j.atherosclerosis.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 4.Anderson TJ, et al. 2012 update of the Canadian Cardiovascular Society guidelines for the diagnosis and treatment of dyslipidemia for the prevention of cardiovascular disease in the adult. Can J Cardiol. 2013;29(2):151–167. doi: 10.1016/j.cjca.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 5.Henderson R, et al. The genetics and screening of familial hypercholesterolaemia. J Biomed Sci. 2016;23(1):39. doi: 10.1186/s12929-016-0256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuchel M, et al. Homozygous familial hypercholesterolaemia: new insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur Heart J. 2014;35(32):2146–2157. doi: 10.1093/eurheartj/ehu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordestgaard BG, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease. Eur Heart J. 2013;34(45):3478–3490. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon CP, et al. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med. 2015;372(25):2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 9.Catapano AL, et al. 2016 ESC/EAS Guidelines for the Management of Dyslipidaemias. Eur Heart J. 2016;37(39):2999–3058. doi: 10.1093/eurheartj/ehw272. [DOI] [PubMed] [Google Scholar]
- 10.Banach M, et al. PoLA/CFPiP/PCS Guidelines for the Management of Dyslipidaemias for Family Physicians 2016. Arch Med Sci. 2017;13(1):1–45. doi: 10.5114/aoms.2017.64712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Goma EM, et al. Treatment gaps in adults with heterozygous familial hypercholesterolemia in the United States: data from the CASCADE-FH Registry. Circ Cardiovasc Genet. 2016;9(3):240–249. doi: 10.1161/CIRCGENETICS.116.001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marais AD, et al. A dose-titration and comparative study of rosuvastatin and atorvastatin in patients with homozygous familial hypercholesterolaemia. Atherosclerosis. 2008;197(1):400–406. doi: 10.1016/j.atherosclerosis.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 13.Raal FJ, et al. Expanded-dose simvastatin is effective in homozygous familial hypercholesterolaemia. Atherosclerosis. 1997;135(2):249–256. doi: 10.1016/S0021-9150(97)00168-8. [DOI] [PubMed] [Google Scholar]
- 14.Gagné C, et al. Efficacy and safety of ezetimibe coadministered with atorvastatin or simvastatin in patients with homozygous familial hypercholesterolemia. Circulation. 2002;105(21):2469–2475. doi: 10.1161/01.CIR.0000018744.58460.62. [DOI] [PubMed] [Google Scholar]
- 15.Momtazi AA, et al. Regulation of PCSK9 by nutraceuticals. Pharmacol Res. 2017;120:157–169. doi: 10.1016/j.phrs.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Consortium EP. The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 17.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19(1):60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam JK, et al. siRNA versus miRNA as therapeutics for gene silencing. Molecular Therapy-Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Y, et al. Therapeutic potentials of gene silencing by RNA interference: principles, challenges, and new strategies. Gene. 2014;538(2):217–227. doi: 10.1016/j.gene.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Davidson BL, McCray PB. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12(5):329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal N, et al. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67(4):657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pecot CV, et al. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Petersen CP, et al. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21(4):533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 26.Schultheis B, et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol. 2014;32(36):4141–4148. doi: 10.1200/JCO.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- 27.Tabernero J, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3(4):406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 28.Bader AG, et al. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18(12):1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momtazi AA et al. (2016) Curcumin as a MicroRNA regulator in cancer: a review [DOI] [PubMed]
- 30.DeVincenzo J, et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci. 2010;107(19):8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra PK, et al. Inhibition of hepatitis C virus replication by intracellular delivery of multiple siRNAs by nanosomes. Mol Ther. 2012;20(9):1724–1736. doi: 10.1038/mt.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Sendi H, et al. miR-122 decreases HCV entry into hepatocytes through binding to the 3′ UTR of OCLN mRNA. Liver Int. 2015;35(4):1315–1323. doi: 10.1111/liv.12698. [DOI] [PubMed] [Google Scholar]
- 33.Erdmann VA, Poller W, and Barciszewski J (2008) RNA technologies in cardiovascular medicine and research. Springer
- 34.Fitzgerald K, et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med. 2017;376(1):41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13(4):644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray KK, et al. (2017) Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N Engl J Med [DOI] [PubMed]
- 37.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110(3):496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 38.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70(18):7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal L, et al. Making sense of antisense. Eur J Cancer. 2005;41(18):2812–2818. doi: 10.1016/j.ejca.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Jackson AL, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12(7):1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birmingham A, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3(3):199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 42.Fedorov Y, et al. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12(7):1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Sall A, Yang D. MicroRNA: an emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9(6):978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goedeke L, et al. miRNA regulation of LDL-cholesterol metabolism. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2016;1861(12):2047–2052. doi: 10.1016/j.bbalip.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Condorelli G, Latronico MV, Dorn GW. microRNAs in heart disease: putative novel therapeutic targets? Eur Heart J. 2010;31(6):649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martino F, et al. Circulating miR-33a and miR-33b are up-regulated in familial hypercholesterolaemia in paediatric age. Clin Sci. 2015;129(11):963–972. doi: 10.1042/CS20150235. [DOI] [PubMed] [Google Scholar]
- 48.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 49.Sahebkar A, et al. Editorial: microRNA-33 inhibition: a potential adjunct to statin therapy? Curr Vasc Pharmacol. 2016;14(4):321–322. doi: 10.2174/1570161114999160513150737. [DOI] [PubMed] [Google Scholar]
- 50.Bouchie A (2013) First microRNA mimic enters clinic, Nature Research [DOI] [PubMed]
- 51.Qiu Z, Dai Y. Roadmap of miR-122-related clinical application from bench to bedside. Expert Opin Investig Drugs. 2014;23(3):347–355. doi: 10.1517/13543784.2014.867327. [DOI] [PubMed] [Google Scholar]
- 52.Van Rooij E and Kauppinen S (2014) Development of microRNA therapeutics is coming of age. EMBO molecular medicine, p. e201100899 [DOI] [PMC free article] [PubMed]
- 53.Soria LF, et al. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci. 1989;86(2):587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soutar AK, Naoumova RP. Mechanisms of disease: genetic causes of familial hypercholesterolemia. Nature clinical practice Cardiovascular medicine. 2007;4(4):214–225. doi: 10.1038/ncpcardio0836. [DOI] [PubMed] [Google Scholar]
- 55.Agarwal V, et al. Predicting effective microRNA target sites in mammalian mRNAs. elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvarez ML, et al. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis. 2015;242(2):595–604. doi: 10.1016/j.atherosclerosis.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goedeke L, et al. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis. 2015;243(2):499–509. doi: 10.1016/j.atherosclerosis.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagschal A, et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med. 2015;21(11):1290–1297. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goedeke L, et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med. 2015;21(11):1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H, et al. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. 2015;243(2):523–532. doi: 10.1016/j.atherosclerosis.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 61.Bai J et al. (2016) A retrospective study of NENs and miR-224 promotes apoptosis of BON-1 cells by targeting PCSK9 inhibition. Oncotarget [DOI] [PMC free article] [PubMed]
- 62.He M, et al. Pro-inflammation NF-kappaB signaling triggers a positive feedback via enhancing cholesterol accumulation in liver cancer cells. J Exp Clin Cancer Res. 2017;36(1):15. doi: 10.1186/s13046-017-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, et al. MicroRNA-132 cause apoptosis of glioma cells through blockade of the SREBP-1c metabolic pathway related to SIRT1. Biomed Pharmacother. 2016;78:177–184. doi: 10.1016/j.biopha.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 64.Zhang H, et al. MicroRNA-449 suppresses proliferation of hepatoma cell lines through blockade lipid metabolic pathway related to SIRT1. Int J Oncol. 2014;45(5):2143–2152. doi: 10.3892/ijo.2014.2596. [DOI] [PubMed] [Google Scholar]
- 65.Li X, et al. MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLoS One. 2013;8(8):e70987. doi: 10.1371/journal.pone.0070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang M, et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res. 2014;55(2):226–238. doi: 10.1194/jlr.M041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Y et al. (2015) A metabolic stress-inducible miR-34a-HNF4 [alpha] pathway regulates lipid and lipoprotein metabolism. Nature communications, 6 [DOI] [PMC free article] [PubMed]
- 68.Soh J, Hussain MM. Supplementary site interactions are critical for the regulation of microsomal triglyceride transfer protein by microRNA-30c. Nutrition & metabolism. 2013;10(1):56. doi: 10.1186/1743-7075-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soh J, et al. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19(7):892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu S-H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Zhang L, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seidah NG. New developments in proprotein convertase subtilisin-kexin 9’s biology and clinical implications. Curr Opin Lipidol. 2016;27(3):274–281. doi: 10.1097/MOL.0000000000000295. [DOI] [PubMed] [Google Scholar]
- 74.Paciullo F, et al. (2017) PCSK9 at the crossroad of cholesterol metabolism and immune function during infections. J Cell Physiol [DOI] [PubMed]
- 75.Momtazi AA, Banach M, Sahebkar A. PCSK9 inhibitors in sepsis: a new potential indication? Expert Opin Investig Drugs. 2017;26(2):137–139. doi: 10.1080/13543784.2017.1272570. [DOI] [PubMed] [Google Scholar]
- 76.Momtazi AA, et al. (2017) PCSK9 and diabetes: is there a link? Drug Discovery Today [DOI] [PubMed]
- 77.Momtazi AA, Banach M, and Sahebkar A (2017) PCSK9 inhibitors in sepsis: a new potential indication?, Taylor & Francis [DOI] [PubMed]
- 78.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 79.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 80.Cohen JC, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 81.Zhao Z, et al. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am J Hum Genet. 2006;79(3):514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hooper AJ, et al. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 2007;193(2):445–448. doi: 10.1016/j.atherosclerosis.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 83.Robinson JG, et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372(16):1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 84.Sabatine MS, et al. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372(16):1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- 85.Sabatine MS, et al. (2017) Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med [DOI] [PubMed]
- 86.Hooper AJ, Burnett JR. Anti-PCSK9 therapies for the treatment of hypercholesterolemia. Expert Opin Biol Ther. 2013;13(3):429–435. doi: 10.1517/14712598.2012.748743. [DOI] [PubMed] [Google Scholar]
- 87.Navarese EP, et al. Effects of proprotein convertase subtilisin/kexin type 9 antibodies in adults with hypercholesterolemia. A systematic review and meta-analysis effects of PCSK9 antibodies in adults with hypercholesterolemia. Ann Intern Med. 2015;163(1):40–51. doi: 10.7326/M14-2957. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X-L, et al. Safety and efficacy of anti-PCSK9 antibodies: a meta-analysis of 25 randomized, controlled trials. BMC Med. 2015;13(1):123. doi: 10.1186/s12916-015-0358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kazi DS, et al. Cost-effectiveness of PCSK9 inhibitor therapy in patients with heterozygous familial hypercholesterolemia or atherosclerotic cardiovascular disease. JAMA. 2016;316(7):743–753. doi: 10.1001/jama.2016.11004. [DOI] [PubMed] [Google Scholar]
- 90.Place RF, et al. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zambrano T, et al. Impact of 3’UTR genetic variants in PCSK9 and LDLR genes on plasma lipid traits and response to atorvastatin in Brazilian subjects: a pilot study. Int J Clin Exp Med. 2015;8(4):5978. [PMC free article] [PubMed] [Google Scholar]
- 92.Cornier M-A, Eckel RH. Non-traditional dosing of statins in statin-intolerant patients—is it worth a try? Current Atherosclerosis Reports. 2014;17(2):475. doi: 10.1007/s11883-014-0475-4. [DOI] [PubMed] [Google Scholar]
- 93.Finkel JB, Duffy D. 2013 ACC/AHA cholesterol treatment guideline: paradigm shifts in managing atherosclerotic cardiovascular disease risk. Trends in Cardiovascular Medicine. 2015;25(4):340–347. doi: 10.1016/j.tcm.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 94.Raal FJ, Blom DJ Anacetrapib in familial hypercholesterolaemia: pros and cons. Lancet 385(9983):2124–2126 [DOI] [PubMed]
- 95.Raal FJ, et al. Pediatric experience with mipomersen as adjunctive therapy for homozygous familial hypercholesterolemia. Journal of clinical lipidology. 2016;10(4):860–869. doi: 10.1016/j.jacl.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 96.McGowan MP, Moriarty PM, Backes JM. The effects of mipomersen, a second-generation antisense oligonucleotide, on atherogenic (apoB-containing) lipoproteins in the treatment of homozygous familial hypercholesterolemia. Clinical Lipidology. 2014;9(5):487–503. doi: 10.2217/clp.14.43. [DOI] [Google Scholar]
- 97.Rader DJ, Kastelein JJP. Lomitapide and mipomersen: two first-in-class drugs for reducing low-density lipoprotein cholesterol in patients with homozygous familial hypercholesterolemia. Circulation. 2014;129(9):1022–1032. doi: 10.1161/CIRCULATIONAHA.113.001292. [DOI] [PubMed] [Google Scholar]
- 98.Gouni-Berthold I, Berthold HK. Mipomersen and lomitapide: two new drugs for the treatment of homozygous familial hypercholesterolemia. Atheroscler Suppl. 2015;18:28–34. doi: 10.1016/j.atherosclerosissup.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 99.Cuchel M, Blom DJ, Averna MR. Clinical experience of lomitapide therapy in patients with homozygous familial hypercholesterolaemia. Atheroscler Suppl. 2014;15(2):33–45. doi: 10.1016/j.atherosclerosissup.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 100.Dai K, Hussain MM. NR2F1 disrupts synergistic activation of the MTTP gene transcription by HNF-4α and HNF-1α. J Lipid Res. 2012;53(5):901–908. doi: 10.1194/jlr.M025130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sheena V, et al. Transcriptional regulation of human microsomal triglyceride transfer protein by hepatocyte nuclear factor-4α. J Lipid Res. 2005;46(2):328–341. doi: 10.1194/jlr.M400371-JLR200. [DOI] [PubMed] [Google Scholar]
- 102.Hirokane H, et al. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J Biol Chem. 2004;279(44):45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- 103.Choi SE, et al. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12(6):1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hussain MM, et al. Multiple functions of microsomal triglyceride transfer protein. Nutrition & metabolism. 2012;9(1):14. doi: 10.1186/1743-7075-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44(1):22–32. doi: 10.1194/jlr.R200014-JLR200. [DOI] [PubMed] [Google Scholar]
- 106.Zhou L, et al. MicroRNAs regulating apolipoprotein B-containing lipoprotein production. Biochimica et Biophysica Acta (BBA)-molecular and cell biology of lipids. 2016;1861(12):2062–2068. doi: 10.1016/j.bbalip.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 107.Soutar AK, Naoumova RP, Traub LM. Genetics, clinical phenotype, and molecular cell biology of autosomal recessive hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2003;23(11):1963–1970. doi: 10.1161/01.ATV.0000094410.66558.9A. [DOI] [PubMed] [Google Scholar]