ABSTRACT
Human parvovirus B19 (B19V) expresses a single precursor mRNA (pre-mRNA), which undergoes alternative splicing and alternative polyadenylation to generate 12 viral mRNA transcripts that encode two structural proteins (VP1 and VP2) and three nonstructural proteins (NS1, 7.5-kDa protein, and 11-kDa protein). Splicing at the second 5′ donor site (D2 site) of the B19V pre-mRNA is essential for the expression of VP2 and the 11-kDa protein. We previously identified that cis-acting intronic splicing enhancer 2 (ISE2) that lies immediately after the D2 site facilitates the recognition of the D2 donor for its efficient splicing. In this study, we report that ISE2 is critical for the expression of the 11-kDa viral nonstructural protein. We found that ISE2 harbors a consensus RNA binding motif protein 38 (RBM38) binding sequence, 5′-UGUGUG-3′. RBM38 is expressed during the middle stage of erythropoiesis. We first confirmed that RBM38 binds specifically with the ISE2 element in vitro. The knockdown of RBM38 significantly decreases the level of spliced mRNA at D2 that encodes the 11-kDa protein but not that of the D2-spliced mRNA that encodes VP2. Importantly, we found that the 11-kDa protein enhances viral DNA replication and virion release. Accordingly, the knockdown of RBM38 decreases virus replication via downregulating 11-kDa protein expression. Taken together, these results suggest that the 11-kDa protein facilitates B19V DNA replication and that RBM38 is an essential host factor for B19V pre-mRNA splicing and for the expression of the 11-kDa protein.
IMPORTANCE B19V is a human pathogen that can cause fifth disease, arthropathy, anemia in immunocompromised patients and sickle cell disease patients, myocarditis, and hydrops fetalis in pregnant women. Human erythroid progenitor cells (EPCs) are most susceptible to B19V infection and fully support viral DNA replication. The exclusive tropism of B19V for erythroid-lineage cells is dependent not only on the expression of viral receptors and coreceptors on the cell surface but also on the intracellular host factors that support B19V replication. Our present study shows that B19V uses a host factor, RNA binding motif protein 38 (RBM38), for the processing of its pre-mRNA during virus replication. Specifically, RBM38 interacts with the intronic splicing enhancer 2 (ISE2) element of B19V pre-mRNA and promotes 11-kDa protein expression, thereby regulating the 11-kDa protein-mediated augmentation of B19V replication. The identification of this novel host-pathogen interaction will provide mechanistic insights into B19V replication and aid in finding new targets for anti-B19V therapeutics.
KEYWORDS: B19, mRNA splicing, parvovirus
INTRODUCTION
Human parvovirus B19 (B19V) belongs to the genus Erythroparvovirus of the Parvoviridae family (1). Productive B19V infection displays a high tropism for burst-forming unit-erythroid (BFU-E) and CFU-erythroid (CFU-E) progenitor cells in human bone marrow and fetal liver (2–5). B19V also infects nonerythroid tissues (6); however, the infection is not productive (7). B19V infection causes human diseases such as erythema infectiosum or fifth disease in children, transient aplastic crises in sickle cell disease patients, pure red blood aplasia in immunocompromised patients, arthropathy, cardiomyopathy in adults, and nonimmune hydrops fetalis in pregnant women (7–9).
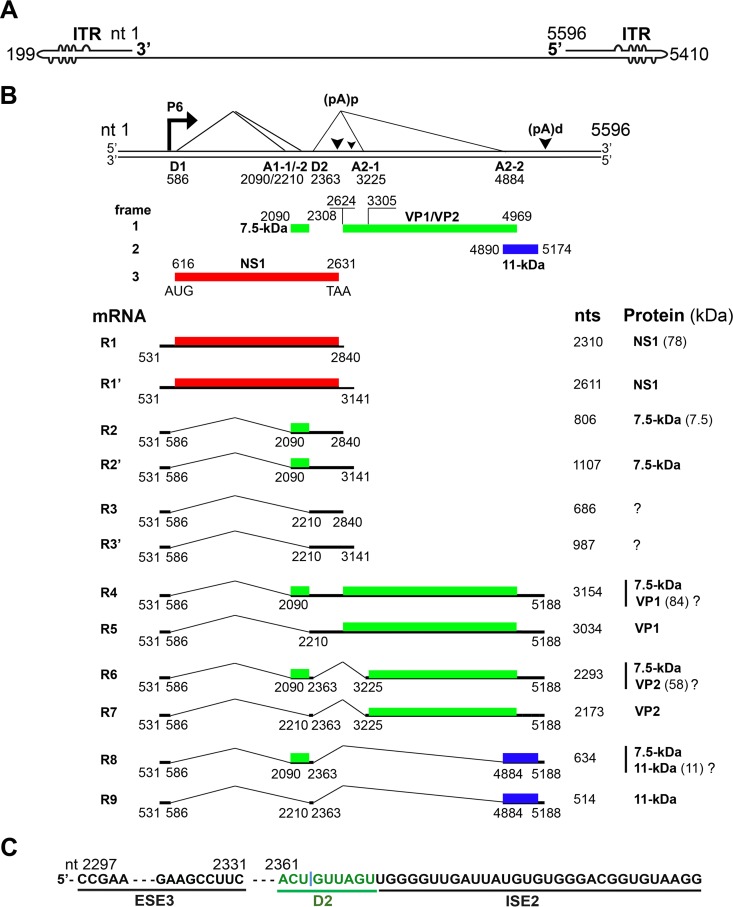
B19V is a small, nonenveloped virus with a single-stranded DNA (ssDNA) genome of 5.6 kb (1). The B19V genome is flanked by two identical inverted terminal repeats (ITRs) at both ends (Fig. 1A) (10). The left portion of the genome encodes the nonstructural NS1 and 7.5-kDa proteins, whereas the right side encodes the structural proteins VP1 and VP2 and an additional nonstructural 11-kDa protein (Fig. 1B) (11–13). The B19V genome has a single promoter at map unit 6 (P6), which transcribes a single precursor mRNA (pre-mRNA) (7). B19V pre-mRNA has two donor sites (D1 and D2) and four acceptor sites (A1-1, A1-2, A2-1, and A2-2), which are used for alternative splicing to generate all species (12) of the viral mRNAs (Fig. 1B). There are two internal (proximal) polyadenylation sites, (pA)p1 and (pA)p2, and one distal (pA)d site (7). mRNAs that encode the NS1 and the 7.5-kDa proteins use the (pA)p1/2 sites, and mRNAs that encode the VP1, VP2, and 11-kDa proteins utilize the (pA)d site for polyadenylation (Fig. 1B) (14).
FIG 1.
B19V transcription map. (A) B19V genome. The linear single-stranded B19V genome is shown in the negative sense, with unpaired or mismatched bases diagrammed as bulges and bubbles. ITR, inverted terminal repeat. (B) Transcription profile. The B19V duplex genome is shown at the top. P6 represents the viral promoter, D1 and D2 denote splice donor sites, and A1-1, A1-2, A2-1, and A2-2 denote splice acceptor sites. Different open reading frames are shown by different colors (red, green, and blue). (pA)p and (pA)d represent polyadenylation sites at the proximal and distal ends, respectively. At the bottom, 9 major RNAs encoding different viral proteins, as indicated, are shown. Question marks indicate that it is unknown whether or not the protein is expressed from the species of mRNA. (C) ESE3, D2, and ISE2 elements. The donor 2 (D2) site of the B19V pre-mRNA is flanked by exon splicing enhancer 3 (ESE3) on the left and intronic splicing enhancer 2 (ISE2) on the right. They act as cis-acting elements for splicing at the D2 site.
In the absence of virus genome replication, most of the viral transcripts are polyadenylated at (pA)p sites in both B19V-permissive and nonpermissive cells, the majority of which encode the NS1 and 7.5-kDa proteins (15, 16). However, virus genome replication overcomes the blockade and enhances the readthrough of the (pA)p sites, generating full-length transcripts that encode the capsid and 11-kDa proteins (15). In the past, our group and others used a B19V replication-competent model, where a nearly full-length B19V genome was amplified by the simian virus 40 (SV40) replication origin in COS-7 cells, to analyze the B19V transcription profile (11, 14, 15, 17, 18). Using that artificial viral DNA replication system, we demonstrated that efficient splicing of the first intron promotes polyadenylation at (pA)p (19). However, splicing within the second intron competes with polyadenylation at (pA)p and stimulates polyadenylation at (pA)d (19). We also showed that the binding of small nuclear U1 RNA to the D2 splice site is sufficient to inhibit polyadenylation at (pA)p (19). Importantly, we previously found that multiple splicing enhancers that define the central exon, which spans the A1-1/A1-2 to D2 sites, determine its inclusion (20). Notably, we identified intronic splicing enhancer 2 (ISE2) after the D2 site of B19V pre-mRNA and demonstrated that the ISE2 sequence (Fig. 1C) is essential for the recognition of the D2 site for splicing (20). The use of a morpholino antisense oligonucleotide for the ISE2 region reduced splicing at the D2 site in B19V-infected ex vivo-expanded human erythroid progenitor cells (EPCs) (20).
In this study, we showed that RNA binding motif protein 38 (RBM38) binds to the B19V ISE2 element, which is essential for the efficient splicing of B19V pre-mRNA at the D2 site. We showed that the knockdown of RBM38 reduced the level of viral mRNA transcripts encoding the 11-kDa protein. Importantly, the 11-kDa protein augmented virus DNA replication and, thereafter, virion release. Overall, our results revealed that RBM38 regulates B19V DNA replication via enhancing B19V pre-mRNA splicing at the D2 site, which ensures the production of viral mRNA transcripts that encode the 11-kDa protein.
RESULTS
ISE2 specifically interacts with the RBM38 protein.
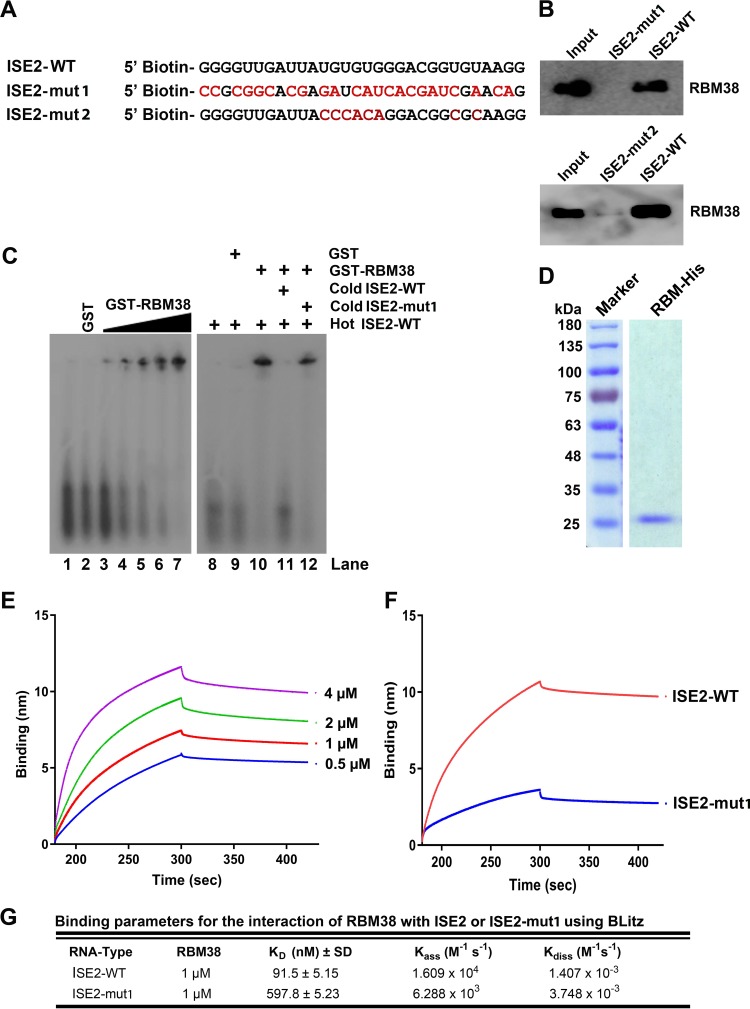
Our previously reported results demonstrated that ISE2 is critical for the recognition of the D2 site for splicing (20). In an in silico analysis of the B19V ISE2 RNA sequence, we found that ISE2 harbors a 5′-UGUGUG-3′ motif, a consensus sequence that binds SUP12, an RBM38 ortholog in Caenorhabditis elegans (21–23). We then asked whether RBM38 interacts with the ISE2 element of the B19V pre-mRNA. We synthesized wild-type ISE2 (ISE2-WT) RNA and a mutant ISE2 (ISE2-mut1) RNA that is identical to the ISEm3 mutation sequence and has been shown to abolish the splicing of B19V pre-mRNA at the D2 site (20) (Fig. 2A) and labeled them with biotin at their 5′ ends. Upon incubation of the two RNA molecules with UT7/Epo-S1 nuclear lysates in the presence of poly(I-C), we performed a pulldown assay using streptavidin-coated beads that bound biotinylated RNA molecules. Upon several washes, the pulled-down proteins were run on SDS-PAGE gels for Western blotting. We found that ISE2-WT pulled down RBM38, whereas ISE2-mut1 did not (Fig. 2B, top). Similarly, we mutated only the RBM38 binding motif within ISE2 (ISE2-mut2) (Fig. 2A) and performed a pulldown assay. Like ISE2-mut1, ISE2-mut2 also did not pull down RBM38 (Fig. 2B, bottom). Next, we purified glutathione S-transferase (GST)-tagged RBM38 for in vitro binding assays. ISE2-WT RNA was synthesized in vitro and radiolabeled with 32P. 32P-labeled hot RNA was incubated with either GST (Fig. 2C, lane 2) or increasing concentrations of GST-RBM38 (Fig. 2C, lanes 3 to 7), and the mixtures were run on a native gel for gel shift assays. It is evident that RBM38 bound ISE2-WT (Fig. 2C). In order to confirm whether the interaction is specific, we coincubated GST-RBM38 and hot ISE2-WT with a molar excess of either cold ISE2-WT or cold ISE2-mut1. Our results showed that only ISE2-WT, but not ISE2-mut1, competed with hot ISE2-WT (Fig. 2C, lane 11 versus lane 12). Furthermore, to determine the binding affinity of RBM38 for ISE2, we purified 6× histidine (His)-tagged RBM38 (Fig. 2D). We synthesized biotinylated ISE2-WT or ISE2-mut1 and performed an in vitro binding assay using biolayer interferometry. The sensograms showed the kinetics binding of RBM38 with ISE2-WT at different protein concentrations (Fig. 2E). Upon comparison of the binding affinities of RBM38 for ISE2-WT and mutant ISE2, the results showed that ISE2-WT bound RBM38 with a high affinity (91.5 ± 5.15 nM) compared to ISE2-mut1 (597.8 ± 5.23 nM) (Fig. 2F). Values for association and dissociation kinetics are also provided (Fig. 2G). In conclusion, our results demonstrated that RBM38 binds specifically to the ISE2 sequence of B19V pre-mRNA but not the mutated ISE2 sequence (ISEm3 or ISE2-mut1) that did not facilitate B19V pre-mRNA splicing at the D2 site in cells (20).
FIG 2.
ISE2 specifically binds the RBM38 protein under in vitro conditions. (A) ISE2 wild-type (ISE2-WT) and ISE2 mutant (ISE2-mut1/2) RNA sequences. The two RNAs shown were used in the binding experiments described below. (B) Pulldown assay. Biotinylated ISE2-WT and ISE2-mut1/2 were incubated with UT7/Epo-S1 nuclear lysates in the presence of poly(I-C) and then pulled down by using streptavidin beads, followed by several washes. The RNA-bound proteins were eluted, run on SDS-PAGE gels for Western blotting, and detected with an anti-RBM38 antibody, as indicated. The UT7/Epo-S1 nuclear lysate was run in lane 1 as a control. (C) Gel shift assay. (Left) Radioactively labeled ISE2-WT was incubated with either purified GST (lanes 2 and 9) or increasing concentrations (5, 10, 50, 100, and 300 nM) of purified GST-RBM38 (lanes 3 to 7, respectively), run on a native gel, and then examined for a shift (left panel). (Right) Similarly, GST-RBM38 was incubated with hot ISE2-WT in the absence (lane 10) or in the presence of excess amounts of cold ISE2-WT (lane 11) and cold ISE2-mut1 (lane 12) for competition assays. Only RNA was run as controls in lanes 1 and 8. (D) RBM38-His purification. RBM38-His was purified as described in Materials and Methods, and 6 μg of protein was run on an SDS–10% PAGE gel and then stained with Coomassie brilliant blue dye. (E to G) Biolayer Interferometry. (E) BLI sensograms showing association and dissociation of the RBM38 protein with ISE2-WT at different concentrations over time. (F) Comparison of binding affinities of the RBM38 protein (1 μM) for the ISE2-WT and ISE2-mut1 RNA molecules. (G) Binding parameters used to calculate KD values (ratio of dissociation and association rate constants). Experiments were repeated at least three times for calculating the means and standard deviations.
RBM38 is expressed during the middle stage of erythroid progenitor development, and knockdown of RBM38 significantly decreases splicing of B19V pre-mRNA at the D2 site.
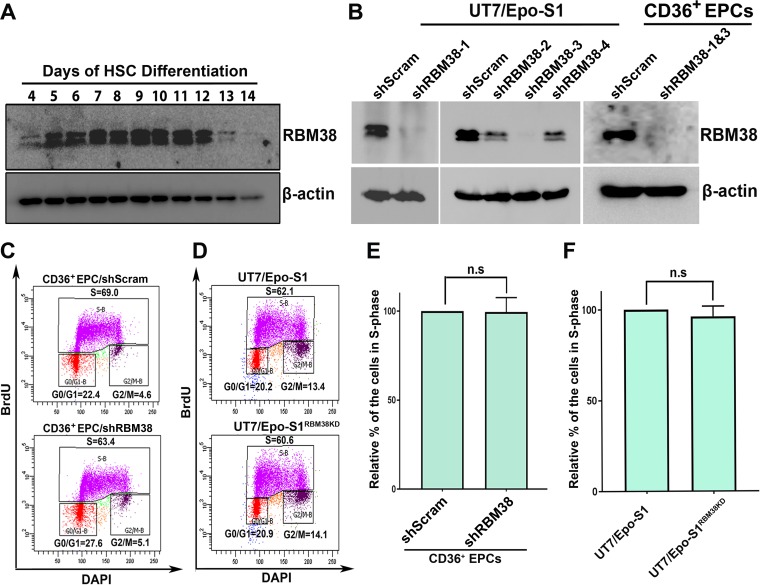
It was reported previously that RBM38 regulates splicing during the erythroid differentiation process (24). We then looked into the expression levels of RBM38 during erythroid developmental stages. Hematopoietic stem cells were cultured from day 4 until day 14, and the cells were collected each day for Western blot analysis of RBM38. As shown in Fig. 3A, The RBM38 expression level increased from day 5 through day 12, which was highly coordinated and consistent with the time course of B19V infection during erythropoiesis (25).
FIG 3.
RBM38 is expressed during the middle phase of erythropoiesis, and its knockdown does not alter cell cycle progression. (A) RBM38 expression during the erythroid differentiation process. Hematopoietic stem cells (HSC) were cultured from day 4 through day 14. The same numbers of cells were collected on each day, lysed, and run for Western blot analysis. The RBM38 protein on the blot was detected by using an anti-RBM38 antibody, and the blot was reprobed for β-actin. (B) RBM38 knockdown. Four lentiviruses expressing shRNAs against RBM38 were transduced into UT7/Epo-S1 cells. CD36+ EPCs were transduced with shRBM38-1 and -3. At 3 days postransduction, the cells were lysed for Western blot analysis using an anti-RBM38 antibody. Blots were reprobed for β-actin. (C to F) Cell cycle analysis. Lentiviruses expressing shRBM38-1 and -3 were used to generate the RBM38-knocked-down UT7/Epo-S1 cell line (UT7/Epo-S1RBM38KD). CD36+ EPCs were transduced with shScram or shRBM38-1 and -3. A BrdU incorporation assay was used to track de novo DNA synthesis as described in Materials and Methods. The cells were processed and analyzed for cell cycle progression by using flow cytometry. (C and D) Representative diagrams showing cell cycle analysis of control and RBM38 knockdown CD36+ EPCs (C) or UT7/Epo-S1 and UT7/Epo-S1RBM38KD cells (D). DAPI, 4′,6-diamidino-2-phenylindole. (E and F) Relative fold changes of the cell population in S phase were calculated for both CD36+ EPCs (E) and UT7/Epo-S1 cells (F). Each experiment was repeated three times for the calculation of means and standard deviations. n.s, no statistical significance (P > 0.05).
Next, using a lentiviral system, we knocked down RBM38 in UT7/Epo-S1 cells using RBM38 short hairpin RNA (shRNA) (shRBM38-1, -2, -3, or -4)-expressing lentiviruses and in CD36+ EPCs using shRBM38-1- and shRBM38-3-expressing lentiviruses (Fig. 3B). RBM38 was successfully knocked down (>90% reduction) in both types of cells. Since B19V replication is dependent on the cellular DNA replication machinery (26, 27), we asked whether the knockdown of RBM38 has any impact on cell cycle progression. Using a bromodeoxyuridine (BrdU) incorporation assay, we determined the percentage of the population of cells in S phase. Our results demonstrated that RBM38 knockdown did not obviously alter the cell proliferation of CD36+ EPCs (Fig. 3C and E) and UT7/Epo-S1 cells (Fig. 3D and F).
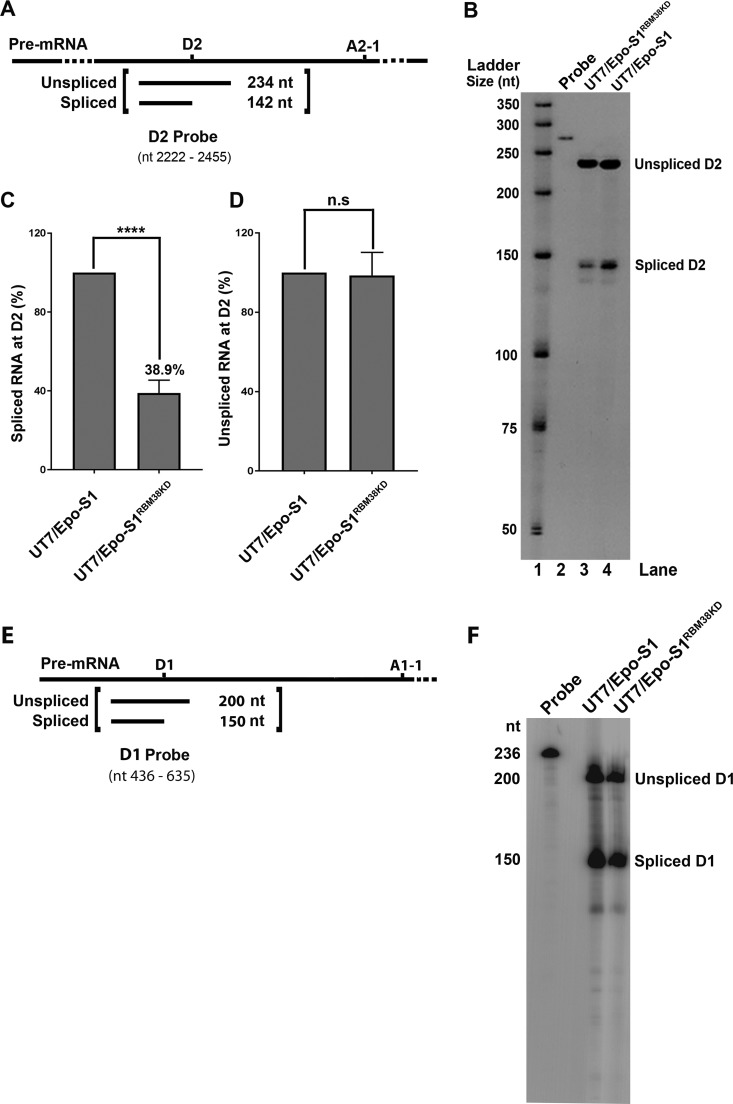
As RBM38 binds ISE2 under in vitro conditions, we next asked whether RBM38 knockdown affects the splicing efficiency at the D2 site. UT7/Epo-S1 and RBM38-knocked-down UT7/Epo-S1RBM38KD cells were transfected with M20 B19V infectious DNA, and the cells were collected after 48 h posttransfection for total RNA extraction, followed by an RNase protection assay. A radiolabeled 234-nucleotide (nt)-long D2 probe was generated and used to detect unspliced and spliced RNAs at the D2 site (Fig. 4A). The results showed that RBM38 knockdown decreased the efficiency of B19V pre-mRNA splicing at the D2 site (Fig. 4B, lane 3). Splicing events at D2 decreased by 61.1% (Fig. 4C), while unspliced RNA remained unaffected (Fig. 4D). Since the D1 site is not flanked by any RBM38 binding motif, as a control, we performed an additional RNase protection assay using a D1 probe (Fig. 4E) to confirm that the RBM38 knockdown did not affect the splicing of B19V pre-mRNA at D1 (Fig. 4F). In conclusion, our results demonstrated that RBM38 is one of the trans-acting factors required for the efficient splicing of B19V pre-mRNA at the D2 site.
FIG 4.
RBM38 promotes splicing at the D2 site. (A) Probe design. A schematic representation of the D2 probe shows the spliced and unspliced RNA species generated by an RNase protection assay. (B) RNase protection assay. Normal UT7/Epo-S1 cells and RBM38-knocked-down cells (UT7/Epo-S1RBM38KD) were transfected with M20. At 2 days posttransfection, the cells were collected for the extraction of total RNA. RNA samples were analyzed by an RNase protection assay using the D2 probe, which was designed to detect spliced/unspliced RNAs from the D2 site of B19V pre-mRNA, as indicated. The RNase protection assay was performed as described in Materials and Methods. (C and D) Quantification. The detected spliced (C) and unspliced (D) RNA bands at the D2 site were quantified in RBM38-knocked-down cell samples and compared to those in normal cell samples (arbitrarily set as 100%). The experiment was repeated at least three times to calculate the means and standard deviations. n.s, no statistical significance (P > 0.05); ****, P < 0.0001. (E and F) D1 splicing. (E) Schematic representation of the D1 probe showing the spliced and unspliced RNA species generated by an RNase protection assay. (F) As described above, RNA samples were subjected to an RNase protection assay using the D1 probe to detect spliced/unspliced RNAs from the D1 site.
RBM38 determines 11-kDa protein expression during B19V replication.
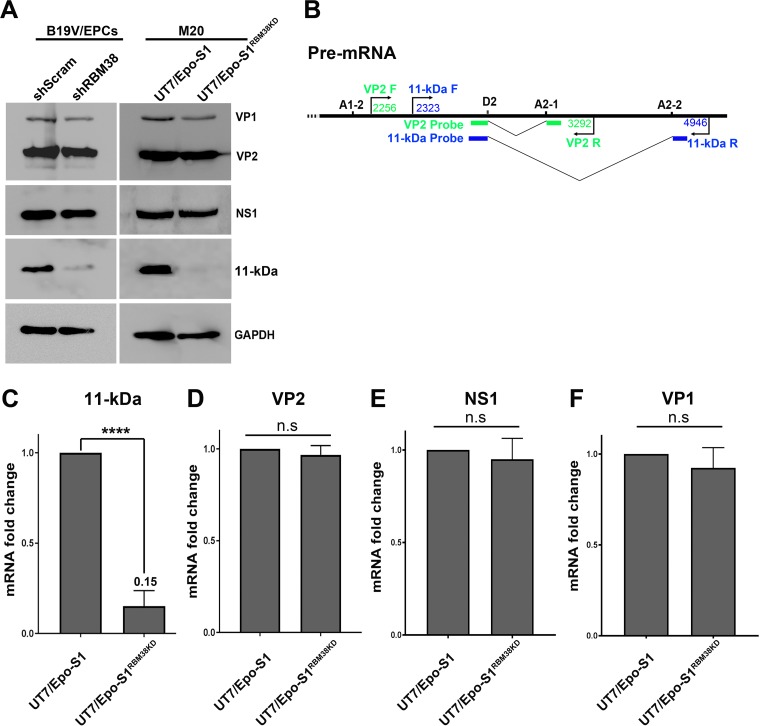
Splicing of B19V pre-mRNA at the D2 site is essential for the generation of viral mRNA transcripts encoding both the VP2 and 11-kDa proteins (Fig. 1B). As RBM38 knockdown decreased the splicing efficiency of B19V pre-mRNA at the D2 site, we asked whether such a decrease affects VP2 and/or 11-kDa protein expression at the protein or the mRNA level. To this end, CD36+ EPCs were infected with B19V, and UT7/Epo-S1 cells were transfected with the M20 infectious clone. The cells were collected at 48 h postinfection or posttransfection for Western blot analysis. The results showed that the level of the 11-kDa protein, but not VP1 and VP2, was significantly decreased in both B19V-infected CD36+ EPCs and M20-transfected UT7/Epo-S1 cells upon RBM38 knockdown (Fig. 5A, left and right, respectively). We then proceeded to analyze viral RNAs extracted as total RNA samples at 48 h posttransfection, using reverse transcription (RT)-quantitative PCR (qPCR) that was specifically designed to detect mRNAs that encode the 11-kDa protein and VP2 (Fig. 5B), as reported previously (28). The RT-qPCR analyses showed that the knockdown of RBM38 in UT7/Epo-S1 cells significantly decreased mRNA levels for the 11-kDa protein by 6.6-fold (Fig. 5C), but the level of VP2 mRNA remained unaffected (Fig. 5D). Also, there was no obvious changes in the levels of NS1- and VP2-encoding mRNAs (Fig. 5E and F). RT-qPCR analyses of viral RNA samples extracted from B19V-infected cells revealed similar results (data not shown).
FIG 5.
RBM38 regulates 11-kDa protein expression. CD36+ EPCs transduced with scramble shRNA (shScram)- and shRBM38-1 and -3 (shRBM38)-expressing lentiviruses were infected with B19V at 2 days postransduction. UT7/Epo-S1 and UT7/Epo-S1RBM38KD cells were transfected with M20. (A) Western blotting. The cells were collected at 2 days postinfection or posttransfection, lysed, and run for Western blotting. Immunoblots were probed for the NS1, VP1/2, and 11-kDa proteins, using their respective antibodies. Blots were reprobed for GAPDH as a loading control. (B) RT-qPCR. Specific primers and probes for the detection of mRNA that encodes the VP2 or 11-kDa protein, as diagrammed, were used for qPCR analysis. (C to F) Detection of mRNAs that encode the 11-kDa, VP2, NS1, and VP1 proteins by RT-qPCR. At 2 days posttransfection, total RNA was extracted from UT7/Epo-S1 and UT7/Epo-S1RBM38KD cells. The RNA was further used to generate cDNA for the quantification of mRNAs that encode the 11-kDa (C), VP2 (D), NS1 (E), and VP1 (F) proteins, using the RT-qPCR approach. Quantified viral mRNA levels were normalized to the level of β-actin mRNA, and mRNAs extracted from M20-transfected control UT7/Epo-S1 cells were used as controls and arbitrarily set as a value of 1. n.s, no statistical significance (P > 0.05); ****, P < 0.0001.
RBM38 knockdown decreases B19V DNA replication and virion release.
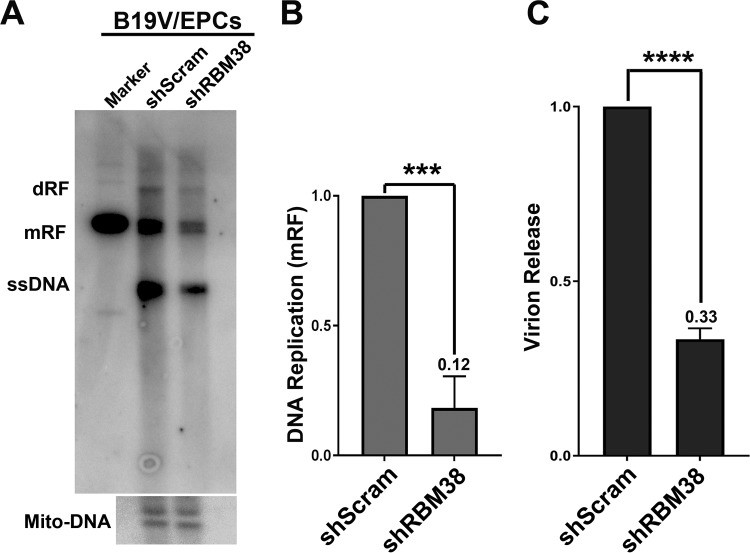
Next, we asked whether RBM38 knockdown affects virus replication. To this end, CD36+ EPCs were transduced with scramble shRNA (shScram)- and shRBM38-expressing lentiviruses and infected with B19V at 48 h postransduction. The cells were collected at 48 h postinfection for the determination of viral DNA replication and virion release. Surprisingly, our results showed that RBM38 knockdown significantly decreased both viral DNA replication (Fig. 6A and B) and virion production (Fig. 6C). Similarly, M20-transfected RBM38 knockdown cells also showed a decrease in virion production (see Fig. 8A, compare groups 1 and 4) and virus DNA replication (see Fig. 8B, compare lanes 1 and 4), compared to those from WT M20-transfected control UT7/Epo-S1 cells.
FIG 6.
RBM38 knockdown decreases viral DNA replication and virion release. CD36+ EPCs were transduced with shScram and shRBM38-1 and -3(shRBM38) lentiviruses. At 2 days postransduction, the cells were infected with B19V. (A and B) Southern blotting. At 2 days postinfection, the cells were collected for Southern blot analysis. (A) The blots were probed with the M20 probe (top) and the mitochondrial DNA probe (Mito-DNA) (bottom). dRF and mRF, double and monomer replicative forms, respectively; ssDNA, single-stranded DNA. (B) The replicated (mRF) DNA bands were quantified and plotted. (C) Virion release. At 3 days postinfection, the supernatants of infected cells were collected for quantification of virions released by using qPCR. Results shown are averages and standard deviations, which were obtained from at least three independent experiments (B and C). ***, P < 0.001; ****, P < 0.0001.
FIG 8.
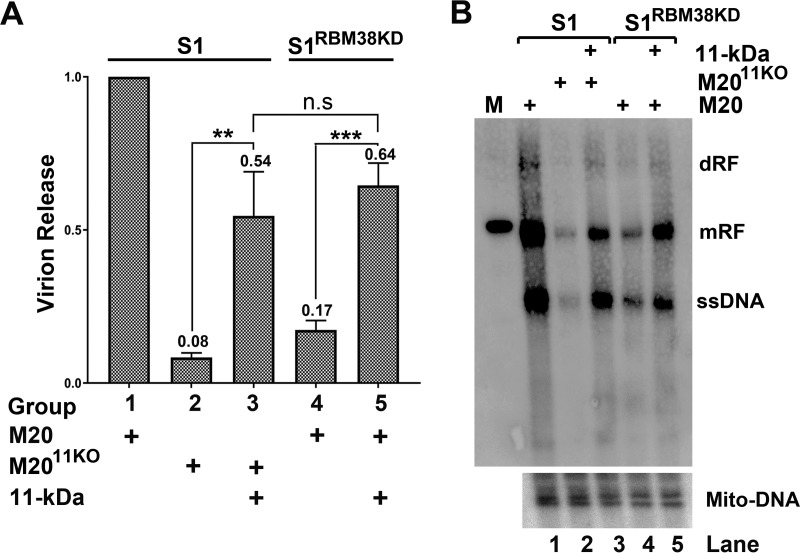
RBM38 knockdown decreases viral DNA replication and virion release via reduction of 11-kDa protein expression. UT7/Epo-S1 cells were transfected with M20 or M2011KO, and 11-kDa-protein-expressing UT7/Epo-S1 cells were transfected with M2011KO, as indicated. UT7/Epo-S1RBM38KD cells or 11-kDa-protein-expressing UT7/Epo-S1RBM38KD cells were transfected with M20. (A) Virion release. At 3 days posttransfection, the supernatants from each transfected sample were collected and used for virion quantification by qPCR. (B) Southern blot analysis. At 3 days posttransfection, cells were collected for Hirt DNA extraction. Hirt DNA samples were subjected to Southern blotting using M20 and Mito-DNA probes. dRF and mRF, double and monomer replicative forms, respectively. n.s, no statistical significance (P > 0.05); **, P < 0.01; ***, P < 0.001.
Thus, our results demonstrated that RBM38 plays an important role in viral DNA replication and virion release during B19V infection of CD36+ EPCs as well as in the transfection of the B19V duplex genome in UT7/Epo-S1 cells.
The 11-kDa protein enhances B19V DNA replication and virion release.
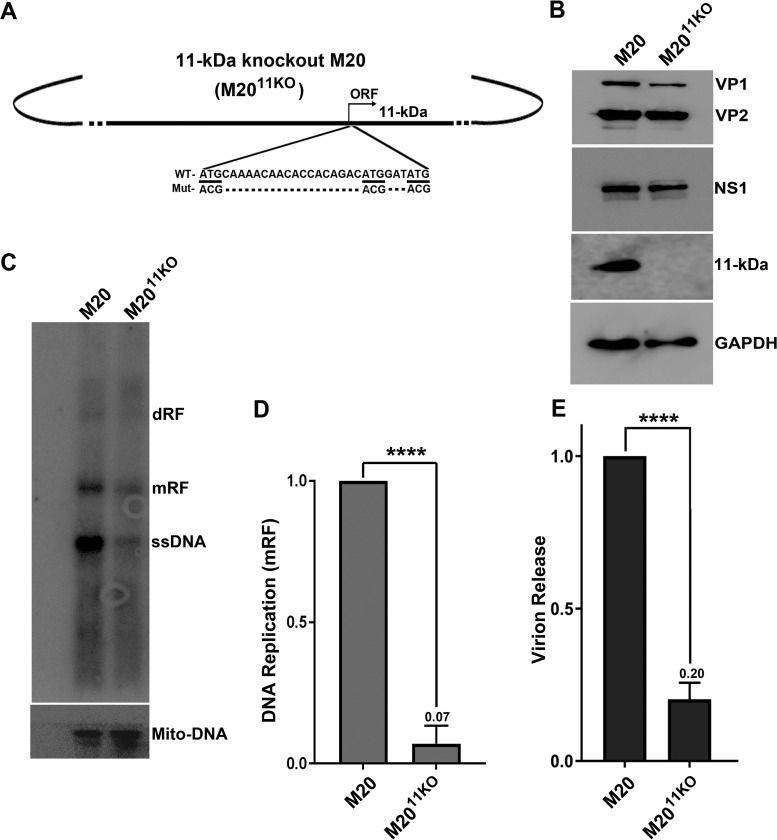
As the knockdown of RBM38 decreases 11-kDa protein expression, we investigated the role of the 11-kDa protein in viral DNA replication and virion release. We first constructed an 11-kDa-protein-null M20 mutant (M2011KO) by silently mutating the three potential AUG sites in the open reading frame (ORF) for the 11-kDa protein (Fig. 7A). We confirmed that M2011KO did not affect the expression of NS1, VP1, and VP2 (Fig. 7B).
FIG 7.
The 11-kDa protein enhances B19V DNA replication and virion release. (A) Diagram of M2011KO. The 11-kDa protein knockout M20 clone (M2011KO) is diagrammed and shown with mutations of three translation initiation codons from ATG to ACG. (B) Western blotting. UT7/Epo-S1 cells were transfected with M20 and M2011KO. At 2 days posttransfection, cells were collected, lysed, and run for Western blot analysis. The proteins on the blots were detected with anti-VP1/2, NS1, 11-kDa protein, and GAPDH antibodies. (C and D) Southern blotting. UT7/Epo-S1 cells were transfected with M20 and M2011KO. At 2 days posttransfection, Hirt DNA samples were extracted from transfected cells and subjected to Southern blotting. (C) The blots were probed by using the M20 probe (top) and the Mito-DNA probe (bottom). mRF, monomer replicative form DNA. (D) mRF DNA bands were quantified, and the means and standard deviations were calculated from three independent experiments. (E) Virion release. At 3 days posttransfection, the supernatants from transfected cells were collected and used for the quantification of virions. Virions released from M20-transfected cells were arbitrarily set as a value of 1. ****, P < 0.0001.
To determine whether 11-kDa protein expression has an impact on virus replication, the M20 and M2011KO clones were transfected into UT7/Epo-S1 cells. At 48 h posttransfection, cells were collected for the extraction of Hirt DNA samples, which were subsequently subjected to Southern blot analysis. As shown in Fig. 7C and D, the level of replication of M2011KO was significantly lower than that of WT M20. At 72 h posttransfection, the media of the transfected cells were collected, subjected to DNase I treatment, and further used to quantify the numbers of released virions. As shown in Fig. 7E, M2011KO-transfected cells had a significant decrease in the amount of released virions, compared with those of WT M20-transfected cells. Collectively, these results strongly supported that the 11-kDa protein augments B19V DNA replication and virion release.
Decreased 11-kDa protein expression in RBM38-knocked-down cells contributes to reduced viral DNA replication and virion release.
In order to confirm whether RBM38 knockdown decreased viral DNA replication via inhibition of the 11-kDa protein, we generated an 11-kDa-protein-expressing lentivirus to transduce UT7/Epo-S1 cells. UT7/Epo-S1 cells were transfected with M20 and M2011KO, and UT7/Epo-S1 cells expressing the 11-kDa protein were transfected with M2011KO. At 72 h posttransfection, the supernatant and cells were collected and examined for virion release and DNA replication, respectively. The results show that 11-kDa protein complementation in M2011KO-transfected cells significantly enhanced virion release (Fig. 8A, compare groups 2 and 3) and virus DNA replication (Fig. 8B, compare lanes 2 and 3). Similarly, RBM38-knocked-down UT7/Epo-S1RBM38KD cells and 11-kDa-protein-expressing UT7/Epo-S1RBM38KD cells were transfected with M20. At 72 h posttransfection, supernatants and cells were collected and examined for virion release and virus DNA replication, respectively. Obviously, we observed that the levels of both virion release (Fig. 8A, compare groups 4 and 5) and virus DNA replication (Fig. 8B, compare lanes 4 and 5) were significantly higher in 11-kDa-protein-expressing UT7/Epo-S1RBM38KD cells than in non-11-kDa-protein-expressing UT7/Epo-S1RBM38KD cells. Overall, the complementation of the 11-kDa protein in UT7/Epo-S1RBM38KD cells recovered ∼60% of the function of the 11-kDa protein in virion release and viral DNA replication during transfection of the M20 clone.
Taken together, these results confirmed that RBM38 plays an important role in B19V DNA replication and virion release through the regulation of 11-kDa protein expression.
DISCUSSION
Parvoviruses with a limited genome size use alternative splicing and alternate polyadenylation to maximize the coding capacity of their genome. B19V pre-mRNA splicing at the D2 site is critical for the production of mRNAs that encode the VP2 and 11-kDa proteins (20). However, the remaining unspliced mRNAs constitute mRNAs that encode the NS1, 7.5-kDa, and VP1 proteins. B19V tightly regulates pre-mRNA splicing at D2 to maintain the variable expression levels of all five viral proteins. Alternative splicing of pre-mRNA is regulated by both cis-acting elements and trans-acting factors that regulate splicing both positively and negatively (29). In the case of B19V pre-mRNA splicing at D2, we previously identified ISE2, a cis-acting element that facilitates the recognition of D2 for splicing (20). In this study, we show that ISE2 is essential for the expression of the viral 11-kDa protein. A cellular trans-acting factor, RBM38, binds ISE2 and regulates splicing at the D2 site. The knockdown of RBM38 decreased the splicing of B19V pre-mRNA at D2, which particularly affects splicing from D2 to A2-2 sites that decreases the levels of mRNAs encoding the 11-kDa protein (Fig. 1B, R8 and R9 mRNA). Importantly, we show that the 11-kDa protein augments viral DNA replication and virion release and that RBM38 promotes B19V replication via 11-kDa protein regulation.
RBM38 binds to ISE2 and regulates splicing at the D2 site and, therefore, 11-kDa protein expression.
During alternative splicing, the splice donor sites are usually weak and are regulated by the interplay of positive and negative cis-acting elements and trans-acting factors (29). The splicing of B19V pre-mRNA at D2 generates mRNAs that encode either the VP2 or 11-kDaprotein, while unspliced mRNAs at D2 encode the NS1, VP1, and 7.5-kDa proteins (Fig. 1, R1, R2, R4, and R5 mRNA). It is obvious that D2 splicing needs to be regulated in such a way that the expression of all five proteins is maintained at an optimal ratio. We identified at least two cis elements, exon splicing enhancer 3 (ESE3) and ISE2, that regulate D2 splicing (20). While looking for other trans-acting factors, we observed that ISE2 carries a 5′-UGUGUG-3′ motif that binds SUP12, an RBM38 ortholog in C. elegans (21–23). RBM38 is expressed during the middle stage of erythroid differentiation, which coincides with the time course of B19V infection (Fig. 3). Using RNA pulldown assays, gel shift assays, and biolayer interferometry (Fig. 2), our results showed that RBM38 specifically binds to ISE2 RNA under in vitro conditions, and the mutated ISE2 sequence (ISEm3 or ISE2-mut1), which did not facilitate the splicing of B19V pre-mRNA at the D2 site (20), also did not specifically bind RBM38. The knockdown of RBM38 in M20-transfected UT7/Epo-S1 cells decreased the splicing efficiency at the D2 site. Surprisingly, the decrease in D2 splicing affected only mRNAs that encode the 11-kDa protein but not VP2-encoding mRNAs (Fig. 5), which implies that RBM38 promotes the splicing of the second intron from the D2 to A2-2 sites. It is possible that another ESE3 or ISE2 binding factor(s) might be responsible for the splicing of the second intron from the D2 to A2-1 sites, which remains to be investigated further in the future. The complex interplay between cis-acting elements surrounding the D2 site and trans-acting factors, at least RBM38, determines the splicing of the second intron and, eventually, the production of the VP2 and 11-kDa proteins. Of note, there are at least five other 5′-UGUGUG-3′ motifs in the B19V pre-mRNA sequence, among which are two sites that follow the cryptic donor D2′. We previously reported that B19V uses a cryptic donor D2′ site, when the D2 site is mutated (20). The cryptic donors might also need RBM38, since it is followed by two similar 5′-UGUGUG-3′ motifs.
The 11-kDa protein enhances B19V DNA replication and virion release from infected cells.
It was reported previously that the 11-kDa protein helps in capsid production and virus infectivity (30). We asked whether the 11-kDa protein plays any other roles in virus replication. Our results show that the 11-kDa protein augments viral DNA replication and virion release (by ∼7- to 10-fold). We previously reported that erythropoietin (Epo) signaling is essential for B19V replication (31). Epo binds Epo receptor and activates the JAK2-STAT5 and MEK/extracellular signal-regulated kinase (ERK) pathways (32). We also demonstrated previously that the ERK pathway negatively regulates B19V replication (33), whereas STAT5 phosphorylation is critical for viral DNA replication (34). The 11-kDa protein has been shown to interact with growth factor receptor-bound protein 2 (Grb2) via its proline-rich region, an Src homology 3 (SH3)-like ligand (35). Grb2 is an adaptor protein that interacts with Ras guanine exchange factor-SOS via its SH3 domain, thereby activating the mitogen-activated protein kinase (MAPK)/ERK signal transduction pathway (36). Since the MEK/ERK pathway impedes B19V replication (33), we speculate that B19V uses the interaction of the 11-kDa protein with Grb2 to disrupt MEK/ERK signaling to promote virus replication. On the other hand, during B19V infection, viral DNA replication (37), NS1 (38), and the 11-kDa protein (39) induce the death of erythroid progenitors. In this study, we examined the role of RBM38 in the production of the 11-kDa protein and its outcomes in the context of virus replication. It is possible that the contribution of the 11-kDa protein to B19V-induced cell death can also be modulated via RBM38 regulation. Whether 11-kDa-protein-mediated cell death plays any role in virion release needs further investigation.
Function of RBM38 as an essential host factor for B19V infection.
RBM38 is an RNA recognition motif (RRM)-containing RNA binding protein of the largest family of RNA binding proteins (40, 41). Diverse roles have been attributed to RBM38, ranging from RNA stability (42) and mRNA translation (43, 44) to the regulation of splicing during erythroid differentiation (24). In the case of B19V, we did not observe any change in the expression levels of NS1, VP1, and VP2 upon the knockdown of RBM38 (Fig. 5); therefore, it is less likely that RBM38 regulates viral mRNA stability and translation during virus infection. The RBM38 ortholog SUP12 regulates the splicing of various genes in C. elegans (21–23). Here we identified RBM38 as one of the trans-acting factors that binds to ISE2, promoting the splicing of the second intron from the D2 to A2-2 sites that favors the expression of the 11-kDa protein. We show mechanistically how B19V uses a cellular host factor (RBM38) for its efficient replication. The expression of RBM38 during the stages when cells are highly susceptible to B19V infection renders it one of the determinants of B19V tropism. Our hypothetical model for D2 splicing of B19V pre-mRNA is that the binding of RBM38 to ISE2 activates the recognition of the D2 site with the A2-2 site (Fig. 9). Since the U2 small nuclear ribonucleoprotein (snRNP) recognizes both the A2-1 and A2-2 sites, we speculate that after binding the ISE2 element, RBM38 interacts with the A2-2 site via an unknown mediator, M3, that specifically binds the A2-2 site (Fig. 9). We are also investigating other host factors that bind the ISE2 region (data not shown) and promote the recognition of D2 with A2-1, which ultimately determines the expression of the VP2 protein. Our overall hypothesis is that the two cis-acting elements flanking the D2 site, i.e., ESE3 and ISE2, with the help of trans-acting factors, regulate the expression of at least two viral proteins (VP2 and 11-kDa protein) during B19V replication. The identification of such trans-acting factors will eventually reveal the entire scenario of how D2 plays a central role in the processing of B19V pre-mRNA, which will identify new targets to inhibit B19V infection.
FIG 9.
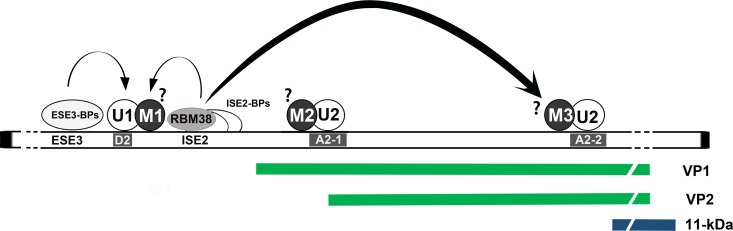
Proposed model for the role of RBM38 in B19V pre-mRNA processing. U1 and U2 represent U1- and U2-small nuclear ribonucleoprotein (snRNP) complexes, and M represents protein mediators associated with U1- and U2-snRNP complexes. RBM38 and ISE2 binding proteins (ISE2-BPs) interact with the ISE2 sequence, whereas ESE3 binding proteins (ESE3-BPs) interact with the ESE3 region of the B19V pre-mRNA. Both ISE2 and ESE3 and their associated trans-acting factors help in the recognition of the D2 site, as shown by arrows. The interaction of RBM38 with ISE2 promotes splicing of the intron from D2 to A2-2. Question marks indicate unknown factors.
MATERIALS AND METHODS
Ethics statement.
Primary human CD133+ hematopoietic stem cells were isolated from bone marrow of healthy donors according to a protocol (protocol 04-H-0179) approved by the National Heart, Lung, and Blood Institute institutional review board. There was no donor information associated with samples when we received the cells.
Primary cells and cell lines.
Human CD133+ hematopoietic stem cells were expanded ex vivo to CD36+ EPCs in Wong medium, as described previously (34, 45). Briefly, CD133+ cells were cultured in Wong medium under normoxia (5% CO2 and 21% O2) up to day 4 and frozen in liquid nitrogen. In each experiment, day 4 cells were seeded under normoxia for 2 to 3 days, prior to incubation under hypoxia (5% CO2 and 1% O2) for 2 days.
UT7/Epo-S1, a human megakaryoblastoid cell line (28, 46), was cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and erythropoietin (2 U/ml; Amgen, Thousand Oaks, CA). UT7/Epo-S1 cells were cultured under hypoxia (1% O2) for 2 to 3 days posttransfection.
UT7/Epo-S1 cells were transduced with lentiviruses expressing shRBM38-1 and -3. The cells were further selected with increasing concentrations of puromycin (1 to 5 μg/ml) and passaged for several weeks to generate the RBM38-knocked-down UT7/Epo-S1 cell line.
Virus and infection.
Plasma sample P430 containing B19V (∼1 ×1012 viral genome copies [vgc]/ml) was obtained from ViraCor Eurofins Laboratories (Lee's Summit, MO). CD36+ EPCs were infected with B19V at a multiplicity of infection (MOI) of 1,000 vgc per cell. The infected cells and media were collected after 2 to 3 days postinfection for Southern/Western blot analyses and for the quantification of virion release, respectively.
Plasmid construction. (i) The 11-kDa protein knockout M20 clone (pM2011KO).
The three ATG codons in the 11-kDa-protein-encoding ORF of pM20 were mutated to ACG codons by using an overlap PCR approach, as described previously (19, 20).
(ii) pLKO constructs.
Lentiviral vector pLKO.1 with an mCherry reporter was used to clone shRNA sequences between the AgeI and EcoRI sites. pLKO.1 containing a scramble shRNA sequence was used as a control (45). The following shRNAs were obtained from Sigma (St. Louis, MO) for knocking down RBM38: shRBM38-1 (5′-CCG GCG GCT TCT CTT TAA TCT AGG TC TCG AGA CCT AGA TTA AAG AGA AGC CGT TTT TTG-3′), shRBM38-2 (5′-CCG GGA CGA CGA TAG TGT TTC TGT ACT CGA GTA CAG AAA CAC TAT CGT CGT CTT TTT TG-3′), shRBM38-3 (5′-CCG GCA ACG TGA ACC TGG CAT ATC TCT CGA GAG ATA TGC CAG GTT CAC GTT GTT TTT TG-3′), and shRBM38-4 (5′-CCG GGA AAC CTG AAA GCA AGA AGT TCT CGA GAA CTT CTT GCT TTC AGG TTT CTT TTT TG-3′) (underlining indicates shRNA sequences).
(iii) pLenti-11-kDa construct.
The pLenti vector was used to clone the optimized ORF encoding the 11-kDa protein between the restriction sites XbaI and SalI, as described previously (31, 45).
Transfection.
UT7/Epo-S1 cells were electroporated with 2 μg of B19V DNA fragments, as reported previously (34, 45), using solution V from the Amaxa Nucleofector system (Lonza, Basel, Switzerland). B19V infectious clone pM20 and the pM2011KO mutant were digested with SalI to release M20 and mutant M2011KO B19V duplex genomes.
RNase protection assay.
Total RNA was extracted from infected or transfected cells by using TRIzol reagent (Invitrogen). RNase protection was performed as described previously (19, 20). Both D1 and D2 probes were generated by using a MAXIscript T7 in vitro transcription kit (Ambion), according to the manufacturer's instructions. The images were captured by using a Typhoon FLA 9000 imager (GE Healthcare) and quantified by using Image Quant TL software (GE Healthcare).
Reverse transcription-quantitative PCR.
cDNA was synthesized by using Moloney murine leukemia virus (M-MLV) kit (Life Technologies, Carlsbad, CA). A multiplex RT-qPCR system was used to detect B19V-specific mRNAs, with β-actin mRNA serving as an internal control (28). For quantification of the levels of the mRNA that encodes the 11-kDa protein, forward primer 5′-GAA GCC TTC TAC ACA CCT TTG G-3′, reverse primer 5′-TGG CTG TCC ACA ATT CTT CAG G-3′, and probe 5′-FAM (6-carboxyfluorescein)-AGA CCA GTT TCG TGA ACT/CTA CAG ATG CA-BHQ (black hole quencher)-3′ were used (28). For quantification of VP2 mRNA levels, forward primer 5′-GAC CAG TTC AGG AGA ATC AT-3′, reverse primer 5′-TTC TGA GGC GTT GTA AGC-3′, and probe 5′-FAM-TCG TGA ACT GTG CAG CTG CCC CTG TG-BHQ1-3′ were used (28). All primers and probes were purchased from Integrated DNA Technologies (Coralville, IA). The primers and probe for quantification of the B19V genome were described previously (34).
RNA pulldown assay.
An RNA pulldown assay was performed as described previously (47, 48), with some modifications. Briefly, ISE2-WT and mutant ISE2 (ISE2-mut1/2) RNA sequences were chemically synthesized by Integrated DNA Technologies (Coralville, IA) with biotinylation at the 5′ end. One hundred microliters of a streptavidin slurry (Gold Biotechnology Inc., St. Louis, MO) was used for each sample, and the samples were washed three times with wash buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM EDTA, 0.5 mM dithiothreitol [DTT], and 1× protease inhibitor cocktail [Sigma]). Beads were finally resuspended in 500 μl of wash buffer, and 10 μl of biotinylated RNA molecules (ISE2-WT or ISE2-mut1/2, at 100 μM) was added to the mixture. The mixtures were rotated for 3 to 4 h at 4°C. The beads were spun down, washed again with wash buffer three times, and resuspended in 500 μl of RNA binding buffer [20 mM Tris-HCl (pH 7.5), 300 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 6 to 10 μg/ml poly(I-C), proteinase inhibitors, and an RNase inhibitor], and 100 μl of the nuclear lysate (at a concentration of 3 to 5 μg/μl) was added to each sample. The UT7/Epo-S1 nuclear extract was prepared as described previously (45). The mixtures were rotated overnight, and the beads were then spun down and washed three times with wash buffer. The bound proteins were eluted by using 60 μl of 2× Laemmli loading buffer and boiled for 5 min. The samples were run for Western blotting.
Electrophoretic mobility shift assay.
ISE2-WT and ISE-mut1 RNA sequences were synthesized by using a MAXIscript T7 in vitro transcription kit (Ambion) and further purified by using an RNA purification kit (catalog no. R1015; Zymo Research, Irvine, CA), according to the manufacturer's instructions. GST and GST-RBM38 proteins were incubated with radiolabeled ISE2-WT RNA in the presence or absence of cold ISE2-WT and cold ISE2-mut1, as indicated for each experiment. A gel shift assay was performed as reported previously (34, 49, 50).
Hirt DNA extraction and Southern blotting.
Hirt DNA (lower-molecular-weight DNA) was extracted from B19V-infected CD36+ EPCs or B19V DNA-transfected UT7/Epo-S1 cells, as reported previously (34, 51). Hirt DNAs extracted from transfected cells were digested with DpnI before being subjected to Southern blot analysis. B19V M20 DNA was used as a template for radioactive labeling to generate the M20 probe. A mitochondrial DNA (Mito-DNA) probe was used as a control for the recovery of Hirt DNA (52).
Western blotting.
Western blotting was performed as reported previously (26, 34). The following primary antibodies were used: anti-RBM38 antibody (catalog no. sc-365898; Santa Cruz, Dallas, TX), anti-VP1/VP2 (catalog no. MAB8293; Millipore, Billerica, MA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog no. A01622; GenScript, Piscataway, NJ), anti-β-actin (catalog no. A5441; Sigma-Aldrich, St. Louis, MO), and anti-BrdU (catalog no. 600-401-C29; Rockland, Limerick, PA). Anti-NS1 and anti-11-kDa-protein antibodies were produced in-house, as described previously (39, 53). Horseradish peroxidase (HRP)-conjugated anti-mouse IgG and anti-rabbit IgG secondary antibodies were purchased from Sigma.
Virion release and quantification.
For the quantification of virions released from transfected UT7/Epo-S1 cells, M20 and M2011KO were electroporated with SalI-linearized DNA. For the quantification of virions released from B19V-infected CD36+ EPCs, shRNA-transduced CD36+ EPCs were infected with B19V. The cells were washed at 3 h postinfection and cultured under hypoxia. At 3 days postelectroporation, or postinfection, 200 μl of the supernatant from each cell sample was collected and treated with Benzonase nuclease (250 U/ml) at 37°C for 2 h. A total of 10 mM EDTA was added to stop the reaction, followed by proteinase K treatment. The nuclease-digestion-resistant viral genome was extracted by using a DNeasy blood and tissue kit (Qiagen, Germantown, MD), according to the manufacturer's instructions. Finally, the recovered viral DNA was resuspended into 200 μl of DNase-free H2O, and vgc were quantified by qPCR, as described previously (28).
Biolayer interferometry.
Biolayer interferometry was used to calculate the binding affinities of RBM38 for ISE2-WT and ISE2-mut1 RNA sequences, as reported previously (54, 55). Briefly, biotinylated RNA was mounted onto streptavidin biosensors (catalog no. 18-5019; Forte Bio Inc., Fremont, CA), equilibrated with RNA binding buffer (20 mM Tris-HCl [pH 7.4], 80 mM NaCl), and then dipped into buffer containing the RBM38 protein at different concentrations to calculate the binding parameters Kass (association rate constant), Kdiss (dissociation rate constant), and KD (equilibrium dissociation constant) by using Blitz Pro software.
Cell cycle analysis.
A BrdU incorporation assay was used to determine the percentage of the cell population in S phase, followed by flow cytometry, as described previously (34, 45).
Lentivirus production.
Lentiviruses were produced according to the instructions provided by Addgene (http://www.addgene.org/tools/protocols/plko) and purified as described previously (31). Cells were transduced with the lentiviral vector at an MOI of ∼5 transduction units/cell, as described previously (31).
Purification of GST- and His-tagged RBM38 proteins.
The bacterially optimized RBM38 ORF was cloned into either the pGEX-4T-3 vector with the GST-encoding sequence at the 5′ end or the pET30a(+) vector between the NdeI and XhoI restriction sites to express the GST-RBM38 and RBM38-His proteins, respectively. The resulting plasmids were transformed into bacterial strain BL21(DE3)/pLysS (Promega, Madison, WI). To induce protein expression, isopropyl-d-1-thiogalactopyranoside (IPTG) was added to the bacterial culture at an optical density at 600 nm (OD600) of 0.6 at a final concentration of 0.5 mM, and the culture was grown for another 2 to 3 h at 37°C. The GST-RBM38 protein was purified as described previously (56). For the purification of RBM38-His, the cells were pelleted down and resuspended in lysis buffer {50 mM Tris (pH 7.4), 150 mM NaCl, 2 mM DTT, 5 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1% Triton X-100, and 1× protease inhibitor cocktail (Sigma)}. The lysate was further sonicated, centrifuged, and passed through a 0.45-μm filter (Thermo). The cleared lysate was mixed with Ni-nitrilotriacetic acid (NTA) beads (Qiagen) and loaded onto a column. The beads were equilibrated with lysis buffer and then washed with lysis buffer, followed by three washes with lysis buffer containing imidazole at concentrations of 20, 30, and 40 mM. During all washing steps, the NaCl concentration was increased to 500 mM. The bound protein was eluted with elution buffer (50 mM Tris, 500 mM NaCl, 0.05% Triton X-100, and 250 mM imidazole). The eluted protein was dialyzed against Tris buffer (20 mM Tris [pH 7.4], 100 mM NaCl), concentrated, quantified, aliquoted, and stored at −80°C.
Statistics.
Statistical analysis was done by using GraphPad Prism version 7.0. Error bars represent means and standard deviations (SD), and statistical significance (P value) was determined by using the Student t test.
ACKNOWLEDGMENTS
We are grateful to members of the Qiu laboratory for technical support and valuable discussions. We are indebted to Susan Wong at the Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, for providing CD133+ human hematopoietic stem cells.
This study was supported in full by PHS grant R01 AI070723 from the National Institute of Allergy and Infectious Diseases to J.Q. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young N, Harrison M, Moore J, Mortimer P, Humphries RK. 1984. Direct demonstration of the human parvovirus in erythroid progenitor cells infected in vitro. J Clin Invest 74:2024–2032. doi: 10.1172/JCI111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young NS, Mortimer PP, Moore JG, Humphries RK. 1984. Characterization of a virus that causes transient aplastic crisis. J Clin Invest 73:224–230. doi: 10.1172/JCI111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozawa K, Kurtzman G, Young N. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233:883–886. doi: 10.1126/science.3738514. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava A, Lu L. 1988. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J Virol 62:3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamson-Small LA, Ignatovich IV, Laemmerhirt MG, Hobbs JA. 2014. Persistent parvovirus B19 infection in non-erythroid tissues: possible role in the inflammatory and disease process. Virus Res 190:8–16. doi: 10.1016/j.virusres.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Qiu J, Soderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young NS, Brown KE. 2004. Parvovirus B19. N Engl J Med 350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 9.Gallinella G. 2013. Parvovirus B19 achievements and challenges. ISRN Virol 2013:898730. doi: 10.5402/2013/898730. [DOI] [Google Scholar]
- 10.Deiss V, Tratschin JD, Weitz M, Siegl G. 1990. Cloning of the human parvovirus B19 genome and structural analysis of its palindromic termini. Virology 175:247–254. doi: 10.1016/0042-6822(90)90205-6. [DOI] [PubMed] [Google Scholar]
- 11.Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol 61:2395–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo W, Astell CR. 1993. A novel protein encoded by small RNAs of parvovirus B19. Virology 195:448–455. doi: 10.1006/viro.1993.1395. [DOI] [PubMed] [Google Scholar]
- 13.St Amand J, Astell CR. 1993. Identification and characterization of a family of 11-kDa proteins encoded by the human parvovirus B19. Virology 192:121–131. doi: 10.1006/viro.1993.1014. [DOI] [PubMed] [Google Scholar]
- 14.Yoto Y, Qiu J, Pintel DJ. 2006. Identification and characterization of two internal cleavage and polyadenylation sites of parvovirus B19 RNA. J Virol 80:1604–1609. doi: 10.1128/JVI.80.3.1604-1609.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan W, Cheng F, Yoto Y, Kleiboeker S, Wong S, Zhi N, Pintel DJ, Qiu J. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J Virol 82:9951–9963. doi: 10.1128/JVI.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JM, Green SW, Shimada T, Young NS. 1992. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J Virol 66:4686–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St Amand J, Beard C, Humphries K, Astell CR. 1991. Analysis of splice junctions and in vitro and in vivo translation potential of the small, abundant B19 parvovirus RNAs. Virology 183:133–142. doi: 10.1016/0042-6822(91)90126-V. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Qiu J, Cheng F, Chu Y, Yoto Y, O'Sullivan MG, Brown KE, Pintel DJ. 2004. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J Virol 78:12929–12939. doi: 10.1128/JVI.78.23.12929-12939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan W, Huang Q, Cheng F, Qiu J. 2011. Internal polyadenylation of the parvovirus B19 precursor mRNA is regulated by alternative splicing. J Biol Chem 286:24793–24805. doi: 10.1074/jbc.M111.227439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan W, Cheng F, Huang Q, Kleiboeker S, Qiu J. 2011. Inclusion of the central exon of parvovirus B19 precursor mRNA is determined by multiple splicing enhancers in both the exon and the downstream intron. J Virol 85:2463–2468. doi: 10.1128/JVI.01708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. 2007. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol 27:8612–8621. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno G, Ono K, Togo M, Watanabe Y, Ono S, Hagiwara M, Kuroyanagi H. 2012. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet 8:e1002991. doi: 10.1371/journal.pgen.1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, Ono S. 2004. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J Cell Biol 167:639–647. doi: 10.1083/jcb.200407085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinicke LA, Nabet B, Shen S, Jiang P, van Zalen S, Cieply B, Russell JE, Xing Y, Carstens RP. 2013. The RNA binding protein RBM38 (RNPC1) regulates splicing during late erythroid differentiation. PLoS One 8:e78031. doi: 10.1371/journal.pone.0078031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong S, Zhi N, Filippone C, Keyvanfar K, Kajigaya S, Brown KE, Young NS. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol 82:2470–2476. doi: 10.1128/JVI.02247-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Kleiboeker S, Deng X, Qiu J. 2013. Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol 87:12766–12775. doi: 10.1128/JVI.02333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y, Qiu J. 2015. Human parvovirus B19: a mechanistic overview of infection and DNA replication. Future Virol 10:155–167. doi: 10.2217/fvl.14.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus b19 can replicate in nonpermissive cells with the help of adenovirus genes and produces infectious virus. J Virol 83:9541–9553. doi: 10.1128/JVI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol 80:5941–5950. doi: 10.1128/JVI.02430-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen AY, Guan W, Lou S, Liu Z, Kleiboeker S, Qiu J. 2010. Role of erythropoietin receptor signaling in parvovirus B19 replication in human erythroid progenitor cells. J Virol 84:12385–12396. doi: 10.1128/JVI.01229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodish HF, Ghaffari S, Socolovsky M, Tong W, Zhang J. 2009. Intracellular signaling by the erythropoietin receptor, p 155–174. In Elliott SG, Foote M, Molineux G (ed), Erythropoietins, erythropoietic factors and erythropoiesis. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 33.Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIFalpha. PLoS Pathog 7:e1002088. doi: 10.1371/journal.ppat.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganaie SS, Zou W, Xu P, Deng X, Kleiboeker S, Qiu J. 2017. Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLoS Pathog 13:e1006370. doi: 10.1371/journal.ppat.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan MM, Tamburic L, Shippam-Brett C, Zagrodney DB, Astell CR. 2001. The small 11-kDa protein from B19 parvovirus binds growth factor receptor-binding protein 2 in vitro in a Src homology 3 domain/ligand-dependent manner. Virology 291:285–291. doi: 10.1006/viro.2001.1217. [DOI] [PubMed] [Google Scholar]
- 36.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. 1992. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 70:431–442. doi: 10.1016/0092-8674(92)90167-B. [DOI] [PubMed] [Google Scholar]
- 37.Guo YM, Ishii K, Hirokawa M, Tagawa H, Ohyagi H, Michishita Y, Ubukawa K, Yamashita J, Ohteki T, Onai N, Kawakami K, Xiao W, Sawada K. 2010. CpG-ODN 2006 and human parvovirus B19 genome consensus sequences selectively inhibit growth and development of erythroid progenitor cells. Blood 115:4569–4579. doi: 10.1182/blood-2009-08-239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffatt S, Yaegashi N, Tada K, Tanaka N, Sugamura K. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J Virol 72:3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen AY, Zhang EY, Guan W, Cheng F, Kleiboeker S, Yankee TM, Qiu J. 2010. The small 11 kDa nonstructural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood 115:1070–1080. doi: 10.1182/blood-2009-04-215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandziulis RJ, Swanson MS, Dreyfuss G. 1989. RNA-binding proteins as developmental regulators. Genes Dev 3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 41.Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou XJ, Wu J, Shi L, Li XX, Zhu L, Sun X, Qian JY, Wang Y, Wei JF, Ding Q. 2017. PTEN expression is upregulated by a RNA-binding protein RBM38 via enhancing its mRNA stability in breast cancer. J Exp Clin Cancer Res 36:149. doi: 10.1186/s13046-017-0620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Xu E, Zhang J, Chen X. 2015. PPM1D phosphatase, a target of p53 and RBM38 RNA-binding protein, inhibits p53 mRNA translation via dephosphorylation of RBM38. Oncogene 34:5900–5911. doi: 10.1038/onc.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez-Dominguez JR, Zhang X, Hu W. 2017. Widespread and dynamic translational control of red blood cell development. Blood 129:619–629. doi: 10.1182/blood-2016-09-741835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu P, Zhou Z, Xiong M, Zou W, Deng X, Ganaie SS, Kleiboeker S. 2017. Parvovirus B19 NS1 protein induces cell cycle arrest at G2-phase by activating the ATR-CDC25C− CDK1 pathway. PLoS Pathog 13:e1006266. doi: 10.1371/journal.ppat.1006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita E, Tada K, Chisaka H, Asao H, Sato H, Yaegashi N, Sugamura K. 2001. Human parvovirus B19 induces cell cycle arrest at G2 phase with accumulation of mitotic cyclins. J Virol 75:7555–7563. doi: 10.1128/JVI.75.16.7555-7563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. 2009. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol 83:167–180. doi: 10.1128/JVI.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu KK. 2006. Analysis of protein-DNA binding by streptavidin-agarose pulldown. Methods Mol Biol 338:281–290. [DOI] [PubMed] [Google Scholar]
- 49.Shen W, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2016. Analysis of cis and trans requirements for DNA replication at the right-end hairpin of the human bocavirus 1 genome. J Virol 90:7761–7777. doi: 10.1128/JVI.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rio DC. 2014. Electrophoretic mobility shift assays for RNA-protein complexes. Cold Spring Harb Protoc 2014:435–440. doi: 10.1101/pdb.prot080721. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2017. Human bocavirus 1 is a novel helper for adeno-associated virus replication. J Virol 91:e00710-. doi: 10.1128/JVI.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. doi: 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol 85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganaie SS, Haque A, Cheng E, Bonny TS, Salim NN, Mir MA. 2014. Ribosomal protein S19-binding domain provides insights into hantavirus nucleocapsid protein-mediated translation initiation mechanism. Biochem J 464:109–121. doi: 10.1042/BJ20140449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salim NN, Ganaie SS, Roy A, Jeeva S, Mir MA. 2016. Targeting a novel RNA-protein interaction for therapeutic intervention of hantavirus disease. J Biol Chem 291:24702–24714. doi: 10.1074/jbc.M116.750729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS, Deng X, Wosen JE, Davulcu O, Yan Z, Engelhardt JF, Brown KE, Chapman MS, Qiu J, Carette JE. 2017. Adeno-associated virus (AAV) serotypes have distinctive interactions with domains of the cellular AAV receptor. J Virol 91:e00391-17. doi: 10.1128/JVI.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]