ABSTRACT
Positive-strand RNA viruses replicate their genomes in membrane-associated structures; alphaviruses and many other groups induce membrane invaginations called spherules. Here, we established a protocol to purify these membranous replication complexes (RCs) from cells infected with Semliki Forest virus (SFV). We isolated SFV spherules located on the plasma membrane and further purified them using two consecutive density gradients. This revealed that SFV infection strongly modifies cellular membranes. We removed soluble proteins, the Golgi membranes, and most of the mitochondria, but plasma membrane, endoplasmic reticulum (ER), and late endosome markers were retained in the membrane fraction that contained viral RNA synthesizing activity, replicase proteins, and minus- and plus-strand RNA. Electron microscopy revealed that the purified membranes displayed spherule-like structures with a narrow neck. This membrane enrichment was specific to viral replication, as such a distribution of membrane markers was only observed after infection. Besides the plasma membrane, SFV infection remodeled the ER, and the cofractionation of the RC-carrying plasma membrane and ER suggests that SFV recruits ER proteins or membrane to the site of replication. The purified RCs were highly active in synthesizing both genomic and subgenomic RNA. Detergent solubilization destroyed the replication activity, demonstrating that the membrane association of the complex is essential. Most of the newly made RNA was in double-stranded replicative molecules, but the purified complexes also produced single-stranded RNA as well as released newly made RNA. This indicates that the purification established here maintained the functionality of RCs and thus enables further structural and functional studies of active RCs.
IMPORTANCE Similar to all positive-strand RNA viruses, the arthropod-borne alphaviruses induce membranous genome factories, but little is known about the arrangement of viral replicase proteins and the presence of host proteins in these replication complexes. To improve our knowledge of alphavirus RNA-synthesizing complexes, we isolated and purified them from infected mammalian cells. Detection of viral RNA and in vitro replication assays revealed that these complexes are abundant and highly active when located on the plasma membrane. After multiple purification steps, they remain functional in synthesizing and releasing viral RNA. Besides the plasma membrane, markers for the endoplasmic reticulum and late endosomes were enriched with the replication complexes, demonstrating that alphavirus infection modified cellular membranes beyond inducing replication spherules on the plasma membrane. We have developed here a gentle purification method to obtain large quantities of highly active replication complexes, and similar methods can be applied to other positive-strand RNA viruses.
KEYWORDS: alphavirus, Semliki Forest virus, replication complex, purification, RNA replication, membrane fractionation
INTRODUCTION
RNA viruses play a major role in emerging and reemerging epidemics, and the most common RNA viruses have a single-stranded genome of messenger RNA polarity, i.e., positive-strand RNA (1, 2). During replication, the genomic RNA serves as a template in the synthesis of a minus strand, which then serves to produce more plus-strand RNAs (3). All eukaryotic positive-strand RNA viruses replicate their genomes in membrane-associated complexes that concentrate replication components, provide a structural framework, and protect viral RNA from host defense mechanisms (4–6). Depending on the virus, the replication membrane originates from the plasma membrane or subcellular organelles such as mitochondria. Two main types of replication-associated membrane modifications have been recognized (5). One type are double-membrane vesicles, in which typically multiple vesicles are interconnected via their outer membrane (7–10). The other type are vesicle-like membrane invaginations called spherules, and the current view is that one or multiple double-stranded RNA (dsRNA) intermediates and, thus, the negative-strand RNA, reside within a spherule, and newly made, mature positive strands are released to the cytoplasm via a narrow neck that is wide enough to allow the import of nucleotides and the export of positive-strand RNAs (11–16).
As a model system to study spherule invaginations, we use an arthropod-borne alphavirus, Semliki Forest virus (SFV). Alphaviruses are found on all continents and are known to cause diseases ranging from mild febrile illnesses to encephalitis and prolonged arthritis (17). SFV is a close relative of the reemerged chikungunya virus (CHIKV) that causes arthralgia, persisting in some cases for years (18, 19). The alphavirus genome encodes two polyproteins. The nonstructural polyprotein P1234 is first cleaved to P123 and nonstructural protein 4 (nsP4), which form an early replication complex (RC) that synthesizes minus-strand RNA using the genome as a template (20, 21). Further cleavage of P123 then yields a late RC consisting of nsP1 to nsP4 and making plus strands from the minus-strand RNA (20). nsP4 is the core RNA-dependent RNA polymerase (RdRp), but RNA synthesis also requires nsP1, which is the capping enzyme and membrane anchor of RCs, nsP2, which is the protease and helicase, and nsP3, which interacts with multiple host proteins (20, 22–31). The second polyprotein, which is translated from a subgenomic RNA, is cleaved to the capsid and envelope proteins (3).
The role of spherules in alphavirus replication was suggested in the 1960s and 1970s, when it was observed that viral RNA synthesis colocalizes with spherule-carrying cytopathic vacuoles (CPVs) in infected cells, and the RNA-synthesizing activity, viral RNA, and CPVs were found to be enriched in the same fraction after isolation (32–34). Spherules (diameter of ∼50 nm) contain electron-dense material and can be immunostained for dsRNA, indicating that the replicative dsRNAs are located inside the spherules (35, 36). Furthermore, nsPs colocalize with CPVs and with dsRNA staining (15, 35, 37), and it has been shown that the template RNA length determines spherule size, strengthening the view of spherules as replication sites (13). Although CPVs revealed the role of spherules in viral RNA replication, spherules are first formed at the plasma membrane. In SFV-infected cells, spherules then are internalized, and after fusion with late endosomes and lysosomes they accumulate in the perinuclear area in CPVs (15). In contrast, Sindbis virus (SINV) and CHIKV spherules predominantly stay on the plasma membrane (35, 38).
Functional in vitro studies on alphavirus replication have to a large extent focused on crude membrane preparations, showing that most of the minus-strand RNA as well as RNA-synthesizing activity are found in a membrane pellet prepared from infected cells (11, 39). Alphaviruses synthesize in cells and in vitro the same RNA species, genomic and subgenomic single-stranded RNA (ssRNA), as well as double-stranded replicative forms (RFs) and replicative intermediates (RIs) (11, 40, 41). In addition, RNA II, corresponding to the 5′ end of the genome up to the subgenomic promoter, is created but its function remains unclear (11, 39, 42). However, only RNA of positive polarity is synthesized in vitro (11, 39). Cytosolic host factors are not required in alphavirus RNA synthesis in vitro, in contrast to positive-strand RNA nidoviruses (11, 39, 43–45). In order to study both the structure and the molecular mechanisms of these membranous RCs, purified and active complexes must be obtained. Alphavirus RCs have been purified in sucrose and glycerol density gradients, indicating that these complexes are indeed membrane associated (32, 40, 41, 45–47). However, purification, which may include detergent solubilization, often yields poor replication activity, and only two to three virus-specific proteins have typically been observed in these complexes. Thus, we still lack an approach to obtain purified, fully functional complexes amenable for structural studies such as cryo-electron microscopy, and there is no structure available for the alphavirus RCs or RdRp.
Here, we report a purification method for SFV RCs, and our aim was to maintain the functionality, integrity, and morphology of the RCs that would enable us to study their biological mechanisms. First, we show that RCs isolated from SFV-infected cells are stable and then further purify them. We demonstrate that the purified RCs are highly active in RNA synthesis and release newly synthesized RNA strands. We also show that SFV infection causes extensive modification of cellular membranes by inducing spherules on the plasma membrane, but the effects reach multiple intracellular membranes.
RESULTS
Isolated SFV RCs are stable.
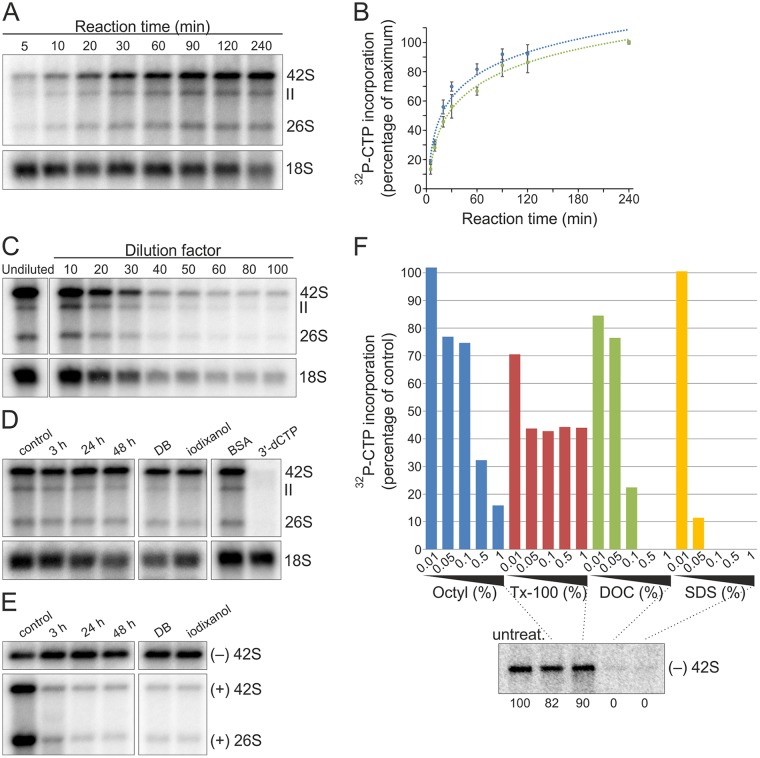
A number of parameters were tested to study if SFV RCs preserve their replication activity under different conditions and thus are amenable for purification. Metabolic labeling has shown that SFV RCs are most active in RNA synthesis in baby hamster kidney (BHK) cells at 4 to 5 h postinfection (p.i.) at a multiplicity of infection (MOI) of 50 (39). Thus, postnuclear supernatant (PNS) was prepared from SFV-infected cells (MOI of 50) at 4 h p.i., and in vitro RNA synthesis was analyzed by measuring the incorporation of [32P]CTP into viral RNA (Fig. 1). Incorporation was readily detected after a 5-min reaction time, and the signal rapidly increased up to 60 min (Fig. 1A and B). Consequently, a 1-h reaction time was routinely used unless otherwise stated. Furthermore, SFV PNS could be diluted at least 100-fold without the loss of replication activity (Fig. 1C).
FIG 1.
SFV RCs are stable and robust in RNA synthesis in vitro. (A) BHK cells were infected with SFV at an MOI of 50, and PNS was prepared at 4 h p.i. In vitro replication was studied by the incorporation of [32P]CTP into SFV RNA, indicated by 26S, II, and 42S. After the incubation times indicated, RNA was isolated and analyzed in a denaturing agarose gel. Ribosomal 18S RNA (18S rRNA) was detected by in-gel hybridization from the same gel. (B) Kinetics of the incorporation of [32P]CTP into 26S (blue) and 42S (green) RNA. Percentages indicate the incorporation compared to the last time point (100%). Data are presented as means ± standard deviations from two independent experiments. (C) Replication assay performed with either undiluted PNS or PNS diluted as indicated. 18S rRNA was detected as described for panel A. For the undiluted sample, 1/10 of the amount of RNA used for other samples was used. (D) Stability of replication activity. PNS was preincubated at 4°C for 3, 24, and 48 h, or PNS was diluted in dilution buffer (DB) or iodixanol and preincubated at 4°C for 24 h before a replication assay. The last two lanes show samples containing 1 mg/ml BSA or 100 μM 3′-dCTP. 18S rRNA was detected by in-gel hybridization from the same gel. (E) Stability of endogenous RNA. To detect endogenous SFV minus and plus strands, total RNA was isolated after the same preincubations as those described for panel D and analyzed by in-gel hybridization with specific probes. (F) Detergent sensitivity. PNS samples containing the indicated detergent concentrations were incubated at 4°C for 1 h and used for either a replication assay or total RNA isolation, followed by in-gel hybridization. After the replication assay, the incorporation of [32P]CTP into 42S RNA was quantified, and percentages indicate the incorporation compared to the untreated PNS. The lower panel shows the presence of endogenous minus-strand RNA in the untreated and 1% detergent-treated samples detected by in-gel hybridization. Numbers below the lanes indicate the percentage of [32P]signal compared to that of the untreated sample. Octyl, n-octylglucoside.
The highly active and robust RNA-synthesizing activity in vitro, which tolerated dilution, indicated that SFV RCs are suitable for purification. We next tested if RCs remain active during the prolonged incubation times that are required to perform the purification procedures. The complexes remained active throughout a 48-h incubation at 4°C (Fig. 1D). In-gel hybridization revealed that the endogenous minus-strand RNA was also stable, while most of the endogenous plus-strand RNA was digested already after 3 h (Fig. 1E). This confirms the idea that in cells most plus strands are released to the cytosol, thus in PNS they are susceptible to cellular RNases, and the minus-strand RNA resides inside the spherules as part of double-stranded RNA molecules in a form that protects them from nucleases. We also determined the stability of RCs in the density gradient medium iodixanol, which is used for subcellular fractionation. After a 24-h incubation, RCs were still highly active and minus-strand RNA was stable (Fig. 1D and E). Bovine serum albumin (BSA; 1 mg/ml) was observed to boost in vitro replication, on average about 120% (Fig. 1D). Thus, in further experiments BSA was routinely used in the reaction mixture. SFV RNA synthesis in vitro could be inhibited by the chain terminator 3′-dCTP (Fig. 1D).
We then determined the tolerance of detergents using PNS and the in vitro replication system. In vitro replication activity of SFV RCs was sensitive to all detergents tested but more sensitive to anionic sodium deoxycholate (DOC) and sodium dodecyl sulfate (SDS) than to nonionic n-octylglucoside and Triton X-100 (Tx-100) (Fig. 1F). One percent DOC and SDS caused the loss of both the minus-strand template and replication activity, indicating that spherules were destroyed and the minus-strand RNA was exposed to cellular nucleases (Fig. 1F). In contrast, about 80% and 90% of the minus-strand RNA present in the untreated sample was recovered after 1% n-octylglucoside and Tx-100 treatments, respectively. Thus, here the partial loss of the activity could be due to disruption of protein-membrane interactions.
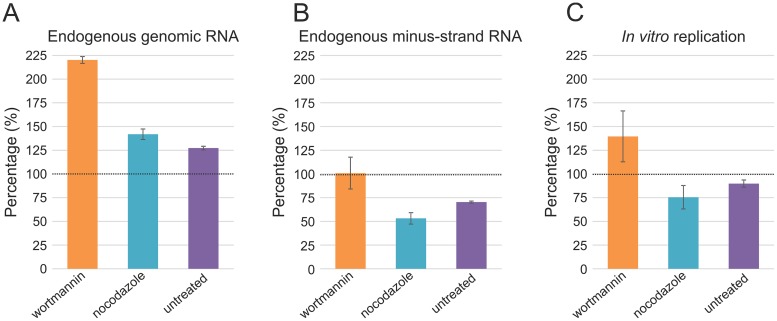
Finally, we tested which subcellular location of spherules is the most suitable for purification by utilizing drug treatments. It has been reported that wortmannin blocks SFV spherules mainly on the plasma membrane, while nocodazole treatment leads to their accumulation in small endocytic vesicles (15). In the absence of drugs, spherules accumulate in CPVs. PNS was prepared from untreated and drug-treated cells infected at an MOI of 500 and collected at 4 h p.i. Quantification of in-gel hybridizations showed that PNS from wortmannin-treated cells contained the largest amount of endogenous minus- and plus-strand RNA, also indicating that the number of RCs was the highest (Fig. 2A and B). Importantly, RCs were highly active in RNA synthesis in vitro independent of their subcellular location, although wortmannin-treated samples gave the highest activity (Fig. 2C). The observed in vitro RNA synthesis was consistent with that obtained in cells using metabolic labeling (15). The wortmannin treatment and an MOI of 500 resulted in about the same amount of the minus-strand RNA as an MOI of 50 without drug treatment (Fig. 2B). However, the amount of plus-strand RNA was doubled (Fig. 2A). This phenomenon was also observed with nocodazole-treated and untreated cells infected at an MOI of 500 compared to untreated cells infected at an MOI of 50 (Fig. 2A and B). If we assume that the minus-strand RNA represents spherules, then at the higher MOI, RCs are formed faster and have more time to synthesize plus strands.
FIG 2.
Spherule location and replication activity. At 4 h p.i., PNS samples were prepared from SFV-infected (MOI of 500) BHK cells, treated with 100 nM wortmannin at 1.5 h p.i. or 5 μM nocodazole at 0 h p.i., or left untreated. These samples were compared to the PNS samples prepared from SFV-infected (MOI of 50), untreated cells to give the percentages indicated in panels A to C. Error bars represent standard deviations from two independent experiments. (A and B) Total RNA was isolated from the PNS samples and plus- and minus-strand RNA were detected by in-gel hybridization with specific probes, followed by quantification of 42S RNA. (C) In vitro incorporation of [32P]CTP into viral RNA. 42S RNA was quantified.
SFV RCs distribute in both the supernatant and the mitochondrial pellet.
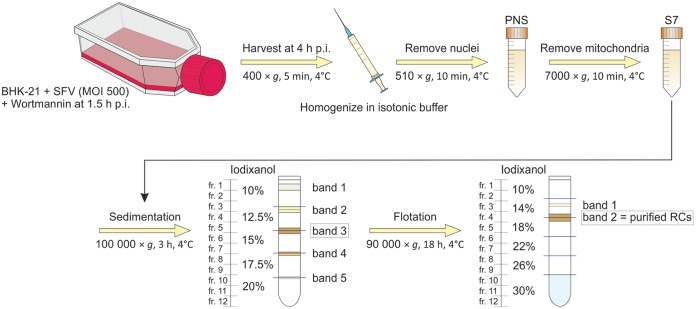
Due to the high abundance and activity, we chose to purify plasma membrane-derived spherules, and the optimized purification method is summarized in Fig. 3. SFV-infected cells (MOI of 500) were treated with wortmannin at 1.5 h p.i. and harvested in an isotonic homogenization buffer at 4 h p.i. Dounce homogenization was used to lyse cells in the previous assays, but it caused disruption of mitochondria and thus cells were lysed for RC purification by passaging through a 22-gauge needle. Nuclei and any remaining intact cells were removed by differential centrifugation, yielding PNS. Further centrifugation to remove mitochondria at 7,000 × g yielded a P7 pellet and S7 supernatant. The same amount of cells was seeded for mock and virus infection, but when cells were collected, mock samples contained twice the amount of cells as that of the infected samples. However, both cell lysates contained similar amounts of proteins (∼13 mg/ml), indicating that there were more proteins in SFV-infected cells than in mock-infected cells. PNS (∼3.2 mg/ml) and S7 (∼2.6 mg/ml) prepared from SFV-infected cells contained more protein than the corresponding samples from mock-infected cells (∼2.0 mg/ml in PNS and ∼1.7 mg/ml in S7), indicating that SFV-infected cells lysed better than mock-infected cells.
FIG 3.
Schematic of RC isolation and purification. Overview of the purification from cell harvesting to iodixanol density gradient centrifugations. From the sedimentation gradient, band 3 was collected for further purification by flotation. Band 2 in the flotation gradient represents the purified RCs.
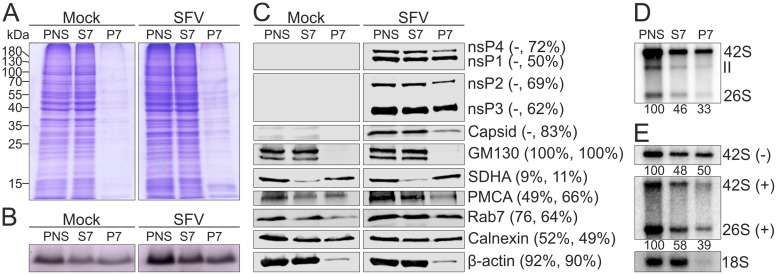
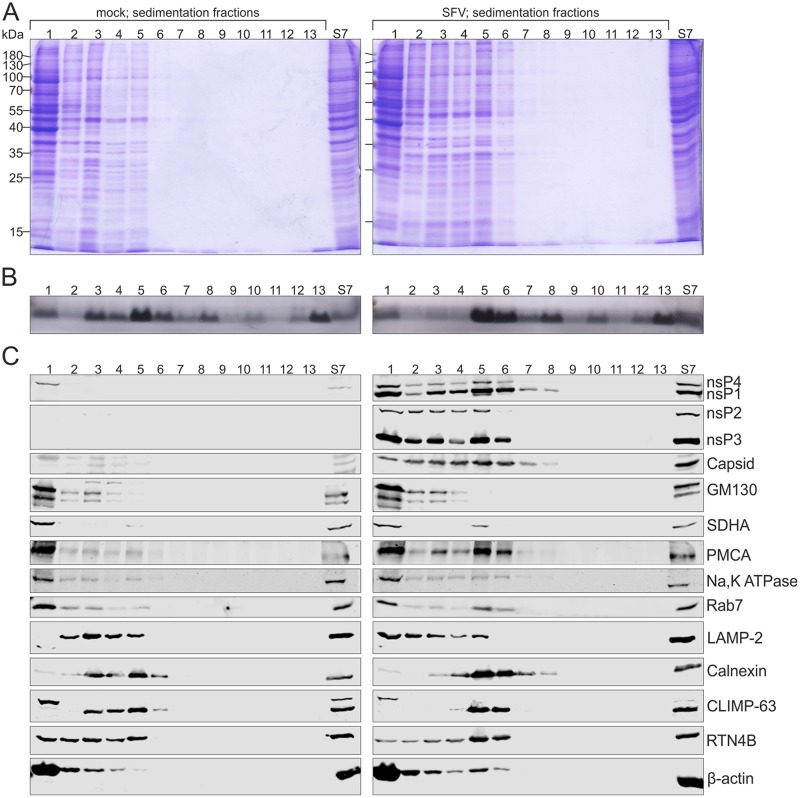
Most of the proteins and about half of the lipids present in PNS from mock- and SFV-infected cells distributed to S7 (Fig. 4A and B). In order to detect lipids, we utilized an approach to stain lipoproteins with Sudan black B after polyacrylamide gel electrophoresis (see Materials and Methods). However, gels were stained after a short electrophoresis to allow detection of species below a 10-kDa marker. Distribution of different viral and cellular markers was further studied by Western blotting (Fig. 4C). Most of nsP2, nsP3, and nsP4, as well as about 50% of nsP1, was found in the viral S7. SFV capsid protein was also mainly found in the S7 fraction. The Golgi matrix protein of 130 kDa (GM130), the endoplasmic reticulum (ER) marker calnexin, the mitochondrial marker succinate dehydrogenase subunit A (SDHA), and the cytosolic marker β-actin showed a similar distribution into S7 and P7 fractions in both mock and virus samples. GM130 was exclusively detected in S7, and β-actin was more prominent in S7 than in P7. Most of SDHA distributed in the P7 pellet, and about half of calnexin was found in S7. About half of the plasma membrane calcium pump ATPase (PMCA) was found in mock S7, while in the virus-infected samples it was more prominent in S7 than in P7. In both mock- and virus-infected samples the late endosomal marker Rab7 was more prominent in S7 than in P7, but the enrichment was stronger in the mock samples.
FIG 4.
Distribution of proteins, lipids, and RNA in the S7 and P7 fractions. PNS was prepared from mock- or SFV-infected (MOI of 500) BHK cells treated with wortmannin and separated into an S7 supernatant and P7 pellet. (A and B) Protein and lipid profiles of PNS, S7, and P7 in an SDS-polyacrylamide gel stained with Coomassie blue (A) or Sudan black B (B). Numbers on the left indicate the molecular masses (kDa) of marker proteins. (C) Distribution of viral and cellular markers as studied by Western blotting. PMCA was used as a marker for the plasma membrane, GM130 for the Golgi membrane, calnexin for the endoplasmic reticulum, SDHA for the mitochondria, Rab7 for the late endosomes, and β-actin for the cytosol. Numbers in parentheses indicate the percentage of the protein present in the S7 fraction compared to that in PNS. The first number is from mock-infected samples and the second one from virus-infected samples. (D) Incorporation of [32P]CTP into SFV RNA in PNS, S7, and P7 prepared from SFV-infected cells. Numbers below the lanes indicate the amount of incorporation into 42S RNA as a percentage of the label incorporated in PNS. (E) Distribution of endogenous minus- and plus-strand RNA analyzed by in-gel hybridization from PNS, S7, and P7 prepared from SFV-infected cells. As a control, a probe against 18S rRNA was used. Numbers below the lanes indicate the amount of 42S RNA as a percentage of that in PNS.
In vitro replication activity present in the viral PNS, S7, and P7 fractions was analyzed by quantifying the incorporation of [32P]CTP into SFV 42S RNA (Fig. 4D). About 50% and 30% of the PNS activity was recovered in S7 and P7, respectively. In-gel hybridization showed that the minus-strand RNA distributed equally to S7 and P7 (Fig. 4E), indicating that differential centrifugation caused a partial loss of replication activity in the pellet fraction. Approximately 60% of the genomic RNA was found in S7.
SFV infection modifies the properties of cellular membranes.
We selected the S7 fraction for further purification in density gradients (Fig. 5 and 6), as more than half of the active RCs remained in this fraction. RCs were first purified from the S7 fractions by sedimentation in a 10 to 20% iodixanol step gradient (Fig. 3) by loading 500 μl of S7 on the top of a 12-ml density gradient, resulting in ∼0.9 and 1.3 mg of protein loaded from mock- and virus-infected samples, respectively. Signals from the cellular markers were comparable in the S7 fractions from mock- and SFV-infected cells (Fig. 5 and 6). After sedimentation, a light-scattering band was detected in every interphase as well as close to the gradient surface. Gradients were fractionated, and the protein pattern in each fraction was determined by SDS-PAGE (Fig. 5A). Most proteins stayed in the top fractions of sedimentation gradients derived from the S7 fraction of mock (labeled mock)- or SFV (labeled SFV)-infected cells, although the latter resulted in the detection of more proteins, especially in fraction (fr.) 5. Lipids were detected in all fractions that correspond to the bands and in the pellet (Fig. 5B). In both gradients, lipids concentrated in fr. 5 and 6 (band 3). However, fr. 3 and 4 (band 2) of the mock gradient contained more lipids than the corresponding fractions of the SFV gradient.
FIG 5.
Infection modulates cellular membranes. S7 was prepared from mock- and SFV-infected (MOI of 500) BHK cells treated with wortmannin, and RCs were purified by a 3-h sedimentation in a 10 to 20% iodixanol step gradient. After centrifugation, gradients were fractionated (fr. 1 to 12, from top to bottom; fr. 13, the pellet) and compared to S7. On the left, fractions are from mock-infected sample, and on the right, fractions are from SFV-infected sample. Protein (A) and lipid (B) patterns were studied in an SDS-polyacrylamide gel stained with Coomassie blue or Sudan black B. Numbers on the left indicate the molecular masses (kDa) of marker proteins. (C) Western blotting shows the distribution of viral and cellular markers as described for Fig. 4. Furthermore, Na,K ATPase was used as an additional marker for the plasma membrane, LAMP-2 for the late endosomes and lysosomes, and CLIMP-63 and RTN4B as additional markers for the endoplasmic reticulum (sheets and tubules, respectively). The sample volumes loaded from the fractions in the Coomassie- or Sudan-stained gels (A and B) and Western blot gels (C) were about five and seven times larger, respectively, than the sample volume loaded from the original S7, shown on the right.
FIG 6.
SFV RCs concentrate with the plasma membrane, ER, and late endosome markers. Light-scattering band 3 from the sedimentation gradient was further purified by an 18-h flotation in a 10 to 30% iodixanol step gradient. After centrifugation, gradients were fractionated (fr. 1 to 12, from top to bottom; fr. 13, the pellet) and compared to S7. Fractions from mock-infected samples are on the left, and fractions from SFV-infected sample are on the right. Protein (A) and lipid (B) patterns were analyzed in an SDS-polyacrylamide gel stained with Coomassie blue or Sudan black B. Numbers on the left indicate the molecular masses (kDa) of marker proteins. (C) Distribution of viral and cellular markers studied by Western blotting as described for Fig. 4. (D) Distribution of Na,K ATPase (plasma membrane), LAMP-2 (late endosomes and lysosomes), and CLIMP-63 and RTN4B (endoplasmic reticulum sheets and tubules, respectively) between flotation fractions 3 to 5 was analyzed and compared to S7 by Western blotting. The sample volumes are the same as those for Fig. 5.
Western blotting revealed the distribution of viral and cellular markers between gradient fractions (Fig. 5C). In the SFV gradient, nsPs concentrated in fr. 1, 3 and 4, and 5 and 6, corresponding to bands 1, 2, and 3, respectively. SFV capsid protein was observed in fr. 1 to 8. The Golgi marker stayed on the top of both gradients, and the remaining mitochondrial marker was mainly on the top and a minor amount was detected in fr. 5. In the mock gradient, the plasma membrane marker PMCA mostly stayed on the top, while in the SFV gradient it concentrated in fr. 1 as well as fr. 5 and 6 (band 3). To further study the distribution of the plasma membrane, another plasma membrane marker, Na,K ATPase, was included, and this marker gave distribution in the sedimentation gradients similar to that of PMCA, although PMCA showed stronger concentration in fr. 5 and 6 (band 3) in the SFV gradient. The late endosomal marker Rab7 predominantly stayed on the top of the mock gradient, while in SFV it concentrated in both fr. 1 as well as fr. 5 and 6 (band 3). Lysosome-associated membrane protein 2 (LAMP-2), which is a marker for late endosomes and lysosomes, showed a different distribution from that of Rab7. In the mock gradient, LAMP-2 concentrated in fr. 2 to 5 and in the SFV gradient in fr. 1 to 5. The ER marker calnexin concentrated in the mock gradient in fr. 3 and 4 (band 2) and fr. 5 and 6 (band 3) and in the SFV gradient in fr. 5 and 6 (band 3). To further examine ER distribution, two additional markers for ER were included. CLIMP-63 (cytoskeleton-linking membrane protein 63), also known as CKAP-4 (cytoskeleton-associated protein 4), is predominantly found in ER sheets (48), and reticulon 4B (RTN4B), also known as neurite outgrowth inhibitor B (Nogo-B), localizes to ER tubules and sheet edges (49). CLIMP-63 showed a distribution similar to that of calnexin, although an additional band was detected in fr. 1. RTN4B was found in fr. 1 to 5 of the mock gradient, while in the SFV gradient RTN4B concentrated in fr. 5 and 6 (band 3), similar to calnexin and CLIMP-63. The cytosolic marker β-actin concentrated in the top fractions, but in the SFV gradient it was also found in fr. 5 and 6 (band 3). We also tested the addition of 150 mM KCl to the cell lysate to determine if a higher salt concentration was required to keep membranes from aggregating, but no effect in sedimentation was observed, indicating that the results were not due to unspecific membrane aggregation.
An in vitro replication assay showed that most SFV replication activity was in fr. 5 and 6 (band 3) of the sedimentation gradient (Fig. 7B), and thus this band, at a density of 1.10 to 1.11 g/ml, was collected for further purification. The final step in the purification was equilibrium centrifugation in a 10 to 30% iodixanol step gradient, as shown in Fig. 3. After flotation, two light-scattering bands were observed and gradients were fractionated. Fractions 10 to 12 represent 30% iodixanol containing band 3 collected from the sedimentation gradient. The upper band in the flotation gradient (Fig. 3, band 1) was considerably weaker than the other band (Fig. 3, band 2), and detectable proteins and lipids concentrated in the area of the band 2 or lower fractions. Coomassie staining revealed very little protein in the mock fractions (Fig. 6A). In the SFV gradient, proteins were mainly observed in fr. 4 (band 2) as well as in fr. 9 to 12. Lipids were detected in both gradients in fr. 4, although SFV gave a much stronger signal (Fig. 6B). In the mock gradient, Western blotting detected only a weak calnexin signal in fr. 3 and 4 (Fig. 6C). In the SFV gradient, nsPs and capsid protein concentrated in fr. 4, except that nsP2 and capsid were also detected in fr. 9 to 12, indicating that nsP2 and capsid protein in these fractions did not float and thus were not membrane associated. The plasma membrane marker PMCA, late endosome marker Rab7, and ER marker calnexin also concentrated in fr. 4. In addition, β-actin and the residual mitochondria were detected in fr. 4. Golgi marker was hardly detectable. The density in fr. 4 was 1.11 g/ml. As viral nsPs and PMCA, Rab7, and calnexin concentrated in fr. 4 in the SFV gradient, fr. 3 to 5 were further studied using additional cellular markers (Fig. 6D). The plasma membrane marker Na,K ATPase, as well as ER markers CLIMP-63 and RTN4B, also concentrated in fr. 4 in the SFV gradient, while these markers were undetectable in the mock fr. 3 to 5. Interestingly, LAMP-2, a marker for the late endosomes and lysosomes, was undetectable in fr. 3 to 5 of both mock and SFV gradients.
FIG 7.
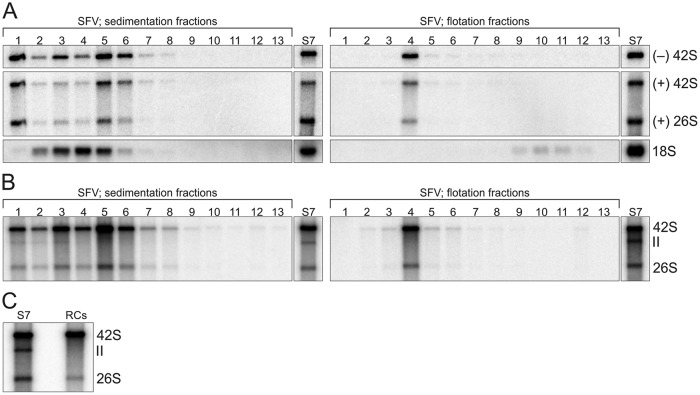
Purified RCs remain active in RNA synthesis. (A) Distribution of endogenous minus- and plus-strand RNAs between SFV sedimentation (left) and flotation (right) fractions studied by in-gel hybridization. 18S rRNA was detected from the same fractions (fr. 1 to 12, from top to bottom; fr. 13 is the pellet), and S7 is shown for comparison. (B) In vitro replication activity of the same fractions as those used for panel A, studied by [32P]CTP incorporation. (C) Comparison of in vitro replication activity of S7 and purified RCs after replication reaction mixtures were incubated 4 h. S7 and RCs indicate [32P]CTP incorporation by the S7 fraction and the purified RCs, respectively. In panel A the volume from the fractions used for RNA isolation was about ∼7.5 times larger than the S7 sample volume, and in panels B and C 20-fold-diluted S7 was used.
We next assayed where SFV minus- and plus-strand RNA as well as 18S rRNA distributed in the sedimentation and flotation gradients (Fig. 7A). 18S rRNA concentrated in sedimentation fr. 4, and only a small amount was detected after flotation in fr. 9 to 12, indicating that the detected rRNA did not float. After sedimentation, the minus- and plus-strand RNA were mainly recovered in fr. 1, 3 and 4, and 5 and 6, and after flotation, both concentrated in fr. 4. Approximately 20% of the minus-strand RNA present in S7 was recovered in flotation fr. 4.
In both sedimentation and flotation gradients, [32P]CTP incorporation colocalized with the endogenous minus-strand RNA (Fig. 7), and high in vitro replication activity was observed after the purification. The RCs in the flotation fr. 4 (band 2) were designated purified RCs and collected for further characterization. The average protein concentration of the purified RCs from three purifications was 0.02 mg/ml, indicating that less than 1% of the proteins present in S7 were remaining. However, the recovery of the activity present in S7 was approximately 20% when the [32P]CTP incorporation into the genomic RNA was compared (Fig. 7C). The purified RCs synthesized both genomic and subgenomic RNA, but RNA II was observed only in sedimentation fr. 1 and 3 (Fig. 7B and C).
Purified RCs display spherule-like structures and release newly made RNA.
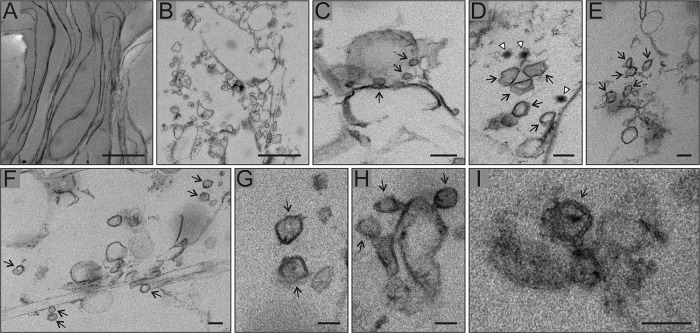
Electron microscopy of the purified RCs and the corresponding membrane fraction from mock-infected cells revealed sheets, vesicles, and some tubule-like membranes (Fig. 8). Sheets, which might be derived from the plasma membrane or ER, were observed in both samples (Fig. 8A). Sheets or vesicles with multiple, spherical invaginated membranes were typical of the purified RCs and could not be detected in the mock samples (Fig. 8B to I). The diameter of these invaginations was similar to that of spherules, about 50 nm, and some of them showed a dark spot in the middle, most likely representing RNA. In addition, membrane sheets displaying smaller round structures, which may represent narrow spherule necks, were observed (Fig. 8C). A few nucleocapsids were also found close to the spherule-like structures (Fig. 8D).
FIG 8.
Purified RC membranes contain spherule-like structures. SFV RCs were purified in subsequent sedimentation and flotation gradients, fixed, and pelleted by differential centrifugation for thin-section electron microscopy. Mock-infected samples served as a control. All micrographs shown are from SFV samples. (A) Membrane sheets observed in both SFV and mock sample. (B) Membranes with spherical invaginations of about 50 nm in diameter typical of SFV only. This most likely represents plasma membrane sheets sectioned in a different orientation than that of panel A, resulting in vesicle-like appearance. (C) Membrane sheet showing smaller round vesicles that may represent neck parts of spherule-like structures, indicated by arrows. (D to I) Close-ups of spherule-like structures, indicated by arrows. In panel D, white arrowheads indicate nucleocapsids. Scale bars, 1,000 nm (A and B), 100 nm (C to F), and 50 nm (G to I).
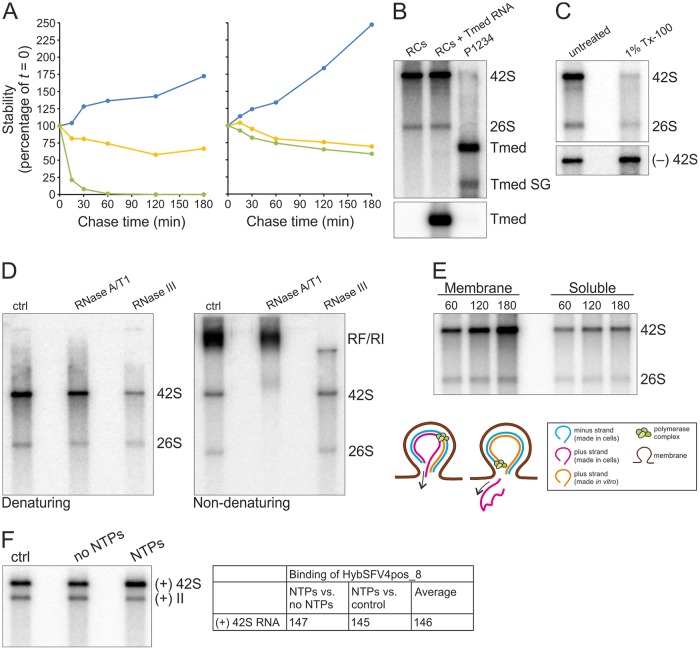
To determine the stability of the newly made RNA, in vitro replication reactions were terminated by adding the nucleoside analogue 3′-dCTP followed by a chase for 3 h (Fig. 9A). In the unterminated control samples, the amount of the [32P]CTP-labeled genomic RNA increased approximately 1.7-fold or 2.5-fold in the reaction mixtures containing S7 or purified RCs, respectively. After termination, the level of the labeled genomic RNA slowly decreased in both S7 and purified RCs at a similar rate. However, the half-life was more than 180 min. In contrast, when a capped in vitro transcript of Tmed RNA, which contains the SFV 5′ and 3′ untranslated regions as well as the subgenomic promoter, was added to the reactions, it was rapidly degraded in the S7 sample and the half-life was less than 15 min. This indicated that the newly synthesized SFV RNA was protected in the S7 sample, and for CHIKV, it has been proposed that membrane or polysome association, RNA structure, or encapsidation protects newly made ssRNA from cellular nucleases (11). In the reaction mixture containing purified RCs, the Tmed RNA was approximately as stable as the viral replication products, suggesting that purification removed most of the cellular nucleases.
FIG 9.
Characterization of the in vitro replication activity of the purified RCs. (A) Stability of newly synthesized SFV genomic RNA. After a 60-min replication assay, [32P]CTP incorporation was blocked by the addition of 100 μM 3′-dCTP, and incubation at 30°C was continued for an additional 180 min. The graph on the left represents an assay with 10-fold-diluted S7 and on the right with the purified RCs. At the indicated chase times radioactivity in 42S was quantified (yellow). As a control, a reaction without 3′-dCTP was quantified (blue). In addition, in vitro transcript of Tmed added in the reaction at a 0-min chase was quantified by in-gel hybridization (green). All values are presented as percentages of the values at the 0-min chase. (B) Purified RCs and additional exogenous template. Replication assay reaction mixtures were incubated for 2 h. RCs indicates [32P]CTP incorporation by the purified RCs. RCs + Tmed RNA shows incorporation by the purified RCs after an exogenous Tmed RNA transcript was added. As a control, [32P]CTP incorporation by the P15 membrane fraction from cells transfected with the P1234 polyprotein and Tmed template plasmids is shown, and genomic and subgenomic (SG) Tmed are indicated. The lower panel shows the presence of Tmed, detected by in-gel hybridization. (C) Detergent stability and sensitivity. Purified RCs were treated with 1% Tx-100, followed by an assay to detect replication (upper) and in-gel hybridization to detect the minus-strand template RNA (lower). (D) ssRNA and RF/RI forms of in vitro-synthesized RNAs. After a 2-h replication assay with the purified RCs, RNA was isolated and treated with RNase A/T1 or III under high-salt conditions to specifically digest ssRNA or dsRNA, respectively, and analyzed under denaturing (left) or nondenaturing (right) conditions. (E) Release of newly made RNA. A replication assay was performed with the concentrated purified RCs, and after 60-, 120-, and 180-min replication reactions, aliquots were removed to prepare pellet and supernatant fractions, followed by RNA isolation and analysis in a denaturing agarose gel. The schematic shows how [32P]CTP is incorporated into viral RNA during in vitro RNA synthesis, resulting in dsRNA containing a plus strand synthesized both in cells (indicated by magenta) and in vitro (indicated by orange). After a round of replication and release of the previous plus strand, an RC contains only the in vitro-synthesized plus strand if replication is semiconservative. (F) Increase in the amount of RNA during in vitro replication. After a 4-h replication assay with the purified RCs and unlabeled NTPs, RNA was isolated and genomic RNA was detected by in-gel hybridization. No NTPs indicates a reaction without added NTPs, and ctrl indicates a sample without any incubations before RNA isolation. 42S RNA was quantified, and average percentages from two independent experiments are shown in the table.
As added RNA remained relatively stable in the purified preparations, we tested if the purified RCs are able to utilize an exogenous template RNA. However, no additional RNA products were synthesized when the Tmed template was provided in addition to the endogenous one (Fig. 9B), indicating that once a functional RC has been formed it is unable to replicate another template. We also tested if RCs remained sensitive to detergents when cellular nucleases were removed by purification. After 1% Tx-100 treatment, the minus-strand template remained stable while replication activity was almost completely lost (Fig. 9C), indicating that the detergent sensitivity was not (solely) due to exposure to nucleases but due to solubilization of the membrane-associated RCs.
We next assessed incorporation of [32P]CTP into ssRNA, RF, and RI by isolating RNA after a replication assay with the purified RCs and treating the isolated RNA with RNases. RF and RI are fully and partially double-stranded RNA molecules, respectively, and under high-salt conditions, RNase A/T1 should digest free ssRNA and single-stranded overhangs of RIs, thus the full-length 32P-labeled RNA observed after the digestion in a denaturing gel is mainly derived from RFs. RNase III should digest RF and double-stranded regions of RIs, and after such a treatment, the full-length 32P-labeled RNA observed in a denaturing gel is mainly derived from ssRNAs. Sixty-four and 19% of the radiolabeled genomic RNA present in the untreated sample was recovered after the RNase A/T1 and III treatments, respectively, in denaturing electrophoresis (Fig. 9D). Thus, approximately 60% of the newly synthesized RNA was in RF, 20% in RI, and 20% in free ssRNAs. However, after both treatments signal might remain that contained an almost full-length 32P-labeled RNA (annealed with the minus-strand template) or an almost full-length 32P-labeled single-stranded overhang. Nondenaturing electrophoresis also supported the presence of both RF and RI, because the RF/RI signal was reduced approximately 32% after the A/T1 digestion. Most likely, single-stranded overhangs in RI were digested and thus the signal was reduced. The untreated and RNase III-treated samples gave equally strong ssRNA signals, whereas after the A/T1 treatment no ssRNA was observed in a nondenaturing gel. The RNase treatments also indicate that SFV replication is semiconservative, because after the A/T1 digestion under high-salt conditions, 32P-labeled genomic RNA was detected in the denaturing gel (Fig. 9D). This is possible only if the labeled RNA was in a dsRNA molecule. If replication were conservative, such a form would not exist.
To determine whether the ssRNAs synthesized by the purified RCs were released, we studied their membrane association. For this, the purified RCs were concentrated to remove iodixanol, and after a replication assay, pellet and supernatant were separated to detect membrane-associated and free RNA. Most of the 32P-labeled RNA stayed associated with the pellet fraction after the replication assay (Fig. 9E). However, 32P-labeled 26S and 42S RNA were also detected in the supernatant, and their amounts increased at the same rate in both pellet and supernatant fraction, indicating that the more RNA was synthesized, the more RNA was found in the supernatant. At every time point, approximately 30% of the labeled genomic RNA was found in the supernatant, which is close to the amount of ssRNA approximated based on the RNase treatments (Fig. 9D and E).
Both the presence of newly made RNA in RIs (Fig. 9D) and release of newly made strands (Fig. 9E) indicated that de novo initiation occurs in vitro. In addition, the linear rate of RNA synthesis in the unterminated RC samples (Fig. 9A) indicated that the purified RCs are able to initiate new strands. Thus, we approached this question by determining how much the amount of viral RNA increases during in vitro replication compared to the amount of RNA present in the RCs after purification. We have previously shown that SFV RCs mainly synthesize RNA of positive polarity in vitro (39), thus here we detected plus-strand RNA using a probe that recognizes an early part of the SFV4 genome (nucleotides 95 to 121). After a 4-h in vitro replication assay at 30°C using unlabeled nucleotide triphosphates (NTPs), RNA was isolated and genomic RNA was detected by in-gel hybridization. In this sample both in-cell-made RNA, present in the purified RCs, as well as in vitro-synthesized, newly made RNA are detected. For comparison, RNA was also isolated from a reaction mixture that contained no added NTPs and from a sample that contained only purified RCs and was not incubated at 30°C. In these samples, only RNA made in cells and present in the purified RCs is detected if we assume that the purified RCs contain no significant amount of cellular NTPs. During in vitro RNA synthesis, the amount of completed genomic RNA increased 50% in the active samples compared to the controls without NTPs or incubation (Fig. 9F). Thus, this result also supports the possibility of de novo initiation during in vitro RNA synthesis and at least indicates active synthesis of very long RNA stretches resulting in complete genomes.
DISCUSSION
Our present study demonstrates that the virus-induced plasma membrane invaginations, i.e., spherules, represent the active replication complexes of alphaviruses. We showed here that all four viral nsPs, minus- and plus-strand RNA, as well as RNA-synthesizing activity were found in the same purified membrane fraction, displaying spherule-like structures on the plasma membrane sheets (Fig. 6 to 8).
Previous purification attempts of alphavirus RCs have mainly focused on the isolation of CPVs (32, 40, 45–47, 50). However, spherules are formed on the plasma membrane and internalization is not typical of all alphaviruses (15, 35, 38). Here, we observed that SFV RCs were most abundant and active when isolated from infected cells treated with wortmannin (Fig. 2), which leads to the retention of the majority of spherules on the plasma membrane (15), thus we chose to purify spherule-carrying plasma membrane sheets. First, we homogenized cells in isotonic buffer using a needle instead of hypotonic buffer and a Dounce homogenizer, such as those used in several previous protocols (32, 40, 41, 45–47). Second, we purified RCs from a postnuclear and postmitochondrial supernatant instead of a membrane pellet (40, 45–47), as differential centrifugation causes a partial loss of activity (Fig. 4) (11, 39). Third, we observed that SFV RCs were sensitive to detergents (Fig. 1F), thus detergent solubilization was excluded. Fourth, we used two consecutive density gradients of iodixanol under isotonic conditions. Our purification protocol yielded RCs that were highly active in synthesizing both genomic and subgenomic RNA as well as releasing newly made strands (Fig. 9). In addition, our purification method produced highly active complexes in large quantities, as from ∼107 cells we purified enough RCs to make at least one hundred standard in vitro replication assays, and it can easily be further scaled up.
The sedimentation analyses revealed that SFV infection extensively modifies cellular membranes (Fig. 5). The plasma membrane had higher density, most likely due to the RNA-containing RCs. Furthermore, major differences in three ER markers were observed, and the ER showed increased density after infection, indicating that SFV causes ER remodeling. Several viruses are known to affect ER organization. For example, herpes simplex virus 1 (HSV-1) infection results in compression of ER around the nuclear membrane, most likely to facilitate nuclear egress of HSV-1 nucleocapsids (51). SFV infection also affected the late endosomal and lysosomal markers. Consequently, SFV has more profound effects on the membrane dynamics of the host cell than merely inducing plasma membrane invaginations.
Purification of the RCs resulted in a replication-active membrane fraction enriched with the markers for the plasma membrane, ER, and late endosomes (Fig. 6). We could remove soluble proteins, the Golgi membranes, and most of the mitochondria. The corresponding membrane fraction from mock-infected cells revealed only a minor signal for the ER marker calnexin, indicating that we purified a virus-specific network of membranes associated with replication. In cells, the peripheral ER tubules form a branched network contacting the cytoskeleton, plasma membrane, and cell organelles (52). The purified membrane preparations revealed ER markers for both ER sheets and tubules, although the marker used for the ER sheets has been reported to be found also at the plasma membrane (48). However, it remains to be studied if the RC-carrying plasma membrane sheets are truly connected to the other membranes or if these membranes have the same density. Clearly, this enrichment was not observed in the mock samples, thus SFV infection modified the studied membranes. However, besides genome replication, expression of SFV structural proteins and host defense responses may affect different membranes. Another possibility is that SFV infection causes the relocalization of some cellular proteins. For example, the late endosomal marker Rab7 was present in the purified RC-membrane fraction, but the endolysosomal marker LAMP-2 could not be detected and thus Rab7 could have been relocalized during infection.
Membrane association of the replicase proteins, mediated mainly by nsP1, is indispensable for alphaviruses (25, 53, 54), and our results showed that detergent solubilization caused a significant loss of replication activity, although the minus-strand template was stable (Fig. 1F and 9C). Thus, the polymerase activity was sensitive to detergents, further highlighting the importance of the membrane association of the RCs. Detergent solubilization has, however, been shown to increase the density of alphaviral RCs while they retain all nsPs and residual RNA synthesis activity (45). It remains to be studied whether the membrane association of SFV nsPs occurs only in the spherule neck or if they are also found inside the membrane invagination. Cryo-electron tomography of Flock House nodavirus (FHV) spherules has revealed a crown structure in the spherule neck with a 12-fold symmetry and a central electron density (12). It was suggested that FHV replicase protein A forms these structures, and if alphavirus RCs establish a similar structure, it may have a different organization, as alphaviruses have four replicase proteins instead of one.
We observed that the purified RCs incorporated [32P]CTP into both ssRNA and dsRNA and released newly made RNA (Fig. 9), and approximately 20 to 30% of the newly made genomic RNA was released as ssRNA. These data indicated that de novo initiation occurs in vitro. Second, we determined that the amount of genomic RNA present in the purified RCs was increased ∼1.5 times during the in vitro replication. As the probe used recognizes an early part of the genome, the results indicate that long RNA strands were produced in vitro, and consequently most in vitro-synthesized strands were either de novo initiated or were reinitiated from a short RNA strand made in cells. Furthermore, the results also showed that large amounts of RNA were produced in vitro. So far, it is unclear how many copies of genomic RNA are contained by SFV spherules. FHV spherules are variable in size, and the authors speculated that some spherules contain more than one dsRNA or positive-strand RNA (12).
In PNS, most of the in vitro-synthesized RNA is single stranded (11, 40), and in vitro-synthesized RF and RI reach the maximal level in about 15 min, after which only the amount of ssRNA increases (55). In alphavirus-infected mammalian cells, CPVs are located close to the ribosome-rich ER, and connecting bridges are formed that may represent RNA strands being translated. Thus, the release of the newly made RNA might require translation or nucleocapsid assembly, and it has been suggested that these steps occur in a matrix between CPVs and the rough ER (36, 56). If the efficient release of the newly made RNA strands requires translation and assembly of the nucleocapsids close to the RCs, this may explain why most of the in vitro-synthesized RNA remained in membrane-associated, double-stranded replicative forms or intermediates. Although we observed SFV capsid protein and nucleocapsids in the purified membrane fraction (Fig. 6 and 8), efficient encapsidation may not be achieved in vitro using purified membranes, as no rRNA was detected.
In conclusion, we showed that alphavirus infection strongly modifies cellular membranes beyond inducing the spherules and that the ER might be connected to the RC-carrying plasma membrane. This is the first step in extending our knowledge of alphavirus membrane-associated RCs, and further studies on these purified complexes should give insights into the host proteins present and required for their activity on the plasma membrane, as well as their structural organization. Currently, alphaviruses, coronaviruses, and arteriviruses represent important groups of pathogenic positive-strand RNA viruses lacking structural information on RdRps (57, 58). The alphaviral core RdRp has been purified in an active form (31), but the structure of the RC might give more information about the active conformation of the polymerase and interactions, as all four nsPs are required for RNA synthesis.
MATERIALS AND METHODS
Cell culture, viruses, and plasmids.
BHK-21 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco), 2 mM l-glutamine (Gibco), 100 U/ml penicillin (Gibco), and 100 μg/ml streptomycin (Gibco). BSR T7/5 cells, derived from BHK stably expressing T7 RNA polymerase (59), were cultured in DMEM supplemented with 10% FBS, 2% (vol/vol) tryptose phosphate broth (Difco), 2 mM l-glutamine, 1% (vol/vol) nonessential amino acids (Gibco), 1 mg/ml G418 (Merck), 100 U/ml penicillin, and 100 μg/ml streptomycin. Both cell lines were grown at 37°C and in 5% CO2. The same conditions were used for virus infection and DNA transfection. SFV4-HA, containing a hemagglutinin (HA) peptide in frame within nsP3 in the XhoI site, has been described and was propagated as done previously (39). Secondary stocks of SFV4-HA were used in all virus infections. The polyprotein construct P123HA4 and template Tmed have been described previously (39, 60).
Isolation of RCs from infected and transfected cells.
About 1 × 107 BHK-21 cells were infected with SFV4-HA at an MOI of 50 or 500 for 1 h or 20 min, respectively, in MEM containing 0.2% BSA and 2 mM l-glutamine. After the adsorption, cells were washed twice and fresh medium was added. Wortmannin (100 nm; Sigma-Aldrich) and nocodazole (5 μM; Calbiochem) treatments were performed as described previously (15). Nocodazole was added at the same time as the virus and wortmannin at 1.5 h p.i. Cells were harvested at 4 h p.i. by trypsinization, and PNS was prepared using a Dounce homogenizer as described in reference 39. BSR cells were cotransfected with the polyprotein and Tmed template plasmids, and 16 h after transfection PNS and P15 membrane fractions were prepared as previously described (39).
Purification of RCs from infected cells.
Approximately 4 × 107 BHK-21 cells were infected with SFV4-HA at an MOI of 500 and treated with wortmannin as described above. Mock-infected cells served as a control and were treated with wortmannin. At 4 h p.i., cells were harvested by scraping in phosphate-buffered saline and washed with homogenization buffer (HB; 250 mM sucrose, 3 mM imidazole [pH 7.4], 2 μg/ml actinomycin D [Sigma-Aldrich], Pierce EDTA-free protease inhibitor [1 tablet per 10 ml; Thermo Fisher Scientific]) and resuspended in 500 μl of HB containing 200 U/ml RiboLock (Thermo Fisher Scientific). Cells were disrupted using a 22-gauge needle and syringe by 24 up-and-down strokes. Unlysed cells and nuclei were removed by centrifugation (510 × g, 10 min, 4°C), yielding PNS, which was diluted 3-fold with HB containing RiboLock. The membrane pellet (P7) and soluble fraction (S7) were prepared by further centrifugation of the diluted PNS (7,000 × g, 10 min, 4°C). The P7 fraction was washed once with and resuspended in HB containing RiboLock.
RCs were purified from S7 fractions by sedimentation (100,000 × g, 3 h, 4°C) in a discontinuous iodixanol (Sigma-Aldrich) gradient consisting of 2.3-ml layers of 20, 17.5, 15, 12.5, and 10% (wt/vol) iodixanol in dilution buffer (DB; 250 mM sucrose, 35 mM HEPES [pH 7.4], 2.5 mM dithiothreitol [DTT], 7 mM KCl) containing Pierce EDTA-free protease inhibitor (1 tablet per 10 ml). Five hundred microliters of S7 was loaded on the top of a gradient. After sedimentation, gradients were fractionated, and protein, lipid, and RNA composition as well as replication activity were analyzed as described below. The density of the fractions was measured by weighing. For further purification, the light-scattering zone between 12.5% and 15% iodixanol layers was collected and purified by equilibrium flotation centrifugation (90,000 × g, 18 h, 4°C) in a discontinuous iodixanol gradient consisting of 3 ml of 30% and 1.8-ml layers of 26, 22, 18, 14, and 10% (wt/vol) iodixanol in DB. A 30% layer contained the light-scattering zone collected from the sedimentation gradient. After flotation, gradients were fractionated and fractions analyzed as the sedimentation fractions. For further experiments, the light-scattering zone between 14% and 18% iodixanol layers was collected, yielding purified RCs. As a control, membranes from wortmannin-treated mock-infected cells were purified using the same protocol.
In vitro replication assays.
A standard 30-μl reaction mixture without BSA contained 25 μl of PNS, 32 mM HEPES (pH 7.4), 225 mM sucrose, 2.3 mM DTT, 6.3 mM KCl, 1.7 μg/ml actinomycin D, and 830 U/ml RiboLock. A standard 30-μl reaction mixture with BSA contained 22 μl of PNS, S7, P7, P15, sedimentation or flotation fraction, or purified RCs, 1 mg/ml BSA, 28 mM HEPES (pH 7.4), 200 mM sucrose, 2 mM DTT, 5.6 mM KCl, 1.5 μg/ml actinomycin D, and 810 U/ml RiboLock. In addition, both reaction mixtures contained 3 mM magnesium acetate, 17 mM creatine phosphate (Sigma-Aldrich), 8.3 U/ml creatine phosphokinase (Sigma-Aldrich), 1 mM ATP, 10 μM UTP, 10 μM GTP, 8.5 μM CTP, and 0.055 μM (5 μCi) [α-32P]CTP (PerkinElmer). If required, samples were diluted with DB, and if samples contained imidazole, 6 mM magnesium acetate was used in the reaction mixture. Reaction mixtures were incubated at 30°C for 1 h unless otherwise stated. After incubation, reactions were terminated by adding LiDS/LET (5% lithium dodecyl sulfate, 20 mM Tris-HCl [pH 7.4], 100 mM LiCl, 2 mM EDTA, and 5 mM DTT) containing 80 μg/ml proteinase K and incubating for 15 min at 37°C. Unincorporated label was removed using RNase-free Micro Bio-Spin P-30 gel columns (Bio-Rad), and the acid phenol method was used to isolate RNA.
To determine stability of RCs, PNS was incubated at 4°C for 3, 24, and 48 h or diluted in DB or 35.5% (wt/vol) iodixanol in DB and incubated at 4°C for 24 h. After incubation, aliquots were removed for total RNA isolation and in-gel hybridization to detect endogenous RNA (see below) and for an in vitro replication assay. To test detergent sensitivity, PNS was treated with 0.01 to 1% (wt/vol) n-octylglucoside, 0.01 to 1% (vol/vol) Tx-100, 0.01 to 1% (wt/vol) DOC, or 0.01 to 1% (wt/vol) SDS in DB at 4°C for 1 h, followed by total RNA isolation and in-gel hybridization to detect endogenous RNA (see below) or a replication assay. To inhibit replication, 100 μM 3′-dCTP (TriLink BioTechnologies) was added to the reaction mixture.
To monitor the stability of newly synthesized RNA in the S7 fraction or purified RCs, 100 μM 3′-dCTP and 220 ng of Tmed in vitro transcript were added to one aliquot and DB to another one after a 1-h replication assay. In vitro transcript was prepared using a SacI-linearized Tmed plasmid and an mMESSAGE mMACHINE T7 transcription kit (Ambion) according to the manufacturer's instructions. After a chase at 30°C, aliquots were removed and reactions were terminated as described above.
The ability of RCs to use an exogenous template RNA was tested by adding 1 μg of Tmed in vitro transcript in a reaction mixture containing purified RCs. As a control, P15 from cells transfected with the polyprotein P123HA4 and template Tmed plasmids was used. Replication assay reaction mixtures were incubated at 30°C for 2 h and terminated as described above. In order to test Tx-100 sensitivity of purified RCs, they were incubated with 1% Tx-100 for 1 h at 25°C. After incubation, aliquots were removed for total RNA isolation and in-gel hybridization to detect endogenous RNA (see below) or for a replication assay.
In order to determine how much the amount of genomic RNA increases during in vitro replication compared to the amount of genomic RNA present in the RCs after purification, replication assays were performed using purified RCs and reaction conditions described above for a standard reaction with BSA, except that 10 μM CTP and no [α-32P]CTP was used. In addition, a reaction devoid of NTPs was performed. After a 4-h incubation at 30°C, RNA was isolated and genomic RNA was detected by in-gel hybridization (see below). As a control, RNA was isolated from the purified RCs without incubation at 30°C.
RNA isolation, denaturing agarose gel electrophoresis, and in-gel hybridization.
Total RNA from replication assay reactions, PNS, S7, and P7 samples, sedimentation and flotation fractions, and purified RCs was isolated using the acid phenol method and analyzed by denaturing formaldehyde agarose gel electrophoresis as described previously (39). If required, spike RNA (Tmed or Tshort in vitro transcript) was added to the samples before RNA isolation to allow normalization. Tshort in vitro transcript was prepared from the Tshort plasmid (60) as described above for Tmed.
In-gel hybridization to detect RNA molecules from dried gels using 32P-labeled oligonucleotides has been described previously (61). SFV4-HA negative-stranded RNA was detected with the probe HybSFV4neg_4, and genomic and subgenomic RNA were detected with the probe HybSFV4pos_5 (39). SFV4-HA genomic RNA and RNA II were detected with the probe HybSFV4pos_8 (5′-GTCAGCCTCAATATCAACATGCACTTT-3′), complementary to nucleotides 95 to 121 of the genome. Tmed RNA was detected with the probe HybTmedpos_2 (39). Tshort RNA was detected with the probe HybTmedpos_3 (5′-AGGTGTATAACAGGTCCTCTCATTAATT-3′), complementary to nucleotides 1270 to 1297. 18S rRNA was detected with the probe 5′-ATGCCCCCGGCCGTCCCTCT-3′.
Dried gels were exposed to PhosphorImager screens to detect 32P label, and the screens were scanned with a Typhoon 9410 imager (GE Healthcare). Quantity One software (Bio-Rad) was used to quantify the incorporated label. Normalization of signals for variations in RNA recovery and loading was done based on quantification of 18S rRNA or spike RNA detected by in-gel hybridization.
RNase treatments of products from an in vitro replication assay.
Replication assays were performed using purified RCs and a 2-h reaction time, and RNA was isolated as described above. Isolated RNA was treated with RNase cocktail enzyme mix (Ambion) containing RNase A and T1 (final concentrations of 2.5 and 100 U/ml, respectively) or RNase III (50 U/ml; NEBNext RNase III RNA fragmentation module) under high-salt conditions. RNase A/T1 reaction mixture contained 4× SSC (600 mM NaCl, 60 mM sodium citrate, pH 7.0), and RNase III reaction mixture contained 2× SSC (300 mM NaCl, 30 mM sodium citrate, pH 7.0) and 1× RNase III reaction buffer. After a 15-min incubation at 37°C, proteinase K was added (80 μg/ml) and incubation was continued for 15 min. RNA was precipitated as described in reference 39. Besides being subjected to denaturing formaldehyde agarose gel electrophoresis, RNA was analyzed without denaturation in a 1.0% (wt/vol) agarose gel in Tris-borate-EDTA buffer after mixing with 2 volumes of gel loading buffer II (Ambion).
RNA release.
To study release of newly synthesized RNA, purified RCs were concentrated and iodixanol was removed by washing with DB in Amicon ultracentrifugal filter units (3,220 × g, 4°C; 50,000 nominal molecular weight limit; Merck Millipore). An in vitro replication assay was performed as described above, and after 1-, 2-, and 3-h reaction times, aliquots were removed and diluted with DB and pellet and supernatant fractions were separated by differential centrifugation (20,600 × g, 15 min, 4°C). The pellet was washed with DB, and the pellet and supernatant were treated with proteinase K (80 μg/ml) for 15 min at 37°C. RNA was isolated using the acid phenol method (39).
Protein and lipid analyses.
Protein concentrations were measured using the Coomassie blue method (62), with BSA as a standard. PNS, S7, P7, and sedimentation and flotation fractions were analyzed by SDS-PAGE with 4% (wt/vol) acrylamide in the stacking gel and 10%, 12%, or 16% (wt/vol) acrylamide in the separation gel. SDS-PAGE (12%) gels were stained with Coomassie blue to show proteins. SDS-PAGE (16%) gels were stained with Sudan black B (Sigma-Aldrich) according to the manufacturer's instructions to detect lipids. After electrophoresis, Sudan black B stains both lipids and lipoproteins. For Western blotting, proteins separated in 10% or 12% SDS-PAGE gels were transferred to Amersham Protran nitrocellulose blotting membrane (GE Healthcare). Membranes were blocked with 5% (wt/vol) nonfat dry milk powder in Tris-buffered saline (TBS), followed by incubation with rabbit polyclonal antibodies against SFV nsP1, nsP2, nsP3, or nsP4 (37) or mouse monoclonal antibody against β-actin (Sigma-Aldrich) in TBS with 5% milk and 0.1% (vol/vol) Tween 20. Alternatively, membranes were blocked with 5% milk in TBS containing 0.1% Tween 20, followed by incubation with rabbit monoclonal antibody against SDHA (Cell Signaling), mouse monoclonal antibody against PMCA (Abcam), rabbit polyclonal antibody against Na,K-ATPase α (Cell Signaling), rabbit polyclonal antibody against calnexin (Abcam), rabbit polyclonal antibody against CLIMP-63 (Abcam), rabbit polyclonal antibody against RTN4A and RTN4B (Abcam), mouse monoclonal antibody against GM130 (BD Biosciences), mouse monoclonal antibody against Rab7 (Abcam), rabbit polyclonal antibody against LAMP-2 (Novus Biologicals), or rabbit polyclonal antibody against SFV capsid protein (63) in TBS with 5% (wt/vol) BSA and 0.1% Tween 20. The primary antibodies were detected with secondary antibodies IRDye800CW donkey anti-rabbit IgG (Li-COR Biosciences) and Alexa Fluor 680 donkey anti-mouse IgG (Invitrogen) and an Odyssey system (Li-COR). Quantification was performed with Image Studio software (Li-COR). The antibody against RTN4A and RTN4B recognized only a band of about 50 kDa, indicating that the detected protein was RTN4B (49), and the result was confirmed using sheep polyclonal antibody against RTN4B (MRC-PPU Reagents).
Electron microscopy.
The light-scattering zone between 14% and 18% iodixanol layers in the flotation gradient (see the purification procedure described above) was collected and fixed with 2% (vol/vol) glutaraldehyde (Sigma-Aldrich) in 100 mM HEPES (pH 7.4) on ice for 30 min. After fixation, membranes were pelleted by differential centrifugation (11,700 × g, 30 min, 4°C). Membrane pellets were washed, osmicated, and embedded into resin (TAAB Laboratories Equipment, Ltd.) using standard procedures, and ultrathin sections (60 nm) were cut, picked on single-slot copper grids, and poststained with lead and uranyl acetate. The homogeneity of the pellet was checked by systematic sectioning through the entire pellet. The micrographs were taken with a Jeol JEM-1400 transmission electron microscope operating at 80 kV at the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki.
ACKNOWLEDGMENTS
We thank Eija Jokitalo for valuable discussions and Arja Välimäki, Mika Lång, and Ilkka Kivistö for excellent technical assistance. We acknowledge the support and the use of resources of the Centre for Virus and Macromolecular Complex Production (ICVIR, University of Helsinki), part of Instruct-FI.
This work was supported by a 3-year research project grant of the University of Helsinki (M.K.P.), an Academy of Finland Postdoctoral Researcher grant (274748; M.K.P.), and Academy of Finland Project grants (265997 and 307802; T.A.).
REFERENCES
- 1.Yang X, Yang H, Zhou G, Zhao GP. 2008. Infectious disease in the genomic era. Annu Rev Genomics Hum Genet 9:21–48. doi: 10.1146/annurev.genom.9.081307.164428. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479-480:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 5.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salonen A, Ahola T, Kääriäinen L. 2005. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol 285:139–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knoops K, Barcena M, Limpens RW, Koster AJ, Mommaas AM, Snijder EJ. 2012. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol 86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Barcena M. 2011. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio 2:e00166-. doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albulescu IC, Tas A, Scholte FE, Snijder EJ, van Hemert MJ. 2014. An in vitro assay to study chikungunya virus RNA synthesis and the mode of action of inhibitors. J Gen Virol 95:2683–2692. doi: 10.1099/vir.0.069690-0. [DOI] [PubMed] [Google Scholar]
- 12.Ertel KJ, Benefield D, Castano-Diez D, Pennington JG, Horswill M, den Boon JA, Otegui MS, Ahlquist P. 2017. Cryo-electron tomography reveals novel features of a viral RNA replication compartment. Elife 6:e25940. doi: 10.7554/eLife.25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallio K, Hellström K, Balistreri G, Spuul P, Jokitalo E, Ahola T. 2013. Template RNA length determines the size of replication complex spherules for Semliki Forest virus. J Virol 87:9125–9134. doi: 10.1128/JVI.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spuul P, Balistreri G, Kääriäinen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J Virol 84:7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 17.Ryman KD, Klimstra WB. 2008. Host responses to alphavirus infection. Immunol Rev 225:27–45. doi: 10.1111/j.1600-065X.2008.00670.x. [DOI] [PubMed] [Google Scholar]
- 18.Schilte C, Staikowsky F, Couderc T, Madec Y, Carpentier F, Kassab S, Albert ML, Lecuit M, Michault A. 2013. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis 7:e2137. doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan AH, Morita K, Parquet Md Mdel C, Hasebe F, Mathenge EG, Igarashi A. 2002. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J Gen Virol 83:3075–3084. doi: 10.1099/0022-1317-83-12-3075. [DOI] [PubMed] [Google Scholar]
- 20.Lemm JA, Rumenapf T, Strauss EG, Strauss JH, Rice CM. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13:2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirako Y, Strauss JH. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol 68:1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahola T, Kääriäinen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A 92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemm JA, Rice CM. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol 67:1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemm JA, Rice CM. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J Virol 67:1905–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spuul P, Salonen A, Merits A, Jokitalo E, Kaariainen L, Ahola T. 2007. Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication. J Virol 81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das PK, Merits A, Lulla A. 2014. Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. doi: 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy WR, Strauss JH. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol 63:4653–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kaariainen L, Ahola T. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem 278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 29.Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI. 2016. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog 12:e1005810. doi: 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, Canard B. 2016. Viral macro domains reverse protein ADP-ribosylation. J Virol 90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubach JK, Wasik BR, Rupp JC, Kuhn RJ, Hardy RW, Smith JL. 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384:201–208. doi: 10.1016/j.virol.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman RM, Levin JG, Grimley PM, Berezesky IK. 1972. Membrane-associated replication complex in arbovirus infection. J Virol 10:504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimley PM, Berezesky IK, Friedman RM. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J Virol 2:1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimley PM, Levin JG, Berezesky IK, Friedman RM. 1972. Specific membranous structures associated with the replication of group A arboviruses. J Virol 10:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol 84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Froshauer S, Kartenbeck J, Helenius A. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol 107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thaa B, Biasiotto R, Eng K, Neuvonen M, Gotte B, Rheinemann L, Mutso M, Utt A, Varghese F, Balistreri G, Merits A, Ahola T, McInerney GM. 2015. Differential phosphatidylinositol-3-kinase-Akt-mTOR activation by Semliki Forest and chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J Virol 89:11420–11437. doi: 10.1128/JVI.01579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietilä MK, Albulescu IC, Hemert MJV, Ahola T. 2017. Polyprotein processing as a determinant for in vitro activity of Semliki Forest virus replicase. Viruses 9:E292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clewley JP, Kennedy SI. 1976. Purification and polypeptide composition of Semliki Forest virus RNA polymerase. J Gen Virol 32:395–411. doi: 10.1099/0022-1317-32-3-395. [DOI] [PubMed] [Google Scholar]
- 41.Sreevalsan T, Yin FH. 1969. Sindbis virus-induced viral ribonucleic acid polymerase. J Virol 3:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wielgosz MM, Huang HV. 1997. A novel viral RNA species in Sindbis virus-infected cells. J Virol 71:9108–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Hemert MJ, de Wilde AH, Gorbalenya AE, Snijder EJ. 2008. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J Biol Chem 283:16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- 44.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barton DJ, Sawicki SG, Sawicki DL. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J Virol 65:1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomatos PJ, Kääriäinen L, Keränen S, Ranki M, Sawicki DL. 1980. Semliki Forest virus replication complex capable of synthesizing 42S and 26S nascent RNA chains. J Gen Virol 49:61–69. [DOI] [PubMed] [Google Scholar]
- 47.Ranki M, Kääriäinen L. 1979. Solubilized RNA replication complex from Semliki Forest virus-infected cells. Virology 98:298–307. [DOI] [PubMed] [Google Scholar]
- 48.Sandoz PA, van der Goot FG. 2015. How many lives does CLIMP-63 have? Biochem Soc Trans 43:222–228. doi: 10.1042/BST20140272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rämö O, Kumar D, Gucciardo E, Joensuu M, Saarekas M, Vihinen H, Belevich I, Smolander OP, Qian K, Auvinen P, Jokitalo E. 2016. NOGO-A/RTN4A and NOGO-B/RTN4B are simultaneously expressed in epithelial, fibroblast and neuronal cells and maintain ER morphology. Sci Rep 6:35969. doi: 10.1038/srep35969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varjak M, Saul S, Arike L, Lulla A, Peil L, Merits A. 2013. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. J Virol 87:10295–10312. doi: 10.1128/JVI.01105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maeda F, Arii J, Hirohata Y, Maruzuru Y, Koyanagi N, Kato A, Kawaguchi Y. 2017. Herpes simplex virus 1 UL34 protein regulates the global architecture of the endoplasmic reticulum in infected cells. J Virol 91:e00271-. doi: 10.1128/JVI.00271-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips MJ, Voeltz GK. 2016. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahola T, Lampio A, Auvinen P, Kääriäinen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J 18:3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salonen A, Vasiljeva L, Merits A, Magden J, Jokitalo E, Kääriäinen L. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J Virol 77:1691–1702. doi: 10.1128/JVI.77.3.1691-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michel MR, Gomatos PJ. 1973. Semliki forest virus-specific RNAs synthesized in vitro by enzyme from infected BHK cells. J Virol 11:900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jose J, Taylor AB, Kuhn RJ. 2017. Spatial and temporal analysis of alphavirus replication and assembly in mammalian and mosquito cells. mBio 8:e02294-. doi: 10.1128/mBio.02294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pietilä MK, Hellström K, Ahola T. 2017. Alphavirus polymerase and RNA replication. Virus Res 234:44–57. doi: 10.1016/j.virusres.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Posthuma CC, Te Velthuis AJW, Snijder EJ. 2017. Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res 234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spuul P, Balistreri G, Hellström K, Golubtsov AV, Jokitalo E, Ahola T. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J Virol 85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scholte FE, Tas A, Martina BE, Cordioli P, Narayanan K, Makino S, Snijder EJ, van Hemert MJ. 2013. Characterization of synthetic Chikungunya viruses based on the consensus sequence of recent E1-226V isolates. PLoS One 8:e71047. doi: 10.1371/journal.pone.0071047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
- 63.Väänänen P, Kääriäinen L. 1979. Haemolysis by two alphaviruses: Semliki Forest and Sindbis virus. J Gen Virol 43:593–601. [DOI] [PubMed] [Google Scholar]