ABSTRACT
Respiratory syncytial virus (RSV) is the main cause of acute lower respiratory tract infection (ALRI) in children worldwide. Virus-host interactions affect the progression and prognosis of the infection. Autophagy plays important roles in virus-host interactions. Respiratory epithelial cells serve as the front line of host defense during RSV infection, However, it is still unclear how they interact with RSV. In this study, we found that RSV induced autophagy that favored RSV replication and exacerbated lung pathology in vivo. Mechanistically, RSV induced complete autophagy flux through reactive oxygen species (ROS) generation and activation of the AMP-activated protein kinase/mammalian target of rapamycin (AMPK-MTOR) signaling pathway in HEp-2 cells. Furthermore, we evaluated the functions of autophagy in RSV replication and found that RSV replication was increased in HEp-2 cells treated with rapamycin but decreased remarkably in cells treated with 3-methylademine (3-MA) or wortmannin. Knockdown key molecules in the autophagy pathway with short hairpinp RNA (shRNA) against autophagy-related gene 5 (ATG5), autophagy-related gene 7 (ATG7), or BECN1/Beclin 1 or treatment with ROS scavenger N-acetyl-l-cysteine (NAC) and AMPK inhibitor (compound C) suppressed RSV replication. 3-MA or shATG5/BECN1 significantly decreased cell viability and increased cell apoptosis at 48 hours postinfection (hpi). Blocking apoptosis with Z-VAD-FMK partially restored virus replication at 48 hpi. Those results provide strong evidence that autophagy may function as a proviral mechanism in a cell-intrinsic manner during RSV infection.
IMPORTANCE An understanding of the mechanisms that respiratory syncytial virus utilizes to interact with respiratory epithelial cells is critical to the development of novel antiviral strategies. In this study, we found that RSV induces autophagy through a ROS-AMPK signaling axis, which in turn promotes viral infection. Autophagy favors RSV replication through blocking cell apoptosis at 48 hpi. Mechanistically, RSV induces mitophagy, which maintains mitochondrial homeostasis and therefore decreases cytochrome c release and apoptosis induction. This study provides a novel insight into this virus-host interaction, which may help to exploit new antiviral treatments targeting autophagy processes.
KEYWORDS: autophagy, RSV, virus replication, ROS, apoptosis
INTRODUCTION
Respiratory syncytial virus (RSV), a member of the Pneumovirus genus in the Paramyxoviridae family, is an enveloped negative-stranded RNA virus. RSV is the most important pathogen causing acute lower respiratory tract infection (ALRI) in infants, preschool children, the elderly, and immunocompromised individuals worldwide (1, 2). RSV infection is the main cause of hospital admission and death from ALRI in children and is associated with high health care costs (3). So far, there is no safe and effective vaccine or specific antiviral drug for RSV. As an intracellular obligate microorganism, virus-host interaction affects the progression and prognosis of the infection. In this study, we focused on RSV-host interaction, especially on how RSV affects autophagy and how autophagy affects RSV replication.
Autophagy plays important roles in virus-host interaction. Autophagy is a highly conservative metabolism process essential for maintaining cellular homeostasis in eukaryotic cells, through degrading redundant or damaged proteins and organelles via the lysosomal degradative pathway and recycling the metabolites (4, 5). Viral infection can induce autophagy, which in turn affects virus infection in different ways. On the one hand, autophagy plays an antiviral role through activating Toll-like receptors (TLRs), participating in virus antigen processing and presenting, and sequestrating and degrading virus directly (6–10). On the other hand, a number of viruses have evolved many strategies to evade or even subvert autophagy for their benefit. They can prevent autophagosome-lysosome fusion, reshape the endomembrane system to create membrane-associated replication “factories,” or suppress antiviral innate immunity to favor virus replication (11–14).
Autophagy can be induced by various viruses; however, how virus induces autophagy remains largely elusive. Cellular stress responses, such as endoplasmic reticulum (ER) stress and oxidative stress, may be induced by viral infection and may trigger autophagy. Accumulating evidence on tumors has highlighted the role of reactive oxygen species (ROS), a key molecule to induce oxidative stress, in autophagy induction (15–18). However, to the best of our knowledge, the relationship between ROS and autophagy regulation during virus infection is not fully understood. Many studies have indicated the causal link between RSV infection and oxidative stress and demonstrated that ROS production plays an important role in RSV pathogenesis through mediating inflammatory responses of lung (19–21). Antioxidant treatment could ameliorate RSV-induced pulmonary inflammation (22). Oxidative stress and autophagy are two different cellular responses to RSV infection. It is intriguing whether these two responses interact with each other.
Besides autophagy, apoptosis also plays a “double-edged sword” role in virus-host interaction. Apoptosis, a programmed cell death controlled by many genes, is required to eliminate misplaced or damaged cells in order to maintain homeostasis. This “sacrifice” of infected cells provides an important host defense mechanism to limit virus replication (23). To obtain a favorable environment, viruses have developed myriad mechanisms to subvert cellular apoptosis to facilitate their replication, assembly, and spreading (24, 25).
Autophagy and apoptosis are completely different physiological process; however, there is plenty of evidence showing that they are closely related. They can regulate each other and even be switched under certain conditions (26, 27). For example, autophagy may limit apoptosis through degrading damaged mitochondria or activated caspase 8 (28–31). Proapoptotic proteins can cleave some autophagy proteins, such as BECN1 and autophagy-related gene 5 protein (ATG5), which might switch those proteins from proautophagic to proapoptotic (32, 33). The dynamic balance between these two pathways affects viral survival and pathogenicity. Though respiratory epithelial cells serve as the front line of host defense, it is still unclear how they interact with RSV. In this study, we investigated the effects of RSV infection on autophagy and the role of autophagy in virus proliferation kinetics both in vivo and in vitro. We discovered a possible interaction between autophagy, apoptosis, and viral replication. This study reveals how RSV affects respiratory epithelial cells and provides a new antiviral option by targeting autophagy.
RESULTS
RSV infection induces autophagy that favors viral replication.
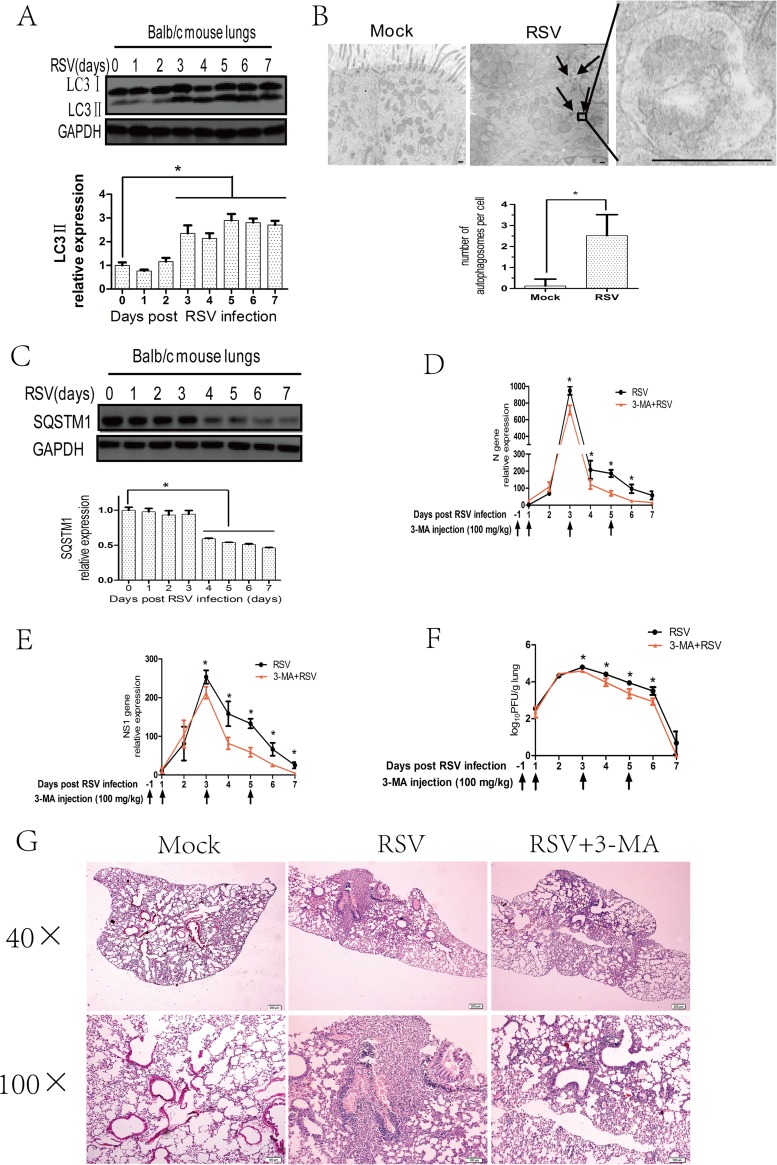
To explore the relationship between RSV infection and autophagy, we first determined whether RSV infection can induce autophagy in vivo. Because the lipidated form of microtubule-associated protein 1 LC3 (referred to as LC3II) is a hallmark of autophagosome (34), we investigated the expression of LC3II in the lungs of mice infected with RSV. The results showed that LC3II expression in RSV-infected mice lungs began to increase at 3 days and was sustained at least up to 7 days after RSV intranasal infection (Fig. 1A), but the total LC3 mRNA level was not affected (data not shown). In addition, we performed transmission electron microcopy (TEM) to observe autophagosomes in the lungs of mice infected with RSV and found that the number of autophagosomes was significantly increased after RSV infection (Fig. 1B).
FIG 1.
RSV-induced autophagy enhances viral replication in the lungs of BALB/c mice. (A) BALB/c mice were mock infected or infected with RSV as described in Materials and Methods. Mouse lungs were harvested at indicated days and the expression of LC3II and GAPDH was analyzed with immunoblotting with specific antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to GAPDH control. The data represent means ± SD for at least 3 mice per group. (B) BALB/c mice were treated as described for panel A for 5 days, and mice lungs were examined by transmission electron microcopy. The black arrows depict double-membraned autophagosomes. The bar graph represents the number of autophagosomes per cell. The data are from 50 cells per sample. Bar, 500 nm. (C) BALB/c mice were treated as described for panel A, and the expression of SQSTM1 and GAPDH of mice lungs was analyzed with immunoblotting with specific antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to GAPDH control. The data represent means ± SD for at least 3 mice per group. (D and E) BALB/c mice were infected with RSV in the presence or absence of 3-MA as described in Materials and Methods. Mouse lungs were harvested at indicated days, and viral N (D) or NS1 (E) genes were detected by qRT-PCR. Gene expression was calculated by comparison to that of one mouse infected with RSV at day 1 postinfection (100%). Results are means ± SD for 5 mice per group. (F) BALB/c mice were treated as described for panels D and E, and lung homogenates at indicated days were prepared to detect viral titers according to the methods described in Materials and Methods. Results are shown as log10 PFU/g lung and represent means ± SD for 5 mice per group. (G) BALB/c mice were treated as described for panels D and E for 5 days, and lung pathology was detected by hematoxylin and eosin (H&E) stain. *, P < 0.05.
The entire process of autophagy (known as autophagic flux) includes not only the increased autophagosome formation but also autophagosome-lysosome fusion and degradation of contents. The expression of polyubiquitin-binding protein SQSTM1, an indicator of autophagic flux (34), can be used to evaluate the effects of RSV infection on autophagic flux. Compared with uninfected control mice, SQSTM1 expression in the lungs of RSV-infected mice was significantly decreased starting at 4 days after RSV infection (Fig. 1C), but its mRNA level was not decreased (data not shown).
To confirm the role of autophagy in RSV replication, we used autophagy inhibitor 3-methylademine (3-MA) to block autophagy and then checked its effects on viral load. The effect of 3-MA was validated by detecting LC3II expression in lungs after RSV infection (data not shown). We used the expression of viral NS1 and N genes in infected lung tissues and viral titers in lung homogenates to represent viral replication. Blocking autophagy by 3-MA reduced viral N and NS1 gene expression starting at 3 days after RSV infection (Fig. 1D and E). A similar pattern of reduction in viral titers was also observed in lung homogenates (Fig. 1F). These results indicated that RSV infection can induce autophagy, which may favor viral replication in vivo.
In addition, we assessed the effects of 3-MA on lung pathology of the RSV-infected mice and found that treatment with 3-MA could ameliorate lung pathology (Fig. 1G). RSV infection caused lymphocyte infiltration, thickened alveolar septum, and induced alveolar hemorrhage in the lungs. 3-MA treatment can ameliorate lymphocyte infiltration and decrease the area of lesions.
RSV infection induces autophagosome formation and complete autophagy flux in HEp-2 cells.
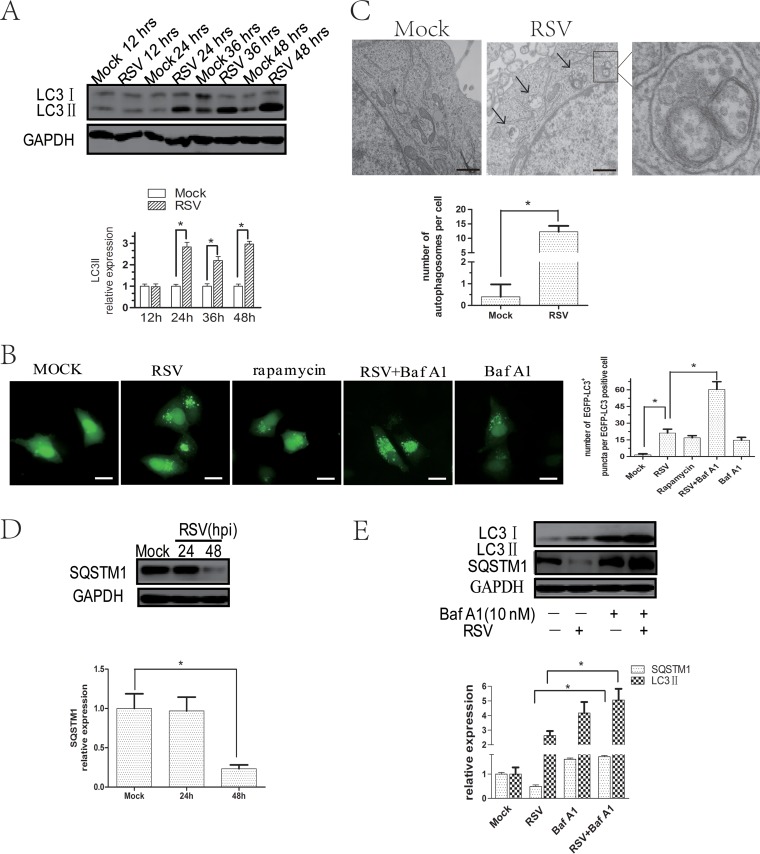
To explore the mechanisms of RSV-induced autophagy, we tested whether RSV could induce autophagy in HEp-2 cells in vitro. The result showed that RSV infection significantly increased LC3II expression in HEp-2 cells from 24 h after infection (Fig. 2A), especially when the multiplicity of infection (MOI) was 5 (data not shown). In addition, the LC3 mRNA level was unaffected by RSV infection (data not shown).
FIG 2.
RSV infection induces complete autophagy flux in HEp-2 cells. (A) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for indicated times. The expression of LC3IIand GAPDH was analyzed with their specific antibodies as described in Materials and Methods. (B) HEp-2 cells transfected with pBABE-puro-EGFP-LC3 plasmid for 24 h were mock infected or infected with RSV in the absence or presence of Baf A1 for 24 h and analyzed by fluorescence microscopy for the presence of fluorescent EGFP-LC3 puncta. Bar, 20 μm. Rapamycin was used as a positive control. The bar graph represents the number of puncta per EGFP-LC3-positive cell. The data are from 50 cells per sample. (C) HEp-2 cells were infected with RSV (MOI = 5) for 24 h and examined by transmission electron microscope. The black arrows depict double-membraned autophagosomes. The bar graph represents the number of autophagosomes per cell. The data are from 50 cells per sample. Bar, 333 nm. (D) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for 24 and 48 h and then analyzed by immunoblotting with anti-SQSTM1 and anti-GAPDH antibodies. (E) Cells were infected with RSV (MOI = 5) for 48 h in the presence or absence of Baf A1 (10 nM). Cell lysates were analyzed by immunoblotting using anti-LC3, anti-SQSTM1, and anti-GAPDH antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to GAPDH control. The data represent means ± SD for 3 independent experiments. *, P < 0.05.
Punctate accumulation of LC3, which represents LC3, is conjugated to phosphatidylethanolamine (PE) and recruited to autophagosome membranes and has been suggested to be a marker of autophagy (35). To further decipher the relationship between RSV and autophagy, we performed fluorescence microscopy to localize LC3 distribution. RSV infection caused a significant change in the distribution of cytoplasmic enhanced green fluorescent protein (EGFP)-LC3 from a diffuse localization to a punctate accumulation, confirming that autophagosome formation was increased (Fig. 2B). Moreover, the number of autophagic vacuoles with double-layered structures was increased in RSV-infected HEp-2 cells as observed by transmission electron microcopy (Fig. 2C). These data indicate that RSV infection induced autophagosome formation.
Compared with mock-infected cells, SQSTM1 expression was significantly decreased at 48 h postinfection in RSV-infected cells, suggesting that autophagic flux was enhanced (Fig. 2D). Additionally, bafilomycin A1 (Baf A1), a vacuolar H+-ATPase inhibitor that can block autophagy flux, significantly increased the number of cytoplasmic EGFP-LC3 puncta (Fig. 2B) and the level of LC3II and SQSTM1 in HEp-2 cells infected with RSV (Fig. 2E). These results further confirmed that RSV infection induced complete autophagy flux.
RSV-induced ROS production favors autophagosome formation through activating AMP-activated protein kinase/mammalian target of rapamycin (AMPK-MTOR) signaling in HEp-2 cells.
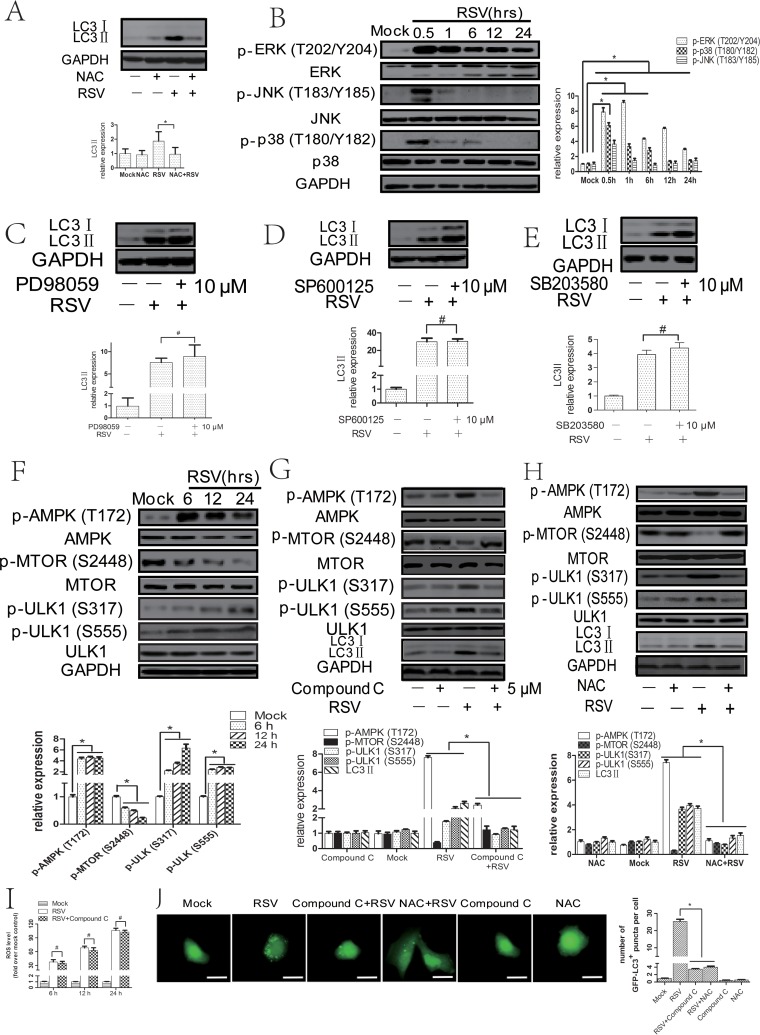
To determine the role of oxidative stress in RSV-induced autophagy, we checked whether RSV infection could induce ROS production in HEp-2 cells. Intracellular ROS levels (probed by DCFH-DA [2′,7′-dichlorodihydrofluorescein diacetate]) in HEp-2 cells were elevated at 6 h, reached a peak at 12 h, and were sustained for up to 24 h after RSV infection (data not shown). To further confirm the involvement of ROS in RSV-induced autophagy, we measured the expression of LC3II and the number of cytoplasmic EGFP-LC3 puncta in infected HEp-2 or A549 cells treated with N-acetyl-l-cysteine (NAC) (a ROS scavenger). We found that 5 mM NAC suppressed RSV-induced ROS levels (data not shown) and decreased RSV-induced LC3II expression and the number of cytoplasmic EGFP-LC3 puncta (Fig. 3A and J), proving the importance of ROS in RSV-induced autophagy. In addition, quantitative real-time PCRs (qPCRs) were performed to rule out transcriptional effects of RSV and NAC on LC3 mRNA levels (data not shown).
FIG 3.
RSV-induced ROS production favors autophagosome formation through activating AMPK-MTOR signaling in HEp-2 cells. (A) HEp-2 cells were mock infected or infected with RSV in the absence or presence of NAC (5 mM) for 24 h. Cell lysates were then analyzed by immunoblotting with anti-LC3 and anti-GAPDH antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to GAPDH control. (B) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for indicated times. The expression of p-ERK (Thr202/Tyr 204), p-JNK (Thr183/Tyr185), and p-p38 (Thr180/Tyr182) was analyzed by immunoblotting. The relative levels of targeted proteins were quantitated by densitometry and normalized to total ERK, JNK, or p38, respectively. (C, D, and E) HEp-2 cells were infected with RSV in the absence or presence of 10 μM PD98059 (C), SP600125 (D), or SB203580 (E) for 24 h. Cell lysates were then analyzed by immunoblotting with anti-LC3 and anti-GAPDH antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to GAPDH control. (F) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for indicated times. The expression of p-AMPK (Thr172), p-MTOR (Ser2448), p-ULK1 (Ser371), and p-ULK1 (Ser555) was analyzed by immunoblotting with specific antibodies as described in Materials and Methods. The relative levels of targeted proteins were quantitated by densitometry and normalized to AMPK, MTOR, or ULK1, respectively. (G and H) HEp-2 cells were mock infected or infected with RSV (MOI = 5) in the absence or presence of compound C (5 μM) (G) or NAC (5 mM) (H) for 24 h. The expression of p-AMPK (Thr172), p-MTOR (Ser2448), p-ULK1 (Ser371), p-ULK1 (Ser555), and LC3II was analyzed by immunoblotting with specific antibodies as described in Materials and Methods. The relative levels of targeted proteins were quantitated by densitometry and normalized to AMPK, MTOR, ULK1, and GAPDH, respectively. (I) HEp-2 cells were mock infected or infected with RSV (MOI = 5) in the absence or presence of compound C for indicated times. The intracellular reactive oxygen species (ROS) level was examined by DCFH-DA assay as described in Materials and Methods. The bar graph represents fold changes in mean fluorescence intensity (MFI) compared to uninfected cells. (J) pBABE-puro-EGFP-LC3-transfected HEp-2 cells were mock infected or infected with RSV (MOI = 5) in the absence or presence of compound C (5 μM) or NAC (5 mM) for 24 h. EGFP-LC3 puncta were visualized by fluorescence microscopy. The bar graph represents the number of puncta per EGFP-LC3 positive cell. The data are from 50 cells per sample. Bar, 20 μm. The data shown represents means ± SD for 3 independent experiments. *, P < 0.05; #, P > 0.05.
Although oxidative stress has been shown to be able to induce autophagy, the precise mechanisms remain poorly elucidated. Some studies reported a link between ROS and mitogen-activated protein kinase (MAPK) or AMPK-MTOR pathways. Therefore, we investigated their roles in autophagy triggered by ROS. As shown in Fig. 3B, the levels of p-Jun N-terminal protein kinase (p-JNK; Thr183/Tyr185), p-extracellular signal-regulated kinase (p-ERK; Thr202/Tyr 204), and p-p38 (Thr180/Tyr182) were increased after RSV infection. Activation of AMPK and inhibition of MTOR were also observed in RSV-infected HEp-2 cells (Fig. 3F). In addition, we detected the phosphorylated ULK1, which is a critical molecule downstream from AMPK and MTOR, and found that RSV infection led to an increase in the level of phosphorylated ULK1 at Ser317 and Ser555 (Fig. 3F). Next, we detected the effects of specific inhibitors for JNK (SP600125), ERK (PD98059), p38 (SB203580), and AMPK (compound C) on LC3II expression in RSV-infected HEp-2 cells. First, we optimized the concentrations of all kinds of inhibitors (data not shown) and verified that RSV-induced LC3II expression was significantly inhibited after blocking the AMPK-MTOR pathway but not the MAPK pathway (Fig. 3C to E and G). In addition, the number of cytoplasmic EGFP-LC3 puncta was decreased in either RSV-infected HEp-2 cells (Fig. 3J) or A549 cells (data not shown) after treatment with compound C. The results indicated that AMPK-MTOR signaling pathways play a major role in RSV-induced autophagy. To explore the relationship between ROS and the AMPK-MTOR signaling pathway, we measured the expression of p-AMPK (Thr172), p-MTOR (Ser2448), p-ULK1 (Ser371), p-ULK1 (Ser555), and LC3II in HEp-2 cells treated with NAC. As shown in Fig. 3H, NAC reversed the expression of p-AMPK (Thr172), p-MTOR (Ser2448), p-ULK1 (Ser371), p-ULK1 (Ser555), and LC3II induced by RSV infection. Inhibition of AMPK by compound C did not affect the intracellular reactive oxygen species (ROS) level in RSV-infected HEp-2 cells (Fig. 3I). The results suggest that ROS production may serve as an upstream signal to activate the AMPK-MTOR signaling pathways, leading to autophagy induction in RSV-infected HEp-2 cells.
Autophagy modulation by using pharmacological regulators affects RSV replication.
To explore the role of autophagy in virus replication, we used validated autophagy modulators to evaluate the impact of autophagy on viral replication.
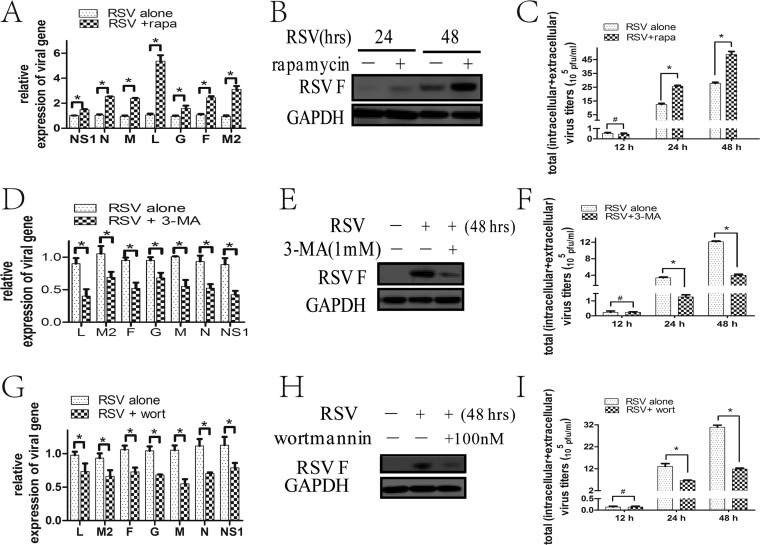
First, we optimized the concentrations of autophagy modulators and verified that 100 nM rapamycin (an MTOR inhibitor) and 1 mM 3-MA and 100 nM wortmannin (phosphatidylinositol 3-kinases [PtdIns3K] inhibitor) promoted and inhibited, respectively, autophagy in RSV-infected HEp-2 cells (data not shown). RSV entry and cell proliferation may affect viral replication. To rule out whether these chemicals affect RSV entry, we pretreated the cells with these chemicals and then infected the cells and examined RSV entry by checking invasive RSV gene expression and mean fluorescence intensity (MFI) of RSV. We found that these chemicals had no effects on RSV entry under these conditions (data not shown). In addition, these pharmacological regulators had no effect on cell proliferation (data not shown).
Then, we tested the effects of autophagy modulators on viral replication at an MOI of 5. The results showed that the expressions of viral M, N, L, G, F, M2, and NS1 genes were significantly enhanced in rapamycin-treated HEp-2 cells (Fig. 4A). Enhanced autophagy by rapamycin was also remarkably associated with augmented RSV F protein expression at 48 h and total viral loads (intracellular and extracellular) at 24 h and 48 h in cells infected with different MOIs (Fig. 4B and C). The proviral effects of rapamycin can be blocked by ATG5 knockdown (data not shown), proving that rapamycin exerts its promoting effects on viral replication through the autophagy pathway. Treating HEp-2 cells with either 3-MA or wortmannin notably decreased viral M, N, L, G, F, M2, and NS1 gene expression and RSV F protein expression at the indicated times and the total viral titers at 24 h and 48 h at different MOIs (Fig. 4D to I). All these results indicated that autophagy may promote virus replication.
FIG 4.
Autophagy modulation by pharmacological regulators affects RSV replication. (A, D, and G) HEp-2 cells were infected with RSV (MOI = 5) in the absence or presence of rapamycin (A), 3-MA (D), or wortmannin (G) for 24 h. Relative expressions of RSV-targeted genes were determined by fluorescence quantitative PCR. (B, E, and H) HEp-2 cells were infected with RSV (MOI = 5) in the absence or presence of rapamycin (B), 3-MA (E), or wortmannin (H) for indicated times. Cell lysates were prepared and analyzed by immunoblotting with anti-RSV F and anti-GAPDH antibodies. (C, F, and I) HEp-2 cells were infected with RSV (MOI = 5) in the absence or presence of rapamycin (C), 3-MA (F), or wortmannin (I) for indicated times. Total virus titers (intracellular + extracellular) were determined as described in Materials and Methods. Data represent means ± SD for 3 independent experiments. rapa, rapamycin; 3-MA, 3-methyladenine; wort, wortmannin; *, P < 0.05; #, P > 0.05.
Silencing autophagy-related genes by short hairpin RNAs (shRNAs) leads to a reduction of RSV replication in HEp-2 cells.
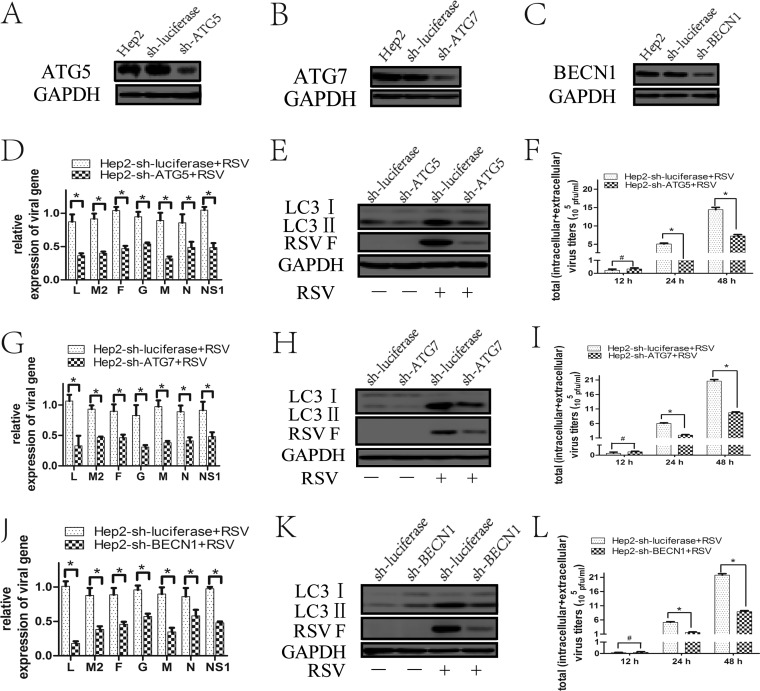
Pharmacological regulators may have off-target effects that might affect the results. Therefore, we used shRNA lentivirus specifically targeting the autophagy-related genes to further confirm their effects on virus replication. Knockdown of these autophagy-related genes (Fig. 5A to C) resulted in a decrease in LC3II expression induced by RSV, demonstrating a successful inhibition of autophagy by the shRNA/lentiviruses targeting ATG5, ATG7, or BECN1 expression (Fig. 5E, H, and K).
FIG 5.
Inhibition of autophagy with specific shRNA targeting ATG5, ATG7, or BECN1 reduces RSV replication. (A to C) HEp-2 cells stably expressing specific shRNA against ATG5 (HEp-2-sh-ATG5), ATG7 (HEp-2-sh-ATG7), BECN1 (HEp-2-sh-BECN1), or luciferase gene (HEp-2-sh-luciferase) were established as described in Materials and Methods. The silencing efficiency of ATG5 shRNA (A), ATG7 shRNA (B), and BECN1 shRNA (C) was validated by immunoblotting with anti-ATG5, anti-ATG7, and anti-BECN1 antibodies, respectively, as well as with anti-GAPDH. (D, G, and J) HEp-2-sh-ATG5 (D), HEp-2-sh-ATG7 (G), HEp-2-sh-BECN1 (J), or HEp-2-sh-luciferase cells were infected with RSV (MOI = 5) for 24 h. Relative expressions of RSV targeted genes were determined by fluorescence quantitative PCR. (E, H, and K) HEp-2-sh-ATG5 (E), HEp-2-sh-ATG7 (H), HEp-2-sh-BECN1 (K) and HEp-2-sh-luciferase (E, H, K) cells were infected with RSV (MOI = 5) for 48 h. The expression of LC3II and RSV F was analyzed by immunoblotting with specific antibodies. (F, I, and L) HEp-2-sh-ATG5 (F), HEp-2-sh-ATG7 (I), and HEp-2-sh-BECN1 (L) cells were infected with RSV for indicated times, and total virus titers (intracellular + extracellular) were determined as described in Materials and Methods. Data represent means ± SD for 3 independent experiments. *, P < 0.05; #, P > 0.05.
Compared with HEp-2 cells transduced with the control shRNA lentivirus, HEp-2 cells transduced with the lentiviral shRNAs targeting ATG5 (HEp-2-sh-ATG5), ATG7 (HEp-2-sh-ATG7), and BECN1 (HEp-2-sh-BECN1) showed a reduction of viral gene expression (M, N, L, G, F, M2, and NS1) and RSV F protein expression at the indicated times as well as a decrease in total viral titers at 24 h and 48 h postinfection (Fig. 5D to L). These results further support the conclusion that autophagy plays an important role in promoting RSV replication.
NAC or compound C decreases RSV replication through an autophagy-dependent mechanism.
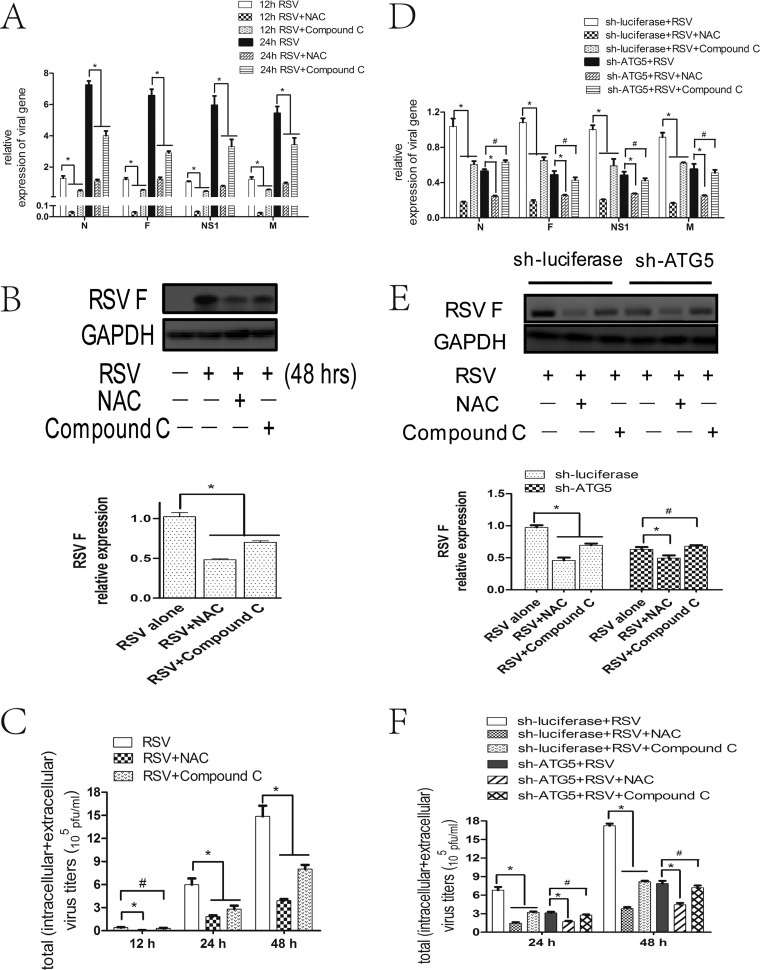
As described above, RSV induced autophagy through the ROS-AMPK-MTOR but not the ROS-MAPK signaling pathways. To further clarify their roles in RSV replication, we tested the effects of NAC, compound C, ERK inhibitor, JNK inhibitor, or p38 inhibitor on viral replication. The results showed that NAC or compound C reduced viral N, F, M, and NS1 gene and RSV F protein expression as well as total viral titers (intracellular and extracellular) in HEp-2 cells, with the former compound having stronger and earlier effects than the latter (Fig. 6A to C). However, ERK/JNK/p38 inhibitor had no effect (data not shown).
FIG 6.
NAC or compound C decreases RSV replication through an autophagy-dependent mechanism. (A, D) HEp-2 (A), HEp-2-sh-ATG5 cells (D), or HEp-2-sh-luciferase cells were infected with RSV (MOI = 5) in the absence or presence of NAC or compound C for indicated times. Relative expressions of RSV targeted genes were determined by fluorescence quantitative PCR. (B, E) HEp-2 (B), HEp-2-sh-ATG5 cells (E), or HEp-2-sh-luciferase cells were treated as described for panel A for 48 h. The expression of RSV F was analyzed with specific antibodies. The relative level of targeted protein was quantitated by densitometry and normalized to GAPDH control. (C, F) HEp-2 (C), HEp-2-sh-ATG5 cells (F), or HEp-2-sh-luciferase cells were treated as described for panel A for indicated times, and total virus titers (intracellular + extracellular) were determined as described in Materials and Methods. Data represent means ± SD for 3 independent experiments. *, P < 0.05; #, P > 0.05.
To further clarify the role of autophagy in the effects of NAC or compound C on RSV replication, we assessed their inhibitory effects in ATG5-deficient HEp-2 cells. The results showed that the inhibitory effects of NAC and compound C on RSV replication disappeared or diminished in ATG5-deficient cells, respectively, indicating that compound C inhibits RSV replication dependent on autophagy, while NAC inhibits RSV replication through both autophagy-dependent and -independent mechanisms (Fig. 6D to F).
Autophagy inhibition decreases cell viability and increases apoptosis of RSV-infected cells at 48 hpi.
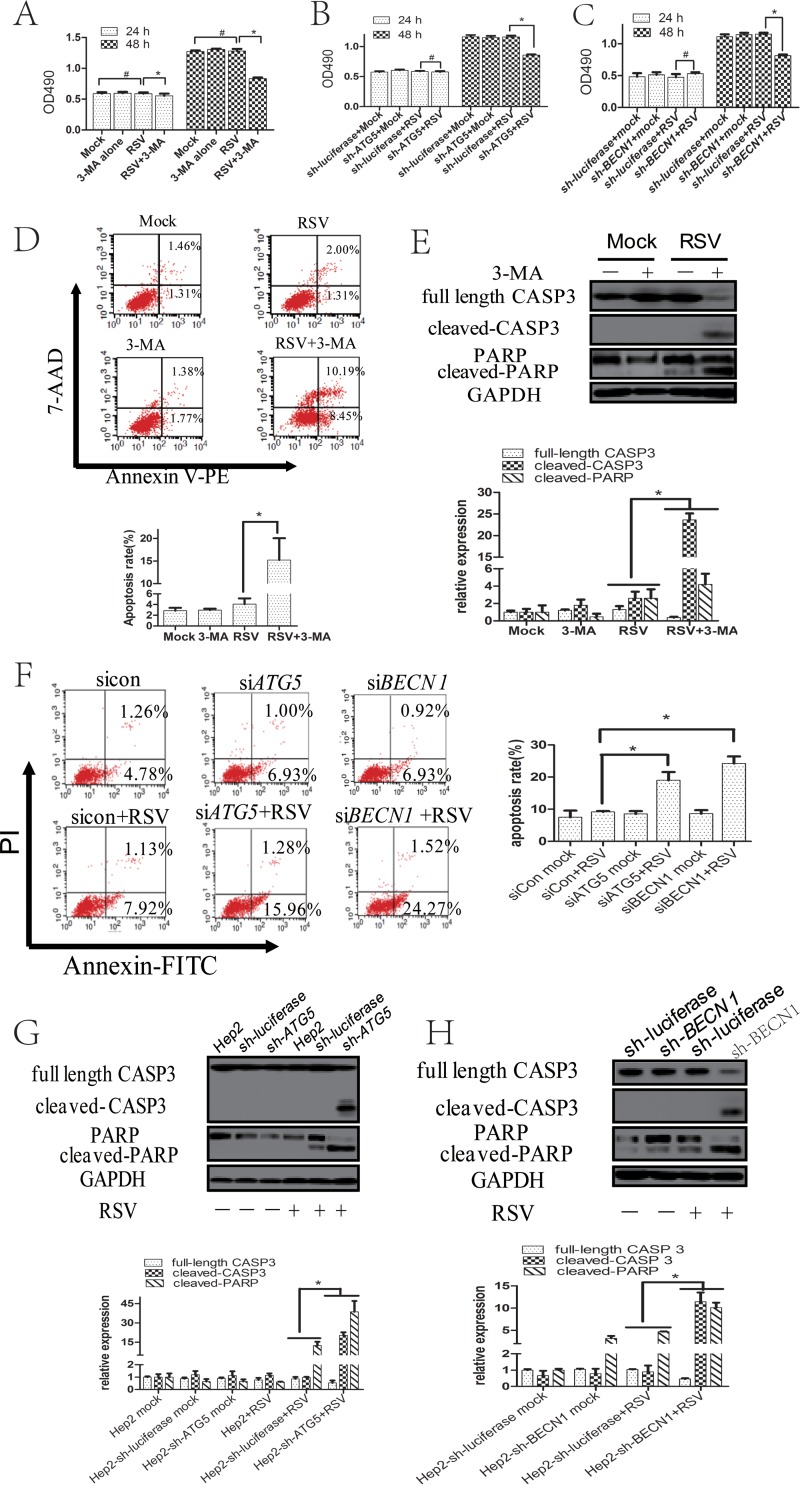
In the above-described experiments, we observed that treatment with 3-MA or shRNAs conferred more floating cells than did lack of treatment at 48 hpi, which led us to explore the effects of 3-MA or shRNAs on cell viability in RSV-infected cells. We performed a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS) assay, and the results showed that 3-MA or sh-ATG5/BECN1 decreased RSV-infected cell viability at 48 hpi but not 24 hpi (Fig. 7A to C). To determine the role of apoptosis, we blocked autophagy with 3-MA or small interfering RNAs (siRNAs) targeting ATG5 or BECN1 and checked the apoptotic rate and the expression of cleaved caspase 3 (CASP3) and cleaved poly(ADP-ribose) polymerase (PARP) in RSV-infected cells. As shown in Fig. 7D and E, 3-MA treatment significantly increased the percentages of apoptosis and the cleavage of CASP3 and PARP at 48 hpi. Similar results were obtained when autophagic activity was inhibited by knocking down ATG5 or BECN1, indicating that autophagy could inhibit apoptosis of RSV-infected HEp-2 cells at 48 hpi (Fig. 7F to H).
FIG 7.
Inhibition of autophagy promotes apoptosis of RSV-infected HEp-2 cells at 48 hpi. (A) HEp-2 cells were uninfected or infected with RSV in the presence or absence of 3-MA for indicated times. Cell viability was determined by MTS assay as described in Materials and Methods. The optical density at 490 nm (OD490) values of sample wells were normalized to that of the blank well. (B, C) HEp-2-sh-ATG5 (B), HEp-2-sh-BECN1 (C), or HEp-2-sh-luciferase cells were uninfected or infected with RSV for indicated times. Cell viability was determined by MTS assay as described in Materials and Methods. The OD490 values of sample wells were normalized to that of the blank well. (D) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for 48 h in the presence or absence of autophagy inhibitor 3-MA. The apoptosis rate was determined by flow cytometry assay using annexin V-PE and 7-aminoactinomycin D (7-AAD) staining as described in Materials and Methods. (E) HEp-2 cells were treated as described for panel D, and the expressions of total CASP3, cleaved CASP3, total PARP, and cleaved PARP were analyzed with their specific antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to the GAPDH control. (F) HEp-2 cells were transfected with negative-control siRNA, ATG5 siRNA, or BECN1 siRNA for 24 h and then uninfected or infected with RSV for another 48 h. The apoptosis rate was determined by flow cytometry assay using annexin V-FITC and PI staining as described in Materials and Methods. (G and H) HEp-2-sh-ATG5 (G), HEp-2-sh-BECN 1 (H), HEp-2-sh-luciferase, or HEp-2 cells were mock infected or infected with RSV (MOI = 5) for 48 h. The expressions of total CASP3, cleaved-CASP3, PARP, and cleaved PARP were analyzed with specific antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to the GAPDH control. *, P < 0.05; #, P > 0.05.
Blocking apoptosis partially reverses the effects of autophagy inhibition on RSV replication at 48 hpi.
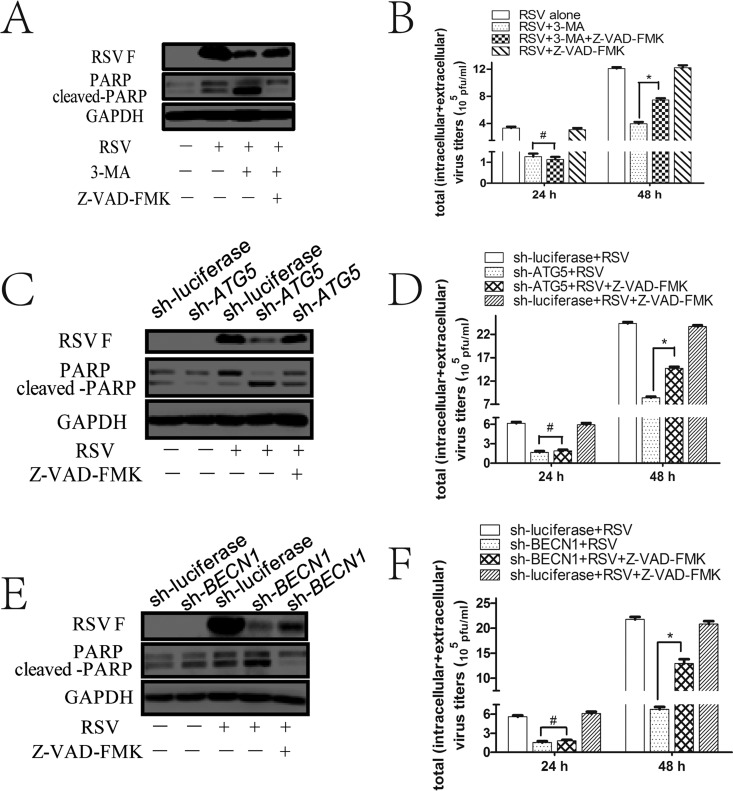
To figure out whether apoptosis is involved in autophagy-mediated viral replication, we blocked apoptosis with a pancaspase inhibitor, carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (Z-VAD-FMK), to see whether it affects virus replication. Interestingly, we found that Z-VAD-FMK did not have any effect on total viral titers at 24 hpi (Fig. 8B, D, and F) but can partially reverse the inhibitory effects of 3-MA or sh-ATG5/BECN1 on RSV F protein expression and total viral titers at 48 hpi (Fig. 8A to F). These results suggest that autophagy-mediated inhibition of apoptosis affects viral replication at 48 hpi only, indicating that other mechanisms may be involved in the proviral role of autophagy at 24 hpi.
FIG 8.
Apoptosis inhibitor can partially reverse the effects of autophagy inhibition on RSV replication at 48 hpi. (A) HEp-2 cells were untreated or pretreated with autophagy inhibitor 3-MA (1 mM) for 2 h and then infected with RSV for another 48 h in the presence or absence of Z-VAD-FMK (50 μM) as described in Materials and Methods. The expression of RSV F, PARP, and cleaved PARP was detected by Western blotting with their specific antibodies. (B) HEp-2 cells were treated as described for panel A for indicated times, and total virus titers were detected as depicted in Materials and Methods. (C and E) HEp-2-sh-ATG5 (C), HEp-2-sh-BECN1 (E), or HEp-2-sh-luciferase cells were untreated or treated with Z-VAD-FMK as described in Materials and Methods during RSV infection for 48 h. The expression of RSV F, PARP, and cleaved PARP was detected by Western blotting with their specific antibodies. (D and F) HEp-2-sh-ATG5 (D), HEp-2-sh-BECN1 (F), or HEp-2-sh-luciferase cells were treated as described for panel C and E for indicated times, and total virus titers were determined as depicted in Materials and Methods. Data represent means ± SD for 3 independent experiments. *, P < 0.05; #, P > 0.05.
RSV subverts mitophagy to reduce cytochrome c release.
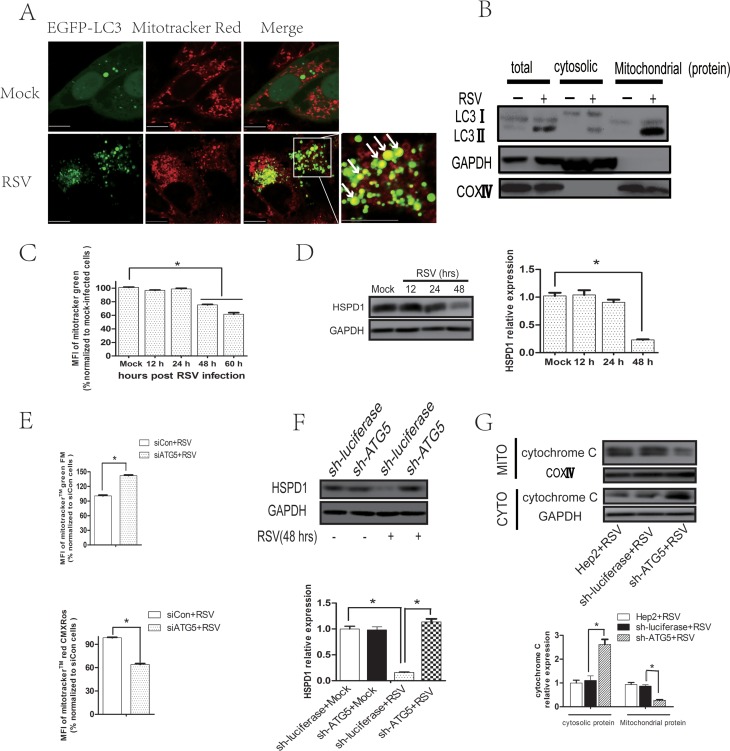
Transmission electron microscopy showed many two- or multilayer membrane structures resembling the mitochondrial structure inside the autophagosome (Fig. 2C). It is well known that mitochondria are at the gateway of the intrinsic mitochondrion-mediated apoptosis pathway. We hypothesized that RSV induced mitophagy, a selective type of autophagy targeting mitochondria to maintain mitochondrial homeostasis and therefore inhibit cell apoptosis. To evaluate mitophagy, we analyzed the localization of EGFP-LC3B and mitochondria labeled with MitoTracker Red CMXRos in RSV-infected cells and found an increase in colocalization of autophagosomes and mitochondria (Fig. 9A). Moreover, we found that enhanced LC3II protein expression was accumulated mainly on mitochondria in RSV-infected cells (Fig. 9B). These observations indicate that RSV induces mitophagy. In agreement with this observation, RSV infection resulted in a decrease of mitochondrial mass analyzed by MitoTracker Green staining (Fig. 9C) and a reduction of expression levels of conservative mitochondrial heat shock protein 60 (HSPD1/HSP60) at 48 h (Fig. 9D). Blocking autophagy by knocking down ATG5 resulted in preservation of HSPD1 expression (Fig. 9F), as well as an increase in total mitochondrial mass (functional mitochondria and dysfunctional mitochondria) and a decrease in functional mitochondrial mass after RSV infection (Fig. 9E). Finally, we observed that the impaired elimination of dysfunctional mitochondria in ATG5 knockdown cells led to an increase in the cytoplasmic cytochrome c level during RSV infection (Fig. 9G), which is a crucial and hallmark event in the mitochondrion-mediated apoptosis pathway (36). These results indicated that RSV infection induces mitophagy, which maintains mitochondrial homeostasis, and therefore decreases cytochrome c release and apoptosis induction.
FIG 9.
RSV subverts mitophagy to reduce cytochrome c release. (A) HEp-2 cells transfected with pBABE-puro-EGFP-LC3 plasmid for 24 h were mock infected or infected with RSV for 48 h. Cells were then stained with MitoTracker Red CMXRos, and colocalization (yellow dots) of mitochondria (red) with autophagosomes (green dots) was observed by confocal microscopy. Bar, 10 μm. (B) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for 48 h. The expression of LC3II, GAPDH, and COXIV of total, cytosolic, or mitochondrial protein was analyzed with their specific antibodies as described in Materials and Methods. (C) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for 12, 24, 48, or 60 h and then stained with MitoTracker Green. Mitochondrial mass was quantified as mean fluorescence intensity, determined by the Tecan Infinite M1000 Pro multimode plate reader, and normalized to that of the mock-infected group. (D) HEp-2 cells were mock infected or infected with RSV (MOI = 5) for 12, 24, or 48 h. The expressions of HSPD1 and GAPDH were analyzed with their specific antibodies as described in Materials and Methods. The relative levels of targeted proteins were quantitated by densitometry and normalized to GAPDH control. (E) HEp-2 cells were transfected with negative-control siRNA or ATG5 siRNA for 24 h and then infected with RSV for another 48 h. Cells were then stained with MitoTracker Red CMXRos and MitoTracker Green. Total (functional + dysfunctional) and functional mitochondrial masses were quantified as mean fluorescence intensity (MFI) of MitoTracker Green or MitoTracker Red CMXRos, respectively. MFI was determined by the Tecan Infinite M1000 Pro multimode plate reader and normalized to that of the control siRNA group. (F) HEp-2-sh-ATG5 or HEp-2-sh-luciferase cells were mock infected or infected with RSV (MOI = 5) for 48 h. The expressions of HSPD1 and GAPDH were analyzed with immunoblotting with specific antibodies. The relative levels of targeted proteins were quantitated by densitometry and normalized to those of the GAPDH control. (G) HEp-2, HEp-2-sh-luciferase, or HEp-2-sh-ATG5 cells were infected with RSV (MOI = 5) for 48 h. The expression of cytochrome c of mitochondrial or cytosolic protein was analyzed with immunoblotting with specific antibodies. The relative level of mitochondrial or cytosolic cytochrome c was quantitated by densitometry and normalized to that of COXIV or GAPDH, respectively. Data represent means ± SD for 3 independent experiments. MITO, mitochondrial protein; CYTO, cytosolic protein. *, P < 0.05.
DISCUSSION
Autophagy plays a key role in pathogenic infection by maintaining the balance between cells and stimuli. Recent studies demonstrate that many RNA and DNA virus infections can induce unidentical autophagy responses. For example, hepatitis C virus (HCV), Sindbis virus, and bluetongue virus (BTV) can enhance autophagy flux (8, 37, 38), while hepatitis B virus (HBV), HIV, respiratory syndrome virus, and herpes simplex virus 1 (HSV-1) inhibit autophagy flux by either inhibiting autophagosome formation or blocking the fusion of autophagosome with lysosome (39–42). Recently, a report even showed that different autophagy flux responses originated from different cells infected with the same strain of rabies virus (43). All these results indicate that autophagy responses are both virus and cell type specific. Recent studies on the relationship between RSV and autophagy focused mainly on the role of RSV-induced dendritic cells (DCs) or macrophage autophagy in regulating DC maturation, cytokine production, and CD4+ T cell responses (44–47). So far, it is not clear whether and how RSV-induced autophagy affects RSV replication. To elucidate this, we first explored the effects of RSV infection on autophagy in vivo. RSV infection resulted in increased LC3II conversion, a decreased SQSTM1 level, and an increased number of autophagosomes in the mice lung, indicating that RSV enhances autophagy flux in vivo. It is worthy of note that the increase in LC3II expression and the decrease in SQSTM1 expression might result from the enhancement of autophagic flux or transcriptional effects of RSV infection. We assessed LC3 or SQSTM1 mRNA levels, and the results showed that they were not regulated at the transcriptional level.
Autophagy inhibition by 3-MA could decrease RSV replication and ameliorate lung pathology in lungs of mice after RSV infection. These results were not consistent with Lukacs lab's findings that autophagy defection led to exacerbated lung pathology resulting from Th2 immune deviation and elevated viral loads. In recent years, researchers have realized that autophagy can exert divergent functions depending on the host microenvironment and stimulus: two genetically different strains of mice (BALB/c and C57BL/6) and two different strains of RSV (strains Long and line 19) elicited different immune responses (48, 49). We used a system different from those used in other studies, which may be the reason why our results are different from theirs. Additionally, we need to be aware that 3-MA is not a specific autophagy inhibitor; further research using mice deficient in key autophagy genes is needed to confirm our findings.
Considering that airway epithelium cells constitute the front line of the host against RSV colonization and replication and that research on the relationship between epithelium cells and autophagy has not been done until now, we used RSV-infected HEp-2 cells as a model to clarify the interaction between virus and autophagy in vitro. In this study, we demonstrated that RSV infection can not only increase autophagosome formation but also enhance autophagy flux in epithelial cells, meaning that RSV affects the whole process of autophagy formation.
To explore the possible mechanisms of RSV-induced autophagy, we found that NAC could remarkably abate RSV-induced LC3II expression and accumulation of EGFP-LC3 puncta, suggesting that ROS plays an essential role in RSV-induced autophagy. ROS is known to be able to induce autophagy, but the precise mechanisms are poorly understood. In this study, we analyzed the signaling pathways involved and found that AMPK-MTOR played a major role in autophagy induction. In addition to MAPK and AMPK-MTOR, we also investigated the PKB-AKT-MTOR signaling pathway, which is considered a key pathway essential for autophagy regulation. However, we did not observe any activation of AKT in the presence or absence of RSV (data not shown), ruling out its possible role in autophagy induction.
To date, the role of autophagy in virus replication has been controversial. Although autophagy commonly serves as a defense mechanism against virus replication, a growing body of evidence pointed out that some viruses have evolved strategies to subvert autophagy in order to favor their replication (50). Our results showed that autophagy inducers and inhibitors can increase and decrease RSV replication, respectively, indicating that autophagy can promote RSV replication. In consideration of nonspecific biological effects of the pharmacological reagents, we verified these results by knocking down essential autophagy-related genes. In addition, we found that NAC or compound C, but not ERK/JNK/p38 inhibitor, reduced viral replication in HEp-2 cells through autophagy-dependent mechanisms. With findings similar to our results, a report has shown that NAC inhibited RSV replication in alveolar II-type epithelial cells (51). These results demonstrate the relationship between ROS, AMPK-MTOR, autophagy, and RSV replication. Importantly, we ruled out the possible effects of chemicals on virus entry and cell viability; both may affect viral replication.
The phenomenon that treatment with 3-MA or shRNAs conferred more floating cells at 48 hpi led us to explore the effects of 3-MA or shRNAs on cell viability. The results showed that treatment with 3-MA or sh-ATG5/BECN1 decreased cell viability in RSV-infected HEp-2 cells only at 48 hpi and not at 24 hpi, which compelled us to probe the role of apoptosis. Apoptosis, a programmed cell death, is another important biological response in host cells during virus infection. Previous studies showed that early-expressed nonstructural proteins NS1 and NS2 of RSV delayed cell apoptosis and boosted viral replication (52). Autophagy and apoptosis, two completely different physiological processes, are closely related. However, the relationship between autophagy, apoptosis, and RSV replication has not been clarified. Our results showed that autophagy inhibition promoted RSV-infected cell apoptosis at 48 h. Z-VAD-FMK can partially reverse the effects of autophagy inhibition on RSV replication at 48 hpi but not 24 hpi. These results indicate that autophagy-mediated inhibition of apoptosis affects viral replication at 48 hpi only and that other mechanisms may be involved in the proviral role of autophagy at 24 hpi. The effects of rapamycin support this conclusion as well. Rapamycin remarkably augmented RSV replication through an autophagy-dependent mechanism. However, rapamycin did not affect cell viability or apoptosis in the cells infected with RSV for 24 h and 48 h (data not shown), indicating that apoptosis may not contribute to the effects of rapamycin on viral replication.
A possible mechanism by which RSV-induced autophagy counteracts apoptosis is that RSV induces mitophagy to eliminate dysfunctional mitochondria and therefore decreases cytochrome c release and apoptosis induction. Generally, autophagy and mitophagy have similar pathways. The autophagy-related genes (ATG) that form a core machinery of autophagy are also essential for mitophagy (53). However, some specific receptors are unique to mitophagy. Further study is required to determine the specific receptors that mediate mitophagy during RSV infection. Besides, other mechanisms may be involved in the proviral role of autophagy at 24 h as described above. Our results showed that autophagy modulation resulted in dramatic changes of almost all RSV gene transcript levels as early as 24 hpi. A likely mechanism is that RSV exploits autophagy for genomic replication and transcription. The processes of RSV replication take place in the context of ribonucleoprotein complexes (RNPs) containing the template RNA associated with nucleoprotein (N) monomers, as well as viral phosphoprotein (P), polymerase, and other accessory proteins (54). The P protein is a factor that is essential for transcription and replication of RSV (54). In this study, we detected subcellular localizations of LC3B and viral phosphoprotein in HEp-2 cells infected with RSV by immunofluorescence assay. The results showed that phosphoproteins were highly colocalized with LC3 (data not shown), indicating that the autophagosome may be involved in viral replication by interacting with RNPs of RSV. More study is needed to determine whether and how RSV exploits autophagy for genomic replication and transcription.
Collectively, this study reveals that autophagy is induced by RSV infection through ROS generation and activation of AMPK-MTOR signaling, which is beneficial for viral replication. Autophagy can inhibit apoptosis of RSV-infected cells to promote virus replication. A possible mechanism by which autophagy inhibits apoptosis is that RSV induces mitophagy to maintain mitochondrial homeostasis and therefore decreases cytochrome c release and apoptosis induction. Our study provides a novel insight into RSV-host interactions and discovery of new antiviral strategies or drugs against RSV infection by targeting autophagy.
MATERIALS AND METHODS
Antibodies and reagents.
The primary antibodies used were anti-LC3B (2775; Cell Signaling), anti-SQSTM1 (5114; Cell Signaling Technology), anti-ATG7 (DF6130; Affinity Biosciences), anti-ATG5 (DF6010; Affinity Biosciences), anti-BECN1 (PD017; MBL), anti-phospho-ERK (Thr202/Tyr204) (4370; Cell Signaling), anti-phospho-JNK (Thr183/Tyr185) (4668; Cell Signaling), anti phospho-p38 (Thr180/Tyr182) (612280; BD), anti-phospho-AMPK (Thr172) (AF3423; Affinity Biosciences), anti-phospho-MTOR (Ser2448) (AF3308; Affinity Biosciences), anti-phospho-ULK1 (Ser317) (12753; Cell Signaling), anti-phospho-ULK1 (Ser555) (5869; Cell Signaling), anti-ULK1 (8054; Cell Signaling), anti-AMPK (AF6423; Affinity Biosciences), anti-MTOR (AF6308; Affinity Biosciences), anti-ERK (BS1112; Bioworld), anti-JNK (BS3630; Bioworld), anti-p38 (ab32142; Abcam), anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; AP0063; Bioworld), anti-PARP (9542; Cell Signaling), anti-CASP3 (14220; Cell Signaling), anti-RSV fusion glycoprotein (anti-RSV F) (ab94968; Abcam), anti-cytochrome c (ab133504; Abcam), anti-HSPD1 (ARG54173; Arigo), anti-COXIV (ab202554; Abcam), anti-RSV phosphoprotein (anti-RSV P; ab94965; Abcam), fluorescein isothiocyanate (FITC)-conjugated anti-RSV nucleoprotein (GTX36623; Gene Tex). Secondary antibodies were horseradish peroxidase (HRP)-labeled goat-anti-mouse (ASS1007; Abgent), HRP-labeled goat-anti-rabbit (ASS1009; Abgent), fluorophore-labeled donkey anti-rabbit IgG (A21206; Invitrogen), and Dylight 594-labeled goat anti-mouse IgG (072-09-18-06; KPL).
The reagents used in the study were N-acetyl-l-cysteine (A7250; Sigma-Aldrich), p38 MAPK inhibitor SB203580 (S1076; Selleckchem), JNK MAPK inhibitor SP600125 (S1460; Selleckchem), ERK MAPK inhibitor PD98059 (S1177; Selleckchem), AMPK inhibitor compound C/dorsomorphin (S7840; Selleckchem), rapamycin (S1039; Selleckchem), bafilomycin A1 (11038; Cayman), 3-methyladenine (S2767; Selleckchem), wortmannin (S2758; Selleckchem), Z-VAD-FMK (S7023; Selleckchem), CellTiter 96 AQueous One Solution kit (G3582; Promega), MitoTracker Red CMXRos (M7512; Invitrogen), MitoTracker Green (M7514; Invitrogen), and mitochondrion isolation kit C1260 (Applygen Technologies Inc.).
Cell lines and virus.
Human laryngeal epithelial cell line HEp-2 from The Institute of Basic Medical Sciences of the Chinese Academy of Medical Sciences (Beijing, China) and HEK 293T cells from Shanghai stem cell bank (Shanghai, China) were maintained in RPMI 1640 medium (31800-022; Gibco) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (P1400; Solarbio), 10 mM HEPES (0511; Biosharp, Amresco), and 10% fetal bovine serum (04-001-1A; Biological Industries) at 37°C with 5% CO2 in a humidified atmosphere. Respiratory syncytial virus A strain Long (The American Type Culture Collection, Rockville, MD, USA) was propagated in HEp-2 cells as previously described (55). Viruses were harvested after 72 to 96 h from the culture supernatant. Viral titers were determined according to the methods described below. RSV was stored at −80°C until use.
Viral infection.
HEp-2 cells were infected with RSV as indicated or mock infected with phosphate-buffered saline (PBS) for 2 h with serum-free RPMI 1640 and shaking every 15 min. After washing with 1× PBS, the cells were cultured with RPMI 1640 medium containing penicillin (100 U/ml), streptomycin (100 μg/ml), and 2% fetal calf serum (FCS) until they were harvested.
Measuring RSV titers in cell lysates and mouse lungs.
RSV titers were determined using a plaque assay (56). Briefly, 10-fold serial dilutions of RSV-infected cell lysates or lung homogenates were incubated with Hep-2 cells in 96-well plates for 2 h with shaking every 15 min. Then, supernatants were removed and replaced with 1% (wt/vol) methylcellulose semisolid culture medium at 37°C in a CO2 incubator for 4 to 7 days. Plaques in Hep-2 monolayers were counted by light microscope. The RSV titer is expressed in PFU/milliliter cell lysates or log10 PFU/gram lung.
ROS assay.
Fluorescent probe DCFH-DA (2′,7′-dichlorodihydrofluorescein diacetate) was utilized to measure intracellular ROS (57). The cells were seeded and infected with RSV for the indicated times. Then, the cells were incubated with 5 μM DCFH-DA for 20 min in the dark, washed three times with PBS, and subjected to fluorescence detection using fluorescence microscopy. Fluorescence intensity was measured using a modular multimode microplate reader (Infinite M1000; Tecan) with excitation and emission wavelengths set at 485 and 525 nm, respectively, to do quantitative analysis. An active oxygen hydrogen donor (48 μM) was used as a positive control.
Western blot analysis.
Mitochondrial and cytoplasmic proteins were separated using a commercially available mitochondrion isolation kit according to the manufacturer's protocol. Total proteins were collected and lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (P0013; Beyotime) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF; BL507A; Biosharp) and phosphatase inhibitors (P1260; Solarbio) on ice for 30 min. The supernatant was collected, and protein concentrations were determined by a NanoDrop 2000c spectrophotometer (EW-83061-12; Thermo Scientific). A total of 30 μg proteins was separated electrophoretically on a 12% SDS-polyacrylamide gel and transferred to a 0.45-μm polyvinylidene difluoride (PVDF) membrane (IPVH00010; Millipore). After being blocked in 5% nonfat milk for 1 h, the membrane was incubated with primary antibodies overnight at 4°C. After 3 10-min washes with Tris-buffered saline (TBS)-Tween, the membrane was incubated with HRP-conjugated secondary antibody for 1 h at room temperature (RT). After washing, the membranes were developed with a Western Lightning plus-ECL reagent (NEL104001EA; PerkinElmer) and detected with a Synoptics Syngene Bioimaging instrument (R114075; Synoptics). The relative expression of proteins was quantified by densitometry using Image J software.
Fluorescence microscopy.
HEp-2 cells were plated on coverslips and transiently transfected with pBABEpuro-EGFP-LC3 plasmid, kindly provided by Jiwu Wei of Nanjing University (22405; Addgene) using Lipofectamine 2000 (11688-019; Invitrogen) for 24 h, followed by RSV infection for another 24 h. Cells were fixed with 4% paraformaldehyde and visualized by an Olympus BX63 fluorescence microscope with an Olympus DP72 color charge-coupled-device (CCD) camera and cellSens standard software to detect autophagic puncta.
Transmission electron microscopy.
HEp-2 cells were seeded in 6-well plates and infected with RSV at a multiplicity of infection (MOI) of 5 for 2 h. After washing with 1× PBS (pH 7.2), cells were incubated for an additional 22 h. Cells were trypsinized and collected by centrifuging at 1,000 rpm for 5 min. The cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4°C. Small parts of the upper right lobe of lung were collected from mice being challenged with PBS or RSV intranasally for 5 days as described below and fixed with 4% glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4°C. Glutaraldehyde-fixed HEp-2 cells or mice lungs were postfixed with 1% osmic acid, dehydrated stepwise with ethanol, and embedded in epoxy resin. Ultrathin sections were cut using a Leica ultramicrotome and stained with uranyl acetate and lead citrate. The above-described procedure was done by the Electron Microscope Center of Hebei Medical College. Cells or lungs were imaged using a Hitachi 7500 transmission electron microscope at 80-kV acceleration voltage. Autophagosomes were defined as double-membrane vacuoles with a diameter of 0.2 to 1.0 μm.
Biochemical intervention.
For detection of autophagic flux, HEp-2 cells were pretreated with 10 nM bafilomycin A1 for 2 h, followed by RSV infection for 24 h in the presence of bafilomycin A1. For autophagy induction and inhibition experiments, HEp-2 cells were pretreated with 100 nM rapamycin, 1 mM 3-MA, or 100 nM wortmannin for 2 h prior to viral infection. Viral adsorption was performed at 37°C for 2 h. The inoculum was removed and washed twice with PBS, and the cells were then incubated in fresh medium containing rapamycin (100 nM), 3-MA (1 mM), or wortmannin (100 nM) until harvesting of the cells. In addition, the same amount of solvent was added to the control group. For apoptosis inhibition, HEp-2 cells were exposed to a general caspase inhibitor, Z-VAD-FMK, in a concentration of 50 μM after virus adsorption for 2 h until harvesting of the cells and supernatants.
shRNA knockdown.
HEp-2 cells stably expressing specific shRNAs against ATG5, ATG7, BECN1, or luciferase gene were established by being transduced with lentiviral particles expressing specific shRNA. Lentiviral particles were packaged by transfecting 293T cells with pSIF-H1-copGFP shRNA Expression Lentivectors (System Biosciences) and packaging vectors using Lipofectamine 2000 according to the manufacturer's instructions. The sequences used in the shRNA targeting ATG5, ATG7, BECN1, and luciferase (the control shRNA) were as follows: 5′-TCATGGAATTGAGCCAATGTT-3′ (58), 5′-GCTGGATGAAGCTCCCAAGGACATT-3′ (59), 5′-GGATGATGAGCTGAAGA GTGTTGAA-3′ (59), and 5′-CTTACGCTGAGTACTTCGA-3′ (60). Western blotting was performed to determine knockdown efficiency.
siRNA knockdown.
Cells were grown to 50% confluence and then transiently transfected with synthetic ATG5 or BECN1 small interfering (siRNA) duplexes (GenePharma) at a concentration of 30 nM using HiperFect Transfection Reagent (301705; Qiagen) according to the manufacturer's instructions. A scrambled siRNA was used as negative control. The silencing efficiency was determined by Western blotting. Twenty-eight hours after transfection, cells were infected with RSV for 48 h as described above to determine the apoptotic rate. Target sites of siRNA duplex were selected according to the methods described in references 58 and 59. The sequences of siRNA duplex used were as follows: ATG5 siRNA (a), 5′AUGGAAUUGAGCCAAUGUUTT3′, and ATG5 siRNA (b), 5′AACAUUGGCUCAAUUCCAUGA3′; BECN1 siRNA (a), ′GGAUGAUGAGCUGAAGAGUGUUGAA3′ (59), and BECN1 siRNA (b), 5′UUCAACACUCUUCAGCUCAUCAUCC3′ (59); scrambled siRNA (a), 5′ UUCUCCGAACGUGUCACGUTT3′, and scrambled siRNA (b), 5′ACGUGACACGUUCGGAGAATT3′.
RNA isolation and quantitative real-time PCR.
Total RNA was extracted from HEp-2 cells or mice lungs using RNAiso Plus Reagent (9109; TaKaRa) according to the manufacturer's protocol. The RNA was reverse transcribed to cDNA using the PrimeScript RT reagent kit with genomic DNA (gDNA) Eraser (047A; TaKaRa). Quantitative real-time PCR (qPCR) was performed with cDNA templates and the PowerUp SYBR green master mix (A25742; Applied Biosystems) and analyzed using the ABI prism 7500 Sequence detection system (Applied Biosystems). The relative expressions of target mRNA were normalized to expression of GAPDH using the 2−ΔΔCT method (where CT is threshold cycle). The primers used for qPCR analysis of RSV amplicons were described in reference 61 and synthetized by Sangon Biotech.
Lung histopathological examination.
Mice lungs were perfused with saline and immediately preserved in 4% paraformaldehyde overnight, dehydrated through a graded alcohol series, embedded in paraffin, cut into five-micrometer-thick sections, and stained with hematoxylin and eosin (H&E). The slides were examined by light microscopy.
Cell viability assay.
Cell viability was assessed by MTS assay [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt; G3582; Promega] according to the manufacturer's instructions. Briefly, cells were seeded in 96-well plates and cultivated for 24 h to adhere. After treatment, cells were incubated with 10 μl CellTiter 96 AQueous One Solution reagent (MTS solution) for 1 h at 37°C in a 5% CO2 incubator. Absorbance was read at a wavelength of 490 nm using the Infinite M1000 PRO microplate reader (Infinite M1000; Tecan).
Apoptosis detection.
Apoptosis was determined by the phycoerythrin (PE)-annexin V apoptosis detection kit (559763; BD Biosciences) or the FITC-annexin V apoptosis detection kit (556547; BD Biosciences) according to the kit guidelines. Briefly, floating and attached cells were collected and resuspended in 1× binding buffer at a concentration of 1 × 106 cells/ml. Then, 100 μl of cell suspension was stained with 5 μl PE-annexin V/FITC-annexin V and 5 μl 7-AAD/propidium iodide (PI) for 15 min at room temperature in the dark. Samples were then analyzed by the FACSCalibur flow cytometer (BD Biosciences) within 1 h to determine the percentages of apoptotic cells (annexin V positive).
Mitochondrial mass assay.
Total mitochondria and functional mitochondria were monitored by fluorescence levels upon staining with 25 nM MitoTracker Green or 20 nM MitoTracker Red CMXRos for 15 min at 37°C, respectively. Mean fluorescence intensity was detected by the Tecan Infinite M1000 Pro multimode plate reader and normalized to mock-infected cells or siCon cells infected with RSV.
Animals and treatment.
Five-week-old female specific-pathogen-free (SPF) BALB/c mice were purchased from the Experimental Animal Center of Hebei Medical University. All mice were housed in temperature-controlled individual ventilated cages (IVC) with 12-h light/12-h dark cycles and were fed with standard chow and sterile tap water. The mice received humane care, and experiments were carried out according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals and with the approval of the Animal Care and Use Committee of Hebei Medical College.
The mice were divided into 3 groups. One group (n = 35) was infected with RSV (5 × 106 PFU per mouse) intranasally. The second group (n = 35) was injected intraperitoneally (i.p.) with the autophagy inhibitor 3-methyladenine (3-MA) prior to RSV intranasal infection, and 3-MA i.p. injection was continued every 2 days (100 mg/kg of body weight) until the mice were sacrificed. The third group was negative-control treated with an equal amount of PBS intranasally (n = 5). The first two groups were randomly divided into 7 subgroups (5 mice/subgroup), and the mice of each subgroup were sacrificed on each day after treatment. LC3II and SQSTM1 expressions in mouse lungs were examined by Western blotting. Viral NS1 and N genes in mouse lungs were detected by qPCR. Lung homogenates were collected to determine viral titers. Autophagosomes and lung pathology in the lung of mice infected with RSV for 5 days were detected by transmission electron microscopy and hematoxylin and eosin (H&E) stain, respectively.
Statistical analysis.
Data are expressed as means ± standard deviations (SD). All experiments were performed in triplicate. SPSS version 16.0 was used for statistical analyses. The significance between two groups was determined using Student's t test. Statistical analyses for multiple groups were performed using one-way analysis of variance (ANOVA). The Mann-Whitney rank sum test was applied for nonnormally distributed data. A P value of <0.05 was considered significant statistically.
ACKNOWLEDGMENTS
We thank Jiwu Wei (Nanjing University, Nanjing, China) for kindly providing pBABEpuro-EGFP-LC3 plasmid. We thank the Department of Electron Microscopy Center, Hebei Medical University, for our TEM work, and we are grateful to Yan Jing, Meng Li, and Zhou Chenming for their help in preparing EM samples and taking and analyzing EM images.
This study was supported by the National Natural Science Foundation of China (grant numbers 31770971, 81671635, and 81500143).
We declare that we have no conflict of interests.
REFERENCES
- 1.Hall CB, Simoes EA, Anderson LJ. 2013. Clinical and epidemiologic features of respiratory syncytial virus. Curr Top Microbiol Immunol 372:39–57. doi: 10.1007/978-3-642-38919-1_2. [DOI] [PubMed] [Google Scholar]
- 2.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. 2009. The burden of respiratory syncytial virus infection in young children. N Engl J Med 360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B. 2010. Autophagy in mammalian development and differentiation. Nat Cell Biol 12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung CS, Taylor GS. 2010. Nuclear shelter: the influence of subcellular location on the processing of antigens by macroautophagy. Autophagy 6:560–561. doi: 10.4161/auto.6.4.11814. [DOI] [PubMed] [Google Scholar]
- 7.Orvedahl A, Sumpter R Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orvedahl A, Macpherson S, Sumpter R, Tallóczy Z, Zou Z, Levine B. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 10.Gorbea C, Makar KA, Pauschinger M, Pratt G, Bersola JL, Varela J, David RM, Banks L, Huang CH, Li H. 2010. A role for Toll-like receptor 3 variants in host susceptibility to enteroviral myocarditis and dilated cardiomyopathy. J Biol Chem 285:23208–23223. doi: 10.1074/jbc.M109.047464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding B, Zhang G, Yang X, Zhang S, Chen L, Yan Q, Xu M, Banerjee AK, Chen M. 2014. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 15:564–577. doi: 10.1016/j.chom.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. Plos Biol 3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. 2012. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem 287:18036–18043. doi: 10.1074/jbc.M111.320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A 104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui KF, Yeung PL, Chiang AK. 2016. Induction of MAPK- and ROS-dependent autophagy and apoptosis in gastric carcinoma by combination of romidepsin and bortezomib. Oncotarget 7:4454–4467. doi: 10.18632/oncotarget.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Zhang T, Sun W, Wang Z, Zuo D, Zhou Z, Li S, Xu J, Yin F, Hua Y, Cai Z. 2016. Erianin induces G2/M-phase arrest, apoptosis, and autophagy via the ROS/JNK signaling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Dis 7:e2247. doi: 10.1038/cddis.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poillet-Perez L, Despouy G, Delage-Mourroux R, Boyer-Guittaut M. 2015. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol 4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewaele M, Maes H, Agostinis P. 2010. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy 6:838–854. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Castro S, Brasier AR, Jamaluddin M, Garofalo RP, Casola A. 2004. Reactive oxygen species mediate virus-induced STAT activation: role of tyrosine phosphatases. J Biol Chem 279:2461–2469. doi: 10.1074/jbc.M307251200. [DOI] [PubMed] [Google Scholar]
- 20.Casola A, Burger N, Liu T, Jamaluddin M, Brasier AR, Garofalo RP. 2001. Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with respiratory syncytial virus. Role in viral-induced interferon regulatory factor activation. J Biol Chem 276:19715–19722. [DOI] [PubMed] [Google Scholar]
- 21.Huang SH. 2010. Inhibitory effect of melatonin on lung oxidative stress induced by respiratory syncytial virus infection in mice. J Pineal Res 48:109–116. doi: 10.1111/j.1600-079X.2009.00733.x. [DOI] [PubMed] [Google Scholar]
- 22.Castro SM, Guerreroplata A, Suarezreal G, Adegboyega PA, Colasurdo GN, Khan AM, Garofalo RP, Casola A. 2006. Antioxidant treatment ameliorates respiratory syncytial virus–induced disease and lung inflammation. Am J Respir Crit Care Med 174:1361–1369. doi: 10.1164/rccm.200603-319OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett H, Mcfadden G. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol 7:160–165. doi: 10.1016/S0966-842X(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 24.Neumann S, Maadidi SE, Faletti L, Haun F, Labib S, Schejtman A, Maurer U, Borner C. 2015. How do viruses control mitochondria-mediated apoptosis? Virus Res 13:45–55. doi: 10.1016/j.virusres.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benedict CA, Norris PS, Ware CF. 2002. To kill or be killed: viral evasion of apoptosis. Nat Immunol 3:1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 26.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. 2007. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 8:741–752. doi: 10.1038/nrn2238,10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 27.Thorburn A. 2008. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis 13:1–9. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. 2007. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 29.Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. 2013. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PloS Pathog 9:e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. 2014. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A 111:6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou W, Jie H, Caisheng L, Goldstein LA, Rabinowich H. 2010. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. 2006. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 33.Wirawan E, Walle LV, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, Rycke RD, Verspurten J, Declercq W. 2010. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green DR. 2000. Apoptotic pathways: paper wraps stone blunts scissors. Cell 102:1–4. doi: 10.1016/S0092-8674(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 37.Ke PY, Chen SSL. 2011. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv S, Xu Q, Sun E, Yang T, Li J, Feng Y, Zhang Q, Wang H, Zhang J, Wu D. 2015. Autophagy activated by bluetongue virus infection plays a positive role in its replication. Viruses 7:4657–4675. doi: 10.3390/v7082838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou T, Jin M, Ding Y, Zhang Y, Sun Y, Huang S, Xie Q, Xu C, Cai W. 2016. Hepatitis B virus dampens autophagy maturation via negative regulation of Rab7 expression. Biosci Trends 10:244–250. doi: 10.5582/bst.2016.01049. [DOI] [PubMed] [Google Scholar]
- 40.Kyei GB, Dinkins CA. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun MX, Huang L, Wang R, Yu YL, Li C, Li PP, Hu XC, Hao HP, Ishag HA, Mao X. 2012. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 8:1434–1447. doi: 10.4161/auto.21159. [DOI] [PubMed] [Google Scholar]
- 42.Lussignol M, Queval C, Bernetcamard MF, Cottelaffitte J, Beau I, Codogno P, Esclatine A. 2013. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol 87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng J, Zhu S, Hu L, Ye P, Wang Y, Tian Q, Mei M, Chen H, Guo X. 2016. Wild-type rabies virus induces autophagy in human and mouse neuroblastoma cell lines. Autophagy 12:1704–1720. doi: 10.1080/15548627.2016.1196315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed M, Morris SH, Jang S, Mukherjee S, Yue Z, Lukacs NW. 2013. Autophagy-inducing protein beclin-1 in dendritic cells regulates CD4 T cell responses and disease severity during respiratory syncytial virus infection. J Immunol 191:2526–2537. doi: 10.4049/jimmunol.1300477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owczarczyk AB, Schaller MA, Reed M, Rasky AJ, Lombard DB, Lukacs NW. 2015. Sirtuin 1 regulates dendritic cell activation and autophagy during respiratory syncytial virus-induced immune responses. J Immunol 195:1637–1646. doi: 10.4049/jimmunol.1500326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris S, Swanson MS, Lieberman A, Reed M, Yue Z, Lindell DM, Lukacs NW. 2011. Autophagy-mediated dendritic cell activation is essential for innate cytokine production and APC function with respiratory syncytial virus responses. J Immunol 187:3953–3961. doi: 10.4049/jimmunol.1100524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokharel SM, Shil NK, Bose S. 2016. Autophagy, TGF-beta, and SMAD-2/3 signaling regulates interferon-beta response in respiratory syncytial virus infected macrophages. Front Cell Infect Microbiol 6:174. doi: 10.3389/fcimb.2016.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howes A, Taubert C, Blankley S, Spink N, Wu X, Graham CM, Zhao J, Saraiva M, Ricciardi-Castagnoli P, Bancroft GJ, O'Garra A. 2016. Differential production of type I IFN determines the reciprocal levels of IL-10 and proinflammatory cytokines produced by C57BL/6 and BALB/c macrophages. J Immunol 197:2838–2853. doi: 10.4049/jimmunol.1501923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore ML, Chi MH, Luongo C, Lukacs NW, Polosukhin VV, Huckabee MM, Newcomb DC, Buchholz UJ, Crowe JE Jr, Goleniewska K, Williams JV, Collins PL, Peebles RS Jr. 2009. A chimeric A2 strain of respiratory syncytial virus (RSV) with the fusion protein of RSV strain line 19 exhibits enhanced viral load, mucus, and airway dysfunction. J Virol 83:4185–4194. doi: 10.1128/JVI.01853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiramel AI, Brady NR, Bartenschlager R. 2013. Divergent roles of autophagy in virus infection. Cells 2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mata M, Morcillo E, Gimeno C, Cortijo J. 2011. N-acetyl-l-cysteine (NAC) inhibit mucin synthesis and pro-inflammatory mediators in alveolar type II epithelial cells infected with influenza virus A and B and with respiratory syncytial virus (RSV). Biochem Pharmacol 82:548–555. doi: 10.1016/j.bcp.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 52.Bitko V, Shulyayeva O, Mazumder B, Musiyenko A, Ramaswamy M, Look DC, Barik S. 2007. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-kappaB-dependent, interferon-independent mechanism and facilitate virus growth. J Virol 81:1786–1795. doi: 10.1128/JVI.01420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanki T, Wang K, Klionsky DJ. 2010. A genomic screen for yeast mutants defective in mitophagy. Autophagy 6:278–280. doi: 10.4161/auto.6.2.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortin J, Martin-Benito J. 2015. The RNA synthesis machinery of negative-stranded RNA viruses. Virology 479-480:532–544. doi: 10.1016/j.virol.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 55.Trudel M, Nadon F, Séguin C, Binz H. 1991. Protection of BALB/c mice from respiratory syncytial virus infection by immunization with a synthetic peptide derived from the G glycoprotein. Virology 185:749–757. doi: 10.1016/0042-6822(91)90546-N. [DOI] [PubMed] [Google Scholar]
- 56.Iqbal M, Lin W, Jabbal-Gill I, Davis SS, Steward MW, Illum L. 2003. Nasal delivery of chitosan–DNA plasmid expressing epitopes of respiratory syncytial virus (RSV) induces protective CTL responses in BALB/c mice. Vaccine 21:1478–1485. doi: 10.1016/S0264-410X(02)00662-X. [DOI] [PubMed] [Google Scholar]
- 57.Myhre O, Andersen JM, Aarnes H, Fonnum F. 2003. Evaluation of the probes 2′,7′-dichlorofluorescin diacetate, luminol, and lucigenin as indicators of reactive species formation. Biochem Pharmacol 65:1575–1582. doi: 10.1016/S0006-2952(03)00083-2. [DOI] [PubMed] [Google Scholar]
- 58.Chen S, Zhou L, Zhang Y, Leng Y, Pei XY, Lin H, Jones R, Orlowski RZ, Dai Y, Grant S. 2014. Targeting SQSTM1/p62 induces cargo loading failure and converts autophagy to apoptosis via NBK/Bik. Mol Cell Biol 34:3435–3449. doi: 10.1128/MCB.01383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. 2008. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat 112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 60.Song X, Yao Z, Yang J, Zhang Z, Deng Y, Li M, Ma C, Yang L, Gao X, Li W. 2016. HCV core protein binds to gC1qR to induce A20 expression and inhibit cytokine production through MAPKs and NF-κB signaling pathways. Oncotarget 7:33796–338808. doi: 10.18632/oncotarget.9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boukhvalova MS, Prince GA, Blanco JC. 2007. Respiratory syncytial virus infects and abortively replicates in the lungs in spite of preexisting immunity. J Virol 81:9443–9450. doi: 10.1128/JVI.00102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]