ZIKV reemerged in recent years, causing outbreaks in many parts of the world. Alarmingly, ZIKV infection has been associated with neurological complications such as Guillain-Barré syndrome (GBS) in adults and congenital fetal growth-associated anomalies in newborns. Host peripheral immune cells are one of the first to interact with the virus upon successful transmission from an infected female Aedes mosquito. However, little is known about the role of these immune cells during infection. In this work, the immune responses of monocytes, known target cells of ZIKV infection, were investigated by high-density transcriptomics. The analysis saw a robust immune response being elicited. Importantly, it also divulged that monocytes prime NK cell activities during virus infection. Removal of monocytes during the infection changed the immune milieu, which in turn reduced NK cell stimulation. This study provides valuable insights into the pathobiology of the virus and allows for the possibility of designing novel targeted therapeutics.
KEYWORDS: NK cells, RNA-seq, Zika virus, immune response, monocytes, transcriptomes
ABSTRACT
Zika virus (ZIKV) has reemerged in the population and caused unprecedented global outbreaks. Here, the transcriptomic consequences of ZIKV infection were studied systematically first in human peripheral blood CD14+ monocytes and monocyte-derived macrophages with high-density RNA sequencing. Analyses of the ZIKV genome revealed that the virus underwent genetic diversification, and differential mRNA abundance was found in host cells during infection. Notably, there was a significant change in the cellular response, with cross talk between monocytes and natural killer (NK) cells as one of the highly identified pathways. Immunophenotyping of peripheral blood from ZIKV-infected patients further confirmed the activation of NK cells during acute infection. ZIKV infection in peripheral blood cells isolated from healthy donors led to the induction of gamma interferon (IFN-γ) and CD107a—two key markers of NK cell function. Depletion of CD14+ monocytes from peripheral blood resulted in a reduction of these markers and reduced priming of NK cells during infection. This was complemented by the immunoproteomic changes observed. Mechanistically, ZIKV infection preferentially counterbalances monocyte and/or NK cell activity, with implications for targeted cytokine immunotherapies.
IMPORTANCE ZIKV reemerged in recent years, causing outbreaks in many parts of the world. Alarmingly, ZIKV infection has been associated with neurological complications such as Guillain-Barré syndrome (GBS) in adults and congenital fetal growth-associated anomalies in newborns. Host peripheral immune cells are one of the first to interact with the virus upon successful transmission from an infected female Aedes mosquito. However, little is known about the role of these immune cells during infection. In this work, the immune responses of monocytes, known target cells of ZIKV infection, were investigated by high-density transcriptomics. The analysis saw a robust immune response being elicited. Importantly, it also divulged that monocytes prime NK cell activities during virus infection. Removal of monocytes during the infection changed the immune milieu, which in turn reduced NK cell stimulation. This study provides valuable insights into the pathobiology of the virus and allows for the possibility of designing novel targeted therapeutics.
INTRODUCTION
Zika virus (ZIKV) gained global attention in 2015 to 2016 when the virus suddenly reemerged in the human population and caused major viral outbreaks across the world with a large disease burden (1). Although ZIKV has been causing sporadic outbreaks since it was first reported in Uganda >60 years ago (2), very little is known about the biology of the virus and the host response to infection. ZIKV is an arthropod-borne flavivirus that causes Zika fever—a disease that for the majority of patients has few or no symptoms (3). However, in severe cases, ZIKV infection may be responsible for neurological complications such as Guillain-Barré syndrome (GBS) in adults (4) and congenital fetal growth-associated anomalies in newborns (5). The host response to ZIKV infection may be one of the main drivers of the different disease phenotypes.
Recent studies have established that ZIKV can infect peripheral blood monocytes (6–9). However, despite ongoing intensive investigative efforts to understand ZIKV-related neuropathogenesis, knowledge regarding the mechanisms of ZIKV infection in peripheral immune cells is lacking. Given that ZIKV is transmitted into the dermis via the bite from a virus-infected mosquito, monocytes would be one of the first immune cells in the blood to interact with the virus when it reaches the circulatory system. Therefore, the interplay between ZIKV and monocytes will be crucial in determining the outcome of infection (10).
This study focused on characterizing the primary ex vivo response of human donor blood monocytes and monocyte-derived macrophages (MDMs) to ZIKV infection. Systematically, RNA sequencing (RNA-seq) was first used to identify and quantify the abundance of host mRNA and characterize viral RNA. This information was subsequently used to map the host response to ZIKV infection in the two different ex vivo cell types. These data also provided insights into the potential adaptation of the virus during viral replication in these cells. Immunophenotyping of peripheral blood cells isolated from patients infected with ZIKV independently was executed to validate the predictions obtained from the differential gene expression analysis. Depletion of CD14+ monocytes in peripheral blood was then performed ex vivo to functionally understand the cross talk between monocytes and priming of NK cells during ZIKV infection. Last, a multiplex assay was carried out to further understand host cell immunoproteomic changes during ZIKV infection. This global analysis of the host immune response provides a novel understanding of the pathobiology of the virus, leading to the possibility of targeted therapeutic interventions in severe cases.
RESULTS
ZIKV targets human peripheral blood monocytes and macrophages.
CD14+ monocytes have been reported to be the main targets of ZIKV during infection (6–9). In this study, human primary CD14+ monocytes were first isolated from fresh peripheral blood mononuclear cells (PBMCs) to enrich this cell type to >90% of the total cell population (Fig. 1A). In addition, isolated monocytes from the same donors were differentiated into monocyte-derived macrophages (MDMs) over 5 days (Fig. 1B). Purified cells were then infected ex vivo with ZIKV, and their permissiveness to ZIKV infection and growth was determined at 24 and 72 h postinfection (hpi) (Fig. 1A). The 24-hpi time point was chosen to represent the acute infection phase, and the 72-hpi time point was chosen to represent a stage by which a substantial host-virus interaction would have taken place (11). Data obtained showed that ZIKV infection of MDMs was more significant than infection of monocytes in all five donors (~40% compared to ~20% at 72 hpi, respectively) (Fig. 1C). A decrease in viral load was observed in the virus-infected MDMs between the two time points, whereas the viral load remained consistent in infected monocytes over time (Fig. 1D). RNA-seq was then performed to assess global mRNA abundance in these cells.
FIG 1 .
Primary human MNCs and MDMs are targets of ZIKV infection. Isolated human primary MNCs and MDMs (2 × 106 cells each) were infected with ZIKV at an MOI of 10 and harvested at 24 and 72 hpi. (A and B) Flow cytometry gating on monocytes (MNCs) (A) and MDMs (B). Gating for positive infection was set using the mock-infected samples. For the dot plots, cells positive for ZIKV antigen (Ag) are shown in red. For the histogram, ZIKV-infected samples (red) were overlaid on mock-infected samples (black). Gating of live cells was performed on single cells. SSC, side scatter; FSC, forward scatter. (C and D) Compiled results for infection (ZIKV Ag) (C) and viral load detected in MNCs and MDMs obtained from five healthy donors (D). All data are presented as means ± standard deviations. **, P < 0.05, by Mann-Whitney U test, two-tailed. Viral load data were not statistically significant between 24 and 72 hpi in MNCs by Mann-Whitney U test, two-tailed.
Genome variation in ZIKV during infection of the peripheral blood.
In order to compare the amounts of virus between the different cell types and determine whether ZIKV underwent genetic diversification during infection, viral sequence reads (obtained from the RNA-seq) were mapped and compared to that of the progenitor virus stock (PF/ZIKV/HPF/2013). These data indicated that for MDMs, 4.53% and 0.43% of total sequence reads (~10 to 140 million reads) mapped to the ZIKV genome at 24 hpi and 72 hpi, respectively, while 24% and 0.8% of sequence reads (~50 to 120 million reads) generated from monocytes mapped to the ZIKV genome at 24 hpi and 72 hpi, respectively.
Due to the inherent error-prone nature of viral RNA replication, nucleotide variants may become established in the viral genome during ZIKV infection in different cell types. To investigate this hypothesis, consensus genome information for each sample and the frequency of minor variants at each nucleotide position in the progenitor stock were determined and compared to the genome of virus present in the infected samples utilizing previously developed workflows (12, 13). The ZIKV consensus genome sequence derived from the progenitor stock was 10,570 nucleotides in length and contained minor variants (as a measure of quasispecies) spread throughout the genome (see Fig. S1A in the supplemental material). Of the 11 valid consensus sequences derived from the virus-infected samples, the virus recovered in cells from five donors (D1 to D5) had the same consensus sequence as the input stock (PF/ZIKV/HPF/2013). However, some donor samples contained viral genomes that had additional nucleotide differences at six different positions (Table S1). These nucleotide differences (Table S1) were visualized as a maximum likelihood phylogenetic tree, where the input stock was used as the reference sample (Fig. S1B). There were only eight high-frequency transition mutations to choose from (log108 = 0.9 [Fig. 2A]), increasing the likelihood of these changes appearing several times. Of these eight transition mutations, six appeared as major variants and thus changed the overall consensus sequence. The nucleotide positions of these six transition mutations (Table S2) indicated that all the changes in the consensus sequence were already present at relatively high frequency as minor variants in the input stock and were subsequently amplified during viral replication. Changes at nucleotide positions 2815 and 4211 were the most common, being found in ~35% of reads mapping to the virus genome. Had these changes been found in ≥50% of reads, they would have been classified as major variants and thus changed the consensus sequence (Table S2).
FIG 2 .
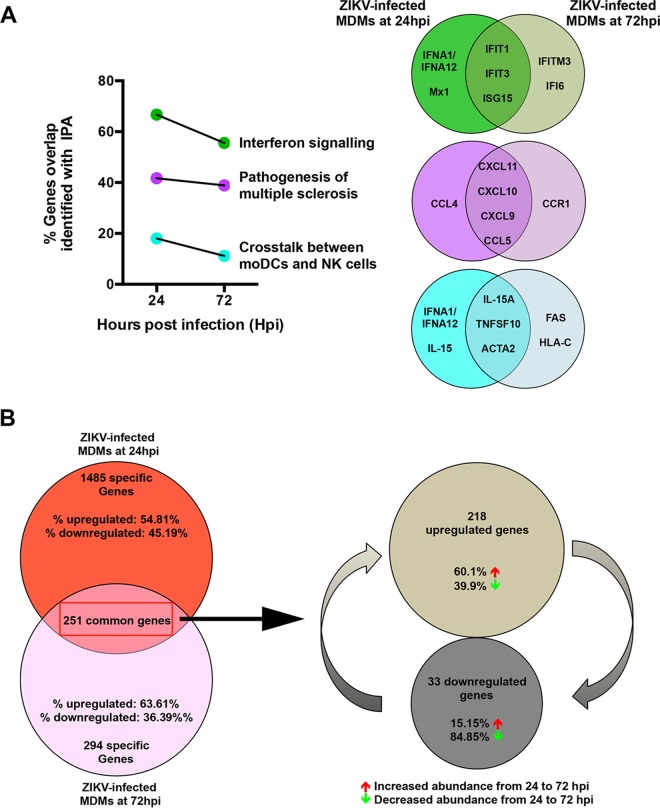
Transcriptomic profiling of host cells during ZIKV infection. Primary human MNCs and MDMs (2 × 106 cells per infection) were infected with ZIKV at an MOI of 10, harvested at 24 and 72 hpi for transcriptomic analysis by RNA-seq, and then compared to mock-infected controls. (A) Venn diagram illustrating the proportion of upregulated signaling pathways identified by IPA in ZIKV-infected MDMs. Upregulation intensities of the 27 common canonical pathways are shown in a heat map. Asterisks within the boxes represent the calculated P values associated with each identified pathway compared to the mock-infected samples. (B) The five most upregulated genes within the top three signaling pathways (interferon pathway, multiple sclerosis pathway, and cross talk between moDCs and NK cells) at 24 and 72 hpi are shown. Data presented were obtained from a total of five donors.
Phylogenetic analyses based on sample consensus sequences. (A) Frequency of ZIKV minor variants (transitions and transversions) recovered from infected human primary MNCs and MDMs isolated from five donors. Bin −3 is where ≤1/1,000 reads show a specific change at an individual nucleotide position. Bin −2 is >1/1,000 and ≤1/100 reads showing a difference. Bin −1 is >1/100 and ≤1/10 reads, and bin >−1 is >1/10 reads showing a change up to a logical limit of just under 1/2. (B) Phylogenetic tree generated from the alignment of consensus sequences of ZIKV RNA recovered from the same samples as described for panel A. All samples included in the tree had a mean sequence coverage of >10 at each nucleotide position. PF/ZIKV/HPF/2013 is the virus strain used for infection and denoted as the reference sample in this analysis. Download FIG S1, PDF file, 0.1 MB (79.2KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of nucleotide differences at specific genome positions. Download TABLE S1, PDF file, 0.1 MB (74.1KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selected nucleotide positions from the minor variant file of the inoculum. Download TABLE S2, PDF file, 0.1 MB (66.7KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptomic profiling reveals key cellular responses to ZIKV infection.
RNA-seq was used to identify and quantify global mRNA abundance in ZIKV-infected peripheral monocytes and MDMs at 24 and 72 hpi. For monocytes, mock- and ZIKV-infected cells at both 24 and 72 hpi exhibited minimal changes in host transcript abundance. For MDMs, the abundances of transcripts that mapped to 1,736 and 545 genes at 24 and 72 hpi, respectively, were significantly different (false discovery rate [FDR] of <0.05) between the mock- and ZIKV-infected samples.
Ingenuity pathway analysis (IPA) was used to interrogate and group the differentially expressed genes into functional pathways (Fig. 2A). Of all the canonical pathways identified, 27 were common in ZIKV-infected MDMs at 24 and 72 hpi (Fig. 2A). This analysis found that genes associated with the interferon response were significantly upregulated at both time points. In addition, signaling pathways involved in the pathogenesis of multiple sclerosis and key pathways involved in monocyte-derived dendritic cell (moDC) and NK cell processes were also shared between the two time points (Fig. 2A). Overall, the top three common pathways activated in MDMs were interferon signaling, multiple sclerosis pathogenic pathways, and cross talk pathways between moDCs and NK cells (Fig. 2A). The specific genes with the most abundant transcripts within these three pathways were analyzed and, compared to the mock-infected controls, were all increased in abundance after ZIKV infection (Fig. 2B).
Virus-infected MDMs exhibit reduced cellular responsiveness.
Transcriptomic profiles of various ZIKV-infected MDMs were compared to evaluate the transition of the cellular host response over the course of ZIKV infection. The percent overlap of the identified transcripts between ZIKV-infected MDMs was assessed at 24 hpi and 72 hpi within the three targeted pathways described above (Fig. 3). Interestingly, the percentage of overlapping transcripts identified at 72 hpi was lower for all three pathways, which may reveal a reduced activation status of these pathways at this stage of the infection. The identification of different transcripts associated with 72 hpi may indicate the different signaling cascades present or the activation status of these cells (Fig. 3A). Global assessment of all identified transcripts revealed that transcripts mapping to 251 genes were in fact present in virus-infected MDMs at both time points. Transcripts that mapped to 1,485 genes were specific to 24 hpi, of which 54.81% exhibited increased abundance compared to the mock controls. By comparison, transcripts that mapped to 294 genes were unique to 72 hpi, with 63.36% of them having increased mRNA abundance compared to the mock controls (Fig. 3B). Within the 251 common genes, transcripts mapping to 218 genes had an increased fold change value compared to the mock-infected controls, indicating that these transcripts were increased in abundance in all ZIKV-infected MDMs. Further inquiry of these transcripts revealed that 60.1% of them were increased in abundance at 72 hpi compared to 24 hpi. Likewise, of the remaining transcripts that mapped to 33 genes and showed decreased abundance, 84.85% were further reduced at 72 hpi.
FIG 3 .
Transition of the host cellular response over the course of ZIKV infection. The host cellular response was analyzed and investigated by RNA sequencing, and significant transcriptomic differences were identified. (A) Transitional analysis (percentage of genes overlapping) of the top three common canonical signaling pathways was performed using IPA of infected MDMs. Venn diagrams indicate the top five common and time-point-specific genes associated with each canonical pathway. (B) Proportion of common and differentially expressed genes within ZIKV-infected MDMs at 24 and 72 hpi. Data presented were obtained from a total of five donors.
NK cells are activated in ZIKV-infected patients.
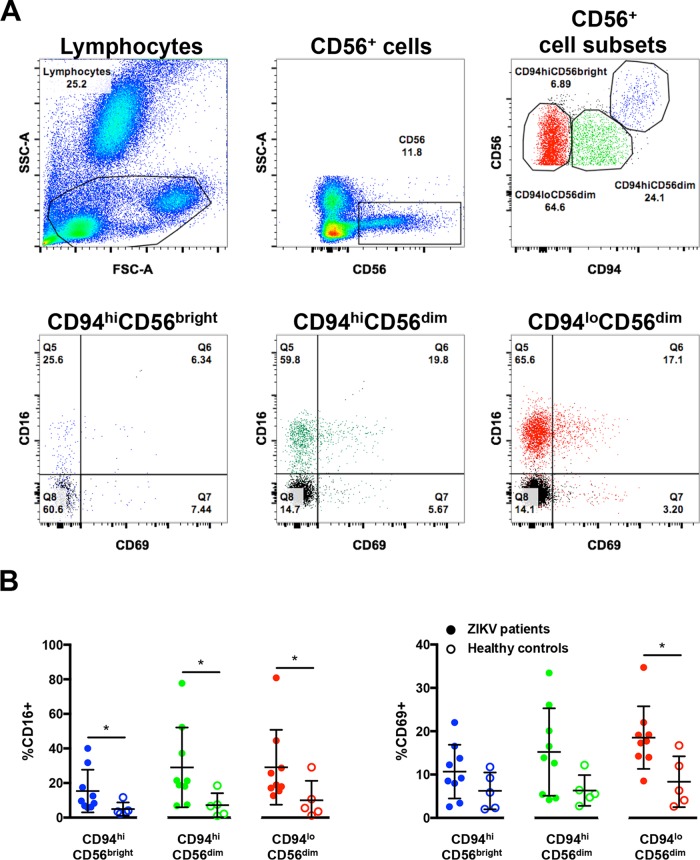
IPA predicted robust cross talk between NK cells and moDCs in peripheral blood upon ex vivo ZIKV infection (Fig. 2 and 3). The IPA prediction that NK cells were activated in the peripheral blood of ZIKV-infected patients was, therefore, investigated by comprehensive immunophenotyping of blood samples taken from ZIKV-infected patients. These patients were recruited from the first endemic ZIKV outbreak in Singapore in 2016 (7, 14). Blood aliquots were obtained from ZIKV-infected patients (n = 9) during the acute disease phase (between 1 and 7 days post-illness onset [PIO]) and were subjected to a whole-blood staining protocol that targeted CD56+ cells, predominantly NK cells (15) (Fig. 4A). Blood from healthy donors (n = 5) was collected and processed in parallel as a control group. Gated cells were further grouped with the C-type lectin receptor CD94, giving three CD56+ populations: CD56bright CD94hi, CD56dim CD94hi, and CD56dim CD94lo (16). The activation status of these populations was then assessed based on the percentage of each subset expressing CD16 and CD69 (Fig. 4B). A higher level of CD16 was observed across all CD56+ subsets in ZIKV-infected patients than in the healthy controls. A higher percentage of the subsets also expressed CD69, a known cellular activation marker (17).
FIG 4 .
Activation of CD56+ cells in patients infected with ZIKV. (A) Gating strategy of CD56+ cell subsets and their expression of CD16 and CD69. Lymphocytes were first gated to exclude the neutrophils. Subsequently, CD56+ cells were identified and further gated into three populations based on the expression of surface marker CD94: CD94hi CD56bright (blue), CD94hi CD56dim (green), and CD94lo CD56dim (red). The data presented correspond to a representative patient infected with ZIKV. Cells from a healthy control are overlaid and depicted as the black population (Q8). CD56 gating was performed on single cells. (B) Compiled data on the percentage of gated subsets that are positive for CD16 (Q5 and Q6) and CD69 (Q6 and Q7). Patients (n = 9) are depicted as filled circles, and healthy controls (n = 5) are depicted as open circles. All data are presented as means ± standard deviations. *, P < 0.05, by Mann-Whitney U test, two-tailed.
CD14+ monocytes prime NK cell activity during ZIKV infection.
Given that peripheral NK cells were activated in ZIKV-infected patients and monocytes are precursors of MDMs, the functional relationship between monocytes and NK cells was assessed. CD14+ monocytes were depleted from human primary PBMCs, with an average efficiency of >95% (Fig. S2A). Lipopolysaccharide (LPS; 10 ng/ml) was used as a positive control to simulate priming of CD56+ CD94+ Lineage− NK cells (Fig. S2B) by monocytes (18). A significant reduction in the activity of NK cells was observed when CD14-depleted PBMCs were stimulated with LPS compared to LPS stimulation of PBMCs containing CD14+ monocytes (Fig. S3A). This effect was evidenced by the reduced levels of the surface markers CD69, CD107a, and intracellular gamma interferon (IFN-γ) in depleted cells, verifying that this approach was an efficient strategy for investigating priming of NK cells by CD14+ monocytes.
Gating strategy of CD56+ CD94+ NK cells. (A) Fresh human PBMCs were subjected to CD14+ monocyte depletion. Flow cytometric plots from one representative donor are shown. A depletion efficiency of >95% was typically obtained. Monocytes are defined as lineage+ cells, with CD14, CD3, CD19, and CD20 included as lineage markers. (B) Live singlets were first gated from the stained PBMCs. CD45+ CD56+ cells were identified, and NK cells were subsequently gated out with CD94 and lineage markers—CD14, CD3, CD19, and CD20. NK cells are defined as CD56+ CD94+ Lineage−. Flow cytometric plots from one representative donor are shown. Download FIG S2, PDF file, 0.5 MB (575.9KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of surface markers by activated NK cells. LPS (10 ng/ml)-stimulated conditions. (A) Compiled percentages of CD69-, CD107a-, and IFN-γ-positive CD56+ CD94+ Lineage− NK cells after LPS (10 ng/ml) stimulation. Expression levels are normalized to the respective mock sample (dotted line). Data shown were derived from seven healthy donors. (B) Total PBMCs and CD14-depleted PBMCs (2 × 106 cells per infection) were infected with ZIKV at an MOI of 10 and harvested at 36 hpi. (B) Compiled percentages of NKG2D and NKG2A-positive CD94+ CD56+ Lineage− NK cells normalized to the respective mock sample. (C) Comparison of the percentage of CD69-positive CD94+ CD56+ Lineage− NK cells between mock-infected and ZIKV-infected full PBMCs and CD14-depleted PBMCs. Data shown were derived from seven donors. Data shown are presented as paired data. (D) The expression levels of CD69, CD107a, and IFN-γ on CD56+ CD94+ Lineage− NK cells at 72 hpi. Expression levels are normalized to respective mock sample. Data were obtained from two donors. Lineage markers CD3, CD19, CD20, and CD14 have been included to rule out the presence of non-NK cells. All data are presented as means ± standard deviations. *, P < 0.05, by Mann-Whitney U test, two-tailed. Download FIG S3, PDF file, 0.2 MB (224.2KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
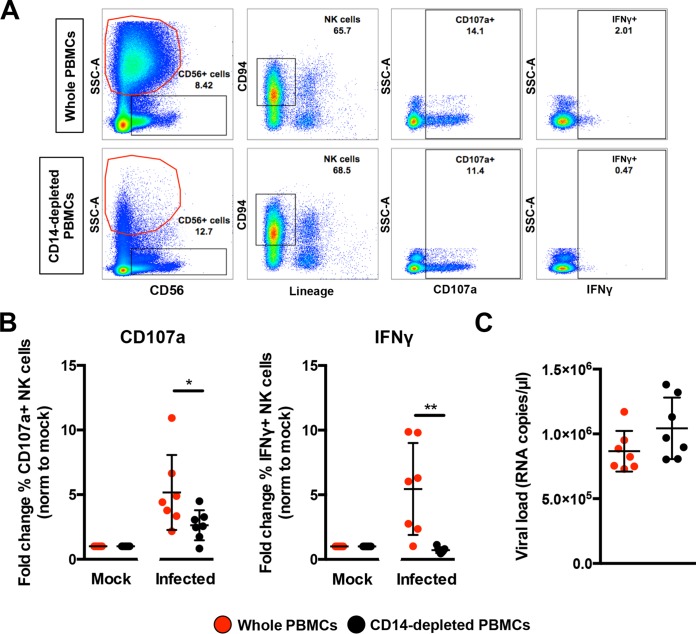
PBMCs were then isolated from seven healthy donors and subjected to CD14 depletion before being either infected with ZIKV or stimulated with LPS in parallel to serve as a control to determine activation of NK cells. ZIKV infection in nondepleted PBMCs resulted in high levels of CD107a and IFN-γ (Fig. 5A) in CD56+ CD94+ Lineage− NK cells at 36 hpi—an optimal time point to detect NK cell priming (19). The opposite effect, however, was observed in ZIKV-infected PBMCs depleted of CD14+ monocytes, as the levels of both CD107a and IFN-γ were significantly reduced (Fig. 5B). Although monocyte depletion did not affect the expression of NK cell activation receptor NKG2A or NKG2D, a general reduction in NKG2D-expressing NK cells was observed during ZIKV infection (Fig. S3B). Surprisingly, the activation marker CD69 was not increased upon ZIKV infection in this study (Fig. S3C). ZIKV viral loads were comparable between the two conditions (Fig. 5C). Interestingly, levels of CD107a and IFN-γ remained high at 72 hpi in nondepleted infected PBMCs compared to depleted infected PBMCs (Fig. S3D).
FIG 5 .
Role of monocytes in NK cell activity. Full PBMCs and CD14-depleted PBMCs (2 × 106 cells per infection) were infected with Zika virus (ZIKV) at an MOI of 10 and harvested at 36 hpi. (A) Gating strategy of CD94+ CD56+ Lineage− NK cells and their expression of CD69, CD107a, and IFN-γ. Plots from one representative donor are shown. The red circle indicates the presence or absence of CD14+ monocytes. (B) Compiled percentages of CD107a- and IFN-γ-positive NK cells (depicted in panel A) as normalized to the respective mock sample. (C) Viral load in the infected cells. Data shown were derived from seven donors. Lineage markers CD3, CD19, CD20, and CD14 have been included to rule out the presence of non-NK cells. All data are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01, by Mann-Whitney U test, two-tailed. Viral load data were not statistically significant between the two conditions by Mann-Whitney U test, two-tailed.
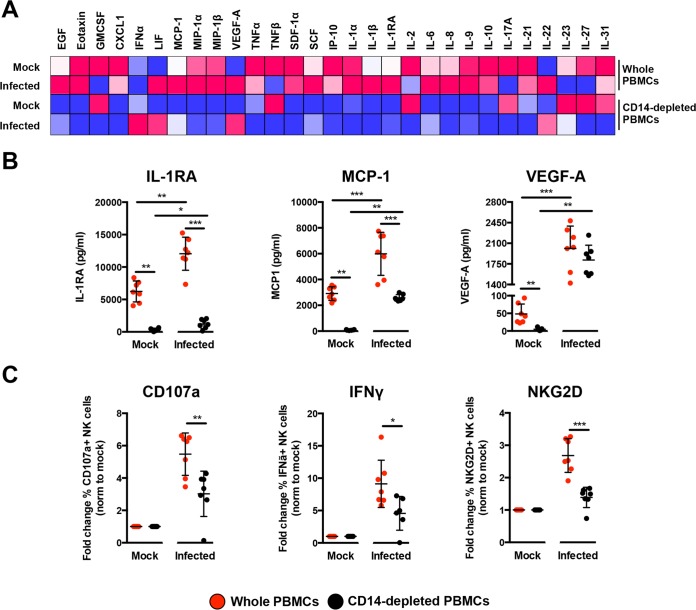
To delve further into the mechanism, the profile of secreted immune mediators from ZIKV-infected PBMCs was quantified using a 45-plex microbead-based immunoassay (20). Levels of 11 mediators were significantly affected by the depletion of CD14+ monocytes (Fig. 6A and S4A), while 8 mediators were affected upon ZIKV infection (Fig. S4B). Interestingly, depletion of CD14+ monocytes and ZIKV infection did not affect the levels of epidermal growth factor (EGF), interleukin 9 (IL-9), IL-17A, macrophage inflammatory protein 1α (MIP-1α), and MIP-1β (Fig. S4C). The effect of CD14+ monocyte depletion was observed in the levels of stem cell factor (SCF) and tumor necrosis factor alpha (TNF-α) only after ZIKV infection (Fig. S4D). Importantly, levels of monocyte chemoattractant protein 1 (MCP-1), IL-1RA, and vascular endothelial growth factor A (VEGF-A) were affected by both CD14+ monocyte depletion and ZIKV infection (Fig. 6B). To further investigate the capacity of the cytokine milieu to prime NK cells, freshly isolated human primary PBMCs were then treated with the same culture supernatants from ZIKV-infected PBMCs and CD14+ monocyte-depleted PBMCs. Stimulation with culture supernatant from ZIKV-infected nondepleted PBMCs led to a significant upregulation in expression of CD107a, IFN-γ, and NKG2D in the CD94+ CD56+ NK cells (Fig. 6C), confirming the importance of monocytes in NK cell priming during ZIKV infection. To rule out priming of NK cells by viruses present in the culture supernatant, a UV treatment procedure was performed to inactivate the virus prior to the stimulation assay. Expectedly, while UV inactivation successfully inactivated ZIKV (Fig. S5A), it also affected the quality of the cytokines and led to reduced priming of NK cells (Fig. S5B).
FIG 6 .
Immune profiling of ZIKV-infected PBMCs. (A) Immune mediators in the culture supernatant of ZIKV-infected PBMCs and CD14-depleted PBMCs were quantified with a 45-plex microbead assay. Concentrations were scaled between 0 and 1. (B) Bar charts of three cytokines, levels of which were significantly affected by both the depletion of CD14+ monocytes and ZIKV infection. (C) Stimulatory capacity of the culture supernatants was further evaluated with freshly isolated PBMCs. Culture supernatant was added in a ratio of 1:10, and cells were harvested at 36 h poststimulation. Compiled percentages of CD107a-, IFN-γ-, and NKG2D-positive CD94+ CD56+ NK cells are shown as normalized to the respective mock sample. Data displayed were derived from seven donors. Lineage markers CD3, CD19, CD20, and CD14 have been included to rule out the presence of non-NK cells. All data are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001, by Mann-Whitney U test, two-tailed.
Quantification of immune mediators. Immune mediators in the culture supernatant of ZIKV-infected PBMCs and CD14-depleted PBMCs were quantified using a 45-plex microbead assay. Quantified immune mediators are grouped into four groups based on their profile: mediators affected by depletion of CD14+ monocytes (A), mediators affected by ZIKV infection (B), mediators not affected by both CD14+ monocyte depletion and ZIKV infection (C), and mediators affected by depletion of CD14+ monocytes only after ZIKV infection (D). Data displayed were derived from seven donors. All data are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001, by Mann-Whitney U test, two-tailed. Download FIG S4, PDF file, 0.5 MB (557.3KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
UV inactivation of ZIKV and proteins. (A) Wild-type (WT) ZIKV was subjected to two different doses (1,000 or 100 mJ/cm2) of UV treatment across different durations. UV-treated ZIKV was subsequently used to infect HEK293T cells, and the amount of viral RNA load was determined at 48 hpi. Levels of viral RNA load are expressed as fold increase relative to the level of viral RNA load detected at 0 hpi with the WT ZIKV. Heat-inactivated (HI) ZIKV was included in parallel as a negative control. (B) Culture supernatants were UV treated (100 mJ/cm2 for 10 min), and their stimulatory capacity was further evaluated with freshly isolated PBMCs in a ratio of 1:10 (with fresh complete medium). Cells were harvested at 36 h poststimulation and evaluated for the percentage of CD107a-; IFN-γ-; and NKG2D-, NKG2A-, and CD69-positive CD94+ CD56+ NK cells. Data shown (n = 2) normalized to the respective mock sample. Lineage markers CD3, CD19, CD20, and CD14 have been included to rule out the presence of non-NK cells. All data are presented as means ± standard deviations. Download FIG S5, PDF file, 0.1 MB (95.6KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Myeloid cells are targets of active ZIKV infection (6–9, 21–23) and can elicit immune responses with detrimental outcomes (6, 8). Both monocytes and macrophages exhibit extensive heterogeneity (24, 25). While it is difficult to obtain tissue-resident macrophages for experimental purposes, human blood is a readily accessible, valuable source of these cells. Transcriptomic profiling of ex vivo human blood monocytes and MDMs has revealed marked differences between these cell types (26, 27). In this study, human primary monocytes were naturally differentiated into MDMs without any bias for an M1 or M2 macrophage phenotype (28). Given that these cells are targets of ZIKV infection (8), investigations into their cellular immune responses during infection will open avenues to exploit their function for therapeutic benefits.
The level of ZIKV infection (as assessed by the amount of ZIKV antigen and genome copy number) was higher in MDMs than monocytes, which corroborates previous observations (8). Transcriptomic differences between monocytes and MDMs (26, 27) would be a plausible explanation for the differential susceptibility of these cells to ZIKV infection. It is also noteworthy that higher ZIKV infection levels were found in purified primary cell populations than in PBMCs, perhaps due to the presence of other immune subsets in PBMCs that may dampen the overall infection level. ZIKV RNA was detected at the two time points, 24 and 72 hpi, and the virus was present as quasispecies postinfection in human primary myeloid cells. The virus consensus sequence and minor variant mapping revealed an overrepresentation of transition mutations at highly variable nucleotide positions in the sequence reads. The proportion of these minor variants indicated a shift toward becoming major variants. A recent study that sequenced ZIKV genomes isolated from infected patients provided important information pertaining to ZIKV transmission (29). These data highlighted the degree of divergence in sequenced genomes and placed further emphasis on understanding virus evolution and transmission effectiveness (30). As not all recovered ZIKV RNA samples contained the same mutations, it will be interesting to determine how different host immune responses can lead to ZIKV quasispecies that acquire different combinations of mutations.
ZIKV infection led to the differential abundance of host transcripts mapping to numerous cellular genes in MDMs but not in monocytes, likely due to higher levels of infection observed in MDMs. Furthermore, it has been reported that different donors could account for significant differences in cellular responses (31, 32). However, this differential effect does not necessarily signify that ZIKV-infected monocytes do not elicit any cellular response to infection but rather that the differences were not measurable by RNA-seq at the read depths used in this analysis. In fact, transcript abundances of numerous genes were different between the mock- and ZIKV-infected monocytes; however, the statistical threshold of an FDR of <0.05 was not reached and thus these findings were excluded from further analyses. Using IPA data mining, these differentially expressed genes were involved in 133 and 63 canonical cellular pathways (27 of them being shared) in MDMs at 24 and 72 hpi, respectively. The reduced number of cellular pathways identified in ZIKV-infected MDMs at the later 72-hpi time point suggests that certain cellular functions may be shut down postinfection (33). This effect could signify that (i) the host cells conserve energy to focus only on essential pathways for survival and/or (ii) the host cells have succumbed to ZIKV infection, which leads to transcriptional shutdown in host cells.
Unsurprisingly, the IFN response was the most highly expressed signaling pathway of these common pathways at both time points because of the virus trigger (34). This observation was further complemented by the presence of few other IFN-related pathways. Observations were found for the next two most expressed pathways—pathogenesis of multiple sclerosis and cross talk between NK cells and moDCs—both of which involve NK cells. Although ZIKV infection has not been previously associated with multiple sclerosis due to the relatively new disease spectrum, other viral infections such as those with Epstein-Barr virus (35) and measles virus (36) have been linked.
CXCL9, CXCL10, CXCL11, and CCL5 (identified as the top genes in the pathway) are chemokines known to stimulate NK cell activation (37, 38). The increased transcript abundance of these immune mediators, coupled with others such as IL-15, is a strong indication that ZIKV-infected macrophages are primed to “communicate” with NK cells. Other recent studies have also provided evidence of cross talk between macrophages and NK cells (18). The increased abundance of TNFSF10 and Fas transcripts in ZIKV-infected MDMs could indicate priming of NK cell-mediated apoptosis (39). Interestingly, levels of typical NK cell-activating cytokines, such as IL-12 (40, 41) and IL-18 (42, 43), were not differentially expressed in this study. However, mRNA levels of IL-23 and IL-27, two cytokines belonging to the family of IL-12 (44) with roles in NK cell activation (45, 46), were increased.
Immunophenotyping of whole-blood samples from ZIKV-infected patients revealed the presence of CD69+ CD56+ immune cells (predominantly the CD56+ NK cells) (15), suggesting the possible priming of NK cells in ZIKV infection. Similar observations were also reported in dengue virus (DENV) patients (47), as well as in volunteers vaccinated with the yellow fever virus (YFV) vaccine (48). The involvement of NK cells was thus explored ex vivo in human primary PBMCs. Interestingly, ex vivo culture alone led to an increase in the basal expression level of CD69 in CD56+ CD94+ NK cells, as previously reported (49). Furthermore, ZIKV infection resulted in reduced levels of CD69, which is a phenomenon also reported for the flavivirus tick-borne encephalitis virus infection in healthy donor NK cells (50). Moreover, NK cells behave differently ex vivo and in vivo (51), which may explain the different levels of CD69 detected in patients and in ex vivo ZIKV-infected NK cells. It was also reported in CD69-deficient mice that the activity of NK cells remains functional (52). High levels of key NK activation markers, including the degranulation marker CD107a and intracellular cytokine IFN-γ, indicate the higher activation status of NK cells. The activity of NK cells was directly dependent on the presence of CD14+ monocytes. ZIKV infection of PBMCs depleted of CD14+ monocytes significantly downregulated the expression of the various NK cell markers, demonstrating the functional role of monocytes as one of the key players for NK cell stimulation. The data presented in this study are further supported by a recent publication in which ZIKV patients had high levels of IL-18, TNF-α, and IFN-γ (20)—immune mediators associated with NK cell function. The use of SJL mice, which lack NK cells (53), as a model of ZIKV infection also suggested a protective role for these immune cells given that these animals succumbed to cortical malformations (54). Likewise, the NK cell-mediated immune response was significantly increased in healthy volunteers receiving a YFV vaccination (55). Thus, the role of NK cells during ZIKV infection should be explored.
Interestingly, multiplex quantification of secreted immune mediators from ex vivo ZIKV-infected PBMCs provided an alternate perspective. IL-18 and IFN-γ, two NK cell-related cytokines, were below the detection limit. However, freshly isolated PBMCs stimulated with culture supernatants from ZIKV-infected PBMCs resulted in increased priming of NK cells, clearly indicating that the concoction of immune mediators is capable of driving NK cell activation.
Nonetheless, the, depletion of CD14+ monocytes would abrogate this activation as observed by the low levels of MCP-1, IL-1RA, VEGF-A, eotaxin, growth-related oncogene alpha (GROα), IFN-α, stromal cell-derived factor 1α (SDF-1α), IFN-γ-induced protein 10 (IP-10), IL-6, IL-1α, IL-1β, IL-8, IL-21, and IL-10. The reduced levels of MCP-1 could also have a detrimental effect on NK cell recruitment and priming (37, 56), although MCP-1 and VEGF-A have been reported to drive the production of each other (57–59). The high levels of secreted IL-1RA from ZIKV-infected PBMCs could also have participated in the increased priming of NK cells, as IL-1RA is known to potentiate the effect of IL-2 stimulation of NK cells (60). Thus, the loss of detectable IL-2 after ZIKV infection in CD14-depleted PBMCs would further reduce NK cell priming. The presence of other immune mediators, such as IL-6, IL-8, IL-10, IP-10, SDF-1α, GROα, IL-1α, and IL-1β, in ZIKV-infected nondepleted PBMCs would further provide an inflammatory condition for cellular activation. While the levels of these immune mediators have been reported to be high in ZIKV patients (20), IL-10 and IP-10 have been demonstrated to contribute to cytolysis and activation of NK cells (37, 61). Levels of leukemia inhibitory factor (LIF) (62), IL-22 (63), and IL-31 (64) were high upon ZIKV infection, indicating their roles in regulating T cells during ZIKV infection (65). T cells could regulate NK cell activity (66), and monocytes could indirectly mediate NK cell functions through the T lymphocytes. Though it was not exhibited in this work, monocytes are known to physically “communicate” with T cells, regulating their activities in the process (67). This is thought to occur via the interaction between CD80/86 and CD28 expressed on the infected monocytes and T cells, respectively (68). Likewise, monocytes could also activate NK cells via physical interactions (69). Interestingly, it was recently reported that NK cells were activated by monocyte-derived dendritic cells in a contact-dependent manner during DENV infection (70). This further highlights the complexity of immune cell interactions in different contexts.
To conclude, through a systematic investigative workflow combining approaches exploring host cell transcriptomes and immunoproteomes, it was demonstrated that monocytes and macrophages do not act alone but in conjunction with other immune cells to orchestrate a series of host immune responses and drive disease progression. As such, a comprehensive understanding of immune cell interaction will have important clinical implications for the design of novel therapeutics that can either dampen down or enhance a response as appropriate.
MATERIALS AND METHODS
Ethics approval and consent to participate.
Whole-blood samples were collected from ZIKV-infected patients who were referred to the Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. Blood was obtained from patients who provided written informed consent. The study protocol was approved by the SingHealth Centralized Institutional Review Board (CIRB; reference no. 2016/2219). Blood samples were collected from healthy donors with written consent in accordance with guidelines from the Health Sciences Authority of Singapore (study approval number NUS IRB 10-250).
Patient whole-blood samples.
This study utilized whole-blood samples obtained from patients (n = 9) admitted to the Communicable Disease Centre at Tan Tock Seng Hospital, Singapore, from 27 August to 18 October 2016. Samples included in this study were collected during the acute phase (1 to 7 days post-illness onset [PIO]) of ZIKV infection. These patients were confirmed to be infected with ZIKV by reverse transcription-PCR (RT-PCR) performed on serum and urine samples obtained during their first visit to the clinic. Patients were screened negative for DENV exposure by RT-PCR and serology. Whole-blood samples were collected in EDTA Vacutainer tubes (Becton, Dickinson). Whole-blood samples were also obtained from healthy volunteers (n = 5) as controls, which were confirmed to be negative for ZIKV RNA by RT-PCR.
Virus preparation.
The ZIKV strain (GenBank accession number KJ776791.2) used in this study was originally isolated from the French Polynesia outbreak in 2013 (71). The virus was propagated as previously described (8). Briefly, the virus was propagated by multiple passages in Vero-E6 cells (ATCC; CRL-1587) and precleared by centrifugation before storage at −80°C. The ZIKV stock contained approximately 1.2 × 107 PFU/ml of infectious virus when titrated on Vero-E6 cells and approximately 3.5 × 109 ZIKV viral RNA copies/ml by the quantitative RT-PCR method described below. The virus titer was determined using standard plaque assays with Vero-E6 cells. Vero-E6 cells were regularly tested for mycoplasma contamination and were grown and passaged in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) supplemented with 10% (vol/vol) fetal bovine serum (FBS). UV inactivation of ZIKV was performed with the CL-1000 UV cross-linker (UVP) at an intensity of 100 mJ/cm2 for 10 min.
Isolation and depletion of monocytes from human PBMCs.
Monocytes were prepared from fresh human PBMCs as previously described (8) and by gradient centrifugation using Ficoll-Paque density gradient medium (GE Healthcare). Subsequently, monocytes were isolated using an indirect magnetic labeling system (monocyte isolation kit II; Miltenyi Biotec). A direct magnetic labeling system (human CD14+ monocyte isolation kit 2; StemCell) was used for depletion of monocytes from PBMCs. The manufacturers’ protocols were strictly adhered to for these procedures.
Differentiation of monocytes into MDMs.
Isolated monocytes were differentiated into MDMs by plating in complete Iscove’s modified Dulbecco’s medium (IMDM) (HyClone) supplemented with 10% (vol/vol) heat-inactivated human serum (HS) (Sigma-Aldrich), which was replaced every 2 days. ZIKV infections were performed on monocytes and MDMs 5 days later, as described below.
Virus infection.
ZIKV infections in monocytes and MDMs (n = 5 each) were performed at a multiplicity of infection (MOI) of 10. Each infection mixture consisted of a virus suspension prepared in serum-free IMDM (HyClone). The cells were incubated with the infection mixture at 37°C and allowed to adsorb for 2 h with intermittent shaking before the virus inoculum was removed and replaced with complete IMDM supplemented with 10% (vol/vol) HS (Sigma-Aldrich). Cells were incubated at 37°C until harvesting at 24 and 72 hpi. The harvested cells for downstream total RNA isolation were stored at −80°C. A total of 140 µl of the infected-cell suspension was used to quantify the viral load. For assessment of monocyte function in NK cell activation during ZIKV infection, total human PBMCs (n = 7) and donor-corresponding CD14-depleted PBMCs (n = 7) were infected with ZIKV at an MOI of 10. In parallel, both PBMC fractions were stimulated with 10 ng/ml lipopolysaccharide (LPS; Sigma) as a positive control to measure NK cell activation. Cells were subsequently treated with 1× brefeldin (eBioscience) and stained with CD107a (BD Pharmingen) 6 h before harvesting at 36 hpi. The viral load was quantified from 140 µl of the infected-cell suspension. Negative controls (n = 7) were cells undergoing the same infection conditions in the absence of infectious ZIKV particles. These controls are referred to as mock-infected samples.
PBMC stimulation assay.
Fresh PBMCs were isolated as described above and subjected to stimulation with ZIKV-infected culture supernatants (n = 7) in a final ration of 1:10 in fresh IMDM (HyClone) supplemented with 10% (vol/vol) HS (Sigma-Aldrich). Cells were subsequently treated with 1× brefeldin (eBioscience) and stained with CD107a (BD Pharmingen) 6 h before harvesting at 36 h for downstream antibody staining.
Viral RNA extraction and viral load analysis.
Viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen), according to the manufacturer’s instructions. Quantification of ZIKV NS5 RNA was determined by quantitative real-time PCR (qRT-PCR) TaqMan assay (72) using a QuantiTect Probe RT-PCR kit (Qiagen) in a 12.5-µl reaction volume. All reactions were performed on a 7900HT Fast real-time PCR system machine (Applied Biosciences).
Total RNA extraction.
Total RNA was extracted using an RNeasy minikit (Qiagen) according to the manufacturer’s instructions. The extracted total RNA was quantified on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific).
Flow cytometry and antibodies.
Detection of ZIKV antigen was carried out in a two-step indirect intracellular labeling process. Briefly, harvested cells were first fixed and permeabilized with fluorescence-activated cell sorting (FACS) lysing solution (BD Biosciences) and FACS permeabilization solution 2 (BD Biosciences), respectively. Antigen staining was then performed with a flavivirus-specific mouse monoclonal antibody (clone 4G2) (Millipore) followed by secondary staining with a goat anti-mouse IgG F(ab′)2 antibody (Invitrogen). Cells were then specifically stained for the surface markers CD45 and CD14 (for ZIKV-infected monocytes and MDMs). Dead cells were excluded by staining with the LIVE/DEAD Fixable Aqua dead cell stain kit (Life Technologies). For PBMCs, surface markers CD45, CD14, CD3, CD19, and CD56 were stained prior to intracellular staining (for ZIKV-infected PBMCs). For patient samples, 100 μl of whole blood was stained for the surface markers CD56, CD94, CD16, CD69, CD107a, NKG2D, and NKG2A. The stained cells were subsequently incubated with FACS lysing solution (BD Biosciences) to lyse the red blood cells. Lymphocytes were first gated to exclude the neutrophils. Subsequently, CD56+ cells were first identified and further defined with the CD94 surface marker to give three other subsets—CD56bright CD94hi, CD56dim CD94hi, and CD56dim CD94lo (16). To specifically assess NK cell activity ex vivo, PBMC fractions were stained for CD107a and various lineage markers (CD3, CD19, CD20, and CD14) (15) in addition to the panel of antibodies used for patient whole-blood staining. The usage of lineage markers excludes the presence of non-NK cells in the ensuing analysis. Stained PBMCs were fixed and permeabilized as described above before intracellular staining of ZIKV antigen and IFN-γ. All gatings were performed on single cells.
All antibodies used were mouse anti-human antibodies and were obtained from BD Pharmingen (CD3, CD19, CD20, CD14, CD69, CD56, CD94, NKG2D, CD107a, and IFN-γ), BioLegend (CD16 and CD45), and Miltenyi Biotec (NKG2A). Data were acquired on a Fortessa flow cytometer (BD Biosciences) with BD FACSDiva software. Data analysis was performed using FlowJo version 9.3.2 software (Tree Star, Inc.).
Cytokine quantification using microbead-based immunoassay and data analyses.
Cytokine levels in supernatant obtained from mock- and ZIKV-infected PBMCs were measured simultaneously using the ProcartaPlex immunoassay (Thermo Fisher Scientific) detecting 45 secreted cytokines, chemokines, and growth factors, including brain-derived neurotropic factor (BDNF); eotaxin/CCL11; epidermal growth factor (EGF); fibroblast growth factor 2 (FGF-2); granulocyte-macrophage colony-stimulating factor (GM-CSF); growth-related oncogene alpha (GROα)/CXCL1; hepatocyte growth factor (HGF); nerve growth factor (NGF) beta (73); leukemia inhibitory factor (10); alpha interferon (IFN-α); IFN-γ; interleukin-1β (IL-1β); IL-1α; IL-1RA; IL-2; IL-4; IL-5; IL-6; IL-7; IL-8/CXCL8; IL-9; IL-10; IL-12p70; IL-13; IL-15; IL-17A; IL-18; IL-21; IL-22; IL-23; IL-27; IL-31; gamma interferon-induced protein 10 (IP-10)/CXCL10; monocyte chemoattractant protein (MCP-1/CCL2); macrophage inflammatory protein 1α (MIP-1α)/CCL3; MIP-1β/CCL4; regulated on activation, normal T cell expressed and secreted (RANTES)/CCL5; stromal cell-derived factor 1α (SDF-1α)/CXCL12; tumor necrosis factor alpha (TNF-α); TNF-β/lymphotoxin alpha (LTA); platelet-derived growth factor (PDGF)-BB; placental growth factor (PLGF); stem cell factor (SCF); vascular endothelial growth factor A (VEGF-A); and VEGF-D. Preparation of samples and reagents and immunoassay procedures were performed according to manufacturers’ instructions. Data were acquired using a Luminex FlexMap three-dimensional (3D) instrument (Millipore) and analyzed using Bio-Plex Manager 6.0 software (Bio-Rad) based on standard curves plotted through a five-parameter logistic curve setting. Levels of BDNF, FGF-2, HGF, nerve growth factor (NGF), IFN-γ, IL-4, IL-5, IL-7, IL-12p70, IL-13, IL-15, IL-18, RANTES, PDGF-BB, PLGF, and VEGF-D were below detection limit and excluded for further analysis. Hierarchical clustering was done using TM4-MeV (http://mev.tm4.org/).
RNA-seq and differential gene expression analysis.
The general approach to RNA-seq and differential expression has been previously described (10, 74) and is detailed in brief below.
RNA-seq.
RNA samples were treated with DNase using an Ambion Turbo DNA-free kit (Ambion) and then purified using AMPure XP beads (Agencourt). The DNase-treated RNA (2 µg) underwent Ribo-Zero treatment using an Epicentre Ribo-Zero Gold kit (human/rat/mouse) (Epicentre) and was repurified on AMPure XP beads. Successful RNA depletion was verified using a Qubit (Thermo Fisher Scientific) and an Agilent 2100 Bioanalyzer (Agilent), and all of the depleted RNA was used as input material for the ScriptSeq v2 RNA-seq library preparation protocol. RNA was amplified for 14 cycles, and the libraries were purified on AMPure XP beads. Each library was quantified using Qubit, and the size distribution was assessed using the AATI fragment analyzer (Advanced Analytical). These final libraries were pooled in equimolar amounts using the Qubit and fragment analyzer data. The quantity and quality of each pool were assessed by the fragment analyzer and subsequently by quantitative PCR (qPCR) using the Illumina library quantification kit (Kapa Biosystems) on a Light Cycler LC480II (Roche) according to the manufacturer’s instructions. The template DNA was denatured according to the protocol described in the Illumina cBot user guide and loaded at a 12 pM concentration. Sequencing was carried out on three lanes of an Illumina HiSeq 2500 with version 4 chemistry, generating 2- by 125-bp paired-end reads.
Bioinformatics analysis.
Briefly, base calling and demultiplexing of indexed reads were performed using CASAVA version 1.8.2 (Illumina) to produce 30 samples from the five lanes of sequence data in fastq format. The raw fastq files were trimmed to remove the Illumina adapter sequences using Cutadapt version 1.2.1 (75). The option “-O 3” was set so that the 3′ end of any read that matched the adapter sequence by ≥3 bp was removed. The reads were further trimmed to remove low-quality bases using Sickle version 1.200 with a minimum window quality score of 20. After trimming, reads of <50 bp were removed. If both reads from a pair passed this filter, each read was included in the R1 (forward reads) or R2 (reverse reads) file. If only one read of a read pair passed this filter, it was included in the R0 (unpaired reads) file. The reference genome used for alignment was the human reference genome assembly GRCh38, downloadable from the Ensembl ftp site. The reference annotation was downloaded from the Ensembl ftp site (ftp://ftp.ensembl.org/pub/release-77/gtf/homo_sapiens/Homo_sapiens.GRCh38.77.gtf.gz). The annotated file contained 63,152 genes. R1/R2 read pairs were mapped to the reference sequence using TopHat2 version 2.1.0 (76), which employs the mapper Bowtie 2 version 2.0.10 (77).
Differential gene expression and functional analysis.
Mapped reads were further analyzed using edgeR version 3.3 (78) to calculate normalized counts per million (CPM), identify genes differentially expressed between infected and mock-infected conditions, and compare infected conditions with each other. Correlation and principal-component analysis (PCA) plots were created in RStudio. Heat maps were generated using GENE-E (Broad Institute; https://software.broadinstitute.org/GENE-E/). IPA was used for gene ontology and pathway analysis. The P value associated with each identified canonical pathway was calculated by Fisher’s exact test (right-tailed). The presence of the 27 common canonical pathways was illustrated in a heat map generated by hierarchical clustering using TM4-MeV (79).
Identification of ZIKV variants.
Bowtie 2 (77) was used to determine the mean sequence coverage. Here, 12 of the 41 samples (including the inoculum) had a mean coverage of >10 following alignment with the ZIKV reference genome (GenBank accession number KJ776791.2) used in this study. The frequencies of minor variants were calculated using QuasiRecomb (80). Sequences of individual viral proteins were compared to the Protein Data Bank using the online NCBI Protein BLAST server (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins).
ACKNOWLEDGMENTS
We thank Siti Naqiah Amrun, Yiu-Wing Kam, Jonathan Cox, Yi-Hao Chan, Guillaume Carissimo, Farhana Abu Bakar, Nicholas Q. R. Kng, Kia-Joo Puan, and Nurhashikin Binte Yusof from SIgN for their help in the processing of patient samples. We also thank Linda Kay Lee from the Communicable Diseases Centre and Ivy Low, Seri Mustafah, and Anis Larbi from the SIgN Flow Cytometry team for their assistance. We are grateful to all patients and healthy volunteers for their participation in the study. Finally, we thank Laurent Rénia and Insight Editing London for comments and proofreading the manuscript prior to submission.
The views expressed are those of the author(s) and not necessarily those of the A*STAR, NHS, the NIHR, the Department of Health, or PHE.
This work was supported by core research grants provided to the Singapore Immunology Network (SIgN) by the Biomedical Research Council (BMRC) and by the BMRC A*STAR-led Zika Virus Consortium Fund (project number 15/1/82/27/001), Agency for Science, Technology and Research (A*STAR), Singapore. This work was also supported by funds provided by the Medical Research Council (MRC) (project number MC_PC_15094) and by the HPRU in Emerging and Zoonotic Infections Cross NIHR Strategic Research Fund (project number ZLKN_11447), United Kingdom.
F.-M.L., D.A.M., J.A.H., and L.F.P.N. designed the study. F.-M.L., D.L., D.A.M., J.A.H., and L.F.P.N. wrote the manuscript. F.-M.L., T.-K.C., C.Y.P.L., J.J.L.T., X.L., W.-X.Y., and Y.F. performed the experiments. F.-M.L., D.L., X.L., Y.F., B.L., N.Y.R., D.A.M., J.A.H., and L.F.P.N. analyzed the data. All other authors were involved in sample collection, processing and analysis, and/or logistical support. All authors read and approved the final manuscript.
Footnotes
This paper was submitted via the mSphereDirect™ pathway.
Contributor Information
Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai.
Anita K. McElroy, University of Pittsburgh.
Mark T. Heise, University of North Carolina at Chapel Hill.
REFERENCES
- 1.Korzeniewski K, Juszczak D, Zwolińska E. 2016. Zika—another threat on the epidemiological map of the world. Int Marit Health 67:31–37. doi: 10.5603/IMH.2016.0007. [DOI] [PubMed] [Google Scholar]
- 2.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 4.Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasil P, Pereira JP Jr, Raja Gabaglia C, Damasceno L, Wakimoto M, Ribeiro Nogueira RM, Carvalho de Sequeira P, Machado Siqueira A, Abreu de Carvalho LM, Cotrim da Cunha D, Calvet GA, Neves ES, Moreira ME, Rodrigues Baião AE, Nassar de Carvalho PR, Janzen C, Valderramos SG, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro—preliminary report. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foo SS, Chen W, Chan Y, Bowman JW, Chang LC, Choi Y, Yoo JS, Ge J, Cheng G, Bonnin A, Nielsen-Saines K, Brasil P, Jung JU. 2017. Asian Zika virus strains target CD14+ blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nat Microbiol 2:1558–1570. doi: 10.1038/s41564-017-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lum FM, Lin C, Susova OY, Teo TH, Fong SW, Mak TM, Lee LK, Chong CY, Lye DCB, Lin RTP, Merits A, Leo YS, Ng LFP. 2017. Sensitive detection of Zika virus antigen in patients’ whole blood as an alternative diagnostic approach. J Infect Dis 216:182–190. doi: 10.1093/infdis/jix276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lum FM, Low DK, Fan Y, Tan JJ, Lee B, Chan JK, Rénia L, Ginhoux F, Ng LF. 2017. Zika virus infects human fetal brain microglia and induces inflammation. Clin Infect Dis 64:914–920. doi: 10.1093/cid/ciw878. [DOI] [PubMed] [Google Scholar]
- 9.Michlmayr D, Andrade P, Gonzalez K, Balmaseda A, Harris E. 2017. CD14+CD16+ monocytes are the main target of Zika virus infection in peripheral blood mononuclear cells in a paediatric study in Nicaragua. Nat Microbiol 2:1462–1470. doi: 10.1038/s41564-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Speranza E, Munoz-Fontela C, Haldenby S, Rickett NY, Garcia-Dorival I, Fang Y, Hall Y, Zekeng EG, Ludtke A, Xia D, Kerber R, Krumkamp R, Duraffour S, Sissoko D, Kenny J, Rockliffe N, Williamson ED, Laws TR, N'Faly M, Matthews DA, Gunther S, Cossins AR, Sprecher A, Connor JH, Carroll MW, Hiscox JA. 2017. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol 18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, Fedorova NB, Puri V, Shabman RS, Pulendran B, Suthar MS. 2017. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog 13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll MW, Matthews DA, Hiscox JA, Elmore MJ, Pollakis G, Rambaut A, Hewson R, García-Dorival I, Bore JA, Koundouno R, Abdellati S, Afrough B, Aiyepada J, Akhilomen P, Asogun D, Atkinson B, Badusche M, Bah A, Bate S, Baumann J, Becker D, Becker-Ziaja B, Bocquin A, Borremans B, Bosworth A, Boettcher JP, Cannas A, Carletti F, Castilletti C, Clark S, Colavita F, Diederich S, Donatus A, Duraffour S, Ehichioya D, Ellerbrok H, Fernandez-Garcia MD, Fizet A, Fleischmann E, Gryseels S, Hermelink A, Hinzmann J, Hopf-Guevara U, Ighodalo Y, Jameson L, Kelterbaum A, Kis Z, Kloth S, Kohl C, Korva M, Kraus A, Kuisma E, Kurth A, Liedigk B, Logue CH, Lüdtke A, Maes P, McCowen J, Mély S, Mertens M, Meschi S, Meyer B, Michel J, Molkenthin P, Muñoz-Fontela C, Muth D, Newman EN, Ngabo D, Oestereich L, Okosun J, Olokor T, Omiunu R, Omomoh E, Pallasch E, Pályi B, Portmann J, Pottage T, Pratt C, Priesnitz S, Quartu S, Rappe J, Repits J, Richter M, Rudolf M, Sachse A, Schmidt KM, Schudt G, Strecker T, Thom R, Thomas S, Tobin E, Tolley H, Trautner J, Vermoesen T, Vitoriano I, Wagner M, Wolff S, Yue C, Capobianchi MR, Kretschmer B, Hall Y, Kenny JG, Rickett NY, Dudas G, Coltart CE, Kerber R, Steer D, Wright C, Senyah F, Keita S, Drury P, Diallo B, de Clerck H, Van Herp M, Sprecher A, Traore A, Diakite M, Konde MK, Koivogui L, Magassouba N, Avšič-Županc T, Nitsche A, Strasser M, Ippolito G, Becker S, Stoecker K, Gabriel M, Raoul H, Di Caro A, Wölfel R, Formenty P, Günther S. 2015. Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature 524:97–101. doi: 10.1038/nature14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowall SD, Matthews DA, Garcia-Dorival I, Taylor I, Kenny J, Hertz-Fowler C, Hall N, Corbin-Lickfett K, Empig C, Schlunegger K, Barr JN, Carroll MW, Hewson R, Hiscox JA. 2014. Elucidating variations in the nucleotide sequence of Ebola virus associated with increasing pathogenicity. Genome Biol 15:540. doi: 10.1186/PREACCEPT-1724277741482641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singapore Zika Study Group 2017. Outbreak of Zika virus infection in Singapore: an epidemiological, entomological, virological, and clinical analysis. Lancet Infect Dis 17:813–821. doi: 10.1016/S1473-3099(17)30249-9. [DOI] [PubMed] [Google Scholar]
- 15.Montaldo E, Del Zotto G, Della Chiesa M, Mingari MC, Moretta A, De Maria A, Moretta L. 2013. Human NK cell receptors/markers: a tool to analyze NK cell development, subsets and function. Cytometry A 83:702–713. doi: 10.1002/cyto.a.22302. [DOI] [PubMed] [Google Scholar]
- 16.Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK, Liu S, McClory S, Marcucci G, Trotta R, Caligiuri MA. 2010. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK cell subsets. Blood 115:274–281. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogel LA, Sun MM, Geurs TL, Carayannopoulos LN, French AR. 2013. Markers of nonselective and specific NK cell activation. J Immunol 190:6269–6276. doi: 10.4049/jimmunol.1202533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel T, Hentges F, Zimmer J. 2012. Consequences of the crosstalk between monocytes/macrophages and natural killer cells. Front Immunol 3:403. doi: 10.3389/fimmu.2012.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marçais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, Rabilloud J, Mayol K, Tavares A, Bienvenu J, Gangloff YG, Gilson E, Vivier E, Walzer T. 2014. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol 15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kam YW, Leite JA, Lum FM, Tan JJL, Lee B, Judice CC, Teixeira DAT, Andreata-Santos R, Vinolo MA, Angerami R, Resende MR, Freitas ARR, Amaral E, Junior RP, Costa ML, Guida JP, Arns CW, Ferreira LCS, Renia L, Proenca-Modena JL, Ng LFP, Costa FTM, Zika-Unicamp Network . 2017. Specific biomarkers associated with neurological complications and congenital central nervous system abnormalities from Zika virus-infected patients in Brazil. J Infect Dis 216:172–181. doi: 10.1093/infdis/jix261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Costa H, Gouilly J, Mansuy JM, Chen Q, Levy C, Cartron G, Veas F, Al-Daccak R, Izopet J, Jabrane-Ferrat N. 2016. Zika virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci Rep 6:35296. doi: 10.1038/srep35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, Cao-Lormeau VM, Choumet V, Briant L, Desprès P, Amara A, Yssel H, Missé D. 2015. Biology of Zika virus infection in human skin cells. J Virol 89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quicke KM, Bowen JR, Johnson EL, McDonald CE, Ma H, O’Neal JT, Rajakumar A, Wrammert J, Rimawi BH, Pulendran B, Schinazi RF, Chakraborty R, Suthar MS. 2016. Zika virus infects human placental macrophages. Cell Host Microbe 20:83–90. doi: 10.1016/j.chom.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon S, Plüddemann A, Martinez Estrada F. 2014. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev 262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon S, Taylor PR. 2005. Monocyte and macrophage heterogeneity. Nat Rev Immunol 5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 26.Martinez FO, Gordon S, Locati M, Mantovani A. 2006. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 27.Ploeger DT, Hosper NA, Schipper M, Koerts JA, de Rond S, Bank RA. 2013. Cell plasticity in wound healing: paracrine factors of M1/M2 polarized macrophages influence the phenotypical state of dermal fibroblasts. Cell Commun Signal 11:29. doi: 10.1186/1478-811X-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Italiani P, Boraschi D. 2014. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol 5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grubaugh ND, Ladner JT, Kraemer MUG, Dudas G, Tan AL, Gangavarapu K, Wiley MR, White S, Thézé J, Magnani DM, Prieto K, Reyes D, Bingham AM, Paul LM, Robles-Sikisaka R, Oliveira G, Pronty D, Barcellona CM, Metsky HC, Baniecki ML, Barnes KG, Chak B, Freije CA, Gladden-Young A, Gnirke A, Luo C, MacInnis B, Matranga CB, Park DJ, Qu J, Schaffner SF, Tomkins-Tinch C, West KL, Winnicki SM, Wohl S, Yozwiak NL, Quick J, Fauver JR, Khan K, Brent SE, Reiner RC Jr, Lichtenberger PN, Ricciardi MJ, Bailey VK, Watkins DI, Cone MR, Kopp EW IV, Hogan KN, Cannons AC, Jean R, Monaghan AJ, Garry RF, Loman NJ, Faria NR, Porcelli MC, Vasquez C, Nagle ER, Cummings DAT, Stanek D, Rambaut A, Sanchez-Lockhart M, Sabeti PC, Gillis LD, Michael SF, Bedford T, Pybus OG, Isern S, Palacios G, Andersen KG. 2017. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 546:401–405. doi: 10.1038/nature22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodpothong P, Auewarakul P. 2012. Viral evolution and transmission effectiveness. World J Virol 1:131–134. doi: 10.5501/wjv.v1.i5.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mamrut S, Avidan N, Staun-Ram E, Ginzburg E, Truffault F, Berrih-Aknin S, Miller A. 2015. Integrative analysis of methylome and transcriptome in human blood identifies extensive sex- and immune cell-specific differentially methylated regions. Epigenetics 10:943–957. doi: 10.1080/15592294.2015.1084462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, Kourilsky P, Wong SC. 2011. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118:e16–e31. doi: 10.1182/blood-2010-12-326355. [DOI] [PubMed] [Google Scholar]
- 33.Carrasco L. 1977. The inhibition of cell functions after viral infection. A proposed general mechanism. FEBS Lett 76:11–15. doi: 10.1016/0014-5793(77)80110-5. [DOI] [PubMed] [Google Scholar]
- 34.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 35.Lünemann JD. 2012. Epstein-Barr virus in multiple sclerosis: a continuing conundrum. Neurology 78:11–12. doi: 10.1212/WNL.0b013e318241f2b3. [DOI] [PubMed] [Google Scholar]
- 36.Tucker WG, Andrew Paskauskas R. 2008. The MSMV hypothesis: measles virus and multiple sclerosis, etiology and treatment. Med Hypotheses 71:682–689. doi: 10.1016/j.mehy.2008.06.029. [DOI] [PubMed] [Google Scholar]
- 37.Robertson MJ. 2002. Role of chemokines in the biology of natural killer cells. J Leukoc Biol 71:173–183. [PubMed] [Google Scholar]
- 38.Thapa M, Welner RS, Pelayo R, Carr DJ. 2008. CXCL9 and CXCL10 expression are critical for control of genital herpes simplex virus type 2 infection through mobilization of HSV-specific CTL and NK cells to the nervous system. J Immunol 180:1098–1106. doi: 10.4049/jimmunol.180.2.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren HS, Smyth MJ. 1999. NK cells and apoptosis. Immunol Cell Biol 77:64–75. doi: 10.1046/j.1440-1711.1999.00790.x. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi M, Fitz L, Ryan M, Hewick RM, Clark SC, Chan S, Loudon R, Sherman F, Perussia B, Trinchieri G. 1989. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med 170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Cao S, Kim S, Chung EY, Homma Y, Guan X, Jimenez V, Ma X. 2005. Interleukin-12: an update on its immunological activities, signaling and regulation of gene expression. Curr Immunol Rev 1:119–137. doi: 10.2174/1573395054065115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.French AR, Holroyd EB, Yang L, Kim S, Yokoyama WM. 2006. IL-18 acts synergistically with IL-15 in stimulating natural killer cell proliferation. Cytokine 35:229–234. doi: 10.1016/j.cyto.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Takeda K, Tsutsui H, Yoshimoto T, Adachi O, Yoshida N, Kishimoto T, Okamura H, Nakanishi K, Akira S. 1998. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity 8:383–390. doi: 10.1016/S1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 44.Kastelein RA, Hunter CA, Cua DJ. 2007. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu Rev Immunol 25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 45.Matsui M, Kishida T, Nakano H, Yoshimoto K, Shin-Ya M, Shimada T, Nakai S, Imanishi J, Yoshimoto T, Hisa Y, Mazda O. 2009. Interleukin-27 activates natural killer cells and suppresses NK-resistant head and neck squamous cell carcinoma through inducing antibody-dependent cellular cytotoxicity. Cancer Res 69:2523–2530. doi: 10.1158/0008-5472.CAN-08-2793. [DOI] [PubMed] [Google Scholar]
- 46.van de Wetering D, de Paus RA, van Dissel JT, van de Vosse E. 2009. IL-23 modulates CD56+/CD3- NK cell and CD56+/CD3+ NK-like T cell function differentially from IL-12. Int Immunol 21:145–153. doi: 10.1093/intimm/dxn132. [DOI] [PubMed] [Google Scholar]
- 47.Azeredo EL, De Oliveira-Pinto LM, Zagne SM, Cerqueira DI, Nogueira RM, Kubelka CF. 2006. NK cells, displaying early activation, cytotoxicity and adhesion molecules, are associated with mild dengue disease. Clin Exp Immunol 143:345–356. doi: 10.1111/j.1365-2249.2006.02996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neves PC, Matos DC, Marcovistz R, Galler R. 2009. TLR expression and NK cell activation after human yellow fever vaccination. Vaccine 27:5543–5549. doi: 10.1016/j.vaccine.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 49.Kim TJ, Kim M, Kim HM, Lim SA, Kim EO, Kim K, Song KH, Kim J, Kumar V, Yee C, Doh J, Lee KM. 2014. Homotypic NK cell-to-cell communication controls cytokine responsiveness of innate immune NK cells. Sci Rep 4:7157. doi: 10.1038/srep07157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krylova NV, Smolina TP, Leonova GN. 2015. Molecular mechanisms of interaction between human immune cells and Far Eastern tick-borne encephalitis virus strains. Viral Immunol 28:272–281. doi: 10.1089/vim.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gotlieb N, Rosenne E, Matzner P, Shaashua L, Sorski L, Ben-Eliyahu S. 2015. The misleading nature of in vitro and ex vivo findings in studying the impact of stress hormones on NK cell cytotoxicity. Brain Behav Immun 45:277–286. doi: 10.1016/j.bbi.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notario L, Alari-Pahissa E, de Molina A, Lauzurica P. 2016. CD69 deficiency enhances the host response to vaccinia virus infection through altered NK cell homeostasis. J Virol 90:6464–6474. doi: 10.1128/JVI.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sellers RS, Clifford CB, Treuting PM, Brayton C. 2012. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet Pathol 49:32–43. doi: 10.1177/0300985811429314. [DOI] [PubMed] [Google Scholar]
- 54.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandão WN, Rossato C, Andrade DG, Faria DDP, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrão-Braga PC. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR III, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sékaly RP. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison BE, Park SJ, Mooney JM, Mehrad B. 2003. Chemokine-mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. J Clin Invest 112:1862–1870. doi: 10.1172/JCI18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eubank TD, Galloway M, Montague CM, Waldman WJ, Marsh CB. 2003. M-CSF induces vascular endothelial growth factor production and angiogenic activity from human monocytes. J Immunol 171:2637–2643. doi: 10.4049/jimmunol.171.5.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuroda T, Kitadai Y, Tanaka S, Yang X, Mukaida N, Yoshihara M, Chayama K. 2005. Monocyte chemoattractant protein-1 transfection induces angiogenesis and tumorigenesis of gastric carcinoma in nude mice via macrophage recruitment. Clin Cancer Res 11:7629–7636. doi: 10.1158/1078-0432.CCR-05-0798. [DOI] [PubMed] [Google Scholar]
- 59.Marumo T, Schini-Kerth VB, Busse R. 1999. Vascular endothelial growth factor activates nuclear factor-kappaB and induces monocyte chemoattractant protein-1 in bovine retinal endothelial cells. Diabetes 48:1131–1137. doi: 10.2337/diabetes.48.5.1131. [DOI] [PubMed] [Google Scholar]
- 60.Conti P, Dempsey RA, Reale M, Barbacane RC, Panara MR, Bongrazio M, Mier JW. 1991. Activation of human natural killer cells by lipopolysaccharide and generation of interleukin-1 alpha, beta, tumour necrosis factor and interleukin-6. Effect of IL-1 receptor antagonist. Immunology 73:450–456. [PMC free article] [PubMed] [Google Scholar]
- 61.Park JY, Lee SH, Yoon SR, Park YJ, Jung H, Kim TD, Choi I. 2011. IL-15-induced IL-10 increases the cytolytic activity of human natural killer cells. Mol Cells 32:265–272. doi: 10.1007/s10059-011-1057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metcalfe SM. 2011. LIF in the regulation of T-cell fate and as a potential therapeutic. Genes Immun 12:157–168. doi: 10.1038/gene.2011.9. [DOI] [PubMed] [Google Scholar]
- 63.Wolk K, Kunz S, Asadullah K, Sabat R. 2002. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol 168:5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Q, Putheti P, Zhou Q, Liu Q, Gao W. 2008. Structures and biological functions of IL-31 and IL-31 receptors. Cytokine Growth Factor Rev 19:347–356. doi: 10.1016/j.cytogfr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elong Ngono A, Vizcarra EA, Tang WW, Sheets N, Joo Y, Kim K, Gorman MJ, Diamond MS, Shresta S. 2017. Mapping and role of the CD8+ T cell response during primary Zika virus infection in mice. Cell Host Microbe 21:35–46. doi: 10.1016/j.chom.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kerdiles Y, Ugolini S, Vivier E. 2013. T cell regulation of natural killer cells. J Exp Med 210:1065–1068. doi: 10.1084/jem.20130960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Charron L, Doctrinal A, Ni Choileain S, Astier AL. 2015. Monocyte:T-cell interaction regulates human T-cell activation through a CD28/CD46 crosstalk. Immunol Cell Biol 93:796–803. doi: 10.1038/icb.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beyersdorf N, Kerkau T, Hünig T. 2015. CD28 co-stimulation in T-cell homeostasis: a recent perspective. Immunotargets Ther 4:111–122. doi: 10.2147/ITT.S61647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welte S, Kuttruff S, Waldhauer I, Steinle A. 2006. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nat Immunol 7:1334–1342. doi: 10.1038/ni1402. [DOI] [PubMed] [Google Scholar]
- 70.Costa VV, Ye W, Chen Q, Teixeira MM, Preiser P, Ooi EE, Chen J. 2017. Dengue virus-infected dendritic cells, but not monocytes, activate natural killer cells through a contact-dependent mechanism involving adhesion molecules. mBio 8:e00741-17. doi: 10.1128/mBio.00741-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baronti C, Piorkowski G, Charrel RN, Boubis L, Leparc-Goffart I, de Lamballerie X. 2014. Complete coding sequence of Zika virus from a French Polynesia outbreak in 2013. Genome Announc 2:e00500-14. doi: 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balm MN, Lee CK, Lee HK, Chiu L, Koay ES, Tang JW. 2012. A diagnostic polymerase chain reaction assay for Zika virus. J Med Virol 84:1501–1505. doi: 10.1002/jmv.23241. [DOI] [PubMed] [Google Scholar]
- 73.Wu D, Wu J, Zhang Q, Zhong H, Ke C, Deng X, Guan D, Li H, Zhang Y, Zhou H, He J, Li L, Yang X. 2012. Chikungunya outbreak in Guangdong Province, China, 2010. Emerg Infect Dis 18:493–495. doi: 10.3201/eid1803.110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bosworth A, Dowall SD, Garcia-Dorival I, Rickett NY, Bruce CB, Matthews DA, Fang Y, Aljabr W, Kenny J, Nelson C, Laws TR, Williamson ED, Stewart JP, Carroll MW, Hewson R, Hiscox JA. 2017. A comparison of host gene expression signatures associated with infection in vitro by the Makona and Ecran (Mayinga) variants of Ebola virus. Sci Rep 7:43144. doi: 10.1038/srep43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, Ming GL. 2016. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 80.Beerenwinkel N, Zagordi O. 2011. Ultra-deep sequencing for the analysis of viral populations. Curr Opin Virol 1:413–418. doi: 10.1016/j.coviro.2011.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analyses based on sample consensus sequences. (A) Frequency of ZIKV minor variants (transitions and transversions) recovered from infected human primary MNCs and MDMs isolated from five donors. Bin −3 is where ≤1/1,000 reads show a specific change at an individual nucleotide position. Bin −2 is >1/1,000 and ≤1/100 reads showing a difference. Bin −1 is >1/100 and ≤1/10 reads, and bin >−1 is >1/10 reads showing a change up to a logical limit of just under 1/2. (B) Phylogenetic tree generated from the alignment of consensus sequences of ZIKV RNA recovered from the same samples as described for panel A. All samples included in the tree had a mean sequence coverage of >10 at each nucleotide position. PF/ZIKV/HPF/2013 is the virus strain used for infection and denoted as the reference sample in this analysis. Download FIG S1, PDF file, 0.1 MB (79.2KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of nucleotide differences at specific genome positions. Download TABLE S1, PDF file, 0.1 MB (74.1KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selected nucleotide positions from the minor variant file of the inoculum. Download TABLE S2, PDF file, 0.1 MB (66.7KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gating strategy of CD56+ CD94+ NK cells. (A) Fresh human PBMCs were subjected to CD14+ monocyte depletion. Flow cytometric plots from one representative donor are shown. A depletion efficiency of >95% was typically obtained. Monocytes are defined as lineage+ cells, with CD14, CD3, CD19, and CD20 included as lineage markers. (B) Live singlets were first gated from the stained PBMCs. CD45+ CD56+ cells were identified, and NK cells were subsequently gated out with CD94 and lineage markers—CD14, CD3, CD19, and CD20. NK cells are defined as CD56+ CD94+ Lineage−. Flow cytometric plots from one representative donor are shown. Download FIG S2, PDF file, 0.5 MB (575.9KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of surface markers by activated NK cells. LPS (10 ng/ml)-stimulated conditions. (A) Compiled percentages of CD69-, CD107a-, and IFN-γ-positive CD56+ CD94+ Lineage− NK cells after LPS (10 ng/ml) stimulation. Expression levels are normalized to the respective mock sample (dotted line). Data shown were derived from seven healthy donors. (B) Total PBMCs and CD14-depleted PBMCs (2 × 106 cells per infection) were infected with ZIKV at an MOI of 10 and harvested at 36 hpi. (B) Compiled percentages of NKG2D and NKG2A-positive CD94+ CD56+ Lineage− NK cells normalized to the respective mock sample. (C) Comparison of the percentage of CD69-positive CD94+ CD56+ Lineage− NK cells between mock-infected and ZIKV-infected full PBMCs and CD14-depleted PBMCs. Data shown were derived from seven donors. Data shown are presented as paired data. (D) The expression levels of CD69, CD107a, and IFN-γ on CD56+ CD94+ Lineage− NK cells at 72 hpi. Expression levels are normalized to respective mock sample. Data were obtained from two donors. Lineage markers CD3, CD19, CD20, and CD14 have been included to rule out the presence of non-NK cells. All data are presented as means ± standard deviations. *, P < 0.05, by Mann-Whitney U test, two-tailed. Download FIG S3, PDF file, 0.2 MB (224.2KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantification of immune mediators. Immune mediators in the culture supernatant of ZIKV-infected PBMCs and CD14-depleted PBMCs were quantified using a 45-plex microbead assay. Quantified immune mediators are grouped into four groups based on their profile: mediators affected by depletion of CD14+ monocytes (A), mediators affected by ZIKV infection (B), mediators not affected by both CD14+ monocyte depletion and ZIKV infection (C), and mediators affected by depletion of CD14+ monocytes only after ZIKV infection (D). Data displayed were derived from seven donors. All data are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001, by Mann-Whitney U test, two-tailed. Download FIG S4, PDF file, 0.5 MB (557.3KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
UV inactivation of ZIKV and proteins. (A) Wild-type (WT) ZIKV was subjected to two different doses (1,000 or 100 mJ/cm2) of UV treatment across different durations. UV-treated ZIKV was subsequently used to infect HEK293T cells, and the amount of viral RNA load was determined at 48 hpi. Levels of viral RNA load are expressed as fold increase relative to the level of viral RNA load detected at 0 hpi with the WT ZIKV. Heat-inactivated (HI) ZIKV was included in parallel as a negative control. (B) Culture supernatants were UV treated (100 mJ/cm2 for 10 min), and their stimulatory capacity was further evaluated with freshly isolated PBMCs in a ratio of 1:10 (with fresh complete medium). Cells were harvested at 36 h poststimulation and evaluated for the percentage of CD107a-; IFN-γ-; and NKG2D-, NKG2A-, and CD69-positive CD94+ CD56+ NK cells. Data shown (n = 2) normalized to the respective mock sample. Lineage markers CD3, CD19, CD20, and CD14 have been included to rule out the presence of non-NK cells. All data are presented as means ± standard deviations. Download FIG S5, PDF file, 0.1 MB (95.6KB, pdf) .
Copyright © 2018 Lum et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.