Abstract
In Drosophila, heterochromatin protein 1 (HP1) suppresses the expression of euchromatic genes that are artificially translocated adjacent to heterochromatin by expanding heterochromatin structure into neighboring euchromatin. The purpose of this study was to determine whether HP1 functions as a transcriptional repressor in the absence of chromosome rearrangements. Here, we show that Drosophila HP1 normally represses the expression of four euchromatic genes in a dosage-dependent manner. Three genes regulated by HP1 map to cytological region 31 of chromosome 2, which is immunostained by anti-HP1 antibodies in the salivary gland. The repressive effect of HP1 is decreased by mutation in Su(var)3–9, whose mammalian orthologue encodes a histone H3 methyltransferase and mutation in Su(var)2–1, which is correlated with histone H4 deacetylation. These data provide genetic evidence that an HP1-family protein represses the expression of euchromatic genes in a metazoan, and that histone modifiers cooperate with HP1 in euchromatic gene repression.
Heterochromatin protein 1 (HP1) first was identified as a component of heterochromatin in Drosophila polytene chromosomes (1). Subsequently, it was shown to mediate variegated silencing of euchromatic genes that are placed abnormally next to constitutive heterochromatin, a phenomenon known as position–effect variegation (PEV; refs. 2–4). In contrast to its role in suppressing genes that are localized abnormally on chromosomes, the only documented role of HP1 in normal gene regulation is in the activation of two Drosophila heterochromatic genes, light and rolled (5).
HP1-family proteins bind to histones and DNA (6–9). Lys-9 of histone H3 is methylated by the modifier proteins SUV39H1 and Suv39H1, the human and murine orthologues of the Drosophila Su(var)3–9 gene product, respectively (10, 11). Histone H3 methylation is thought to promote transcriptional silencing by creating a binding site for HP1-family proteins to form heterochromatic subdomains (6–8). Histone deacetylation also is thought to promote silencing. Mutations in the Drosophila histone deacetylase (HDAC) 1 suppress PEV, indicating that HDAC1 normally promotes silencing (12). Similarly, Su(var)2–1 mutations suppress PEV silencing and cause hyperacetylation of histones (13).
Although HP1 is involved in heterochromatin structure and position–effect silencing, there are no examples of normal transcriptional repression of genes by HP1. Among HP1-family proteins, only Swi6p in fission yeast has been shown to be involved in normal gene repression, silencing the mating type cassettes (14, 15). Here, we describe a previously unreported role for HP1 in the repression of euchromatic genes in Drosophila. We have identified genes residing in the euchromatin of chromosome 2 that are repressed normally by HP1 and other heterochromatin-associated factors. In contrast to previous examples of euchromatic gene silencing by heterochromatin, these HP1-repressed genes are located well away from constitutive heterochromatin. The genes show a graded, inverse response in transcription proportionate to HP1 dosage. In addition, reduced doses of other PEV modifiers, including Su(var)3–9 and Su(var)2–1, also increase expression of these HP1-regulated genes. Three HP1-regulated genes that we have identified span 145 kb in region 31, a region that is immunostained brightly in the salivary gland polytene chromosomes with anti-HP1 antibodies, suggesting that HP1 directly regulates the repression of a broad chromatin domain. These results provide genetic evidence for an HP1-dependent heterochromatin-repression mechanism operating in the normal suppression of euchromatic genes in a metazoan.
Materials and Methods
Drosophila Stocks.
Drosophila melanogaster stocks were maintained in a background of Df(1)w,y1w67c23 with the Su(var) mutation carried over either CyO, y+ or TM3, Ser y+. The Su(var) alleles Su(var)2–101, Su(var)2–401, Su(var)3–601, Su(var)3–903, Su(var)2–504, Su(var)2–505, and Su(var)2–5149 have been described (3, 5, 16). Heterozygous flies carry a mutation in one of their genes, giving one functional dose of their proteins. HP1(−/−) larvae were produced by crossing Df(1)w,yw; Su(var)2–504/CyO, y+ and Df(1)w,yw; Su(var)2–505/CyO, y+ or Df(1)w,yw; Su(var)2–504/CyO, y+ and Df(1)w,yw; Su(var)2–5149/CyO, y+, and larval genotypes were determined by mouth-hook pigmentation as described (5). Dp(2;2)P90/CyO, y+ larvae (black mouth hook) carry a duplication of the HP1 gene on one chromosome, and Dp(2;2)P90/Dp(2;2)P90 larvae (yellow mouth hook) carry a duplication of the HP1 gene on both chromosomes. Larvae were harvested at early third instar stage, flash-frozen on dry ice, and stored at −70°C until use.
Representational Difference Analysis (RDA).
RDA of cDNA was performed essentially as described (17, 18). Messenger RNA was extracted from early-stage third instar HP1+/+ and HP1−/− larvae and used for cDNA synthesis. Representative cDNA fragments (representations) were generated by DpnII restriction endonuclease digestion of double-stranded cDNAs followed by annealing of R-oligomers and PCR amplification. Representations from wild-type and mutant larvae were used as driver and tester, respectively, in subtractive hybridization after the removal of R-oligomers from both representations and the annealing of J-oligomers to the tester. Difference products were obtained from unhybridized cDNA fragments of the tester by PCR-amplification by using annealed oligomer as the primer. Driver to tester ratios of 100:1, 1,000:1, and 10,000:1 were used in the first, second, and third rounds of subtractive hybridization, respectively. Oligomers annealed to tester were changed between consecutive subtractive hybridizations. Difference products from second and third round hybridizations were analyzed by agarose gel electrophoresis and DNA sequencing.
Northern Blot Analysis.
Total RNA was isolated from third instar larvae. Frozen larvae (100 mg) were homogenized in 1 ml of TRIzol reagent (Life Technologies, Grand Island, NY) at 4°C with a Dounce homogenizer. Homogenized samples were incubated for 10 min at room temperature and mixed vigorously with 0.2 ml of chloroform. The colorless upper aqueous phase was isolated after centrifuging the sample at 12,000 × g for 15 min at 4°C. Isolated samples were mixed with 0.5 ml of isopropyl alcohol followed by incubation for 10 min at room temperature. RNA was precipitated by spinning the mixed sample at 12,000 × g for 10 min at 4°C and quantitated by spectrophotometry several times to ensure the exact amount was present in each sample after purification by acidic phenol extraction and ethanol precipitation. Ethidium bromide-stained gels also were analyzed to assure equal RNA loading before blotting. Gel electrophoresis, blotting, radiolabeling, and hybridization were carried out by standard methods (19). cDNA fragments isolated by RDA were used for the detection of Ser4 and CG13135 transcripts, and PCR-amplified fragments of an exon were used for the detection of the other transcripts. Plasmid clones of Su(var)2–5 (an HP1 cDNA clone) and Rp49 (a ribosomal protein 49 genomic clone) were used to detect their respective RNAs. To compare transcript levels in a particular genotype, the same blot was reprobed after stripping. The density of each RNA band was measured by the NIH IMAGE program after scanning autoradiograms. The intensity of each band compared with wild type was calculated after deduction of the average background signal of each blot.
Immunofluorescence Microscopy.
Immunofluorescent staining of Drosophila polytene chromosomes was performed as described (20). Briefly, salivary glands dissected from wild-type third instar larvae were fixed in formaldehyde buffer and squashed on glass slides. Chromosomes were incubated with rabbit anti-HP1 antibodies (gift of S. C. R. Elgin, Washington University, St. Louis, MO) at room temperature for 1 to 2 hr. Fluorescein isothiocyanate-conjugated goat anti-rabbit IgG was used for secondary staining for 30 min at room temperature. The slides were mounted in 90% glycerol-PBS and examined by fluorescence and phase-contrast microscopy.
Results
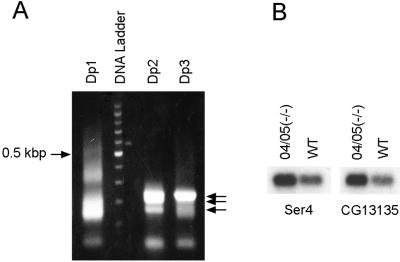
We used RDA (17, 18) to identify Drosophila genes repressed by HP1. In this technique, RNA samples from two different sources are compared to identify specific RNAs that are overrepresented in one sample relative to the other. We compared RNA from third instar larvae lacking functional HP1 (HP1−/−; ref. 5) to RNA from wild-type larvae (HP1+/+) to look for RNAs overexpressed in the HP1−/− sample. We obtained four prominent bands after three rounds of subtractive hybridization. After cloning and sequencing of DNA fragments from these four bands, we identified two euchromatic genes, Ser4 and CG13135, whose expression levels were anticipated to be higher in HP1(−/−) larvae than in wild type (Fig. 1A). Subsequently, we confirmed that the expression of these two genes is repressed 2.5- to 3-fold in wild-type compared with HP1(−/−) larvae by Northern blot analysis (Fig. 1B).
Figure 1.
Detection of the genes overexpressed in HP1-null larvae [04/05(−/−)] relative to wild type [WT(+/+)] by RDA. (A) Ethidium bromide-stained agarose gel showing difference products generated by PCR-amplification after the first (Dp1), second (Dp2), and third (Dp3) rounds of subtractive hybridization in RDA. Arrows indicate prominent bands of Dp2 and Dp3. DNA ladder indicates 100-bp differences, and the 0.5-kb band (0.5 kbp) is indicated. (B) Northern blot analysis of RNA from HP1-null and wild-type larvae with radiolabeled Ser4 and CG13135 cDNA fragments isolated from Dp2 and Dp3. 04/05(−/−) represents HP1-null larvae generated by crossing between heterozygous Su(var)2–504 and Su(var)2–505 parents.
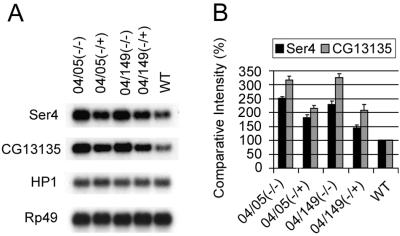
To ensure that the effects of HP1 mutations on the expression of Ser4 and CG13135 are caused by HP1 dosage and not by specific alleles or linked modifiers, we generated larvae that carried different HP1 mutations by crossing Su(var)2–504/+ to Su(var)2–505/+ or Su(var)2–504/+ to Su(var)2–5149/+. Su(var)2–504 and Su(var)2–5149 encode truncated HP1 proteins, whereas Su(var)2–505 encodes a frame-shift mutation at codon 10 (3, 5). Each allele was isolated in a separate mutational screen in a different genetic background. Expression of Ser4 and CG13135 was elevated in all HP1-mutant larvae, whereas their expression in sibs decreased in proportion to the number of functional copies of the HP1 gene present (Fig. 2). In larvae with one functional dose of HP1, expression of both genes was increased 1.5- to 2-fold, whereas in HP1-null larvae, the average expression levels of both genes are 2.5- to 3-fold that observed in wild type. These results show that loss of Ser4 and CG13135 repression is caused by reduced functional HP1 gene dosage and not by genetic background.
Figure 2.
Expression of Ser4 and CG13135 is regulated by HP1. (A) Northern blot analysis of Ser4 and CG13135 RNA in larvae with different HP1 heterozygous (+/−) or null (−/−) mutant backgrounds. (B) Comparative intensities of bands are normalized to wild type (WT) and expressed in percentages. Bars indicate standard errors from three independent experiments. 04/05(−/−) and 04/05(−/+) larvae were generated from Su(var)2–504/CyO, y+ and Su(var)2–505/CyO, y+ parents. 04/149(−/−) and 04/149(−/+) were produced by crossing between Su(var)2–504/CyO, y+ and Su(var)2–5149/CyO, y+ parents. Transcripts of HP1 and Rp49 are shown as loading controls. Note that these Su(var)2–5 alleles have no effect on steady-state HP1 RNA message but yield no normal HP1 protein (refs. 3 and 5).
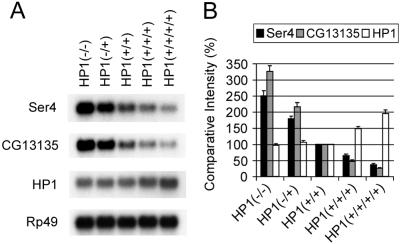
To test whether CG13135 and Ser4 are repressed further by HP1 overexpression, we generated flies with three or four copies of functional HP1 by using the Dp(2;2)P90 chromosome, which carries a tandem duplication including the HP1 gene (7). The expression of CG13135 and Ser4 decreased progressively with the increased HP1 gene dosage across the entire range of HP1 dosage examined (Fig. 3). The repressive effect of HP1 on the expression of CG13135 (13-fold from 0–4 doses) was greater than on Ser4 (6-fold from 0–4 doses). HP1 transcripts increased in HP1(+/+/+) and (+/+/+/+) larvae, as expected.
Figure 3.
Transcriptional repression of Ser4 and CG13135 depends on functional HP1 dosage. (A) Northern blot analysis of Ser4, CG13135, and HP1 RNA. Genotypes giving different HP1 doses are: HP1(−/−), Su(var)2–504/Su(var)2–505; HP1(−/+), Su(var)2–504/CyO, y+ or Su(var)2–505/CyO, y+; HP1 (+/+), Su(var)2–5+/Su(var)2–5+; HP1(+/+/+), Dp(2;2)P90/CyO, y+; and HP1 (+/+/+/+), Dp(2;2)P90/Dp(2;2)P90. Transcripts of Rp49 are shown as loading control. (B) Comparative intensities of bands are normalized to wild type [HP1(+/+)] and expressed in percentages. Bars indicate standard errors from three independent experiments. Note that HP1 RNA levels increase, as expected, to 1.5 and 2 times wild-type levels in HP1(+/+/+) and HP1(+/+/+/+).
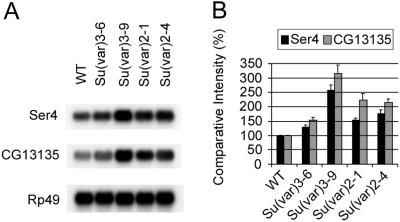
Several other modifiers of PEV have been reported, including Su(var)2–1, 2–4, 3–6, and 3–9 (5, 16). The aggregate histone 4 acetylation level is increased in Su(var)2–1 mutants and this mutation displays a lethal interaction with the histone deacetylase inhibitor N-butyrate (13). Human SUV39H1 and murine Suv39H1, mammalian orthologues of Drosophila Su(var)3–9, encode histone H3-specific methyltransferases that selectively methylate Lys-9 of the amino terminus of histone H3 in vitro (10). Methylated Lys-9 on histone H3 creates a binding site for HP1 proteins in yeast and mammals (6–8). Su(var)2–4 and Su(var)3–6 are strong dominant suppressors of PEV, although the mechanisms of these suppressors are unknown (16, 21). Su(var)3–6 encodes a protein phosphatase (21) that might be essential for modification of chromosomal proteins such as Su(var)3–7 (22). We investigated the effects of these modifiers on the expression of CG13135 and Ser4. Most of the PEV-modifier mutations tested significantly elevate the levels of Ser4 and CG13135 expression (Fig. 4). In particular, the increased expression of HP1-regulated genes caused by Su(var)3–9 mutation parallels the effect of HP1 mutations, which is consistent with recent findings that histone H3 methylation promotes HP1 binding (6–8).
Figure 4.
Elevated expression of Ser4 and CG13135 in larvae with mutations of Su(var)3–601, Su(var)3–903, Su(var)2–101, and Su(var)2–401. (A) Northern blot analysis of Ser4 and CG13135 RNA. Larvae were heterozygous for the indicated Su(var) mutation. Rp49 is a loading control. (B) Comparative intensities of bands are normalized to wild type (WT) and expressed in percentages. Bars indicate standard errors from three independent experiments.
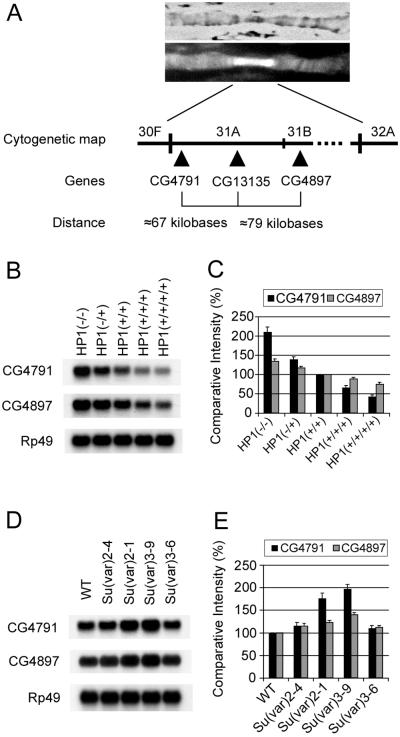
CG13135 and Ser4 are localized in cytological regions 31A and 25B of chromosome 2L, respectively. Cytological region 31 is a prominent euchromatic site of HP1 binding on the left arm of chromosome 2 (Fig. 5A and ref. 20). We picked two other genes (CG4791 and CG4897) in cytological region 31, which are positioned within 100 kb proximal and distal to CG13135 (Fig. 5A), and tested whether those two genes also are regulated by HP1. Like CG13135 and Ser4, the expression level of CG4791 is progressively decreased in response to increasing HP1 copy number (Fig. 5 B and C). CG4897 expression also is decreased, although it seems to be less sensitive to HP1 dosage. The expression profiles of CG4791 and CG4897 in PEV-suppressor mutations differ in magnitude from those of CG13135 (compare Fig. 4 to Fig. 5 D and E). Importantly, however, the Su(var)3–9 and Su(var)2–1 mutations significantly enhance the expression of all three genes (Figs. 4 and 5 D and E). Thus, three genes in the euchromatic region 31 are similarly repressed by HP1, and this repression depends on the dosage of histone modifiers.
Figure 5.
Expression profiles of CG4791 and CG4897 located proximal and distal to CG13135. (A) Immunofluorescence micrograph showing labeling with anti-HP1 antibodies (Upper, phase-contrast image; Lower, fluorescence image) and diagram around cytological region 31 of Drosophila polytene chromosome 2L. Symbols of three genes and approximate distances are illustrated. (B) Northern blot analysis of CG4791 and CG4897 RNA from larvae with different HP1 copy numbers. Functional doses of HP1 are indicated in parentheses. (C) Comparative intensities of bands in B are normalized to wild type [HP1(+/+)] and expressed in percentages. (D) Northern blot analysis of CG4791 and CG4897 RNA from larvae with different Su(var) mutant backgrounds. (E) Comparative intensities of bands in D are normalized to wild type (WT) and expressed in percentages. Bars in C and E indicate standard errors from three independent experiments.
Discussion
These results provide evidence that HP1 represses genes at their endogenous euchromatic locations. All previous examples of HP1-dependent repression involved artificial repression of normally active euchromatic genes when they were translocated close to heterochromatin by chromosome rearrangements. Hints at connections between HP1 and euchromatic gene regulation have come from yeast 2-hybrid protein screens and coimmunoprecipitation assays in which HP1-family proteins were found to associate with transcription corepressors (23–26). Although HP1 has been known to bind at several euchromatic sites by chromosome immunostaining for a long time (20), this study provides evidence that euchromatic HP1-binding sites represent domains of HP1-dependent gene repression. Although HP1 could be acting indirectly by regulating the regulators of euchromatic genes, the linear inverse response to HP1 from 0–4 doses (across two levels of underexpression and two levels of overexpression) and the observable binding of HP1 to the chromosomal interval containing HP1 target genes suggest that HP1 repression is direct. Furthermore, the Su(var)3–9 and Su(var)2–1 proteins are required for the HP1-dependent repression of euchromatic genes. These in vivo data implicate specific covalent modifications of histones as prerequisites for higher-order euchromatin structure organized by HP1 and, further, suggest that the mechanism of HP1-mediated repression in euchromatin shares features with HP1-dependent heterochromatin-mediated silencing in PEV.
Three of the HP1-repressed genes map to region 31, which is one of the most prominent HP1-binding euchromatic regions in the Drosophila genome. Is region 31 of Drosophila chromosome 2 a domain of intercalary heterochromatin? Region 31 is a well banded interval in polytene nuclei, lacking the disorganized, attenuated appearance of pericentric heterochromatin. Although a 2-fold under-replication of the interval cannot be ruled out, region 31 is neither dramatically under-replicated nor significantly late replicating in polytene chromosomes, nor does it contain easily broken regions (weak points) or ectopic pairing sites characteristic of intercalary heterochromatin (27). Meiotic recombination is not suppressed significantly across region 31 (28) in contrast to the pericentric heterochromatin, where recombination is absent. Sequence analysis reveals no significant homology to any of the major satellite DNA sequences characteristic of pericentric heterochromatin (29) or any significant amount of repetitious DNA sequences in tandem. The density of ORFs in region 31 (128 ORFs) is not significantly lower than the adjacent numbered euchromatic segments (108 ORFs in region 30; 106 ORFs in region 32). In contrast, gene density is thought to be much lower per unit length of DNA in heterochromatin than in euchromatin. Taken together, these observations strongly suggest that region 31 is not simply an island of heterochromatin in a sea of euchromatin. Instead, we believe that region 31 represents a euchromatin domain subject to repression by HP1.
Silencing of euchromatic genes by PEV results in mosaic expression of such genes in tissues where the genes are normally expressed uniformly. We do not know whether HP1-mediated repression of region 31 genes is similarly mosaic or whether HP1 reduces expression of target genes uniformly in all cells. Additionally, we do not know whether HP1 is targeted directly to down-regulated region 31 genes or whether such targeting is general or tissue specific.
HP1-family proteins are highly conserved from fission yeast to humans. Human orthologues of HP1 enhance PEV when expressed in Drosophila (30), suggesting functional conservation between human and Drosophila HP1 proteins. Expression of HP1Hsα is down-regulated at the RNA and protein levels in metastatic, compared with nonmetastatic, breast cancer, and HP1 antisense oligonucleotides induce increased invasiveness in breast cancer cell lines, suggesting that decreased HP1Hsα expression can influence the expression of genes involved in tumor-cell invasion (31). Natural target genes of human HP1 proteins are unknown, but chromosome immunolocalization studies have revealed numerous sites of human HP1-family protein binding in euchromatin (32). Our results suggest that some of these sites may represent targets of authentic HP1-mediated transcriptional regulation in euchromatin.
Acknowledgments
We thank P. Harte for critical comments on the manuscript. This work was supported by National Institutes of Health Grants CA66974 (to H.J.W.) and GM57005 (to J.C.E.). H.J.W. is an Irma T. Hirschl scholar.
Abbreviations
- HP1
heterochromatin protein 1
- PEV
position–effect variegation
- RDA
representational difference analysis
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.James T C, Elgin S C. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C R. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eissenberg J C, Morris G D, Reuter G, Hartnett T. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eissenberg J C, Elgin S C. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 5.Lu B Y, Emtage P C R, Duyf B J, Hilliker A J, Eissenberg J C. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature (London) 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Bannister A J, Zegerman P, Partridge J F, Miska E A, Thomas J O, Allshire R C, Kouzarides T. Nature (London) 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 8.Nakayama J-I, Rice J C, Strahl B D, Allis C D, Grewal S I S. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 9.Zhao T, Heyduk T, Allis C D, Eissenberg J C. J Biol Chem. 2000;275:28332–28338. doi: 10.1074/jbc.M003493200. [DOI] [PubMed] [Google Scholar]
- 10.Rea S, Eisenhaber F, O'Carroll D, Strahl B D, Sun Z, Schmid M, Opravil S, Mechtler K, Ponting C P, Allis C D, et al. Nature (London) 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 11.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mottus R, Sobel R E, Grigliatti T A. Genetics. 2000;154:657–668. doi: 10.1093/genetics/154.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn R, Heymann S, Lindigkeit R, Reuter G. Chromosoma. 1986;93:398–403. [Google Scholar]
- 14.Lorentz A, Heim L, Schmidt H. Mol Gen Genet. 1992;233:436–442. doi: 10.1007/BF00265441. [DOI] [PubMed] [Google Scholar]
- 15.Lorentz A, Ostermann K, Fleck O, Schmidt H. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- 16.Wustmann G, Szidonya J, Taubert H, Reuter G. Mol Gen Genet. 1989;217:520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]
- 17.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lisitsyn N, Lisitsyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.James T C, Eissenberg J C, Craig C, Dietrich V, Hobson A, Elgin S C R. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 21.Dombradi V, Axton J M, Glover D M, Cohen P T W. FEBS Lett. 1989;247:391–395. doi: 10.1016/0014-5793(89)81377-8. [DOI] [PubMed] [Google Scholar]
- 22.Reuter G, Spierer P. BioEssays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 23.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 24.Seeler J-S, Marchio A, Sitterlin D, Transy C, Dejean A. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehming N, Le Saux A, Schüller J, Ptashne M. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolshakov V N, Zharkikh A A, Zhimulev I F. Chromosoma. 1985;92:200–208. [Google Scholar]
- 28.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. 454. [Google Scholar]
- 29.Lohe A R, Hilliker A J, Roberts P A. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J, Hwang K-K, Worman H J, Courvalin J-C, Eissenberg J C. Chromosoma. 2001;109:536–544. doi: 10.1007/s004120000113. [DOI] [PubMed] [Google Scholar]
- 31.Kirschmann D A, Lininger R A, Gardner L M G, Seftor E A, Odero V A, Ainsztein A M, Earnshaw W C, Wallrath L L, Hendrix M J C. Cancer Res. 2000;60:3359–3363. [PubMed] [Google Scholar]
- 32.Minc E, Allory Y, Worman H J, Courvalin J-C, Buendia B. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]