Abstract
Locus control regions (LCRs) are defined by their ability to confer high-level tissue-specific expression to linked genes in transgenic assays. Previously, we reported that, at its native site, the murine β-globin LCR is required for high-level β-globin gene expression, but is not required to initiate an open chromatin conformation of the locus. To further investigate the mechanism of LCR-mediated transcriptional enhancement, we have analyzed allele-specific β-globin expression and the pattern of histone acetylation in the presence and absence of the LCR. In single cells from mice heterozygous for a deletion of the LCR, β-globin expression from the LCR-deleted allele is consistently low (≈1–4% of wild type). Thus, the endogenous LCR enhances globin gene expression by increasing the rate of transcription from each linked allele rather than by increasing the probability of establishing transcription per se. Furthermore, in erythroid cells from mice homozygous for the highly expressing wild-type β-globin locus, hyperacetylation of histones H3 and H4 is localized to the LCR and active genes. In mice homozygous for the LCR deletion reduced histone hyperacetylation is observed in LCR proximal sequences; however, deletion of the LCR has no effect on the localized hyperacetylation of the genes. Together, our results suggest that, in its native genomic context, the LCR follows the rate model of enhancer function, and that the developmentally specific hyperacetylation of the globin genes is independent of both the rate of transcription and the presence of the LCR.
A number of eukaryotic gene loci are regulated by locus control regions (LCRs). These elements have been defined by their ability to confer high-level tissue-specific expression to linked genes, as well as to overcome position effects in transgenic assays. Thus, LCRs are believed to not only enhance transcription but also to have dominant chromatin opening activity (reviewed in refs. 1–3), distinguishing them functionally from classical enhancers. At present, the most fully characterized multigene clusters containing LCRs are the mouse and the human β-globin loci. By using homologous and site-specific recombination techniques, we deleted the LCRs of the mouse and human globin loci in tissue culture models to determine whether these elements mediate chromatin opening and transcriptional activation in their native genomic locations. Surprisingly, although these deletions resulted in the significant reduction or elimination of globin gene expression, deletion of neither the human nor mouse LCR resulted in a closed chromatin structure of the endogenous β-globin locus (4, 5). Most recently we deleted the LCR in mice and demonstrated that, after germ-line transmission, the locus resides in an open chromatin conformation as measured by DNaseI sensitivity, and that the genes are expressed at very low levels (6). These findings indicate that elements outside of the LCR are capable of mediating an open chromatin structure in the absence of the LCR, and that the primary function of the LCR in its endogenous location is to enhance transcription of the β-like globin genes.
Two contrasting models of enhancer-mediated activation of transcription have been proposed. One view is that enhancers function by increasing the rate at which a gene is transcribed (rate or rheostat model). An alternate view suggests enhancers function primarily by increasing the probability that a linked gene will establish transcriptional activity (binary model), rather than to increase the rate of transcription (1, 7). Although most experimental data are consistent with either model, only a few studies have attempted to distinguish between these two possibilities, as such distinction requires quantitative assays of expression in single cells. Recent analyses of transgenes integrated at defined genomic locations in erythroid cell lines have provided evidence for both models. For example, investigation of control constructs and constructs containing the β-globin LCR 5′HS2 enhancer integrated at the same genomic sites in human erythroleukemia K562 cells revealed a role for enhancers in the localization of genes into nuclear compartments that favor stable gene expression (8). These studies provided a potential mechanism for the binary model: the “off” state may reflect the location of a gene in a heterochromatic nuclear compartment, and the “on” state may reflect enhancer-mediated relocation of the gene to a compartment permissive for expression. In contrast, study of the same 5′HS2 enhancer, as well as other components of the β-globin LCR, at defined genomic sites in murine erythroleukemia (MEL) cells revealed enhancer-mediated affects on the rate of transcription rather than on the probability of achieving the active state (9). The different conclusions from these studies may reflect the nature of the genomic sites investigated.
Histone acetylation has been associated with chromatin opening and gene regulation in many systems. Localized hyperacetylation is generally linked to transcriptional activation (10–12). Furthermore, studies of the sequential events during promoter activation suggest that histone-acetyl transferase (HAT) recruitment occurs before polymerase initiation and that histone acetylation can be a rate-limiting step in this process (13–16). In addition, studies of histone acetylation throughout multigene loci have suggested that one of the functions of LCRs is to recruit HAT activity (12, 17–20). It has been hypothesized that increased histone acetylation could result in chromatin decondensation and accessibility to trans-activating factors at the distal promoter, linking LCR function and promoter activation.
To investigate the molecular events leading to LCR-mediated activation in its native genomic context, we have performed single-cell analyses of β-globin gene expression and have determined the extent of histones H3 and H4 acetylation in globin loci of erythroid cells from wild-type (wt) mice and from mice with a germ-line deletion of the endogenous LCR (ΔLCR). Our single-cell analysis of mice heterozygous for wt and ΔLCR alleles reveals homogeneous low-level expression from the ΔLCR allele in every erythroid cell analyzed, suggesting that the absence of the LCR reduces the rate of expression from linked genes. Moreover, analysis of mice homozygous for the wt locus reveals high levels of hyperacetylation localized to the LCR and the active genes. Although acetylation of sequences proximal to the LCR is markedly reduced in homozygous ΔLCR mice, hyperacetylation of the genes is unaffected by the deletion of the LCR.
Taken together, our results suggest that LCR-mediated transcriptional enhancement at the native locus follows the rate model, and that hyperacetylation of the globin genes in erythroid cells is independent of both the rate of transcription and the presence of the LCR.
Materials and Methods
Single-Cell Allele-Specific Expression Analysis.
Analysis of transcription was performed on mice heterozygous for the HbbS (S) and HbbD (D) alleles. Mouse blood was heparinized and washed with PBS. The equivalent of 0.2 μl of blood was resuspended in 1 ml of Retic-Count (Becton Dickinson) and was incubated for 30–90 min at room temperature. Reticulocytes were identified and isolated by using a fluorescence-activated cell sorter (FACS) Vantage Flow Cytometer and single-cell deposition unit (Becton Dickinson) according to the manufacturer's recommendations with the exception of using linear scales for measuring light scatter. A group of cells were sorted to confirm their identity as reticulocytes by staining with new methylene blue. Stream alignment was confirmed before each collection. To minimize the risk of sorting more than one cell per well, a narrow forward scatter pulse width was gated on, and hardware was checked by deposition of single 10-μm fluorescent beads into tubes. Of all 576 wells checked, each contained only 1 bead.
Reticulocytes were sorted into PCR tubes containing 5 μl of lysis buffer [10 mM Tris, pH 7.4/5 mM DTT/0.4% Nonidet P-40/0.5 units Rnasin (Promega)]. After mixing, tubes were briefly centrifuged at 1,000 × g and stored at −80°C. For reverse transcription (RT), primer BM2R (GGTTTAGTGGTACTTGTGACGG) was added, samples were heated to 70°C for 10 min, cooled on ice, briefly centrifuged, and warmed to 42°C before adding the RT mix. RT was done at a primer concentration of 100 nM with Superscript II RT (GIBCO) according to the manufacturer except that the reaction contained 5 units/μl of RT enzyme and Rnasin at 1 unit/μl. Reverse transcriptase was inactivated at 70°C, samples cooled to 37°C, and RNaseH (GIBCO) was added at 0.15 units/μl and incubated for 20 min. Additional reverse primer and the forward primer BMAJinF (CAAGCTGCATGTGGATCCTGA) were added to a concentration of 80 nM in the presence of PCR buffer and Taq polymerase (Perkin–Elmer). PCR was performed as described except that after 6 cycles, samples were aliquoted into 3 tubes, additional PCR mix containing α-labeled [32P]CTP was added, and the aliquots from each cell were amplified an additional 17, 19, and 21 cycles. The amplification products were digested with BstXI, and the resulting restriction fragments were separated on a 6% acrylamide gel and quantified as described (21, 22).
To be included in the quantitative analysis, samples needed to demonstrate (i) linear amplification of the more abundant product from the S allele in all three aliquots, (ii) a signal to noise ratio over two, and (iii) that the D/S ratio (ratio of RT-PCR products from the D and S alleles) was not decreasing with increasing cycles of PCR, as this indicates heteroduplex formation. Zero cell controls were routinely run and yielded no product. Ten-cell pools from S/S mice were analyzed and never yielded bands that migrated at the position of the D product. Ten percent of the mutant ΔLCR-D/S single cells had considerably less signal from the control S allele and therefore had to be excluded from our analysis. A mix of a single wt cell with 9 heterozygous mutant cells resulted in S/D ratios of 0.09 and 0.11, comparable to the predicted value of 0.12 and clearly different from the 0.03 seen only in mutant cells, confirming that a single “jackpot” cell would be detected in a 10-cell pool.
Chromatin Immunoprecipitation (IP).
A single cell suspension of predominantly erythroid mouse spleen cells was generated as described (6, 23). Formaldehyde cross-linking and chromatin purification were done as described (12) with the exception that the dialyzed chromatin was sonicated to reduce the average size to 500 bp. IP with Abs against panacetylated histones H4 and H3 acetylated at lysines 9 and 14 (Upstate Biotechnology, Lake Placid, NY) were performed as described (12), but by using 10 μg of input chromatin.
Duplex PCR Analysis of Immunoprecipitated DNA.
Quantitative PCR of input and bound chromatin was performed by using 1–2 ng of DNA. Primers were designed and shown to be mouse-specific and gave products between 280 and 400 bp. Primers to the amylase gene were used in duplex PCR assays with the globin primer sets as described (12). For each sequence, PCR reactions were performed in parallel under conditions of linear amplification, as described previously, for 27 cycles by using identical temperature profiles for all primer pairs. Quantitation was performed as described (12).
Primer Sequences.
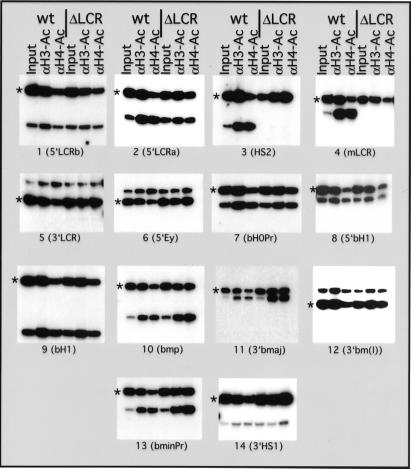
The positions of amplicons used in the mouse β-globin locus are shown in Fig. 4 and primer sequence and product size are listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Figure 4.
Quantification of duplex PCR results. Position of amplicons in the β-globin locus and quantification of chromatin IP results. The ratios of products obtained with the β-globin and amylase primers were determined for each input and Ab-bound sample for the wt and ΔLCR locus. The ratio for the bound fraction is normalized to the input to determine the relative level of enrichment. Enrichment of the globin sequences is reflected by a number >1. The x axis is drawn at 1, reflecting no enrichment. Shown are the averages of enrichment with standard deviation from three independent IPs.
Results
Analysis of Globin Expression in Individual Erythroid Cells from Mice.
Our previous analysis of pools of erythroid cells from mice heterozygous for a germ-line deletion of the endogenous LCR revealed that the ΔLCR allele expresses β-globin at a level only 1–4% of that of the wt allele (6). The binary model would predict that the low-level expression observed in bulk RNA results from a minority of cells which express at normal levels, whereas the majority of cells are transcriptionally silent. In contrast, if the LCR affects the rate of transcription, each allele would express at a similar low rate and reflect the level we previously observed in the analysis of bulk mRNA. To distinguish between these two possibilities, we developed a sensitive assay that allows the quantitative comparison of expression from a mutant allele to that from a wt control allele in individual erythroid cells (reticulocytes). This RT-PCR assay exploits polymorphisms between the two major β-globin alleles in mice, and was designed to assure quantitation in the linear range of amplification, minimize cycles of amplification, avoid heteroduplex formation, and minimize the manipulation and transfer of small amounts of cellular material and nucleic acids, all of which can lead to errors in quantitation (see Materials and Methods). Targeted deletion of the endogenous LCR was performed on the HbbD (D) allele, and heterozygotes with a wt HbbS (S) allele were generated, allowing comparison of expression from the D to the S alleles in individual cells.
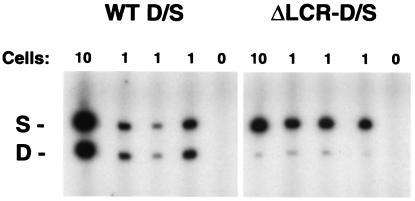
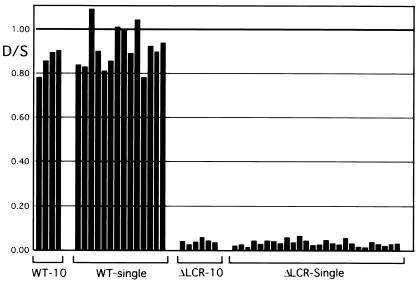
To determine whether deletion of the LCR leads to a heterogeneous population of high expressing jackpot cells and nonexpressing cells or to a decrease in expression in all cells, a combination of 10-cell pools and single reticulocytes from wt D/S and mutant ΔLCR-D/S mice were analyzed for adult mouse β-globin expression. Representative samples are shown in Fig. 1, and the results of quantitation are shown in graphic form in Fig. 2. The ratio of expression from the D allele to the S allele in the wt cells was close to 1 for both 10-cell pools and single cells [0.86 ± 0.06 (n = 4) for the pools and 0.91 ± 0.09 (n = 14) for the single cells]. A similar analysis of mice heterozygous for the LCR deletion (ΔLCR-D/S) revealed ratios of 0.04 ± 0.01 (n = 6) for the 10-cell pools and 0.03 ± 0.01 (n = 26) for the single cells, respectively. These results are remarkably similar to those observed in our prior analysis of bulk RNA (6). An additional 35 single cells were analyzed, all of which showed a comparably weak signal for the D allele. None had an intense band consistent with being a jackpot cell and none were nonexpressers. The signal to noise ratio observed in these 35 cells did not meet our stringent criteria, thus they were not used for quantitation (see Materials and Methods).
Figure 1.
Transcription from individual β-globin alleles in ΔLCR and wt mice. RNA from mice heterozygotic for the HbbD allele (D), and the HbbS allele (S) was analyzed from individual reticulocytes by RT-PCR. The targeted deletion of the LCR deletion is on the D allele, whereas the S allele is wt. Representative samples of 10-cell pools (10), single cells (1), and no-cell controls (0) from wt D/S mice and ΔLCR-D/S mice are shown. WT D/S, wt D/S animals; ΔLCR-D/S, mutant ΔLCR-D/S mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
Figure 2.
Quantitation of transcription from individual β-globin alleles in ΔLCR and wt mice. The ratio of expression from the D allele to that from the S allele for wt D/S animals (WT) or mutant ΔLCR-D/S mice (ΔLCR). Data from 10-cell pools and individual cells are shown.
In summary, all 61 evaluated ΔLCR-D/S single cells demonstrated a low level of expression from the D allele. No jackpot and no nonexpressing cells were observed. Although the existence of jackpot or nonexpressing cells cannot be ruled out, statistical analysis of the single-cell results reveals a 95% confidence that the true frequency of jackpots or nonexpressers is less than 5% (binomial exact confidence interval). In theory, the LCR could be regulating transcription in a binary manor, by increasing the probability that an allele is expressed at a rate of 100% rather than a basal rate of 4%. Such a mechanism is unlikely because partial LCR deletions in mice reduce expression to 30% of normal in both bulk- and single-cell analyses, rather than resulting in 30% of the cells expressing at a rate of 100% and nonexpressers (or cells expressing at a rate of 4%) comprising 70% of the population (M.A.B., unpublished observation). Thus, we conclude that removal of the LCR reduces the rate of transcription from each individual allele in vivo but not the probability that the allele is expressed.
Analysis of Histone Acetylation of the Mouse β-Globin Locus.
Previously, we observed LCR-independent formation of DNaseI-sensitive chromatin and remodeling of the β-globin gene promoter in erythroid cells from mice carrying a germ-line deletion of the LCR (6). Thus, the open chromatin state and promoter remodeling are not sufficient for high-level β-globin gene expression, and high-level expression requires the LCR. These findings provide an opportunity to address the question of which epigenetic modifications are LCR dependent and which are associated with high level of expression of the linked β-like globin genes. Modifications of histones, primarily acetylation and deacetylation, have been implicated in gene activation and repression (24, 25). Thus, the LCR could activate transcription by mediating HAT recruitment to the designated promoters. In this case, deletion of the LCR would lead to reduced levels of acetylation, which in turn would lead to the low and homogeneous basal expression observed in our single-cell analysis (see above).
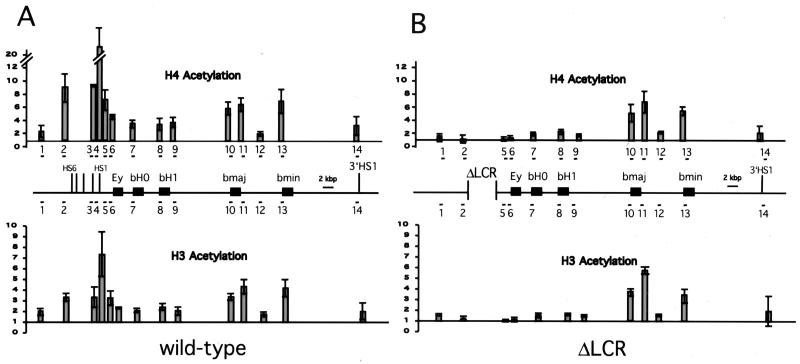
To determine how the histone acetylation of a multigene locus is influenced by the LCR, we examined the acetylation status of core histones H3 and H4 in wt and LCR-deletion alleles in the mouse β-globin locus in vivo. Mice were treated with phenylhydrazine as described (6) and, during recovery of the resultant hemolytic anemia, spleens were isolated at a time when erythroid cells are predominant. Single-cell suspensions were generated, and protein and DNA were cross-linked with formaldehyde. Cross-linked chromatin was isolated by isopycnic centrifugation and subsequently immunoprecipitated with Abs specific for acetylated histones H3 or H4. Ab-bound material enriched for each specific acetylated histone was analyzed for the abundance of sequences from the mouse globin locus and compared with a transcriptionally inactive control gene (amylase) by a duplex PCR approach as described (12). The ratio of the globin sequence to the control gene in the Ab-bound fraction was quantified and normalized to that of the input material for the IP. To generate a detailed picture of this multigene locus, a total of 14 sequences throughout the mouse β-globin locus were analyzed, spanning 103 kbp of genomic DNA (Figs. 3 and 4).
Figure 3.
Analysis of histones H3 and H4 acetylation across the wt and ΔLCR mouse β-globin locus. Chromatin was immunoprecipitated with an Ab that detects all acetylated variants of histone H4 [aH4-Ac] and an Ab that detects acetylated lysines 9 and 14 of histone H3 [aH3-Ac]. Abundance of the mouse globin sequence was detected relative to the mouse amylase gene, which is hypoacetylated in erythroid cells (12). Duplex PCR was performed on the input and Ab-bound fraction from the chromatin IPs by using amylase- and βglobin-specific primers. In each set, the amylase amplification product is marked with an asterisk. The β-globin amplicons are numbered in 5′ to 3′ direction of the locus. Locations of the amplicons in the locus are shown in Fig. 4 and primer sequences are listed in Table 1. Quantification of the ratio of globin to amylase is pictured in Fig. 4 and reveals up to 25-fold enrichment of globin sequences in the bound fraction. The figure shows the amplification products from one of three independent experiments.
Histone Acetylation of the wt β-Globin Locus.
Fig. 3 shows the acetylation analysis of the locus and Fig. 4A shows the relative enrichment for acetylated histones H4 and H3 for sequences in the normal mouse β-globin locus. The analysis reveals regions with different levels of acetylation for both H3 and H4 throughout the locus. Sequences at and flanking the LCR (amplicons 2–5) show a high level of enrichment indicative of histone hyperacetylation and the recruitment of HATs to this control element. The increased acetylation of the LCR is also present at two sequences flanking the LCR. The regions downstream of the LCR, including the inactive genes Ey and bH1, as well as the pseudogene bH0, are modestly enriched compared with the control amylase gene (amplicons 6–9), suggesting that they are in an intermediate level of acetylation—less than the hyperacetylated LCR but higher than the amylase gene, which is hypoacetylated in erythroid cells (12).
The actively transcribed β-major and β-minor genes show stronger enrichment of histone acetylation than the inactive genes, suggesting that they reside in chromatin that is hyperacetylated for histones H3 and H4 at levels comparable to the LCR. This localized hyperacetylation was detected at the major and minor promoters (amplicons 10 and 13) and at the 3′ end of the major gene (amplicon 11), indicating that the promoter and the gene are hyperacetylated. However, this hyperacetylation seems to be limited to the genes themselves, because the intergenic region between major and minor (amplicon 12) displays only a modest level of acetylation. In summary, our analysis of the wt murine β-globin locus from anemic spleen reveals hyperacetylation for histones H3 and H4 localized to the LCR and the active genes, whereas the intervening regions show a reduced level of acetylation.
Histone Acetylation of the ΔLCR β-Globin Locus.
To determine whether the acetylation pattern of the wt locus depends on the LCR, we analyzed mice containing a targeted deletion of the LCR. Mice homozygous for the LCR deletion die during embryogenesis but can be rescued by breeding in a human β-globin yeast artificial chromosome (YAC) transgene (6, 26). The acetylation pattern of these ΔLCR homozygous mice was analyzed at the identical set of sequences used in the analysis of wt mice, with the exception of two (amplicons 3 and 4), which reside in the deleted LCR (Figs. 3 and 4B). The remaining LCR-proximal amplicons reside immediately upstream or downstream of the deletion. Although both of these LCR sequences displayed hyperacetylation in the wt mice, they displayed a low level of acetylation on deletion of the LCR, suggesting that the hyperacetylation of LCR-proximal chromatin depends on sequences at the LCR.
As in the wt locus, the region between the LCR and the major and minor genes shows a lower level of enrichment. However, the relative enrichment at all four sequences (amplicons 6–9) is modestly reduced (2-fold) in the LCR deletion compared with the normal allele, indicating that the absence of the LCR leads to a reduced level of acetylation in this region.
Remarkably, even in the β-globin LCR-deletion allele, the chromatin at the major and minor genes displays a localized peak of acetylation indistinguishable from the highly expressing wt allele. The localization of the hyperacetylated subregion is similar, as is the relative level of enrichment for histones H3 and H4. This result has two implications: (i) The LCR, although required for full transcriptional activation of the appropriate promoters in the mouse locus, does not establish the degree of histone acetylation of these genes, and (ii) the level of acetylation at the mouse major and minor genes does not correlate with an activated level of transcription, but rather with transcription per se.
Discussion
Little is known about how distal cis-acting sequences such as enhancers and LCRs influence the sequential changes in chromatin structure and promoter activation leading to high levels of gene expression (27). Previously, we showed that the endogenous mouse β-globin LCR is not required for an open chromatin structure of the globin locus, but is necessary for high expression of the globin genes (6). To understand the mechanism by which the LCR activates expression at the endogenous mouse β-globin locus in vivo, we have performed an analysis of core histones H3 and H4 acetylation throughout the locus and analyzed the expression from individual alleles in the presence and absence of the LCR.
Histone Acetylation and LCR Function.
The modification of histones, especially acetylation and deacetylation, is involved in regulating chromatin structure and gene expression (24, 25). The association of histone hyperacetylation and gene activity in several systems has led to the suggestion that the recruitment of HAT activity to a promoter is a rate-limiting step in transcriptional activation. Consistent with this suggestion, we observe localized hyperacetylation at the LCR and at the highly expressed genes of the wt locus, indicating the recruitment of HAT activity to the LCR and the active genes. In contrast, the initial examination of acetylation of the avian β-globin locus with a non-histone Ab against acetylated lysine revealed a locuswide and uniform level of hyperacetylation (28). However, a more recent analysis of the avian locus with Abs specific for acetylated histones H3 and H4 revealed localized hyperacetylation of regulatory elements and active genes (18), similar to the mouse locus.
We and others have proposed that the LCR is required for hyperacetylation of the β-globin genes (12, 20). Yet, our analysis of the mouse LCR deletion allele reveals that core histone hyperacetylation of the active β-globin genes is LCR independent. Because the Abs used in the analysis of the murine locus are not directed against specific H3 or H4 lysine residues, we cannot exclude the possibility that deletion of the LCR affects the pattern of acetylated lysines without changing the overall level of H3 and/or H4 hyperacetylation.
We have shown previously that β-globin-promoter remodeling occurs in the absence of the LCR (6). Thus, promoter hyperacetylation and remodeling do not correlate with the level of transcription, and the LCR is not necessary for the recruitment of HATs to the promoter and transcribed sequences. These processes must therefore depend on specific non-LCR sequences, or are a direct consequence of an open chromatin domain in an erythroid milieu. Such sequences may reside upstream of the LCR, potentially in the regions of homology with the human locus that are absent in the various deletion thalassemias (29). It has been suggested that the reduced acetylation in the region between the LCR and the active genes in the wt mouse globin locus is indicative of hypoacetylated chromatin (20). Our data suggest rather that these sequences are in an intermediate or “basal” state, lower than the hyperacetylated genes and the LCR but still enriched compared with a gene that is not expressed in erythroid cells (amylase). Although deletion of the LCR does not affect acetylation at the β-major and -minor promoters, this deletion does result in a strong reduction in acetylation proximal to the LCR and a small but reproducible reduction at the sequences further downstream. Thus, the LCR might affect not only histone acetylation at sequences proximal to the LCR proximal sequences, but also at those between the LCR and the active genes.
In our recent study of the human β-globin locus, we also observed localized hyperacetylation of the LCR and the active gene in the wt locus, but only for histone H3 (12). Our results from the mouse locus do not suggest a histone H3 preference of the recruited HAT activity, and these results are consistent with a recent analysis of murine fetal liver at a developmental stage, in which the adult genes are expressed (day 14.5 after conception; ref. 20). This consistency of results during different stages of development suggests that the acetylation we observe is typical of mouse erythroid cells expressing the adult β-globin genes.
In our analysis of the human locus in cell lines, promoter hyperacetylation was not observed in the absence of the LCR (12). Of note, no transcription is detected in the absence of the LCR in the human locus, whereas the mouse locus shows low-level transcription in the absence of the LCR (4–6). Thus, promoter hyperacetylation might reflect transcription per se, rather than correlate with the level of transcription. In the case of the mouse locus, gene proximal sequences may be sufficient to mediate this low-level transcription. Our results do not exclude the possibility that histone acetylation can be a rate-limiting step in the process of transcriptional activation of the mouse globin genes, but indicate that the LCR acts subsequent to promoter hyperacetylation.
Rate vs. Binary Enhancement of Transcription.
Work from several systems, including targeted deletion of cis-acting elements from endogenous loci (30, 31), suggested that enhancers and LCRs work primarily by affecting the probability that a gene is expressed (binary model), leading to the prediction that deletion of the LCR would result in a decrease in the number of alleles that are expressed. The binary model predicts that the low level of expression detected in the absence of the LCR would result from a low percentage of alleles expressing at wt levels (jackpots), whereas the majority of alleles are transcriptionally silent. Previous analysis, using a chromosome-transfer system, determined that a targeted deletion of the murine β-globin LCR led to moderate and variable levels of expression in a human cell line, and no jackpot cells were observed (4). However, this trans-species chromosome transfer system does not allow for passage through the germline, and the pattern of epigenetic modifications of the locus in tissue culture cells may not reflect those seen in a physiologically normal situation. Thus, we examined reticulocytes of mice after the LCR deletion was passed through the germline for several generations. Expression from the ΔLCR allele is low in single cells and is comparable to that observed in the analysis of bulk populations. Expression is remarkably uniform, and no jackpot cells or nonexpressing cells are observed. Although statistically it is impossible to rule out the possibility of rare jackpot cells or nonexpressing cells, the vast majority of alleles homogeneously express a low level of β-globin mRNA, which accounts for the low basal level of expression observed in the bulk RNA analysis. Our single-cell analysis, in conjunction with our hyperacetylation and chromatin data, is consistent with the majority of cells having a homogeneous phenotype in regard to transcription, histone acetylation, and chromatin structure: (i) low level basal transcription, (ii) hyperacetylation and promoter remodeling of the appropriately developmentally expressed genes, and (iii) an open chromatin structure of the β-globin domain. Thus, the primary effect of the LCR at the endogenous locus in vivo is to stimulate the rate of transcription rather than to increase the probability that a specific allele is expressed.
Our results do not contradict a binary model for LCR or enhancer function, but rather suggest that these elements have several activities that may be context-dependent. Several studies leading to the binary model involved transgenes stably integrated at random ectopic genomic sites (32). At sites where the transgene integrates into a silenced chromatin domain, the enhancer must lead to the opening of the domain, and this property may occur in a binary manner. For example, enhancer activation of transgenes is associated with a relocalization away from areas of pericentric heterochromatin in the nucleus, and this may lead to an open DNaseI-sensitive and hyperacetylated chromatin structure (8). Once an open active structure is achieved, enhancers and LCRs may act primarily to alter the rate of expression. Consistent with this model, the addition of enhancer constructs at ectopic genomic sites, which are in an open chromatin conformation in the absence of an enhancer, affects only the rate of transcription (9). Thus, at the endogenous mouse β-globin locus, where an open structure is present even in the absence of the LCR, the major role for the LCR is to convert basal transcription to activated transcription.
A Model for β-Globin Activation.
Taken together, these results suggest a model of sequential activation of the β-globin locus in which promoter remodeling and gene hyperacetylation lead to uniform low-level (basal) transcription and precede LCR-mediated high-level (activated) transcription. The interdependence of this acetylation and chromatin remodeling is not known, but neither is sufficient to enhance expression. The temporal order of molecular events leading to transcriptional activation is still poorly understood; however, in the few cases in which an endogenous locus was studied, HAT recruitment to a promoter has been shown to be critical for transcription and to occur before polymerase initiation (13–16). The LCR may be necessary for modifying or recruiting new components to the remodeled and hyperacetylated promoter, or for additional chromatin modifications of the locus not detected here.
Alternatively, LCR activity might occur subsequent to transcriptional initiation. Transcriptional pausing is a general rate-limiting step after polymerase initiation, and various activators, enhancers, and LCRs are known to increase elongation efficiency. For example, we have shown previously that the 3′Cα IgH LCR activates c-myc gene expression by overcoming promoter-proximal polymerase pausing (33, 34). Thus, the β-globin LCR may convert expression from a basal to an activated state by increasing the elongation efficiency of transcription initiated at the β-globin promoters. Unfortunately, the viability of homozygous ΔLCR mice depends on the presence of a human β-globin transgene, and the high sequence similarity between the mouse and human β-globin genes excludes the use of standard techniques to detect paused polymerases. However, regardless of the potential mechanisms of LCR-dependent gene activation, our results indicate that this activation is independent of HAT recruitment to the promoter. Further elucidation of how the LCR elevates the rate of transcription will require the determination of the components and modifications of the proteins bound to the promoter in the presence and absence of the LCR.
Supplementary Material
Acknowledgments
We thank Jessica Halow for expert technical assistance; the Fred Hutchinson Cancer Research Center Shared Resources Flow Cytometry Laboratory, Image Analysis Laboratory, and Core Biotech Facility; the members of the Groudine laboratory for helpful suggestions; Mike Bulger for sequence data; and Claire Francastel, Matthew Lorincz, and Mike Bulger for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants DK44746 and HL65440 (to M.G.), and by fellowships from the Deutsche Forschungsgemeinschaft (DFG) and the Rett Syndrome Research Foundation (RSRF) (to D.S.). M.A.B. is a J. S. McDonnell Foundation Scholar.
Abbreviations
- LCR
locus control region
- HAT
histone-acetyl transferase
- RT
reverse transcription
- D
HbbD
- S
HbbS
- wt
wild type
- IP
immunoprecipitation
References
- 1.Bulger M, Groudine M. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 2.Engel J D, Tanimoto K. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 3.Higgs D R. Cell. 1998;95:299–302. doi: 10.1016/s0092-8674(00)81761-4. [DOI] [PubMed] [Google Scholar]
- 4.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I, Kennedy M, Keller G, Groudine M. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 5.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender M A, Bulger M, Close J, Groudine M. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 7.Fiering S, Whitelaw E, Martin D I. BioEssays. 2000;22:381–387. doi: 10.1002/(SICI)1521-1878(200004)22:4<381::AID-BIES8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Francastel C, Walters M C, Groudine M, Martin D I. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- 9.Molete J M, Petrykowska H, Bouhassira E E, Feng Y Q, Miller W, Hardison R C. Mol Cell Biol. 2001;21:2969–2980. doi: 10.1128/MCB.21.9.2969-2980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 11.Parekh B S, Maniatis T. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 12.Schubeler D, Francastel C, Cimbora D M, Reik A, Martin D I, Groudine M. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 13.Cosma M P, Tanaka T, Nasmyth K. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 14.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 15.Krebs J E, Kuo M H, Allis C D, Peterson C L. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang Y, Hu X, DiRenzo J, Lazar M A, Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 17.Elefant F, Su Y, Liebhaber S A, Cooke N E. EMBO J. 2000;19:6814–6822. doi: 10.1093/emboj/19.24.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litt M D, Simpson M, Recillas-Targa F, Prioleau M, Felsenfeld G. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurry M T, Krangel M S. Science. 2000;287:495–498. [PubMed] [Google Scholar]
- 20.Forsberg E C, Downs K M, Christensen H M, Im H, Nuzzi P A, Bresnick E H. Proc Natl Acad Sci USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender M A, Reik A, Close J, Telling A, Epner E, Fiering S, Hardison R, Groudine M. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 22.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 23.Reitman M, Lee E, Westphal H, Felsenfeld G. Mol Cell Biol. 1993;13:3990–3998. doi: 10.1128/mcb.13.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 25.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 26.Gaensler K M, Kitamura M, Kan Y W. Proc Natl Acad Sci USA. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 28.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weatherall D J. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Nienhus A W, Majerus P W, Varmus H, editors. Philadelphia: Saunders; 1994. pp. 157–206. [Google Scholar]
- 30.Xu Y, Davidson L, Alt F W, Baltimore D. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 31.Ronai D, Berru M, Shulman M J. Mol Cell Biol. 1999;19:7031–7040. doi: 10.1128/mcb.19.10.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 33.Madisen L, Groudine M. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- 34.Madisen L, Krumm A, Hebbes T R, Groudine M. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.