Abstract
The gene encoding the rice transcription factor, REB (rice endosperm bZIP) was cloned from a bacterial artificial chromosome library of rice. The cloned 6,227-bp-long Reb gene is composed of six exons and five introns and is flanked by a 1.2-kb 5′ promoter and a 1.2-kb 3′ terminator region. The function of the Reb gene was explored by a transient assay by using a rice immature endosperm system. The effector constructs containing the native gene or fusion genes linking Reb to the rice actin (Act) or globulin (Glb) gene promoters and the reporter gene construct Glb-β-glucuronidase (GUS) were used in this study. When these effector constructs were cotransferred with the reporter uidA gene encoding GUS under the control of the Glb promoter into immature rice endosperm cells, the Glb promoter was activated. The transient GUS expression was 2.0 to 2.5-fold higher with the effector construct than without. When the upstream activation sequence containing the GCCACGT(A/C)AG motifs of the Glb promoter was deleted, the activation by REB was abolished. On the other hand, a gain-of-function experiment showed that inserting the upstream activation sequence into the glutelin-1 (Gt1) promoter made it responsive to activation by REB. When cotransformed with Reb gene, mature transgenic rice grains containing the human lysozyme gene driven by the Glb promoter produced 3.7-fold more lysozyme. Accumulation of recombinant lysozyme in mature seed ranged from 30.57 to 279.61 μg⋅mg−1 total soluble protein in individual transformants from 30 independent transformation events. Thus, our results show that REB is not only a transcriptional activator, it can also be used to increase the expression of recombinant protein in transgenic rice grains.
Keywords: Reb gene, lysozyme expression, transgenic plant, gain-of-function, loss-of-function
The developing endosperm of cereal grains and the developing cotyledons of legume grains are highly suitable as bioreactors for the production of recombinant proteins (1–3). Among the tested recombinant proteins are a protein-engineered enzyme (1), recombinant antibodies in the form of single chain antibody fragments (scFvs) (2, 3), the human milk protein beta casein (4), the antimicrobial protein lactoferrin (5), and thioredoxin (6). Although expression levels of 53 ± 2.8 μg⋅mg−1 total soluble protein (TSP) (range 37–77 μg⋅mg−1) have been obtained with recombinant (1,3-1,4)-β-glucanase in the barley endosperm (1), the production level of single chain antibody fragments reached only 32 ng⋅mg−1 TSP in rice endosperm and 1.4 ng⋅mg−1 TSP in wheat endosperm (3). A high level of expression in transgenic grains becomes an important issue when the commercial production, application, or purification requires a high concentration of recombinant protein in the grain. Successful increases of recombinant protein synthesis and storage have been achieved by codon optimization of the transgene to a C + G content of more than 60% (1), by selection of a stronger promoter (7), and by using a signal peptide to target the protein into the endoplasmic reticulum for deposition into protein bodies (1, 2). A further or alternative way of increasing the amount of recombinant protein might be the transgenic expression of transcription factors that have the ability to transactivate the target genes by binding to nucleotide sequence motifs in the gene promoter (8–11). This possibility has been investigated with the experiments reported in the present communication.
In cereals, bZIP proteins have been identified as transcriptional factors for genes encoding storage proteins in the endosperm (10–20). The basic amino acids containing domain of these proteins bind to a recognition nucleotide sequence in the promoter, whereas their leucine repeat domain interacts with the transcriptional machinery, leading to a dramatic increase of transcription initiation of the gene in the endosperm of the cereal grain (11, 15). Nakase et al. (21) have cloned a bZIP protein gene from rice. This protein, named REB for rice endosperm bZIP protein, binds specifically to the sequences GCCACGTAAG and GCCACGTCAG in the distal part of the rice globulin (Glb) gene promoter (21). Its function as a transcriptional activator or suppressor, however, remains to be elucidated.
We have isolated a rice Reb gene from a bacterial artificial chromosome (BAC) library and shown with transient expression analyses that the REB protein can specifically enhance the expression of the β-glucuronidase (GUS) reporter gene under the control of the Glb gene promoter. The enhancement requires the presence of the REB upstream activation sequence (UAS) in the Glb promoter. It is further demonstrated that REB can increase the expression of human lysozyme in developing transgenic rice grains.
Materials and Methods
Cloning of Reb Gene.
Screening of BAC library.
With reference to the Reb gene sequence (21), PCR primers were designed and used to screen a rice BAC library (22). The forward primer had the sequence 5′-CTGATATGTGCCCATGTTCCAAAC-3′, and the reverse primer was 5′-CCTTGCTGAATGCAGATGTTTCAC-3′. The screening strategy of three-dimensional DNA pools of the BAC library has been described (23). PCR was carried out with 100 ng of pooled BAC DNA/10 mM Tris (pH 8.3)/1.5 mM MgCl2/50 mM KCl/0.5 μM dNTP and used a program of denaturing at 94°C for 5 min followed by 30 cycles of 94°C for 45 s, 60°C for 45 s, and 72°C for 1 min. The plasmid DNA of a positive BAC clone was prepared as described (22), the BAC DNA was digested with HindIII, and the presence of the Reb gene was confirmed by Southern analysis (24).
Subcloning and sequencing.
The Reb gene was retrieved from the BAC by subcloning two fragments into the pBluescript KS+ vector (Stratagene). First, the promoter and partial coding region was obtained as a KpnI–HindIII fragment (Fig. 1). In a second step, a HindIII fragment containing the remaining coding region and the 3′ terminator region was obtained by shotgun cloning (Fig. 1). The two fragments were ligated at the internal HindIII site, generating an intact Reb gene. The DNA was sequenced with an automatic ABI 371 instrument, and alignment of the sequence to the cDNA (21) established the structure of the gene (Fig. 1). The plasmid containing the gene is designated as pAPI266 (Native-Reb).
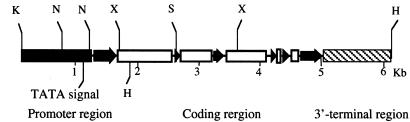
Figure 1.
Restriction map of the rice Reb gene isolated from BAC clone 42B9. The gene sequence of 6.227 kb consists of five introns (open bar) and six exons (solid arrow) flanked by 1.2 kb of the 5′ promoter (solid bar) and 1.2 kb of the 3′ region (striped bar). The restriction enzymes indicate K (KpnI), N (NruI), X (XhoI), S (StuI), and H (HindIII).
Construction of expression plasmids.
Plasmids API212 (Glb-GUS), carrying the rice globulin gene promoter fused to the GUS reporter gene, and API142 (Gt1-GUS), containing the rice glutelin 1 gene promoter fused to the GUS reporter gene, were used for transient expression assays. The globulin promoter (GenBank accession no. X63990) and glutelin 1 promoter (GenBank accession no. Y00687) were obtained from M202 (Oryza sativa Joponica subsp.) by PCR amplification. Plasmid API267 (Glb-Reb) was prepared by cloning the Reb coding region (NruI–SacI fragment, Fig. 1) into the Glb-GUS plasmid after removal of the GUS gene by digestion with SmaI–SacI, thus replacing the GUS gene with the Reb gene. Using the same strategy, plasmid API277 (Actin-Reb) was made by replacing the GUS gene of plasmid Act1-D-GUS (7) with the complete Reb gene (NruI–SacI fragment). A gene encoding the mature polypeptide of human lysozyme (EC 3.2.1.17) with a G + C content of 68.4% was synthesized (Operon Technologies, Alameda, CA) based on the sequence of GenBank accession no. J03801. The DNA was digested with DraI–XhoI and ligated into the NaeI–XhoI sites of the expression cassette in plasmid API241, which contains the rice globulin gene promoter and signal peptide (GenBank accession no. X63990). The resulting plasmid was named pAPI264.
Transient Expression Assays.
Details of the transient expression assays developed by Y. S. Hwang will be presented elsewhere. Briefly, rice spikelets with immature endosperm (7–9 days after pollination) of M202 were collected from plants grown in the greenhouse at 30°C. The spikelets were sterilized with 70% ethanol for 10 min. After evaporation of residual ethanol, the endosperm was dissected, and 10 immature endosperms were placed on a filter paper in a Petri dish containing AA medium (25) supplemented with 20 mM ammonium nitrate. Fifty microliters of gold particles (60 mg⋅ml−1 at 1:1 ratio of 1.0 and 1.5–3.0 μm diameter gold particle) were coated with 5 μg of DNA consisting of a mixture of the reporter gene, the effector gene, and the internal control gene at a molar ratio of 1:1:1. DNA coating was carried out as described in the instruction manual of the Biolistic PDS system (Bio-Rad). As internal control, pAHC18 containing the luciferase gene driven by an ubiquitin promoter (26) was used. In tests without the effector gene, effector DNA was replaced by pBluescript DNA. Particle bombardment was carried out with a Biolistic helium gun device at 1100 psi (Biolistic PDS 1000/He system, Bio-Rad). After bombardment, the immature endosperms were incubated at 25°C for 24 h in 5 ml of AA medium supplemented with 20 mM ammonium nitrate, 50 μg⋅ml−1 cefotaxin, and 50 μg⋅ml−1 timentin (SmithKline Beecham) to prevent bacterial growth. The endosperms were then harvested and ground with 55 μl of extraction buffer (0.1 M potassium phosphate, pH 8.0/1 mM EDTA/10 mM DDT/5% glycerol/0.2 mM leupeptin/0.2 μM phenylmethylsulfonyl fluoride). The extract was centrifuged at 25,000 × g for 5 min at 4°C. From the resulting supernatant, a 20-μl aliquot was added to 180 μl of luciferase assay buffer (0.25 M Tricine, pH 7.8/150 mM magnesium chloride/10 mM ATP/1 mM DDT/100 μg⋅ml−1 BSA). Another 20-μl aliquot was added to 200 μl of GUS assay buffer (Tropix, Bedford, MA). Luciferase activity was measured after incubation at 25°C for 20 min and GUS activity at 37°C for 1 h with a Monolight 2010 chemi-illuminometer according to the manufacturer's instructions (Analytical Luminescence Laboratory, San Diego). GUS activity was normalized to the luciferase activity and expressed relative to the activity of Glb-GUS. All data are averaged from at least six assays of two independent experiments.
Transformation and Plant Regeneration.
Microprojectile-mediated transformation of rice was carried out according to the procedure described in ref. 25. Calli were derived from the hypocotyls of germinating mature seeds of the cultivar TP309 (Oryza sativa, subsp. Japonica). Calli with a diameter of 2–4 mm were selected and placed on an N6 medium (Sigma) supplemented with 0.3 M mannitol and 0.3 M sorbitol for about 20 h before bombardment. Bombardment was carried out with the Biolistic PDC-1000/He instrument (Bio-Rad). Fifty microliters of gold particles (60 mg⋅ml−1 at 1:1 ratio of 1.0 and 1.5–3.0 μm diameter gold particle) were coated with effector DNA, target DNA, and selection marker DNA in a ratio of 3:3:1 (wt/wt) and accelerated with a helium pressure of 1,100 psi. After a 2-day incubation, calli were transferred to N6 selection media containing 35 mg⋅l−1 hygromycin B and allowed to grow in dark at 26°C for 45 days. Calli resistant to hygromycin B were transferred to regeneration media to generate plantlets as described in ref. 25. After shoots had reached a height of 1–3 cm, the plantlets were transferred to rooting media (MS plus 0.05 mg⋅l−1 α-naphthaleneactic acid, Sigma). After 2 weeks, the plantlets were transferred to soil and grown in a greenhouse to maturity.
PCR Analysis of Transgenic Plants.
Genomic DNA samples were prepared from transgenic plant leaves as described in ref. 27 and used as template for amplification with two pairs of primers for identification of the transgenes. For the rice Reb gene, the forward primer 5′-CCATCCAATCCAATCCACTCCAAC-3′ is based on a 3′ untranslated terminal sequence of the gene, and the reverse primer covers the vector sequence 5′-AGGCGATTAAGTTGGGTAACG-3′ (see Fig. 5). For the human lysozyme transgene, the 5′-CCTAGCCAAAGTCTTCGAGCGGTG-3′ forward primer hybridizes to the 5′ end of the gene's ORF and the reverse primer 5′-GCGATGTTGTCTTGCAGC-3′ to its 3′ end (Fig. 5). The PCR mixture contained 100 ng of genomic DNA/10 mM Tris (pH 8.3)/1.5 mM MgCl2/50 mM KCl/0.5 μM dNTP. Amplification used a program of denaturing at 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s. The PCR products were resolved by electrophoresis in a 1.2% agarose gel.
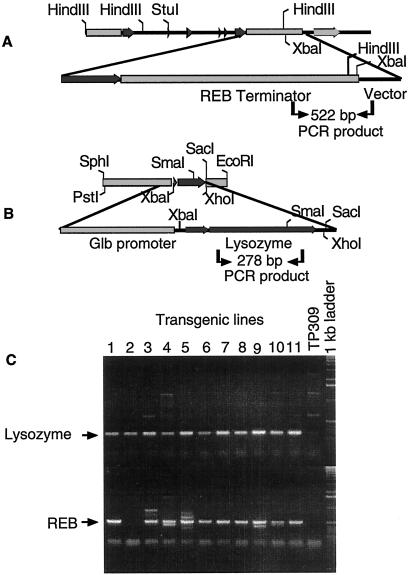
Figure 5.
PCR analysis of T0 transgenic plants containing the Reb and the human lysozyme gene. (A) The plasmid construct, API266 (Native-Reb), the primer positions (indicated by arrows), and amplified fragment size. One primer was designed from the vector region and the other from the Reb terminator. Only transformants can be amplified. (B) The diagram shows construct API264 (Glb-Lys), the primer positions, and the amplified fragment size. The primers hybridize to an internal sequence of the human lysozyme gene. (C) PCR analysis of Native-Reb/Glb-Lys cotransformed plants. Arrows mark the 522-bp fragment of the Reb/vector region and the 278-bp fragment from internal sequence of the human lysozyme gene.
Antibody Preparation and Western Blotting.
A peptide with 15 aa, PPPQSAAAAQQQGGG, from the position 19–23 of rice Reb gene was synthesized, and antiserum were prepared from rabbit by Alpha Diagnostic (San Antonio, TX). The total protein was extracted from immature seeds of transgenic lines at 14 days after pollination by using the extraction buffer (66 mM Tris, pH 6.8/2% SDS/2% β-mercaptoethanol). The resulting extract of total protein was run on a 12% polyacrylamide gel and blotted onto a nitrocellulose membrane according to the manufacturer's instructions (Bio-Rad). The blot was blocked with blocking solution (5% nonfat milk powder/0.02% sodium azide in PBS, pH 7.4) at 4°C overnight. Then, the blot was incubated with anti-REB antibody with 1:2,000 dilution of reaction solution (5% nonfat milk powder/0.05% Tween 20 in PBS, pH 7.4) at room temperature for 2 h. After washing with PBS for three times, 5 min each time, the blots were incubated with mice anti-rabbit IgG-conjugated alkaline phosphatase (1:2,500) in incubation solution (5% nonfat milk powder in PBS, pH 7.4) at room temperature for 2 h. After washing three times with TBS (pH 7.4), the blots were developed with 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium (Sigma).
Lysozyme Activity Assay.
The lysozyme assay used a procedure developed by Jianmin Huang (Applied Phytologics, Sacramento, CA). Briefly, 20 individual seeds from each T1 transgenic plant were ground in 1 ml of chilled extraction buffer (PBS plus 0.35 M NaCl). After centrifugation at 25,000 × g at 4°C for 5 min, the resulting supernatant was recovered. A series of dilutions was placed in a 96-well microtiter plate containing 250 μl of 0.015% Micrococcus letus cells in each well (Sigma). Human lysozyme (EC 3.2.1.17, Sigma) was used as standard. Lysozyme activity was measured as a decrease in turbidity by using Microplate Reader 3550 (Bio-Rad). The lysozyme concentration in the samples was read from a standard curve for different concentrations of human lysozyme. The lysozyme expression level in a given transgenic plant was computed as the average for the lysozyme content of all seeds of that plant. Total soluble protein in seed extracts was estimated with the Bradford protein assay (Bio-Rad).
Results
Cloning of the Reb Gene from a Rice BAC Library.
A rice BAC library was screened with the three-dimensional pool PCR strategy (23) by using PCR primers based on the cDNA sequence of the Reb gene (21). One positive clone, 42B9, was obtained. Restriction site mapping of 42B9 indicated that there was no convenient restriction sites for subcloning the entire Reb gene in a single fragment. The complete Reb gene was therefore generated by ligating a 1,775-bp fragment containing the promoter and the 5′ coding region to a 4,452-bp fragment containing the 3′ coding and terminator region (Fig. 1). DNA sequencing of the Reb gene revealed five introns, six exons, 1.16 kb of the 5′ promoter sequence, and 1.2 kb of the 3′ region totaling 6,227 bp (Fig. 1). Comparison of the ORF of our gene with the Reb cDNA sequence (Accession no. AB021737) revealed 99.97% DNA sequence identity and 99.99% amino acid identity resulting from two amino acid changes: Ile165→Asn and Glu215→Lys. These differences are likely to be because of polymorphisms among rice varieties.
Transcriptional Activation of the Globulin Promoter by REB.
To determine whether binding of the REB protein to the motif [GCCACGT(A/C)AG] in the globulin (Glb) gene promoter activates transcription of this promoter, plasmids containing fusions of the Reb coding region with the Glb promoter and the rice actin (Act) gene promoter were prepared (Fig. 2A). These plasmids as well as the expression plasmid containing the native Reb gene (pAPI266) were cobombarded into the rice endosperm with the plasmid containing the GUS reporter gene driven by the Glb promoter and the internal control plasmid containing the luciferase gene driven by the ubiquitin promoter.
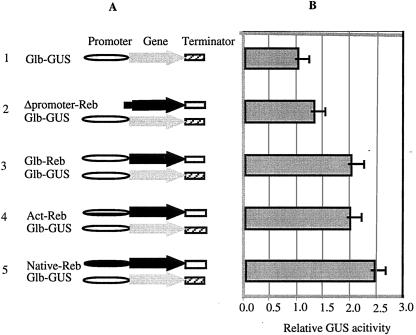
Figure 2.
The trans-activation function of the Reb gene toward the Glb promoter as analyzed with the GUS transient expression assay. (A) Schematic diagram of the constructs used for the transient assays. 1, GUS reporter construct with the Glb promoter and the Nos terminator; 2, Δpromoter-Reb-Term cobombarded with Glb-GUS-Nos reporter gene construct; 3, Glb-Reb-native-Term cobombarded with Glb-GUS-Nos reporter gene construct; 4, Act-Reb-Term cobombarded with Glb-GUS-Nos reporter gene construct; and 5, Native-Reb-Term cobombarded with Glb-GUS-Nos reporter gene construct. (B) Relative GUS activity corresponding to effector/reporter combinations shown in A. The error bar indicates the standard variation. The symbols indicate the Glb promoter (open bar), the Act promoter (horizontal striped bar), the Reb native promoter (black bar), the Reb gene (black arrow), the GUS gene (gray arrow), the Reb terminator (gray bar), and the Nos (striped bar).
Setting the level of GUS expression by the Glb promoter as 1, the codelivery of the plasmids containing the Reb gene increased GUS expression irrespective of whether the gene was driven by its own promoter, the Glb promoter, or the Act promoter (Fig. 2B). The increases were 2.43-, 2.01-, and 1.98-fold, respectively. The activation of GUS expression was abolished when a promoterless Reb construct was cobombarded with Glb-GUS (Fig. 2B). These results suggested that the REB protein functions as a transcriptional activator of the Glb promoter.
Identification of the UAS for REB.
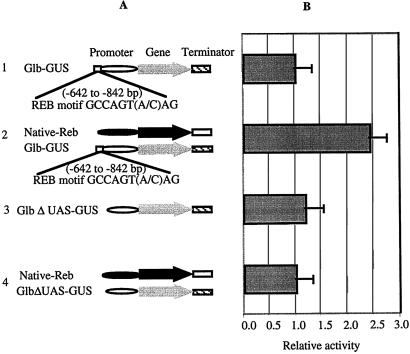
Using a band-shift assay, Nakase et al. (21) have shown that the binding motifs of REB are GCCACGTAAG or GCCACGTCAG. An analysis of the Glb promoter sequences revealed two copies of GCCACGTAAG and one copy of GCCACGTCAG clustered around −700 bp distal to the TATA box of the Glb promoter (Fig. 3A). If the binding motifs for REB signify a UAS, deletion of this sequence would abolish the transactivation by REB (experiment 3 in Fig. 3A). On the other hand, insertion of the binding motif into a promoter that cannot be activated by REB might introduce transactivation responsiveness. Accordingly, the 200 bp with the three motifs located at positions −642 to −842 from the TATA box were deleted from the Glb promoter (Fig. 3A). The deletion has no effect on the expression of GUS, as both Glb-GUS and GlbΔUAS-GUS show the same level of background GUS expression (experiments 1 and 3 in Fig. 3). When Native-Reb was cobombarded with GlbΔUAS-GUS (experiment 4), transcriptional activation by REB was lost (experiment 4 in Fig. 3). These data indicate that REB activation of Glb-GUS occurs through this 200-bp fragment containing REB binding motifs.
Figure 3.
The loss-of-function analysis of the REB trans-activation by transient assay. (A) Schematic diagram of constructs used for loss-of-function analysis. 1, the GUS reporter construct with the Glb promoter and the Nos terminator; 2, Native-Reb-native terminator construct cobombarded with Glb-GUS-Nos reporter construct; 3, GlbΔUAS-GUS-Nos construct; 4, Native-Reb-Term cobombarded with the Glb reporter construct in which REB UAS motifs of the Glb promoter are deleted. (B) Relative GUS activity corresponding to effector/reporter combinations shown in A. The error bar indicates the standard variation. The symbols indicate the Glb promoter (open bar), the Reb native promoter (black bar), the Reb gene (black arrow), the GUS gene (gray arrow), the Reb terminator (gray bar), and the Nos (striped bar).
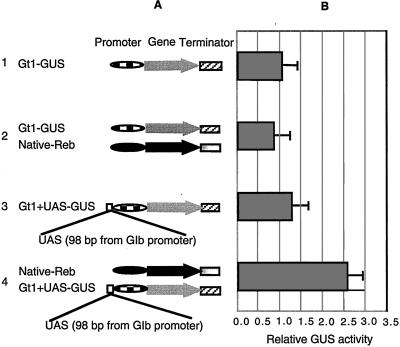
Scanning the rice glutelin1 (Gt1) promoter sequence does not reveal the presence of REB binding motifs. This suggested that the Gt1 promoter could serve as a candidate for constructing a gene that gains the REB response function through addition of the UAS from the Glb promoter. A construct containing the Gt1 promoter linked with the GUS gene was tested, and the results established that cobombardment of developing endosperm with the native Reb gene and the Gt1-GUS does not activate the Gt1 promoter (experiments 1 and 2 in Fig. 4). The 98-bp Glb fragment (from −626 to −724 distal to TATA box of the Glb promoter) containing three copies of GCCACGT(C/A)AG (Fig. 3) was inserted into the Gt1 promoter at a position −630 bp distal to TATA box of the Gt1 promoter to generate Gt1+UAS-GUS (Fig. 4A). Addition of the 98-bp fragment to the Gt1 promoter does not significantly increase its capacity for GUS expression (experiment 3 in Fig. 4). The small increase of GUS activity observed could be because of the presence of some endogenous REB protein in immature rice endosperm. However, when Gt1+UAS-GUS construct was cotransformed into the endosperm with the native Reb gene, a 2.5-fold increase in GUS activity was obtained (experiment 4 in Fig. 4).
Figure 4.
The gain-of-function analysis of the REB trans-activation by transient assay. (A) Schematic diagram of plasmid constructs used for the gain-of-function analysis. 1, Gt1-GUS-Nos; 2, Gt1-GUS-Nos with effector Native-Reb-Term; 3, modified Gt1-UAS-GUS-Nos; 4, modified Gt1+UAS-GUS-Nos with effector Native-Reb-Term. (B) Relative GUS activity corresponding to reporter constructs, Gt1-GUS-Nos, Gt1-UAS-GUS-Nos, and REB effector constructs shown in A. The error bar indicates the standard variation. The symbols indicate the Gt1 promoter (box bar), the Reb native promoter (black bar), the Reb gene (solid bar), the GUS gene (gray arrow), the Reb terminator (gray bar), the Nos terminator (striped bar), and the Reb UAS (open box).
The loss of activation function, when the 200-bp fragment (from −642 to −842 distal to TATA box of the Glb promoter) was removed from the Glb gene promoter, and the gain of this function, when the 98-bp fragment (from −626 to −724 distal to TATA box of the Glb promoter) was added to the Gt1 promoter, establishes the 98 bp of the fragment as a UAS.
The Reb Gene Increases Human Lysozyme Expression in Transgenic Rice Seed.
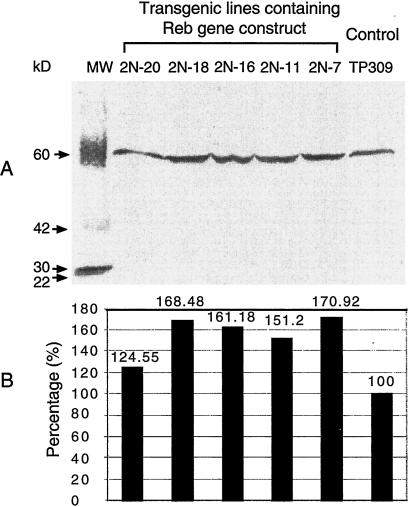
Because transient gene expression of REB in rice immature endosperm trans-activates the Glb promoter, transgenic plants expressing a target gene driven by the Glb promoter might be expected to produce more of the target protein when additional REB protein is present. A synthetic human lysozyme gene was linked with the Glb promoter to generate Glb-Lys, which was used to transform rice with or without Native-Reb. Additional transformants were made by cobombardment of Glb-Lys with the Act-Reb construct as well as with the Act-Reb construct alone. Eight individual transgenic rice plants were obtained from independent transformation events with Act-Reb construct. All of them were late flowering and displayed high sterility and broad leaves (data not shown). Analysis of these plants was not pursued. In contrast, normal plant phenotypes were obtained among transformants containing Glb-Lys alone or both Glb-Lys and native Reb. To determine whether the Reb gene and Glb-Lys are present in the transgenic rice genome, one primer designed from vector sequence and another designed from the Reb gene 3′ terminator were used to identify these lines. In this case, only the recombinant Reb gene could be amplified (Fig. 5A). PCR analysis confirmed the presence of transgenes into the rice genome. As shown in Fig. 5C, 10 of 11 plants from independent transformation events contained both Reb and the lysozyme transgenes. The REB protein of immature seeds from five randomly selected transgenic lines was detected by Western blotting. The expression level of the REB protein in transgenic lines ranged from 25% to 71% higher than that in untransformed TP309 (Fig. 6). This demonstrated that the transgenic Reb gene was active in transgenic plants. Increased REB expression provides the potential in enhancing target gene expression in transgenic seeds.
Figure 6.
The detection of the REB protein expression in immature seed of the transgenic plants by Western blotting. (A) Western blot of five transgenic lines containing Glb-lys/Native-Reb. TP309 is used as base-line control. (B) The quantitative analysis of the REB protein based on A by the Kodak 1D Program.
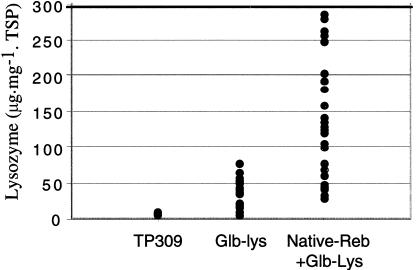
Seeds of confirmed transgenic rice plants were harvested at maturity, and the lysozyme activity was analyzed. Lysozyme expression in the seeds from 30 independent transformation events containing both the Native-Reb and the Glb-Lys ranged from 30.57 to 279.61 μg⋅mg−1 TSP with an average of 125.75 ± 68.65 μg⋅mg−1 TSP (Fig. 7). Seeds of 17 transgenic events containing the Glb-Lys gene alone expressed lysozyme in amounts ranging from 7 to 76 μg⋅mg−1 TSP with an average of 33.95 ± 20.55 μg⋅mg−1 TSP (Fig. 7). No lysozyme activity was detected in untransformed rice seeds (Fig. 7). The results showed that the expression level of the lysozyme increased an average of 3.7-fold when the seeds were transgenic for both the Reb gene and Glb-Lys. Statistical analysis (t test) showed that the amount of lysozyme in seeds from the plants transgenic for the Reb gene and Glb-Lys is significantly higher than in the plants transgenic for the Glb-Lys alone (P < 0.001).
Figure 7.
The expression of human lysozyme in mature seed of T0 transgenic plants. Seeds of 30 plants containing the Reb and lysozyme genes and seeds from 17 plants containing only the lysozyme gene were analyzed for lysozyme activity. The lysozyme gene was under the control of the Glb promoter, and the Reb gene was under the control of its own promoter. Individual seeds from each plant were analyzed. Transgenic seeds lacking detectable amounts of lysozyme were excluded. The activities of 20 lysozyme-positive seeds per plant, including both hemizygous and homozygous seeds, were averaged and plotted on the graph.
Discussion
Several lines of evidence indicate that REB is a transcriptional activator that can increase the expression of recombinant protein as demonstrated here with β-glucuronidase in the developing endosperm and human lysozyme in the transgenic rice seeds. With transient gene expression analysis, we showed that expression of the Reb gene driven by either its own promoter or by the promoters of the actin or the globulin genes can double the expression of the GUS protein (Fig. 2). Transcriptional activation by REB is through an interaction with the UAS located in the 5′-distal region of the Glb promoter (Fig. 3). Addition of the UAS to a nonresponsive promoter provided the transactivation function of the REB (Fig. 4). Production of human lysozyme from Glb-Lys in developing transgenic rice seeds was 3.7-fold higher when coexpressed with REB. The increasing dosage of REB protein with the lysozyme expression level is consistent in the detected transgenic plants.
The increased expression of human lysozyme in transgenic plants carrying the Native-Reb gene is unlikely to be because of other factors, such as the position of integration. All transgenic plants with and without the Native-Reb gene were obtained with the same procedure and at the same time. The majority of both types of transgenic plants grew normally in the greenhouse. Thus, they are assumed to share the same random variation, and any difference in expression between the means of the two groups should be attributed to the presence or absence of additional REB.
The present experiments demonstrate that the use of a limiting transcriptional activator for enhancement of the expression of recombinant protein needs to be expressed specifically in the tissue used for production of the recombinant protein. In our experiments, the transgenic rice plants containing the Reb gene driven by the Act gene promoter show mutant phenotypes and are sterile. Presumably, this aberrant phenotype is because of the constitutive expression of the transcriptional activator REB in all plant cells, causing disturbances of the normal gene expression program during rice development. Comparable abnormalities of transgenic plants expressing a transcription factor have been observed in Arabidopsis (28). In both cases, it is concluded that a constitutive promoter is not suitable to drive a transcription factor gene with the aim to increase recombinant protein expression in the plants. The constitutive expression leads to untimely expression of the genes, as the same transcription factor is used in different cells and at different developmental stages. Its action is pleiotropic (28). To avoid this complication, we have used endosperm-specific promoters in the transgenic plants for both the recombinant protein-producing gene and for the REB transcription factor. Both the globulin gene and the native Reb gene are expressed in endosperm cells (21, 29); thus, their promoters are compatible for cell-specific interaction.
Acknowledgments
We thank Ray Wu for Act-1 D promoter construct, Frank E. Hagie for encouragement, Diter von Wettestein and Robert J. Schemidt for helpful discussion and critical review of the manuscript, and Jianmin Huang, Dorice Yalda, and Cass McCullar for technical assistance.
Abbreviations
- TSP
total soluble protein
- BAC
bacterial artificial chromosome
- GUS
βglucuronidase
- UAS
upstream activation sequence
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF395819).
References
- 1.Horvath H, Huang J, Wong O, Kohl E, Okita T, Kannangara C G, von Wettstein D. Proc Natl Acad Sci USA. 2000;97:1914–1919. doi: 10.1073/pnas.030527497. . (First Published February 4, 2000; 10.1073/pnas.030527497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrin Y, Vaquero C, Gerrard I, Sack M, Drossard J, Stoger E, Christou P, Fischer R. Mol Breeding. 2000;6:345–352. [Google Scholar]
- 3.Stoger E, Vaquero C, Torres E, Sack M, Nicholson L, Drossard J, Williams S, Keen D, Perrin Y, Christou P, Fischer R. Plant Mol Biol. 2000;42:583–590. doi: 10.1023/a:1006301519427. [DOI] [PubMed] [Google Scholar]
- 4.Chong D K X, Roberts W, Arakawa T, Illes K, Bagi G, Slattery C W, Langridge W H R. Transgenic Res. 1997;6:289–296. doi: 10.1023/a:1018410712288. [DOI] [PubMed] [Google Scholar]
- 5.Chong D K X, Langridge W H R. Transgenic Res. 2000;9:71–78. doi: 10.1023/a:1008977630179. [DOI] [PubMed] [Google Scholar]
- 6.Cho M J, Wong L H, Marx C, Jiang W, Lemaux P G, Buchanan B B. Proc Natl Acad Sci USA. 1999;96:14641–14646. doi: 10.1073/pnas.96.25.14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McElroy D, Zhang W G, Cao J, Wu R. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Waston J D. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 9.Wu C Y, Suzuki A, Washida H, Takaiwa F. Plant J. 1998;14:673–683. doi: 10.1046/j.1365-313x.1998.00167.x. [DOI] [PubMed] [Google Scholar]
- 10.Vicente-Carbajosa J, Onate L, Lara P, Diaz I, Carbonero P. Plant J. 1998;13:629–640. doi: 10.1111/j.1365-313x.1998.00068.x. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt R J, Ketudat M, Aukerman M J, Hoschek G. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlan R S, Hammond-Kosack M, Bevan M. Plant J. 1999;19:173–181. doi: 10.1046/j.1365-313x.1999.00522.x. [DOI] [PubMed] [Google Scholar]
- 13.Holdsworth M J, Munoz-Blanco J, Hammond-Kosack M, Colot V, Schuch W, Bevan M W. Plant Mol Biol. 1995;29:711–720. doi: 10.1007/BF00041162. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mena M, Vicente-Carbajosa J, Schmidt R J, Carbonero P. Plant J. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- 16.Onate L, Vicente-Carbajosa J, Lara P, Diaz I, Carbonero P. J Biol Chem. 1999;274:9175–9182. doi: 10.1074/jbc.274.14.9175. [DOI] [PubMed] [Google Scholar]
- 17.Schwechheimer C, Bevan M. Trends Plant Sci. 1998;3:378–383. [Google Scholar]
- 18.Schwechheimer C, Zourelidou M, Bevan M W. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:127–150. doi: 10.1146/annurev.arplant.49.1.127. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z D, Ueda T, Messing J. Gene. 1998;223:321–332. doi: 10.1016/s0378-1119(98)00244-3. [DOI] [PubMed] [Google Scholar]
- 20.Yunes J A, Neto G C, Dasilva M J, Leite A, Ottoboni L M M, Arruda P. Plant Cell. 1994;6:237–249. doi: 10.1105/tpc.6.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakase M, Aoki N, Matsuda T, Adachi T. Plant Mol Biol. 1997;33:513–522. doi: 10.1023/a:1005784717782. [DOI] [PubMed] [Google Scholar]
- 22.Yang D, Parco A, Nandi S, Subudhi P, Zhu Y, Wang G, Huang N. Theor Appl Genet. 1997;95:1147–1154. doi: 10.1007/s001220050379. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Yang D, Domingo J, Ni J, Huang N. Proc Natl Acad Sci USA. 1998;95:5661–5666. doi: 10.1073/pnas.95.10.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Chen L, Zhang S, Beachy R N, Fauquet C M. Plant Cell Rep. 1998;18:25–31. doi: 10.1007/BF00232975. [DOI] [PubMed] [Google Scholar]
- 26.Christensen A H, Quail P H. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 27.Dellaporta S L, Wood J, Kicks J B. Plant Mol Biol Rep. 1983;1:19–20. [Google Scholar]
- 28.Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 29.Wu C Y, Adachi T, Hatano T, Washida H, Suzuki A, Takaiwa F. Plant Cell Physiol. 1998;39:885–889. doi: 10.1093/oxfordjournals.pcp.a029404. [DOI] [PubMed] [Google Scholar]